Interaction of a Novel Alternatively Spliced Variant of HSD11B1L with Parkin Enhances the Carcinogenesis Potential of Glioblastoma: Peiminine Interferes with This Interaction
Abstract
1. Introduction
2. Materials and Methods
2.1. GBM and Other Cancer Cell Lines, Primary Cells, Culture Medium and Reagents
2.2. RT-PCR
2.3. Western Blot
2.4. Construction and Transfection of Plasmid
2.5. Cell Proliferation Assay
2.6. Scratch Test for Cell Migration
2.7. Cell Invasion Assay
2.8. Transient Transfection of Small Interfering RNA of Parkin
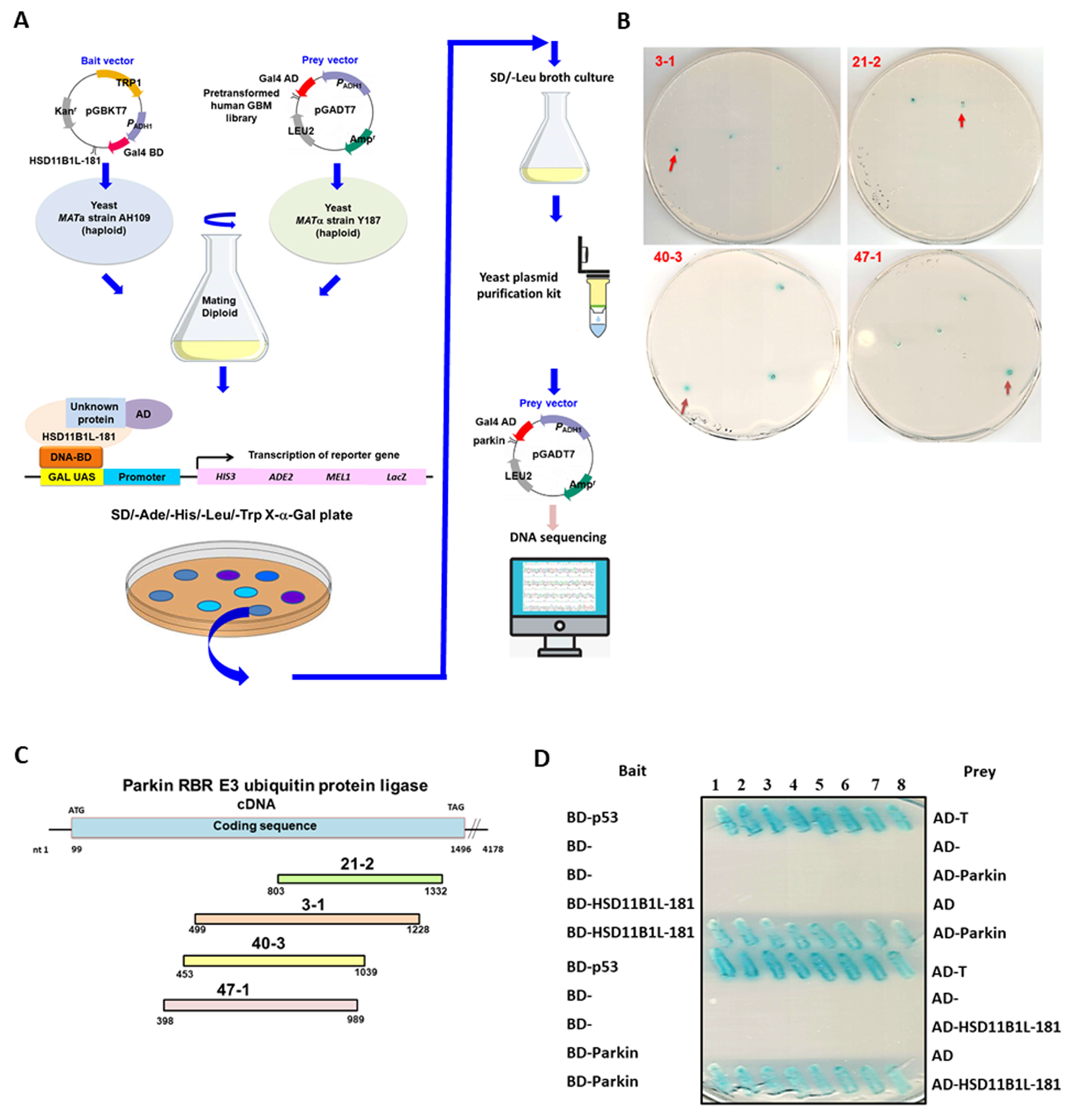
2.9. Interaction Screening by Yeast Two-Hybrid
2.10. Yeast Two-Hybrid Assays
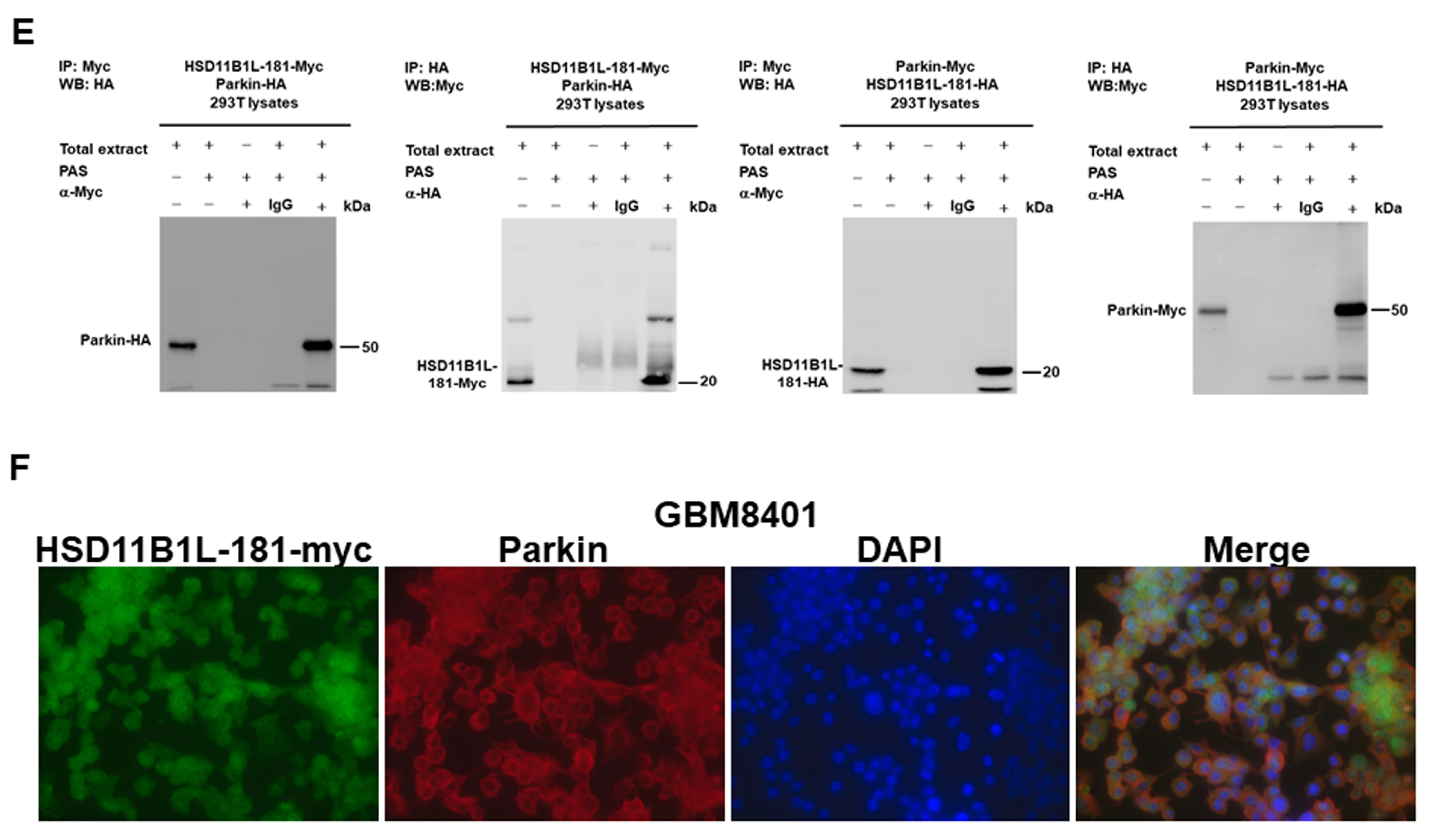
2.11. Co-Immunoprecipitation for HSD11B1L-181 and Parkin
2.12. Immunofluorescence Staining Analysis
2.13. PMN Treatment, Yeast Two-Hybrid Based Growth Assay
2.14. TUNEL Assay
2.15. Subcutaneous Tumor Xenograft Model of Mouse
2.16. Intracranial Xenograft Model of Mouse
2.17. Luminescence Imaging of Intracranial Glioblastoma
2.18. Statistical Analysis Methods
3. Results
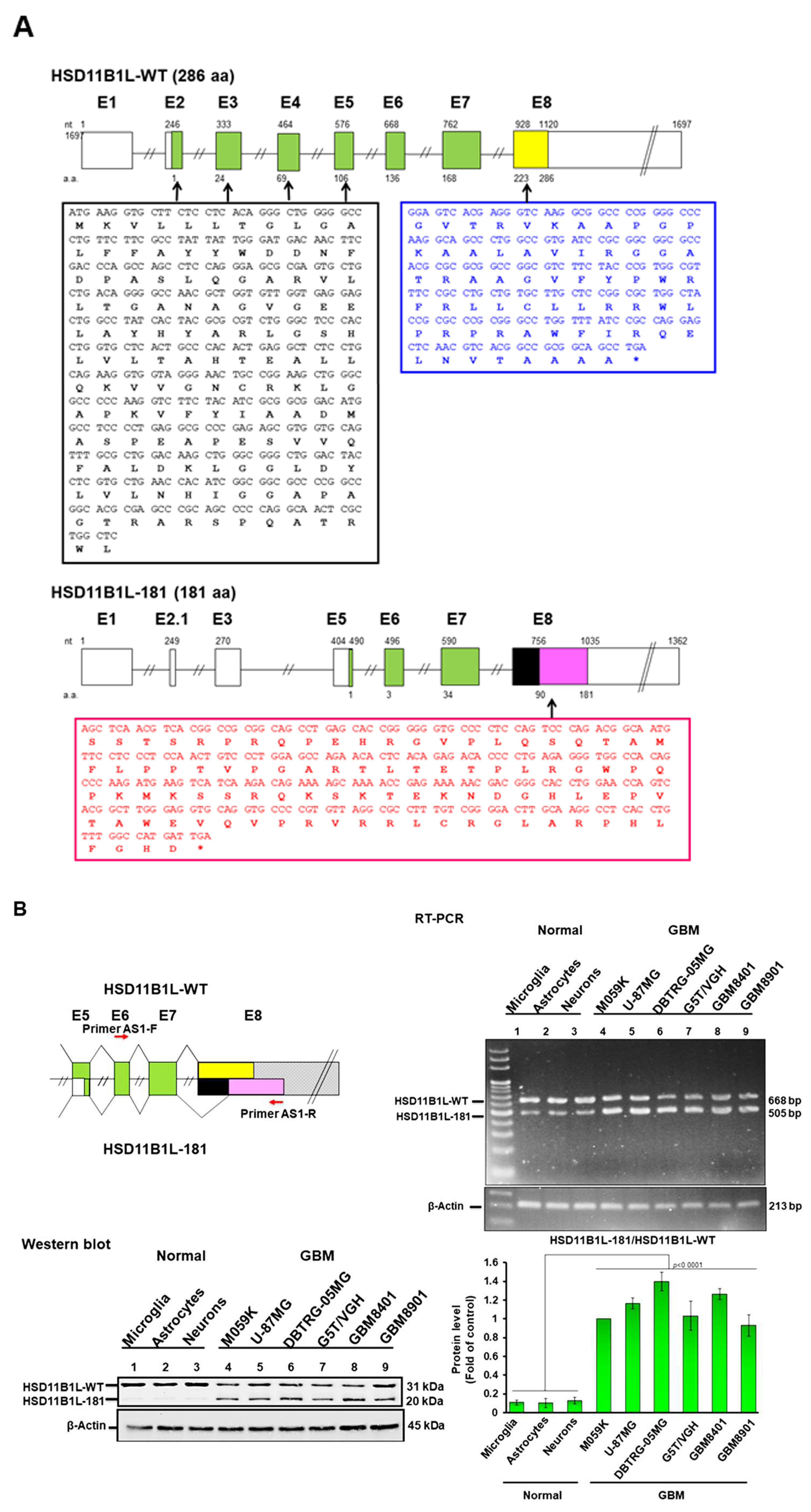
3.1. Characterization of Alternatively Spliced Variants of the Hydroxysteroid 11-Beta Dehydrogenase 1 like Gene (HSD11B1L) in GBM
3.2. HSD11B1L-181 Mature Transcript Is Predominantly Expressed in GBM
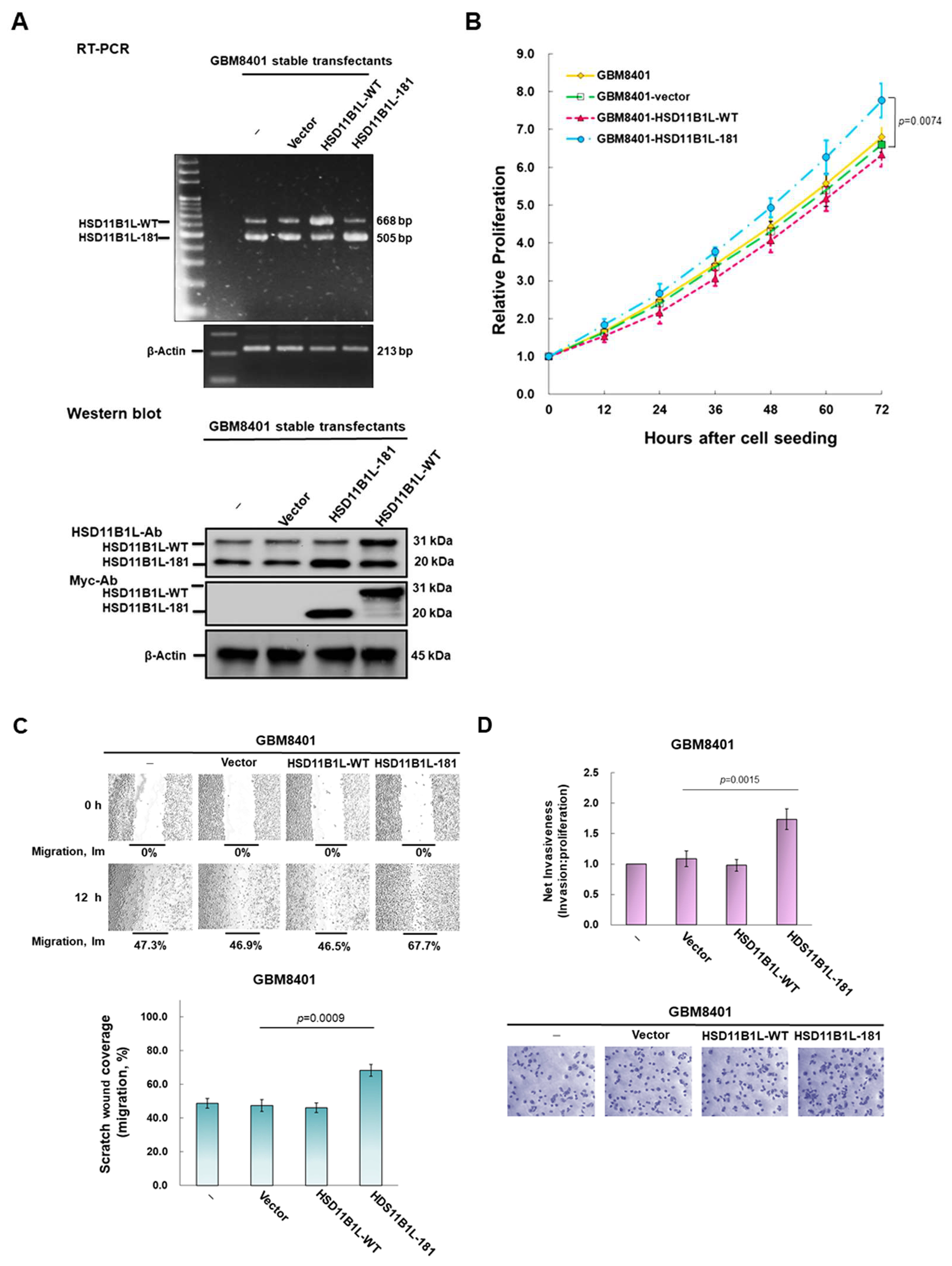
3.3. HSD11B1L-181 but Not HSD11B1L-WT Can Promote GBM Cell Proliferation, Migration and Invasion
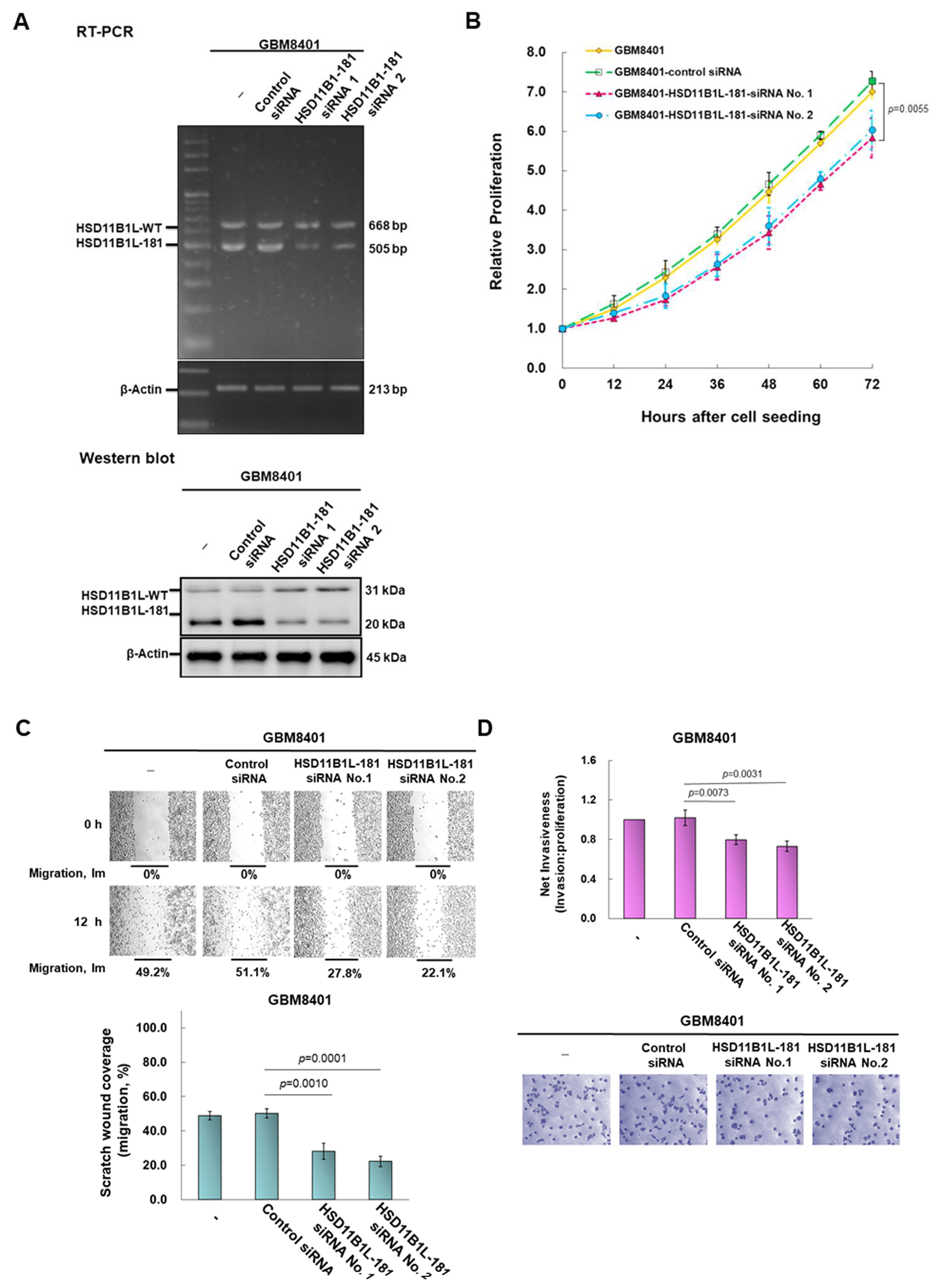
3.4. Downregulation of the Expression of HSD11B1L-181 via RNAi Suppress Characteristics of Proliferation, Migration and Invasion of GBM Cells
3.5. Specific Interaction between HSD11B1L-181 and Parkin
3.6. HSD11B1L-181 Uses the C-Terminal Amino Acid Fragment to Interact with the Middle Amino Acid Fragment of Parkin
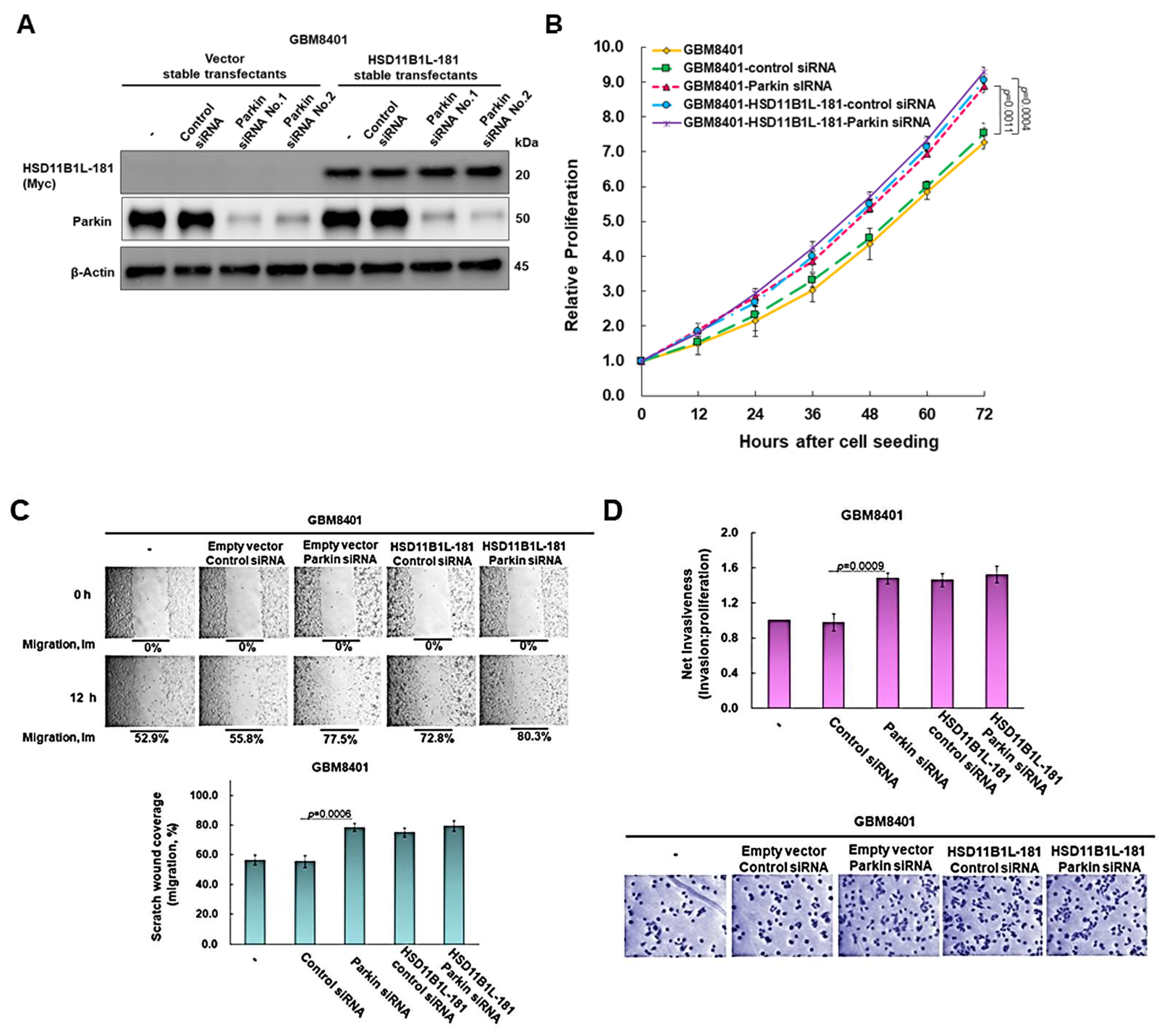
3.7. The Proliferation, Migration and Invasion Ability of GBM Cells Will Be Significantly Augmented by Parkin Knockdown
3.8. Parkin Knockdown Does Not Enhance the Ability of HSD11B1L-181 to Promote the Proliferation, Migration and Invasion of GBM Cells
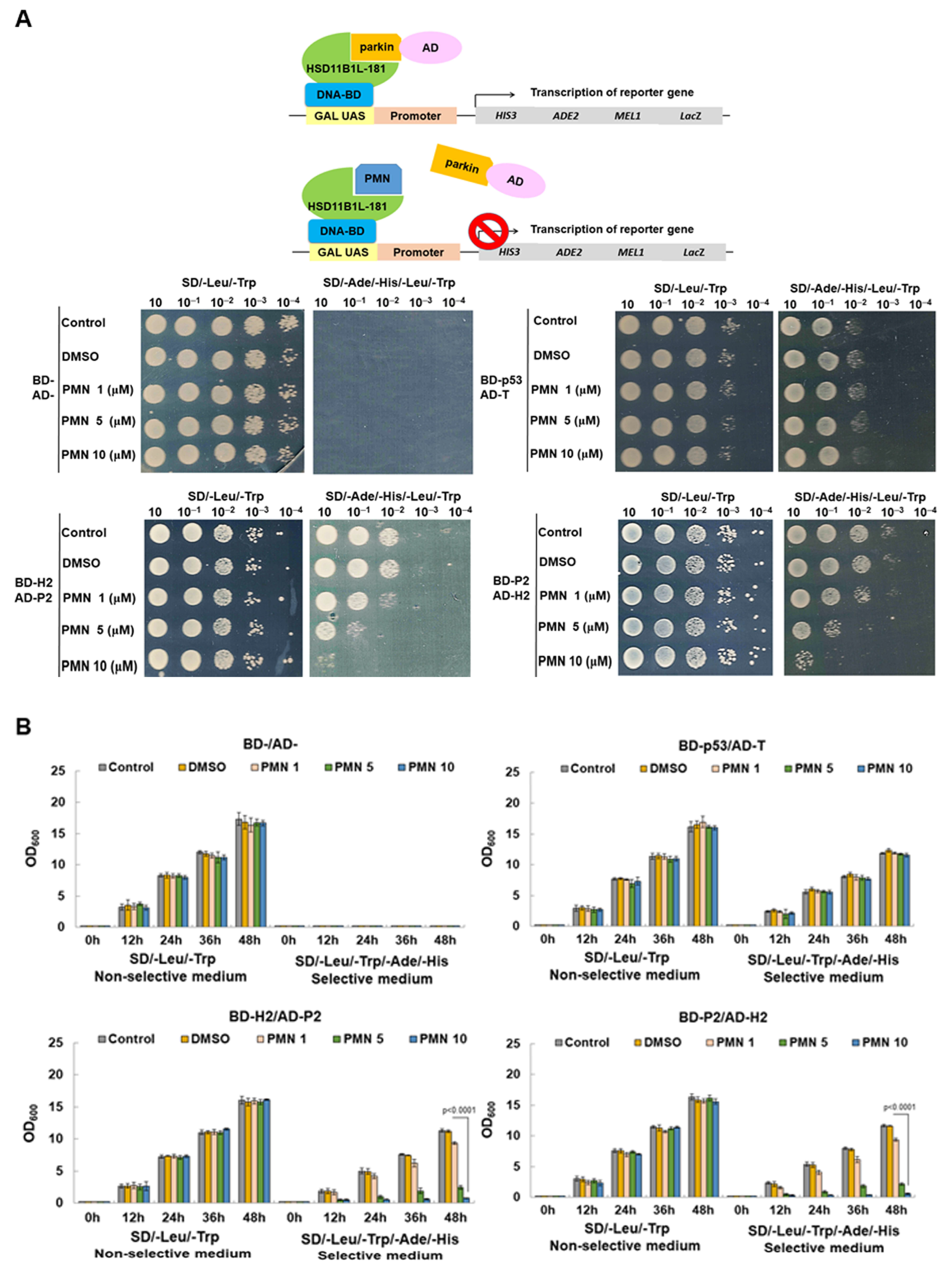
3.9. PMN Can Interfere with the Interaction between HSD11B1L-181 and Parkin in the Yeast Two-Hybrid Model
