Microencapsulated Hepatocytes Differentiated from Human Induced Pluripotent Stem Cells: Optimizing 3D Culture for Tissue Engineering Applications
Abstract
1. Introduction
2. Materials and Methods
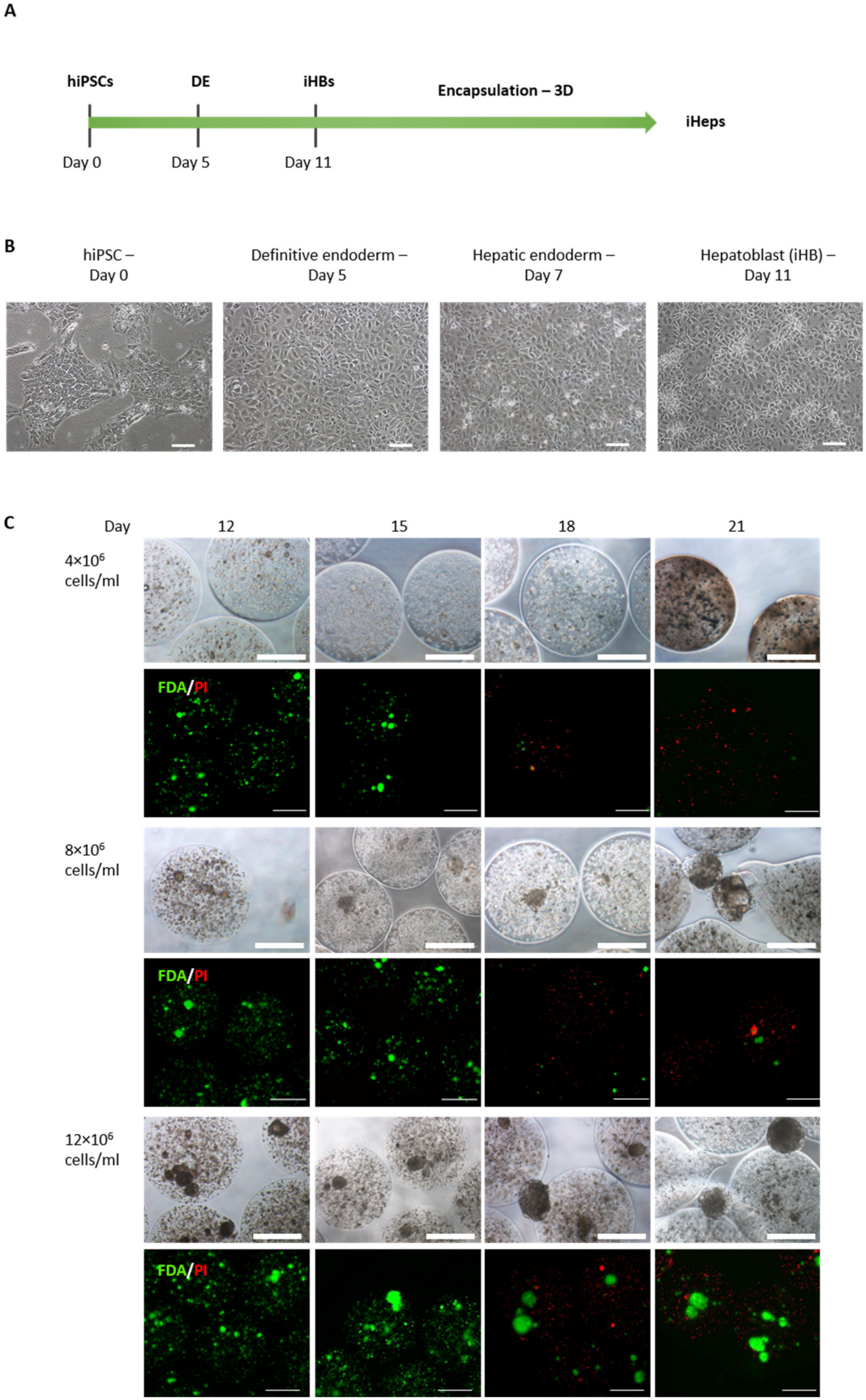
2.1. Hepatocyte Differentiation from hiPSCs (iHeps)
2.2. Encapsulation of iHBs as Single Cells in Alginate
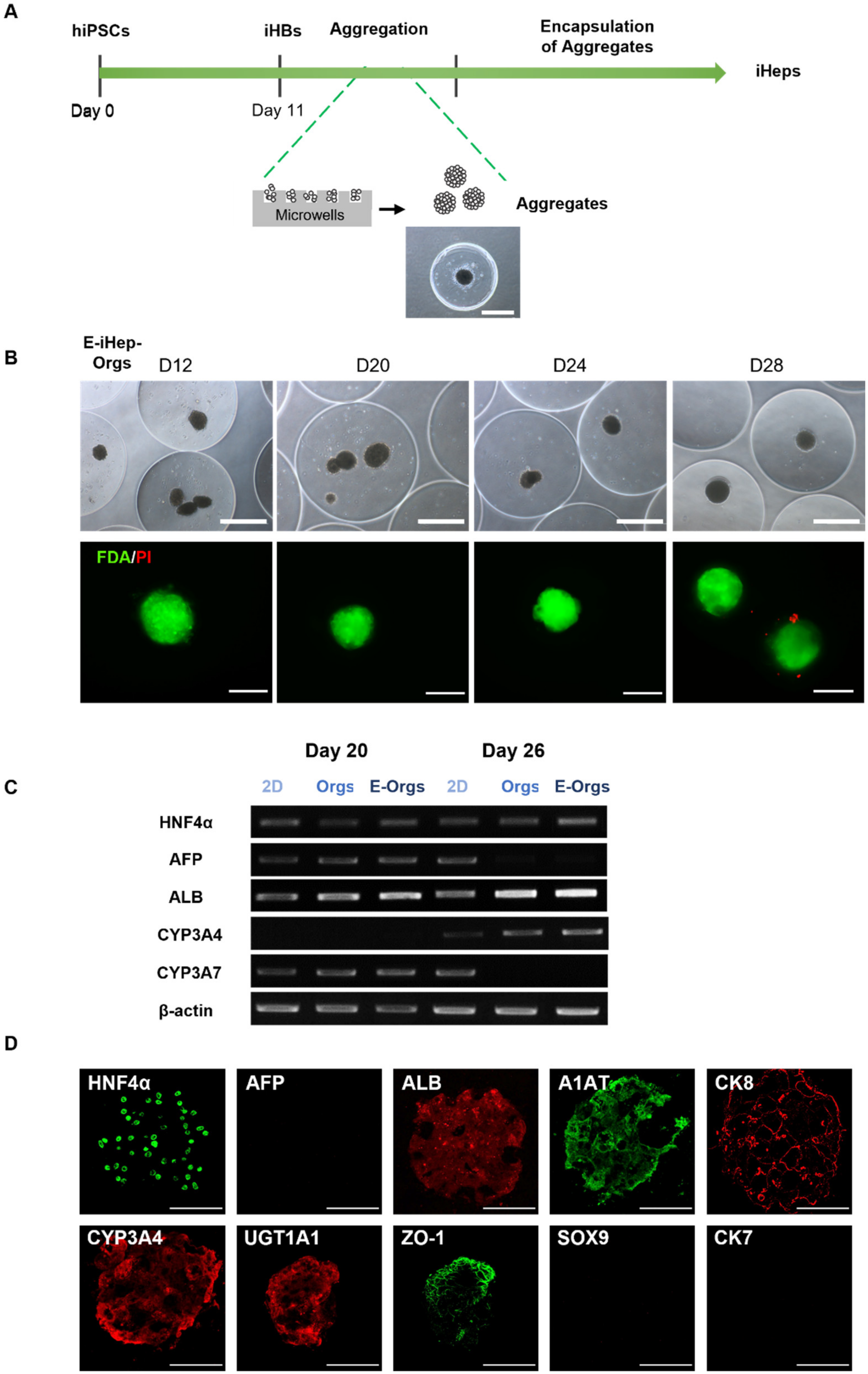
2.3. iHB Aggregation Followed by Encapsulation in Alginate
2.4. Cell Viability Assay
2.5. Assessment of Hepatic Functions In Vitro
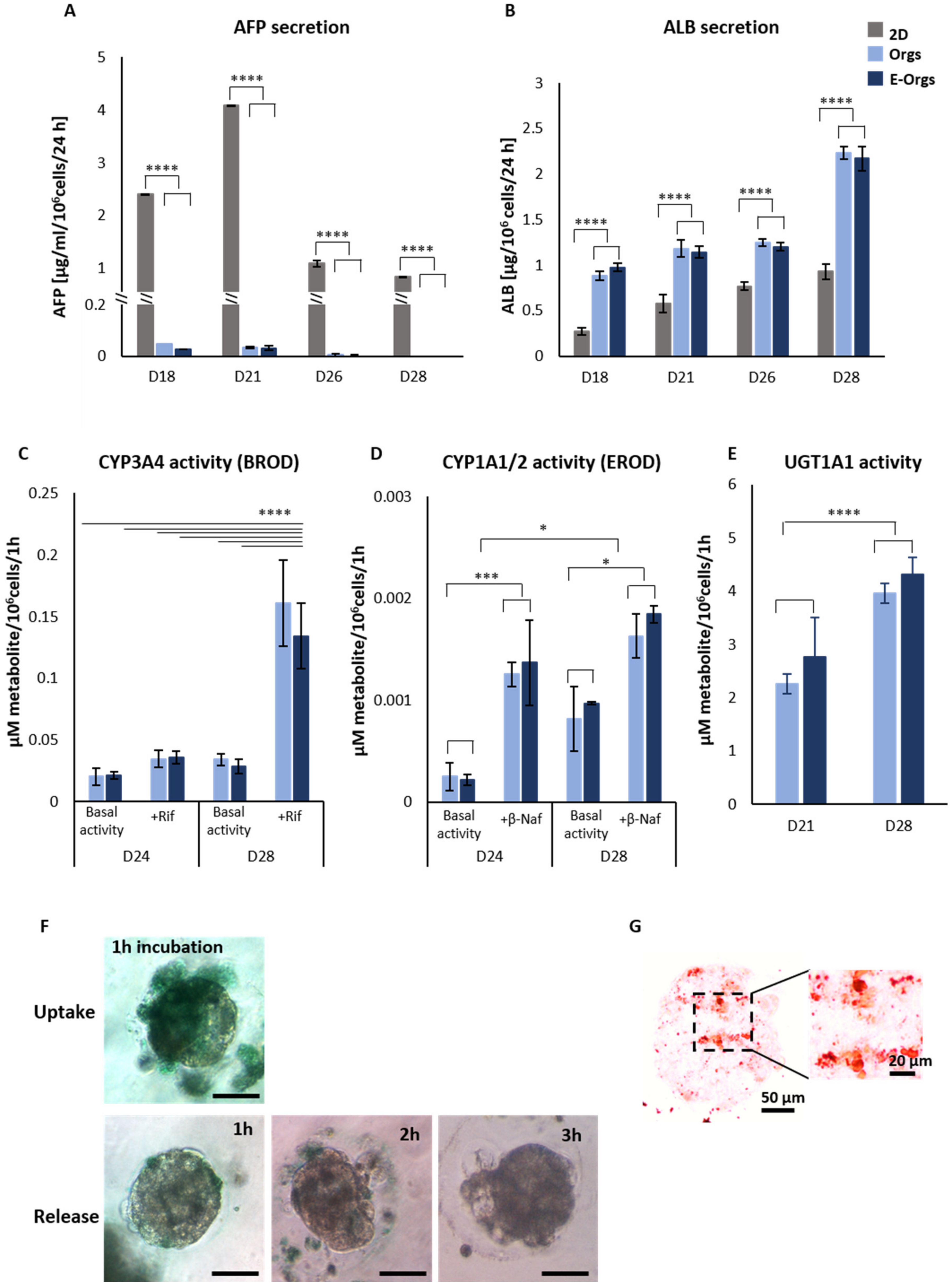
2.5.1. Albumin and α-Fetoprotein Synthesis
2.5.2. Biotransformation Activity—Phase I Metabolism
2.5.3. Biotransformation Activity—Phase II Metabolism
2.5.4. Uptake and Release of Indocyanine Green (ICG)
2.5.5. Lipid Storage—Oil Red O’ Staining
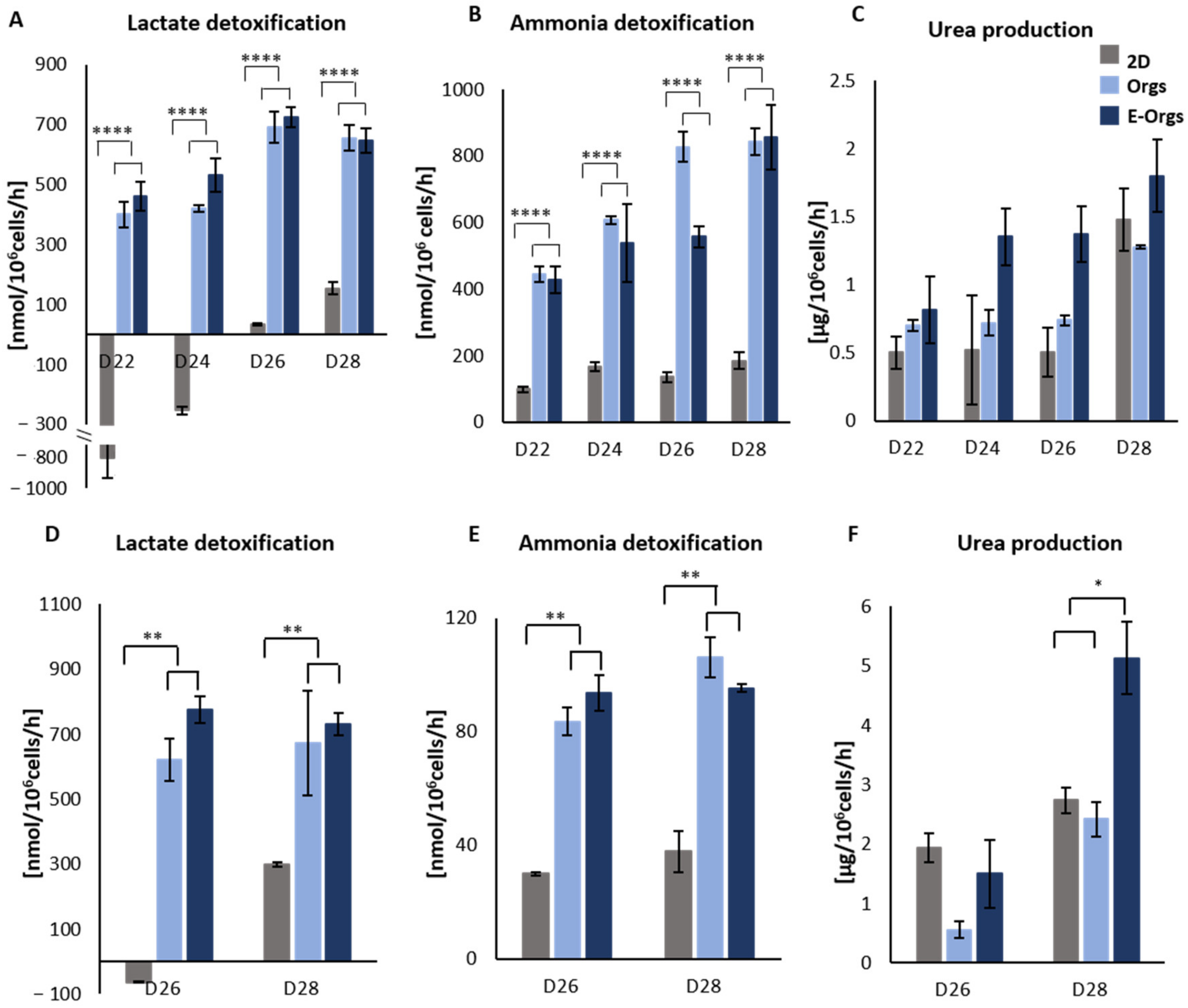
2.5.6. Urea Production and Lactate-Ammonia Detoxification
2.5.7. Data Normalization and Statistical Analysis
3. Results
3.1. iHB Generation and Encapsulation
3.2. Cell Aggregation of iHBs Prior to Alginate Encapsulation
3.3. Functional Assessment of E-iHep-Orgs
3.4. Functional Assessment of E-iHeps under Pathological Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Akamatsu, N.; Sugawara, Y.; Kokudo, N. Acute Liver Failure and Liver Transplantation. Intractable Rare Dis. Res. 2013, 2, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Vacanti, J.P.; Kulig, K.M. Liver Cell Therapy and Tissue Engineering for Transplantation. Semin. Pediatr. Surg. 2014, 23, 150–155. [Google Scholar] [CrossRef]
- Hewitt, N.J.; Lechón, M.J.G.; Houston, J.B.; Hallifax, D.; Brown, H.S.; Maurel, P.; Kenna, J.G.; Gustavsson, L.; Lohmann, C.; Skonberg, C.; et al. Primary Hepatocytes: Current Understanding of the Regulation of Metabolic Enzymes and Transporter Proteins, and Pharmaceutical Practice for the Use of Hepatocytes in Metabolism, Enzyme Induction, Transporter, Clearance, and Hepatotoxicity Studies. Drug Metab. Rev. 2007, 39, 159–234. [Google Scholar] [CrossRef]
