Age-Dependent Dysregulation of APP in Neuronal and Skin Cells from Fragile X Individuals
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Fibroblasts and iPSCs Cultures
2.2. iPSC Neural Differentiation
2.3. Human Forebrain-Specific Organoid Cultures
2.4. SUnSET Assay
2.5. Ammonium Sulfate Precipitation
2.6. Peptide Treatment
2.7. Western Blot
2.8. RT-qPCR
- hFMR1 Forward 5′-TGT CAG ATT CCC ACC TCC TG-3′
- hFMR1 Reverse 5′-TAA CCA CCA ACA GCA AGG CT-3′
- hHPRT1 Forward 5′-TGC TGA GGA TTT GGA AAG GGT-3′
- hHPRT1 Reverse 5′-TCG AGC AAG ACG TTC AGT CC-3′
- hGAPDH Forward 5′-CTC AAC TAC ATG GTT TAC ATG-3′
- hGAPDH Reverse 5′-CCA TTG ATG ACA AGC TTC CCG-3′
2.9. Statistics
3. Results
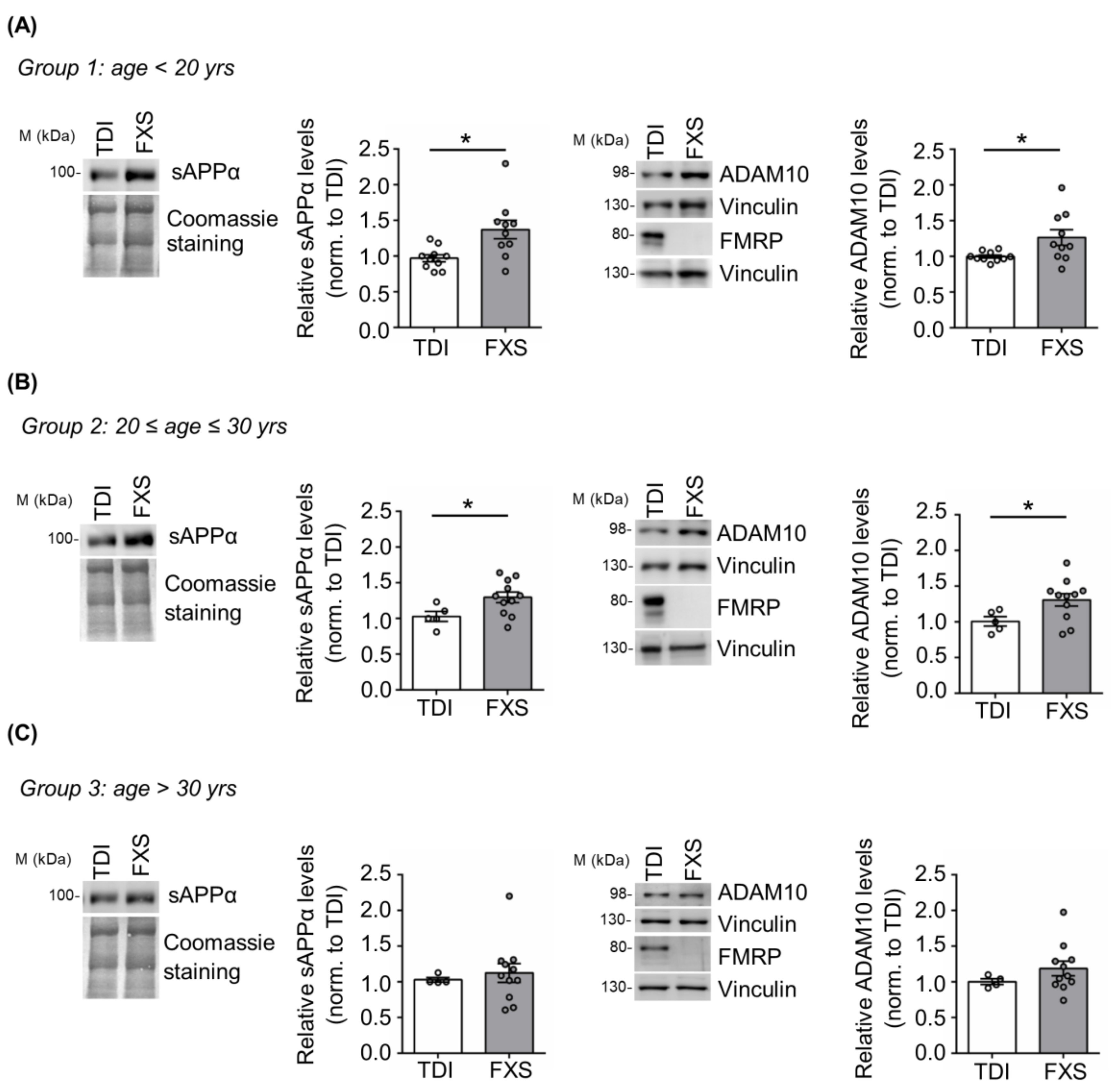
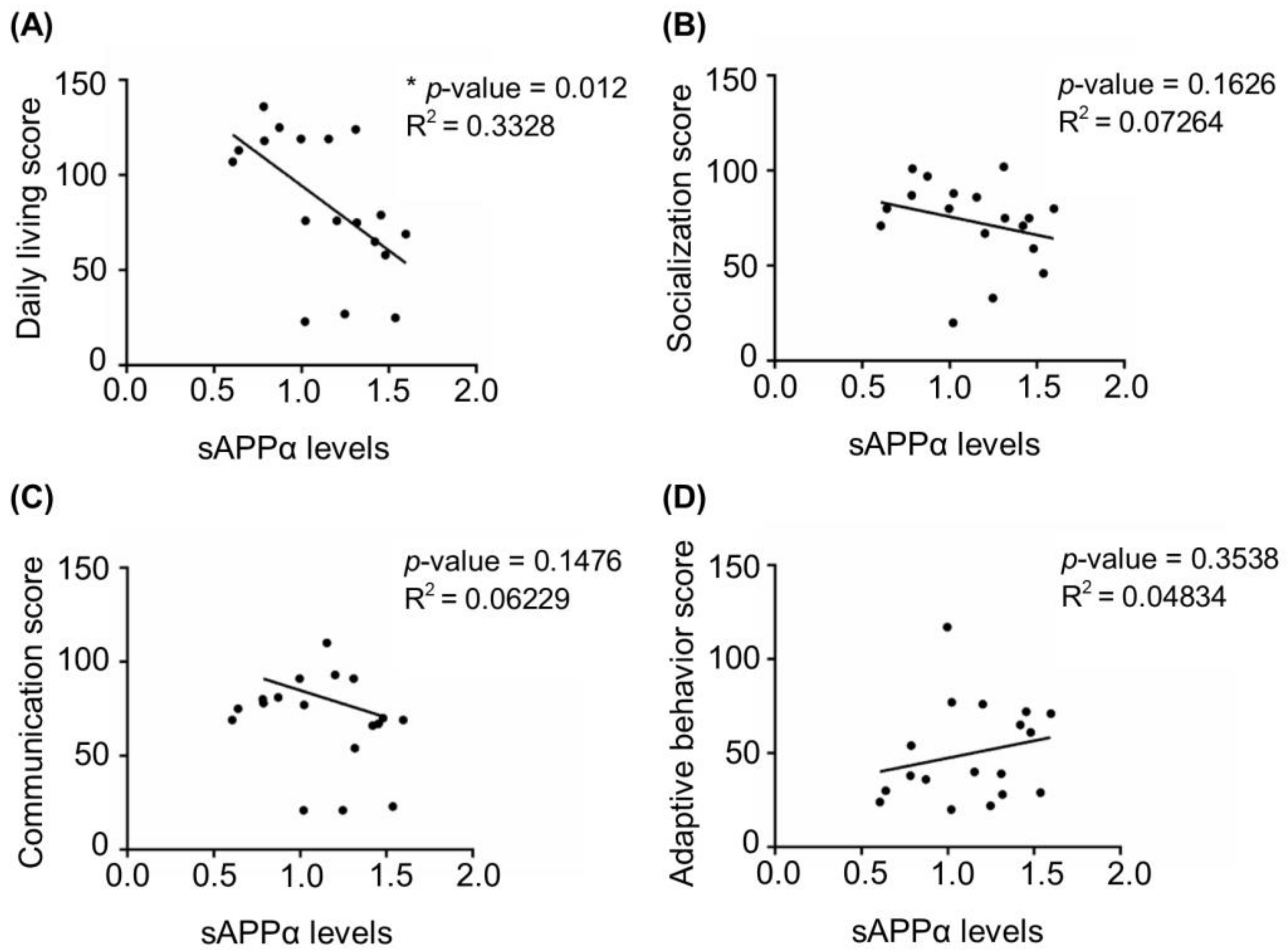
3.1. Age-Dependent Dysregulation of APP Processing in Human FXS Fibroblasts
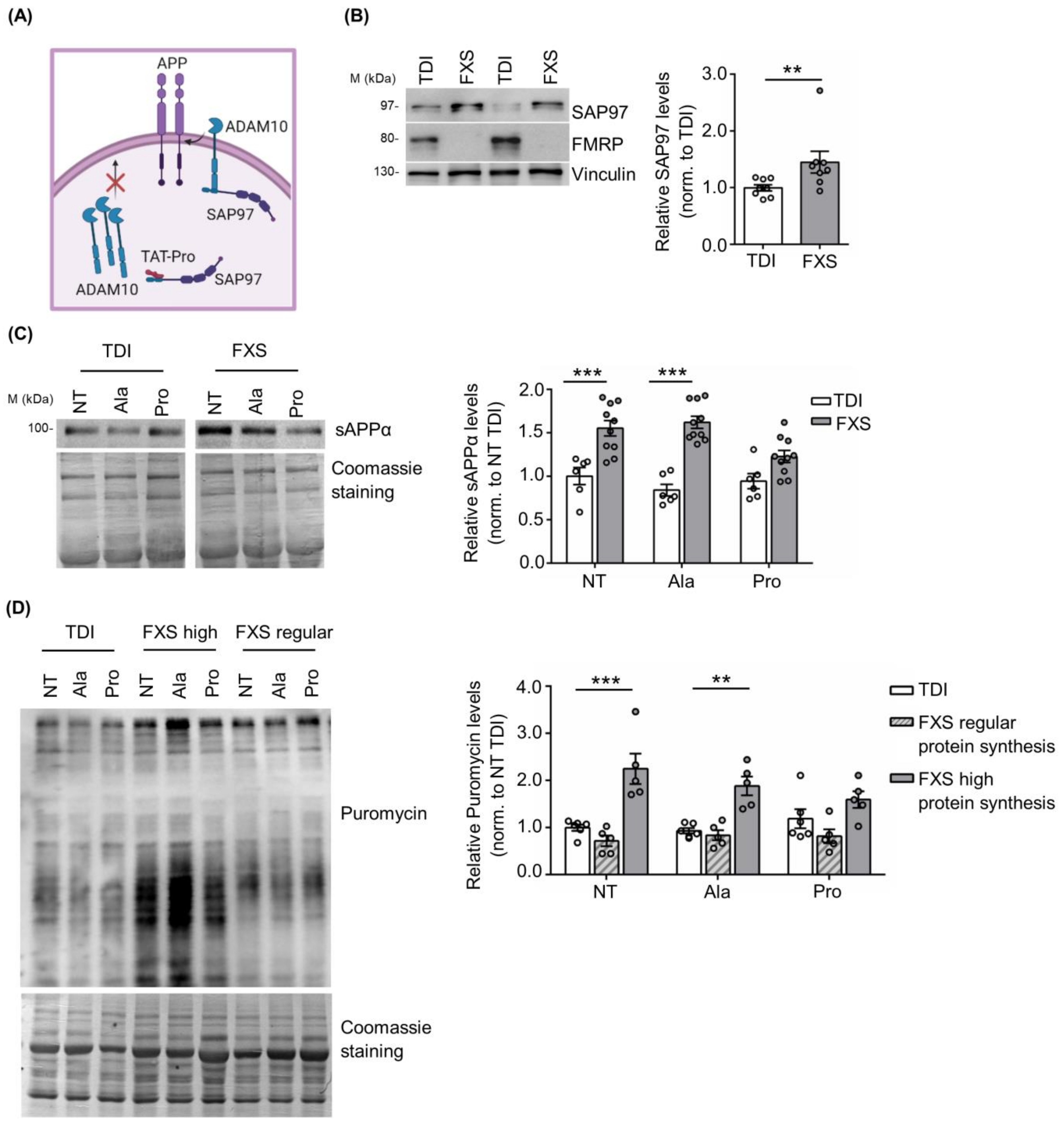
3.2. Peptide-Based Cellular Therapy Reduces sAPPα Release and Exaggerated Protein Synthesis
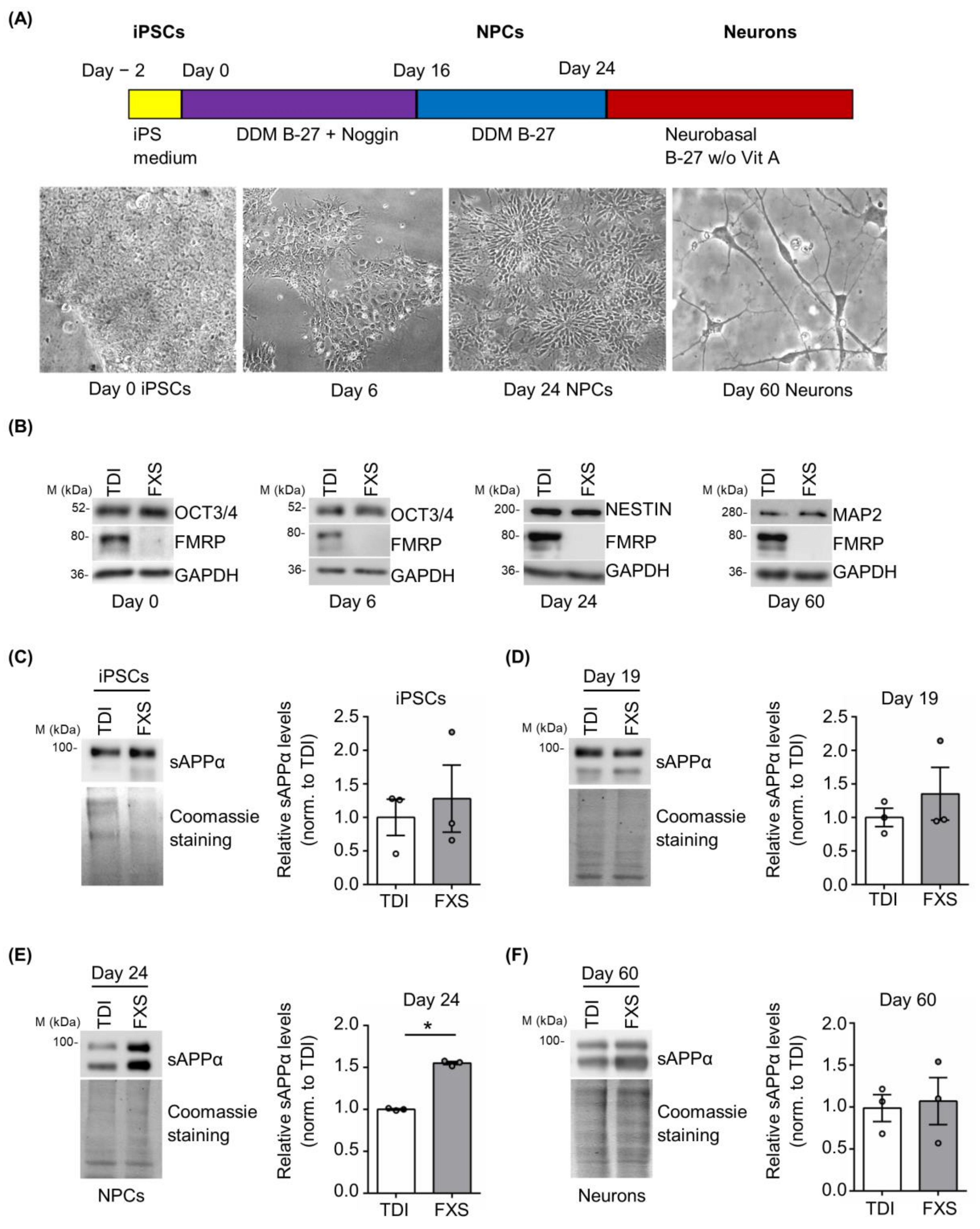
3.3. Excess of sAPPα Is Detected in Differentiating FXS iPSCs and Forebrain Organoids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bagni, C.; Tassone, F.; Neri, G.; Hagerman, R. Fragile X syndrome: Causes, diagnosis, mechanisms, and therapeutics. J. Clin. Investig. 2012, 122, 4314–4322. [Google Scholar] [CrossRef]
- Bagni, C.; Zukin, R.S. A Synaptic Perspective of Fragile X Syndrome and Autism Spectrum Disorders. Neuron 2019, 101, 1070–1088. [Google Scholar] [CrossRef]
- Roberts, J.E.; Ezell, J.E.; Fairchild, A.J.; Klusek, J.; Thurman, A.J.; McDuffie, A.; Abbeduto, L. Biobehavioral composite of social aspects of anxiety in young adults with fragile X syndrome contrasted to autism spectrum disorder. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2018, 177, 665–675. [Google Scholar] [CrossRef]