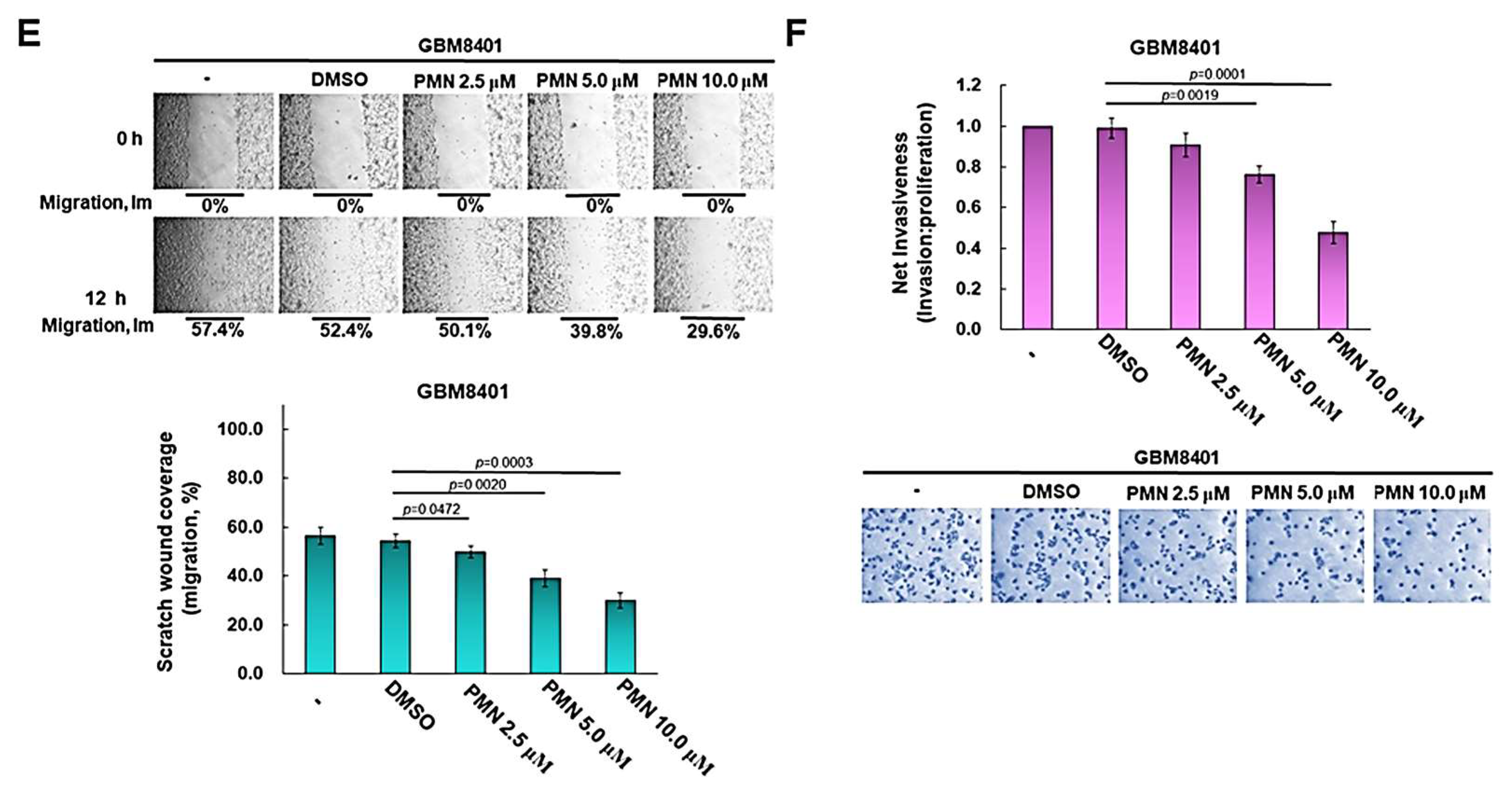
3.10. PMN Treatment Inhibits the Proliferation, Migration and Invasion of GBM Cells in Vitro
3.11. PMN Treatment Prevents Tumor Growth in a Nude Mice Model of Subcutaneous Xenograft of GBM
3.12. PMN Treatment Inhibits Tumor Growth in Orthotopic GBM Xenograft Immunodeficient Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lim, M.; Xia, Y.; Bettegowda, C.; Weller, M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 2018, 15, 422–442. [Google Scholar] [CrossRef]
- Weller, M.; van den Bent, M.; Preusser, M.; Le Rhun, E.; Tonn, J.C.; Minniti, G.; Bendszus, M.; Balana, C.; Chinot, O.; Dirven, L.; et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat. Rev. Clin. Oncol. 2021, 18, 170–186. [Google Scholar] [CrossRef]
- Noorani, I.; Mischel, P.S.; Swanton, C. Leveraging extrachromosomal DNA to fine-tune trials of targeted therapy for glioblastoma: Opportunities and challenges. Nat. Rev. Clin. Oncol. 2022, 19, 733–743. [Google Scholar] [CrossRef]
- Marasco, L.E.; Kornblihtt, A.R. The physiology of alternative splicing. Nat. Rev. Mol. Cell Biol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qian, J.; Gu, C.; Yang, Y. Alternative splicing and cancer: A systematic review. Signal Transduct. Target. Ther. 2021, 6, 78. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.; Xiao, L.; Zhou, W.; Jin, X.; Xu, Z.; Xu, C.; Wang, P.; Luo, M.; Wang, M.; Ma, K.; et al. A pan-cancer analysis of alternative splicing of splicing factors in 6904 patients. Oncogene 2021, 40, 5441–5450. [Google Scholar] [CrossRef] [PubMed]
- Frankiw, L.; Baltimore, D.; Li, G. Alternative mRNA splicing in cancer immunotherapy. Nat. Rev. Immunol. 2019, 19, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; McIntosh, C.S.; Mastaglia, F.L.; Wilton, S.D.; Aung-Htut, M.T. Neurodegenerative diseases: A hotbed for splicing defects and the potential therapies. Transl. Neurodegener. 2021, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Iyoda, T.; Fujita, M.; Fukai, F. Biologically Active TNIIIA2 Region in Tenascin-C Molecule: A Major Contributor to Elicit Aggressive Malignant Phenotypes From Tumors/Tumor Stroma. Front. Immunol. 2020, 11, 610096. [Google Scholar] [CrossRef] [PubMed]