- Elaut, G.; Henkens, T.; Papeleu, P.; Snykers, S.; Vinken, M.; Vanhaecke, T.; Rogiers, V. Molecular Mechanisms Underlying the Dedifferentiation Process of Isolated Hepatocytes and Their Cultures. Curr. Drug Metab. 2006, 7, 629–660. [Google Scholar] [CrossRef]
- Castell, J.V.; Jover, R.; Martnez-Jimnez, C.P.; Gmez-Lechn, M.J. Hepatocyte Cell Lines: Their Use, Scope and Limitations in Drug Metabolism Studies. Expert Opin. Drug Metab. Toxicol. 2006, 2, 183–212. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, R.E.; Fleming, H.E.; Bhatia, S.N. Pluripotent Stem Cell-Derived Hepatocyte-Like Cells. Biotechnol. Adv. 2014, 32, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Behbahan, I.S.; Duan, Y.; Lam, A.; Khoobyari, S.; Ma, X.; Ahuja, T.P.; Zern, M.A. New Approaches in the Differentiation of Human Embryonic Stem Cells and Induced Pluripotent Stem Cells toward Hepatocytes. Stem Cell Rev. 2011, 7, 748–759. [Google Scholar] [CrossRef]
- Messina, A.; Luce, E.; Hussein, M.; Dubart-Kupperschmitt, A. Pluripotent-Stem-Cell-Derived Hepatic Cells: Hepatocytes and Organoids for Liver Therapy and Regeneration. Cells 2020, 9, 420. [Google Scholar] [CrossRef]
- Vasconcellos, R.; Alvarenga, É.C.; Parreira, R.C.; Lima, S.S.; Resende, R.R. Exploring the Cell Signalling in Hepatocyte Differentiation. Cell. Signal. 2016, 28, 1773–1788. [Google Scholar] [CrossRef]
- Cho, C.H.; Parashurama, N.; Park, E.Y.H.; Suganuma, K.; Nahmias, Y.; Park, J.; Tilles, A.W.; Berthiaume, F.; Yarmush, M.L. Homogeneous Differentiation of Hepatocyte-like Cells from Embryonic Stem Cells: Applications for the Treatment of Liver Failure. FASEB J. 2008, 22, 898–909. [Google Scholar] [CrossRef] [PubMed]
- Hannoun, Z.; Steichen, C.; Dianat, N.; Weber, A.; Dubart-Kupperschmitt, A. The Potential of Induced Pluripotent Stem Cell Derived Hepatocytes. J. Hepatol. 2016, 65, 182–199. [Google Scholar] [CrossRef] [PubMed]