- Verdura, E.; Pérez-Cano, L.; Sabido-Vera, R.; Guney, E.; Hyvelin, J.-M.; Durham, L.; Gomez-Mancilla, B. Heterogeneity in Fragile X Syndrome Highlights the Need for Precision Medicine-Based Treatments. Front. Psychiatry 2021, 12, 722378. [Google Scholar] [CrossRef]
- Protic, D.D.; Aishworiya, R.; Salcedo-Arellano, M.J.; Tang, S.J.; Milisavljevic, J.; Mitrovic, F.; Hagerman, R.J.; Budimirovic, D.B. Fragile X Syndrome: From Molecular Aspect to Clinical Treatment. Int. J. Mol. Sci. 2022, 23, 1935. [Google Scholar] [CrossRef]
- Hagerman, R.J.; Berry-Kravis, E.; Hazlett, H.C.; Bailey, D.B.; Moine, H.; Kooy, R.F.; Tassone, F.; Gantois, I.; Sonenberg, N.; Mandel, J.L.; et al. Fragile X syndrome. Nat. Rev. Dis. Prim. 2017, 3, 17065. [Google Scholar] [CrossRef]
- Cregenzán-Royo, O.; Brun-Gasca, C.; Fornieles-Deu, A. Behavior Problems and Social Competence in Fragile X Syndrome: A Systematic Review. Genes 2022, 13, 280. [Google Scholar] [CrossRef]
- Kaufmann, W.E.; Kidd, S.A.; Andrews, H.F.; Budimirovic, D.B.; Esler, A.; Haas-Givler, B.; Stackhouse, T.; Riley, C.; Peacock, G.; Sherman, S.L.; et al. Autism Spectrum Disorder in Fragile X Syndrome: Cooccurring Conditions and Current Treatment. Pediatrics 2017, 139, S194–S206. [Google Scholar] [CrossRef]
- Salcedo-Arellano, M.J.; Dufour, B.; McLennan, Y.; Martinez-Cerdeno, V.; Hagerman, R. Fragile X syndrome and associated disorders: Clinical aspects and pathology. Neurobiol. Dis. 2020, 136, 104740. [Google Scholar] [CrossRef]