- Tiek, D.M.; Khatib, S.A.; Trepicchio, C.J.; Heckler, M.M.; Divekar, S.D.; Sarkaria, J.N.; Glasgow, E.; Riggins, R.B. Estrogen-related receptor beta activation and isoform shifting by cdc2-like kinase inhibition restricts migration and intracranial tumor growth in glioblastoma. FASEB J. 2019, 33, 13476–13491. [Google Scholar] [CrossRef]
- Voss, D.M.; Sloan, A.; Spina, R.; Ames, H.M.; Bar, E.E. The Alternative Splicing Factor, MBNL1, Inhibits Glioblastoma Tumor Initiation and Progression by Reducing Hypoxia-Induced Stemness. Cancer Res. 2020, 80, 4681–4692. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wu, Q.; Cheng, S.; Li, W. Systematic Profiling of mRNA Splicing Reveals the Prognostic Predictor and Potential Therapeutic Target for Glioblastoma Multiforme. J. Oncol. 2021, 2021, 4664955. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, J.; Liu, Z.; Wang, Y.; Xuan, S.; Zhao, P. Comprehensive Characterization of Alternative mRNA Splicing Events in Glioblastoma: Implications for Prognosis, Molecular Subtypes, and Immune Microenvironment Remodeling. Front. Oncol. 2020, 10, 555632. [Google Scholar] [CrossRef]
- Qiu, J.; Wang, C.; Hu, H.; Chen, S.; Ding, X.; Cai, Y. Transcriptome analysis and prognostic model construction based on splicing profiling in glioblastoma. Oncol. Lett. 2021, 21, 138. [Google Scholar] [CrossRef]
- Wang, L.; Shamardani, K.; Babikir, H.; Catalan, F.; Nejo, T.; Chang, S.; Phillips, J.J.; Okada, H.; Diaz, A.A. The evolution of alternative splicing in glioblastoma under therapy. Genome Biol. 2021, 22, 48. [Google Scholar] [CrossRef]
- Wang, Z.; Gao, L.; Guo, X.; Feng, C.; Lian, W.; Deng, K.; Xing, B. Development of a Nomogram With Alternative Splicing Signatures for Predicting the Prognosis of Glioblastoma: A Study Based on Large-Scale Sequencing Data. Front. Oncol. 2020, 10, 1257. [Google Scholar] [CrossRef]
- Pan, Y.B.; Wang, S.; Yang, B.; Jiang, Z.; Lenahan, C.; Wang, J.; Zhang, J.; Shao, A. Transcriptome analyses reveal molecular mechanisms underlying phenotypic differences among transcriptional subtypes of glioblastoma. J. Cell Mol. Med. 2020, 24, 3901–3916. [Google Scholar] [CrossRef]