- Espejel, S.; Roll, G.R.; McLaughlin, K.J.; Lee, A.Y.; Zhang, J.Y.; Laird, D.J.; Okita, K.; Yamanaka, S.; Willenbring, H. Induced Pluripotent Stem Cell–Derived Hepatocytes Have the Functional and Proliferative Capabilities Needed for Liver Regeneration in Mice. J. Clin. Investig. 2010, 120, 3120–3126. [Google Scholar] [CrossRef] [PubMed]
- Baxter, M.; Withey, S.; Harrison, S.; Segeritz, C.-P.; Zhang, F.; Atkinson-Dell, R.; Rowe, C.; Gerrard, D.T.; Sison-Young, R.; Jenkins, R.; et al. Phenotypic and Functional Analyses Show Stem Cell-Derived Hepatocyte-like Cells Better Mimic Fetal Rather than Adult Hepatocytes. J. Hepatol. 2015, 62, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Godoy, P.; Hewitt, N.J.; Albrecht, U.; Andersen, M.E.; Ansari, N.; Bhattacharya, S.; Bode, J.G.; Bolleyn, J.; Borner, C.; Böttger, J.; et al. Recent Advances in 2D and 3D in Vitro Systems Using Primary Hepatocytes, Alternative Hepatocyte Sources and Non-Parenchymal Liver Cells and Their Use in Investigating Mechanisms of Hepatotoxicity, Cell Signaling and ADME. Arch. Toxicol. 2013, 87, 1315–1530. [Google Scholar] [CrossRef] [PubMed]
- Messina, A.; Luce, E.; Benzoubir, N.; Pasqua, M.; Pereira, U.; Humbert, L.; Eguether, T.; Rainteau, D.; Duclos-Vallée, J.-C.; Legallais, C.; et al. Evidence of Adult Features and Functions of Hepatocytes Differentiated from Human Induced Pluripotent Stem Cells and Self-Organized as Organoids. Cells 2022, 11, 537. [Google Scholar] [CrossRef] [PubMed]
- Gieseck III, R.L.; Hannan, N.R.F.; Bort, R.; Hanley, N.A.; Drake, R.A.L.; Cameron, G.W.W.; Wynn, T.A.; Vallier, L. Maturation of Induced Pluripotent Stem Cell Derived Hepatocytes by 3D-Culture. PLoS ONE 2014, 9, e86372. [Google Scholar] [CrossRef]
- Glicklis, R.; Shapiro, L.; Agbaria, R.; Merchuk, J.C.; Cohen, S. Hepatocyte Behavior within Three-Dimensional Porous Alginate Scaffolds. Biotechnol. Bioeng. 2000, 67, 344–353. [Google Scholar] [CrossRef]