- Klusek, J.; O’Connor, S.L.; Hickey, A.; Hills, K.J.; Abbeduto, L.; Roberts, J.E. Attention/Deficit Hyperactivity Disorder in Adolescent and Young Adult Males With Fragile X Syndrome. Am. J. Intellect. Dev. Disabil. 2022, 127, 213–230. [Google Scholar] [CrossRef]
- Davis, J.K.; Broadie, K. Multifarious Functions of the Fragile X Mental Retardation Protein. Trends Genet. 2017, 33, 703–714. [Google Scholar] [CrossRef]
- Pasciuto, E.; Bagni, C. SnapShot: FMRP mRNA Targets and Diseases. Cell 2014, 158, 1446.e1. [Google Scholar] [CrossRef]
- Richter, J.D.; Zhao, X. The molecular biology of FMRP: New insights into fragile X syndrome. Nat. Rev. Neurosci. 2021, 22, 209–222. [Google Scholar] [CrossRef]
- Banerjee, A.; Ifrim, M.F.; Valdez, A.N.; Raj, N.; Bassell, G.J. Aberrant RNA translation in fragile X syndrome: From FMRP mechanisms to emerging therapeutic strategies. Brain Res. 2018, 1693, 24–36. [Google Scholar] [CrossRef]
- Kurosaki, T.; Mitsutomi, S.; Hewko, A.; Akimitsu, N.; Maquat, L.E. Integrative omics indicate FMRP sequesters mRNA from translation and deadenylation in human neuronal cells. Mol. Cell 2022, 82, 4564–4581.e11. [Google Scholar] [CrossRef]
- Sawicka, K.; Hale, C.R.; Park, C.Y.; Fak, J.J.; Gresack, J.E.; Van Driesche, S.J.; Kang, J.J.; Darnell, J.C.; Darnell, R.B. FMRP has a cell-type-specific role in CA1 pyramidal neurons to regulate autism-related transcripts and circadian memory. Elife 2019, 8, e46919. [Google Scholar] [CrossRef]
- Maurin, T.; Lebrigand, K.; Castagnola, S.; Paquet, A.; Jarjat, M.; Popa, A.; Grossi, M.; Rage, F.; Bardoni, B. HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic Acids Res. 2018, 46, 6344–6355. [Google Scholar] [CrossRef]
- Westmark, C.J.; Malter, J.S. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLoS Biol. 2007, 5, e52. [Google Scholar] [CrossRef]