- Li, Y.; Guo, D. Genome-wide profiling of alternative splicing in glioblastoma and their clinical value. BMC Cancer 2021, 21, 958. [Google Scholar] [CrossRef] [PubMed]
- Bird, A.D.; Greatorex, S.; Reser, D.; Lavery, G.G.; Cole, T.J. Hydroxysteroid dehydrogenase HSD1L is localised to the pituitary-gonadal axis of primates. Endocr. Connect 2017, 6, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Q.; Wang, X.; Xue, B.H.; Zhao, Y.; Xie, F.; Wang, S.D.; Xue, C.; Wang, Y.; Zhang, Y.S.; Qian, L.J. Chronic stress promotes glioma cell proliferation via the PI3K/Akt signaling pathway. Oncol. Rep. 2021, 46, 202. [Google Scholar] [CrossRef] [PubMed]
- Murillo-González, F.E.; García-Aguilar, R.; Vega, L.; Elizondo, G. Regulation of Parkin expression as the key balance between neural survival and cancer cell death. Biochem. Pharmacol. 2021, 190, 114650. [Google Scholar] [CrossRef]
- Agarwal, S.; Muqit, M.M.K. PTEN-induced kinase 1 (PINK1) and Parkin: Unlocking a mitochondrial quality control pathway linked to Parkinson’s disease. Curr. Opin. Neurobiol. 2021, 72, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Wahabi, K.; Perwez, A.; Rizvi, M.A. Parkin in Parkinson’s Disease and Cancer: A Double-Edged Sword. Mol. Neurobiol. 2018, 55, 6788–6800. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, Y.; Yuan, S.; Cao, Y.; Bi, Z. Peiminine Induces G0/G1-Phase Arrest, Apoptosis, and Autophagy via the ROS/JNK Signaling Pathway in Human Osteosarcoma Cells in Vitro and in Vivo. Front. Pharmacol. 2021, 12, 770846. [Google Scholar] [CrossRef]
- Li, J.; Yang, D.; Li, Z.; Zhao, M.; Wang, D.; Sun, Z.; Wen, P.; Dai, Y.; Gou, F.; Ji, Y.; et al. PINK1/Parkin-mediated mitophagy in neurodegenerative diseases. Ageing Res. Rev. 2023, 84, 101817. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Panicker, N.; Dawson, V.L.; Dawson, T.M. Cell Biology of Parkin: Clues to the Development of New Therapeutics for Parkinson’s Disease. CNS Drugs 2022, 36, 1249–1267. [Google Scholar] [CrossRef]
- Ham, S.J.; Lee, D.; Yoo, H.; Jun, K.; Shin, H.; Chung, J. Decision between mitophagy and apoptosis by Parkin via VDAC1 ubiquitination. Proc. Natl. Acad. Sci. USA 2020, 117, 4281–4291. [Google Scholar] [CrossRef]