- Capone, S.H.; Dufresne, M.; Rechel, M.; Fleury, M.-J.; Salsac, A.-V.; Paullier, P.; Daujat-Chavanieu, M.; Legallais, C. Impact of Alginate Composition: From Bead Mechanical Properties to Encapsulated HepG2/C3A Cell Activities for in Vivo Implantation. PLoS ONE 2013, 8, e62032. [Google Scholar] [CrossRef]
- David, B.; Barbe, L.; BarthèS-Biesel, D.; Legallais, C. Mechanical Properties of Alginate Beads Hosting Hepatocytes in a Fluidized Bed Bioreactor. Int. J. Artif. Organs 2006, 29, 756–763. [Google Scholar] [CrossRef]
- David, B.; Dufresne, M.; Nagel, M.-D.; Legallais, C. In Vitro Assessment of Encapsulated C3A Hepatocytes Functions in a Fluidized Bed Bioreactor. Biotechnol. Prog. 2004, 20, 1204–1212. [Google Scholar] [CrossRef]
- Pasqua, M.; Pereira, U.; Messina, A.; de Lartigue, C.; Vigneron, P.; Dubart-Kupperschmitt, A.; Legallais, C. HepaRG Self-Assembled Spheroids in Alginate Beads Meet the Clinical Needs for Bioartificial Liver. Tissue Eng. Part A 2020, 26, 613–622. [Google Scholar] [CrossRef]
- Tostões, R.M.; Leite, S.B.; Miranda, J.P.; Sousa, M.; Wang, D.I.C.; Carrondo, M.J.T.; Alves, P.M. Perfusion of 3D Encapsulated Hepatocytes—A Synergistic Effect Enhancing Long-Term Functionality in Bioreactors. Biotechnol. Bioeng. 2011, 108, 41–49. [Google Scholar] [CrossRef]
- Andersen, T.; Auk-Emblem, P.; Dornish, M. 3D Cell Culture in Alginate Hydrogels. Microarrays 2015, 4, 133–161. [Google Scholar] [CrossRef]
- Mahler, S.; Desille, M.; Frémond, B.; Chesné, C.; Guillouzo, A.; Campion, J.-P.; Clément, B. Hypothermic Storage and Cryopreservation of Hepatocytes: The Protective Effect of Alginate Gel against Cell Damages. Cell Transplant. 2003, 12, 579–592. [Google Scholar] [CrossRef]
- Hang, H.; Shi, X.; Gu, G.X.; Wu, Y.; Gu, J.; Ding, Y. In Vitro Analysis of Cryopreserved Alginate-Poly-L-Lysine-Alginate-Microencapsulated Human Hepatocytes. Liver Int. Off. J. Int. Assoc. Study Liver 2010, 30, 611–622. [Google Scholar] [CrossRef]