- Lee, E.K.; Kim, H.H.; Kuwano, Y.; Abdelmohsen, K.; Srikantan, S.; Subaran, S.S.; Gleichmann, M.; Mughal, M.R.; Martindale, J.L.; Yang, X.; et al. HnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat. Struct. Mol. Biol. 2010, 17, 732–739. [Google Scholar] [CrossRef]
- Pasciuto, E.; Ahmed, T.; Wahle, T.; Gardoni, F.; D’Andrea, L.; Pacini, L.; Jacquemont, S.; Tassone, F.; Balschun, D.; Dotti, C.G.; et al. Dysregulated ADAM10-Mediated Processing of APP during a Critical Time Window Leads to Synaptic Deficits in Fragile X Syndrome. Neuron 2015, 87, 382–399. [Google Scholar] [CrossRef]
- Zhang, T.; Pang, P.; Fang, Z.; Guo, Y.; Li, H.; Li, X.; Tian, T.; Yang, X.; Chen, W.; Shu, S.; et al. Expression of BC1 Impairs Spatial Learning and Memory in Alzheimer’s Disease Via APP Translation. Mol. Neurobiol. 2018, 55, 6007–6020. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gu, B.J.; Masters, C.L.; Wang, Y.J. A systemic view of Alzheimer disease–insights from amyloid-β metabolism beyond the brain. Nat. Rev. Neurol. 2017, 13, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Delport, A.; Hewer, R. The amyloid precursor protein: A converging point in Alzheimer’s disease. Mol. Neurobiol. 2022, 59, 4501–4516. [Google Scholar] [CrossRef] [PubMed]
- Deyts, C.; Thinakaran, G.; Parent, A.T. APP Receptor? To Be or Not to Be. Trends Pharmacol. Sci. 2016, 37, 390–411. [Google Scholar] [CrossRef]
- Müller, U.C.; Deller, T.; Korte, M. Not just amyloid: Physiological functions of the amyloid precursor protein family. Nat. Rev. Neurosci. 2017, 18, 281–298. [Google Scholar] [CrossRef]