- Rouland, L.; Duplan, E.; Ramos Dos Santos, L.; Bernardin, A.; Katula, K.S.; Manfioletti, G.; Idbaih, A.; Checler, F.; Alves da Costa, C. Therapeutic potential of parkin as a tumor suppressor via transcriptional control of cyclins in glioblastoma cell and animal models. Theranostics 2021, 11, 10047–10063. [Google Scholar] [CrossRef]
- Aksoy Yasar, F.B.; Shingu, T.; Zamler, D.B.; Zaman, M.F.; Chien, D.L.; Zhang, Q.; Ren, J.; Hu, J. Quaking but not parkin is the major tumor suppressor in 6q deleted region in glioblastoma. Front. Cell Dev. Biol. 2022, 10, 931387. [Google Scholar] [CrossRef]
- Viotti, J.; Duplan, E.; Caillava, C.; Condat, J.; Goiran, T.; Giordano, C.; Marie, Y.; Idbaih, A.; Delattre, J.Y.; Honnorat, J.; et al. Glioma tumor grade correlates with parkin depletion in mutant p53-linked tumors and results from loss of function of p53 transcriptional activity. Oncogene 2014, 33, 1764–1775. [Google Scholar] [CrossRef]
- Maugeri, G.; D’Amico, A.G.; Magro, G.; Salvatorelli, L.; Barbagallo, G.M.; Saccone, S.; Drago, F.; Cavallaro, S.; D’Agata, V. Expression profile of parkin isoforms in human gliomas. Int. J. Oncol. 2015, 47, 1282–1292. [Google Scholar] [CrossRef] [PubMed]
- Rampioni Vinciguerra, G.L.; Sonego, M.; Segatto, I.; Dall’Acqua, A.; Vecchione, A.; Baldassarre, G.; Belletti, B. CDK4/6 Inhibitors in Combination Therapies: Better in Company Than Alone: A Mini Review. Front. Oncol. 2022, 12, 891580. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Park, M. Molecular crosstalk between cancer and neurodegenerative diseases. Cell. Mol. Life Sci. 2020, 77, 2659–2680. [Google Scholar] [CrossRef]
- Gong, Y.; Zack, T.I.; Morris, L.G.; Lin, K.; Hukkelhoven, E.; Raheja, R.; Tan, I.L.; Turcan, S.; Veeriah, S.; Meng, S.; et al. Pan-cancer genetic analysis identifies PARK2 as a master regulator of G1/S cyclins. Nat. Genet. 2014, 46, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Clemm von Hohenberg, K.; Muller, S.; Schleich, S.; Meister, M.; Bohlen, J.; Hofmann, T.G.; Teleman, A.A. Cyclin B/CDK1 and Cyclin A/CDK2 phosphorylate DENR to promote mitotic protein translation and faithful cell division. Nat. Commun. 2022, 13, 668. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, C.; Zhao, Y.; Yue, X.; Wu, H.; Huang, S.; Chen, J.; Tomsky, K.; Xie, H.; Khella, C.A.; et al. Parkin targets HIF-1alpha for ubiquitination and degradation to inhibit breast tumor progression. Nat. Commun. 2017, 8, 1823. [Google Scholar] [CrossRef]