- Syanda, A.M.; Kringstad, V.I.; Blackford, S.J.I.; Kjesbu, J.S.; Ng, S.S.; Ma, L.; Xiao, F.; Coron, A.E.; Rokstad, A.M.A.; Modi, S.; et al. Sulfated Alginate Reduces Pericapsular Fibrotic Overgrowth on Encapsulated CGMP-Compliant HPSC-Hepatocytes in Mice. Front. Bioeng. Biotechnol. 2022, 9, 816542. [Google Scholar] [CrossRef] [PubMed]
- Caron, J.; Pène, V.; Tolosa, L.; Villaret, M.; Luce, E.; Fourrier, A.; Heslan, J.-M.; Saheb, S.; Bruckert, E.; Gómez-Lechón, M.J.; et al. Low-Density Lipoprotein Receptor-Deficient Hepatocytes Differentiated from Induced Pluripotent Stem Cells Allow Familial Hypercholesterolemia Modeling, CRISPR/Cas-Mediated Genetic Correction, and Productive Hepatitis C Virus Infection. Stem Cell Res. Ther. 2019, 10, 221. [Google Scholar] [CrossRef]
- Gautier, A.; Carpentier, B.; Dufresne, M.; Vu Dinh, Q.; Paullier, P.; Legallais, C. Impact of Alginate Type and Bead Diameter on Mass Transfers and the Metabolic Activities of Encapsulated C3A Cells in Bioartificial Liver Applications. Eur. Cell. Mater. 2011, 21, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Leite, S.B.; Teixeira, A.P.; Miranda, J.P.; Tostões, R.M.; Clemente, J.J.; Sousa, M.F.; Carrondo, M.J.T.; Alves, P.M. Merging Bioreactor Technology with 3D Hepatocyte-Fibroblast Culturing Approaches: Improved in Vitro Models for Toxicological Applications. Toxicol. In Vitro 2011, 25, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Pasqua, M.; Pereira, U.; de Lartigue, C.; Nicolas, J.; Vigneron, P.; Dermigny, Q.; Legallais, C. Preclinical Characterization of Alginate-Poly-L-Lysine Encapsulated HepaRG for Extracorporeal Liver Supply. Biotechnol. Bioeng. 2021, 118, 453–464. [Google Scholar] [CrossRef]