- Steubler, V.; Erdinger, S.; Back, M.K.; Ludewig, S.; Fässler, D.; Richter, M.; Han, K.; Slomianka, L.; Amrein, I.; von Engelhardt, J.; et al. Loss of all three APP family members during development impairs synaptic function and plasticity, disrupts learning, and causes an autism-like phenotype. EMBO J. 2021, 40, e107471. [Google Scholar] [CrossRef]
- Lazarov, O.; Demars, M.P. All in the Family: How the APPs Regulate Neurogenesis. Front. Neurosci. 2012, 6, 81. [Google Scholar] [CrossRef]
- Cho, Y.; Bae, H.G.; Okun, E.; Arumugam, T.V.; Jo, D.G. Physiology and pharmacology of amyloid precursor protein. Pharmacol. Ther. 2022, 235, 108122. [Google Scholar] [CrossRef]
- Southam, K.A.; Stennard, F.; Pavez, C.; Small, D.H. Knockout of Amyloid β Protein Precursor (APP) Expression Alters Synaptogenesis, Neurite Branching and Axonal Morphology of Hippocampal Neurons. Neurochem. Res. 2019, 44, 1346–1355. [Google Scholar] [CrossRef]
- Corbett, N.J.; Hooper, N.M. Soluble amyloid precursor protein α: Friend or foe? Adv. Exp. Med. Biol. 2018, 1112, 177–183. [Google Scholar] [CrossRef]
- Jorissen, E.; Prox, J.; Bernreuther, C.; Weber, S.; Schwanbeck, R.; Serneels, L.; Snellinx, A.; Craessaerts, K.; Thathiah, A.; Tesseur, I.; et al. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J. Neurosci. 2010, 30, 4833–4844. [Google Scholar] [CrossRef]
- Kuhn, P.H.; Wang, H.; Dislich, B.; Colombo, A.; Zeitschel, U.; Ellwart, J.W.; Kremmer, E.; Rossner, S.; Lichtenthaler, S.F. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010, 29, 3020–3032. [Google Scholar] [CrossRef] [PubMed]
- Musardo, S.; Marcello, E.; Gardoni, F.; Di Luca, M. ADAM10 in synaptic physiology and pathology. Neurodegener. Dis. 2014, 13, 72–74. [Google Scholar] [CrossRef] [PubMed]