- Maugeri, G.; D’Amico, A.G.; Reitano, R.; Saccone, S.; Federico, C.; Cavallaro, S.; D’Agata, V. Parkin modulates expression of HIF-1alpha and HIF-3alpha during hypoxia in gliobastoma-derived cell lines in vitro. Cell Tissue Res. 2016, 364, 465–474. [Google Scholar] [CrossRef]
- Xu, H.; Zong, H.; Ma, C.; Ming, X.; Shang, M.; Li, K.; He, X.; Du, H.; Cao, L. Epidermal growth factor receptor in glioblastoma. Oncol. Lett. 2017, 14, 512–516. [Google Scholar] [CrossRef]
- Peng, W.; Shi, S.; Zhong, J.; Liang, H.; Hou, J.; Hu, X.; Wang, F.; Zhang, J.; Geng, S.; Sun, X.; et al. CBX3 accelerates the malignant progression of glioblastoma multiforme by stabilizing EGFR expression. Oncogene 2022, 41, 3051–3063. [Google Scholar] [CrossRef]
- Liu, K.; Li, F.; Han, H.; Chen, Y.; Mao, Z.; Luo, J.; Zhao, Y.; Zheng, B.; Gu, W.; Zhao, W. Parkin Regulates the Activity of Pyruvate Kinase M2. J. Biol. Chem. 2016, 291, 10307–10317. [Google Scholar] [CrossRef]
- Spratt, D.E.; Walden, H.; Shaw, G.S. RBR E3 ubiquitin ligases: New structures, new insights, new questions. Biochem. J. 2014, 458, 421–437. [Google Scholar] [CrossRef]
- Liu, C.; Zhen, D.; Du, H.; Gong, G.; Wu, Y.; Ma, Q.; Quan, Z.S. Synergistic anti-inflammatory effects of peimine, peiminine, and forsythoside a combination on LPS-induced acute lung injury by inhibition of the IL-17-NF-kappaB/MAPK pathway activation. J. Ethnopharmacol. 2022, 295, 115343. [Google Scholar] [CrossRef]
- Chen, Z.K.; Zhao, D.; Feng, S.X.; Xu, J. Pharmacodynamics and Cellular Uptake of Peimine and Peiminine in Inflammatory Model Non-Small-Cell Lung Cancer Epithelial Cells (A549). Evid. Based Complement. Alternat. Med. 2022, 2022, 2946201. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.L.; Hung, H.S.; Tsai, C.W.; Liu, S.P.; Chiang, Y.T.; Kuo, Y.H.; Shyu, W.C.; Lin, S.Z.; Fu, R.H. Peiminine Reduces ARTS-Mediated Degradation of XIAP by Modulating the PINK1/Parkin Pathway to Ameliorate 6-Hydroxydopamine Toxicity and alpha-Synuclein Accumulation in Parkinson’s Disease Models In Vivo and In Vitro. Int. J. Mol. Sci. 2021, 22, 10240. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Xu, W.; Jiang, J.; Wang, Y.; Guo, Y.; Yang, R.; Chang, Y.; Zhao, B.; Wang, Z.; Zhang, J.; et al. Peiminine Suppresses RANKL-Induced Osteoclastogenesis by Inhibiting the NFATc1, ERK, and NF-kappaB Signaling Pathways. Front. Endocrinol. 2021, 12, 736863. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cai, H.; Ren, L.; Yang, Y.; Yang, H.; Liu, J.; Li, S.; Zhang, Y.; Zheng, X.; Tan, W.; et al. Sphingosine kinase 1 promotes growth of glioblastoma by increasing inflammation mediated by the NF-kappaB /IL-6/STAT3 and JNK/PTX3 pathways. Acta Pharm. Sin. B 2022, 12, 4390–4406. [Google Scholar] [CrossRef]
- Orlicka-Plocka, M.; Fedoruk-Wyszomirska, A.; Gurda-Wozna, D.; Pawelczak, P.; Krawczyk, P.; Giel-Pietraszuk, M.; Framski, G.; Ostrowski, T.; Wyszko, E. Implications of Oxidative Stress in Glioblastoma Multiforme Following Treatment with Purine Derivatives. Antioxidants 2021, 10, 950. [Google Scholar] [CrossRef]
- Zhao, B.; Shen, C.; Zheng, Z.; Wang, X.; Zhao, W.; Chen, X.; Peng, F.; Xue, L.; Shu, M.; Hou, X.; et al. Peiminine Inhibits Glioblastoma in Vitro and in Vivo Through Cell Cycle Arrest and Autophagic Flux Blocking. Cell Physiol. Biochem. 2018, 51, 1566–1583. [Google Scholar] [CrossRef]
- Fuentes-Fayos, A.C.; Vazquez-Borrego, M.C.; Jimenez-Vacas, J.M.; Bejarano, L.; Pedraza-Arevalo, S.; F, L.L.; Blanco-Acevedo, C.; Sanchez-Sanchez, R.; Reyes, O.; Ventura, S.; et al. Splicing machinery dysregulation drives glioblastoma development/aggressiveness: Oncogenic role of SRSF3. Brain 2020, 143, 3273–3293. [Google Scholar] [CrossRef]
- Huang, R.; Li, Z.; Li, C.; Wang, G.; Yan, P.; Peng, L.; Wang, J.; Zhu, X.; Hu, P.; Zhang, J.; et al. Germ Cell-Specific Gene 1-Like Protein Regulated by Splicing Factor CUGBP Elav-Like Family Member 5 and Primary Bile Acid Biosynthesis are Prognostic in Glioblastoma Multiforme. Front. Genet. 2019, 10, 1380. [Google Scholar] [CrossRef]
- Li, F.; Yi, Y.; Miao, Y.; Long, W.; Long, T.; Chen, S.; Cheng, W.; Zou, C.; Zheng, Y.; Wu, X.; et al. N(6)-Methyladenosine Modulates Nonsense-Mediated mRNA Decay in Human Glioblastoma. Cancer Res. 2019, 79, 5785–5798. [Google Scholar] [CrossRef] [PubMed]