- Rebelo, S.P.; Costa, R.; Estrada, M.; Shevchenko, V.; Brito, C.; Alves, P.M. HepaRG Microencapsulated Spheroids in DMSO-Free Culture: Novel Culturing Approaches for Enhanced Xenobiotic and Biosynthetic Metabolism. Arch. Toxicol. 2015, 89, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Jitraruch, S.; Dhawan, A.; Hughes, R.D.; Filippi, C.; Soong, D.; Philippeos, C.; Lehec, S.C.; Heaton, N.D.; Longhi, M.S.; Mitry, R.R. Alginate Microencapsulated Hepatocytes Optimised for Transplantation in Acute Liver Failure. PLoS ONE 2014, 9, e113609. [Google Scholar] [CrossRef]
- Kilbride, P.; Lamb, S.; Gibbons, S.; Bundy, J.; Erro, E.; Selden, C.; Fuller, B.; Morris, J. Cryopreservation and Re-Culture of a 2.3 Litre Biomass for Use in a Bioartificial Liver Device. PLoS ONE 2017, 12, e0183385. [Google Scholar] [CrossRef] [PubMed]
- Maguire, T.; Davidovich, A.; Wallenstein, E.; Novik, E.; Sharma, N.; Pedersen, H.; Androulakis, I.; Schloss, R.; Yarmush, M. Control of Hepatic Differentiation via Cellular Aggregation in an Alginate Microenvironment. Biotechnol. Bioeng. 2007, 98, 631–644. [Google Scholar] [CrossRef]
- Fang, S.; Qiu, Y.; Mao, L.; Shi, X.; Yu, D.; Ding, Y. Differentiation of Embryoid-Body Cells Derived from Embryonic Stem Cells into Hepatocytes in Alginate Microbeads in Vitro. Acta Pharmacol. Sin. 2007, 28, 1924–1930. [Google Scholar] [CrossRef]
- Blackford, S.J.I.; Ng, S.S.; Segal, J.M.; King, A.J.F.; Austin, A.L.; Kent, D.; Moore, J.; Sheldon, M.; Ilic, D.; Dhawan, A.; et al. Validation of Current Good Manufacturing Practice Compliant Human Pluripotent Stem Cell-Derived Hepatocytes for Cell-Based Therapy. STEM CELLS Transl. Med. 2019, 8, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zhou, X.; Liu, T.; Zhong, Z.; Zhou, Q.; Iqbal, W.; Xie, Q.; Wei, C.; Zhang, X.; Chang, T.M.S.; et al. Direct Differentiation of Human Embryonic Stem Cells to 3D Functional Hepatocyte-like Cells in Alginate Microencapsulation Sphere. Cells 2022, 11, 3134. [Google Scholar] [CrossRef]