- Dar, N.J.; Glazner, G.W. Deciphering the neuroprotective and neurogenic potential of soluble amyloid precursor protein alpha (sAPPα). Cell. Mol. Life Sci. 2020, 77, 2315–2330. [Google Scholar] [CrossRef]
- Coronel, R.; Palmer, C.; Bernabeu-Zornoza, A.; Monteagudo, M.; Rosca, A.; Zambrano, A.; Liste, I. Physiological effects of amyloid precursor protein and its derivatives on neural stem cell biology and signaling pathways involved. Neural Regen. Res. 2019, 14, 1661–1671. [Google Scholar] [CrossRef] [PubMed]
- Westmark, C.J. Fragile X and APP: A Decade in Review, a Vision for the Future. Mol. Neurobiol. 2019, 56, 3904–3921. [Google Scholar] [CrossRef] [PubMed]
- Westmark, C.J.; Sokol, D.K.; Maloney, B.; Lahiri, D.K. Novel roles of amyloid-beta precursor protein metabolites in fragile X syndrome and autism. Mol. Psychiatry 2016, 21, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Ray, B.; Sokol, D.K.; Maloney, B.; Lahiri, D.K. Finding novel distinctions between the sAPPα-mediated anabolic biochemical pathways in Autism Spectrum Disorder and Fragile X Syndrome plasma and brain tissue. Sci. Rep. 2016, 6, 26052. [Google Scholar] [CrossRef]
- Lahiri, D.K.; Sokol, D.K.; Erickson, C.; Ray, B.; Ho, C.Y.; Maloney, B. Autism as early neurodevelopmental disorder: Evidence for an sAPPα-mediated anabolic pathway. Front. Cell. Neurosci. 2013, 7, 94. [Google Scholar] [CrossRef]
- Westmark, C.J.; Westmark, P.R.; O’Riordan, K.J.; Ray, B.C.; Hervey, C.M.; Salamat, M.S.; Abozeid, S.H.; Stein, K.M.; Stodola, L.A.; Tranfaglia, M.; et al. Reversal of fragile X phenotypes by manipulation of AβPP/Aβ levels in Fmr1KO mice. PLoS ONE 2011, 6, e26549. [Google Scholar] [CrossRef]