- Paz, I.; Kosti, I.; Ares, M., Jr.; Cline, M.; Mandel-Gutfreund, Y. RBPmap: A web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014, 42, W361–W367. [Google Scholar] [CrossRef] [PubMed]
- Sergeeva, O.V.; Shcherbinina, E.Y.; Shomron, N.; Zatsepin, T.S. Modulation of RNA Splicing by Oligonucleotides: Mechanisms of Action and Therapeutic Implications. Nucleic Acid Ther. 2022, 32, 123–138. [Google Scholar] [CrossRef]
- Ma, W.K.; Voss, D.M.; Scharner, J.; Costa, A.S.H.; Lin, K.T.; Jeon, H.Y.; Wilkinson, J.E.; Jackson, M.; Rigo, F.; Bennett, C.F.; et al. ASO-Based PKM Splice-Switching Therapy Inhibits Hepatocellular Carcinoma Growth. Cancer Res. 2022, 82, 900–915. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, R.-H.; Hong, S.-Y.; Tsai, C.-W.; Liu, S.-P.; Chiu, S.-C.; Wu, M.-Z.; Shyu, W.-C.; Lin, S.-Z. Interaction of a Novel Alternatively Spliced Variant of HSD11B1L with Parkin Enhances the Carcinogenesis Potential of Glioblastoma: Peiminine Interferes with This Interaction. Cells 2023, 12, 894. https://doi.org/10.3390/cells12060894
Fu R-H, Hong S-Y, Tsai C-W, Liu S-P, Chiu S-C, Wu M-Z, Shyu W-C, Lin S-Z. Interaction of a Novel Alternatively Spliced Variant of HSD11B1L with Parkin Enhances the Carcinogenesis Potential of Glioblastoma: Peiminine Interferes with This Interaction. Cells. 2023; 12(6):894. https://doi.org/10.3390/cells12060894
Chicago/Turabian StyleFu, Ru-Huei, Syuan-Yu Hong, Chia-Wen Tsai, Shih-Ping Liu, Shao-Chih Chiu, Meng-Zhen Wu, Woei-Cherng Shyu, and Shinn-Zong Lin. 2023. "Interaction of a Novel Alternatively Spliced Variant of HSD11B1L with Parkin Enhances the Carcinogenesis Potential of Glioblastoma: Peiminine Interferes with This Interaction" Cells 12, no. 6: 894. https://doi.org/10.3390/cells12060894
APA StyleFu, R.-H., Hong, S.-Y., Tsai, C.-W., Liu, S.-P., Chiu, S.-C., Wu, M.-Z., Shyu, W.-C., & Lin, S.-Z. (2023). Interaction of a Novel Alternatively Spliced Variant of HSD11B1L with Parkin Enhances the Carcinogenesis Potential of Glioblastoma: Peiminine Interferes with This Interaction. Cells, 12(6), 894. https://doi.org/10.3390/cells12060894