- Haque, T.; Chen, H.; Ouyang, W.; Martoni, C.; Lawuyi, B.; Urbanska, A.; Prakash, S. Investigation of a New Microcapsule Membrane Combining Alginate, Chitosan, Polyethylene Glycol and Poly-L-Lysine for Cell Transplantation Applications. Int. J. Artif. Organs 2005, 28, 631–637. [Google Scholar] [CrossRef]
- Deng, S.; Zhu, Y.; Zhao, X.; Chen, J.; Tuan, R.S.; Chan, H.F. Efficient Fabrication of Monodisperse Hepatocyte Spheroids and Encapsulation in Hybrid Hydrogel with Controllable Extracellular Matrix Effect. Biofabrication 2021, 14, 015002. [Google Scholar] [CrossRef]
- Liu, W.-M.; Zhou, X.; Chen, C.-Y.; Lv, D.-D.; Huang, W.-J.; Peng, Y.; Wu, H.-P.; Chen, Y.; Tang, D.; Guo, L.-N.; et al. Establishment of Functional Liver Spheroids From Human Hepatocyte-Derived Liver Progenitor-Like Cells for Cell Therapy. Front. Bioeng. Biotechnol. 2021, 9, 738081. [Google Scholar] [CrossRef]
- Song, W.; Lu, Y.-C.; Frankel, A.S.; An, D.; Schwartz, R.E.; Ma, M. Engraftment of Human Induced Pluripotent Stem Cell-Derived Hepatocytes in Immunocompetent Mice via 3D Co-Aggregation and Encapsulation. Sci. Rep. 2015, 5, 16884. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Wu, J.; Qiu, W.-L.; Yang, L.; Deng, X.; Zhou, Y.; Chen, Y.; Li, X.; Yu, L.; Li, H.; et al. Large-Scale Generation of Functional and Transplantable Hepatocytes and Cholangiocytes from Human Endoderm Stem Cells. Cell Rep. 2020, 33, 108455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, Y.; Chen, L.; Liu, T.; Li, Y.; Wang, Y.; Geng, Y. Efficient Large-Scale Generation of Functional Hepatocytes from Mouse Embryonic Stem Cells Grown in a Rotating Bioreactor with Exogenous Growth Factors and Hormones. Stem Cell Res. Ther. 2013, 4, 145. [Google Scholar] [CrossRef] [PubMed]
- Steichen, C.; Luce, E.; Maluenda, J.; Tosca, L.; Moreno-Gimeno, I.; Desterke, C.; Dianat, N.; Goulinet-Mainot, S.; Awan-Toor, S.; Burks, D.; et al. Messenger RNA- versus Retrovirus-Based Induced Pluripotent Stem Cell Reprogramming Strategies: Analysis of Genomic Integrity. Stem Cells Transl. Med. 2014, 3, 686–691. [Google Scholar] [CrossRef]