- Westmark, C.J.; Chuang, S.-C.; Hays, S.A.; Filon, M.J.; Ray, B.C.; Westmark, P.R.; Gibson, J.R.; Huber, K.M.; Wong, R.K.S. APP Causes Hyperexcitability in Fragile X Mice. Front. Mol. Neurosci. 2016, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Jacquemont, S.; Pacini, L.; Jønch, A.E.; Cencelli, G.; Rozenberg, I.; He, Y.; D’Andrea, L.; Pedini, G.; Eldeeb, M.; Willemsen, R.; et al. Protein synthesis levels are increased in a subset of individuals with fragile X syndrome. Hum. Mol. Genet. 2018, 27, 2039–2051. [Google Scholar] [CrossRef] [PubMed]
- Berry-Kravis, E.M.; Lindemann, L.; Jønch, A.E.; Apostol, G.; Bear, M.F.; Carpenter, R.L.; Crawley, J.N.; Curie, A.; Des Portes, V.; Hossain, F.; et al. Drug development for neurodevelopmental disorders: Lessons learned from fragile X syndrome. Nat. Rev. Drug Discov. 2017, 4, 280–299. [Google Scholar] [CrossRef]
- Berry-Kravis, E. Fragile X Syndrome: Supportive Treatment, Unmet Needs, and Paths to Novel Interventions and Disease-Targeted Therapies. Am. J. Intellect. Dev. Disabil. 2022, 127, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Brick, D.J.; Nethercott, H.E.; Montesano, S.; Banuelos, M.G.; Stover, A.E.; Schutte, S.S.; O’Dowd, D.K.; Hagerman, R.J.; Ono, M.; Hessl, D.R.; et al. The Autism Spectrum Disorders Stem Cell Resource at Children’s Hospital of Orange County: Implications for Disease Modeling and Drug Discovery. Stem Cells Transl. Med. 2014, 3, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Zhou, Y.; Li, Y.; Han, Y.; Xu, J.; Niu, W.; Li, Z.; Liu, S.; Feng, H.; Huang, W.; et al. A human forebrain organoid model of fragile X syndrome exhibits altered neurogenesis and highlights new treatment strategies. Nat. Neurosci. 2021, 24, 1377–1391. [Google Scholar] [CrossRef] [PubMed]