- Rahman, T.M.; Selden, C.; Khalil, M.; Diakanov, I.; Hodgson, H.J.F. Alginate-Encapsulated Human Hepatoblastoma Cells in an Extracorporeal Perfusion System Improve Some Systemic Parameters of Liver Failure in a Xenogeneic Model. Artif. Organs 2004, 28, 476–482. [Google Scholar] [CrossRef]
- Cheng, N.; Wauthier, E.; Reid, L.M. Mature Human Hepatocytes from Ex Vivo Differentiation of Alginate-Encapsulated Hepatoblasts. Tissue Eng. Part A 2008, 14, 1–7. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussein, M.; Pasqua, M.; Pereira, U.; Benzoubir, N.; Duclos-Vallée, J.-C.; Dubart-Kupperschmitt, A.; Legallais, C.; Messina, A. Microencapsulated Hepatocytes Differentiated from Human Induced Pluripotent Stem Cells: Optimizing 3D Culture for Tissue Engineering Applications. Cells 2023, 12, 865. https://doi.org/10.3390/cells12060865
Hussein M, Pasqua M, Pereira U, Benzoubir N, Duclos-Vallée J-C, Dubart-Kupperschmitt A, Legallais C, Messina A. Microencapsulated Hepatocytes Differentiated from Human Induced Pluripotent Stem Cells: Optimizing 3D Culture for Tissue Engineering Applications. Cells. 2023; 12(6):865. https://doi.org/10.3390/cells12060865
Chicago/Turabian StyleHussein, Marwa, Mattia Pasqua, Ulysse Pereira, Nassima Benzoubir, Jean-Charles Duclos-Vallée, Anne Dubart-Kupperschmitt, Cecile Legallais, and Antonietta Messina. 2023. "Microencapsulated Hepatocytes Differentiated from Human Induced Pluripotent Stem Cells: Optimizing 3D Culture for Tissue Engineering Applications" Cells 12, no. 6: 865. https://doi.org/10.3390/cells12060865
APA StyleHussein, M., Pasqua, M., Pereira, U., Benzoubir, N., Duclos-Vallée, J.-C., Dubart-Kupperschmitt, A., Legallais, C., & Messina, A. (2023). Microencapsulated Hepatocytes Differentiated from Human Induced Pluripotent Stem Cells: Optimizing 3D Culture for Tissue Engineering Applications. Cells, 12(6), 865. https://doi.org/10.3390/cells12060865