- Espuny-Camacho, I.; Michelsen, K.A.; Gall, D.; Linaro, D.; Hasche, A.; Bonnefont, J.; Bali, C.; Orduz, D.; Bilheu, A.; Herpoel, A.; et al. Pyramidal Neurons Derived from Human Pluripotent Stem Cells Integrate Efficiently into Mouse Brain Circuits In Vivo. Neuron 2013, 77, 440–456. [Google Scholar] [CrossRef]
- Gaspard, N.; Bouschet, T.; Herpoel, A.; Naeije, G.; van den Ameele, J.; Vanderhaeghen, P. Generation of cortical neurons from mouse embryonic stem cells. Nat. Protoc. 2009, 4, 1454–1463. [Google Scholar] [CrossRef]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef]
- Schmidt, E.K.; Clavarino, G.; Ceppi, M.; Pierre, P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 2009, 6, 275–277. [Google Scholar] [CrossRef]
- Wingfield, P. Protein precipitation using ammonium sulfate. Curr. Protoc. Protein Sci. 2001, 3, psa03fs13. [Google Scholar] [CrossRef]
- Pedini, G.; Buccarelli, M.; Bianchi, F.; Pacini, L.; Cencelli, G.; D’Alessandris, Q.G.; Martini, M.; Giannetti, S.; Sasso, F.; Melocchi, V.; et al. FMRP modulates the Wnt signalling pathway in glioblastoma. Cell Death Dis. 2022, 13, 719. [Google Scholar] [CrossRef] [PubMed]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- Epis, R.; Marcello, E.; Gardoni, F.; Vastagh, C.; Malinverno, M.; Balducci, C.; Colombo, A.; Borroni, B.; Vara, H.; Dell’Agli, M.; et al. Blocking ADAM10 synaptic trafficking generates a model of sporadic Alzheimer’s disease. Brain 2010, 133, 3323–3335. [Google Scholar] [CrossRef]
- Marcello, E.; Gardoni, F.; Mauceri, D.; Romorini, S.; Jeromin, A.; Epis, R.; Borroni, B.; Cattabeni, F.; Sala, C.; Padovani, A.; et al. Synapse-associated protein-97 mediates alpha-secretase ADAM10 trafficking and promotes its activity. J. Neurosci. 2007, 27, 1682–1691. [Google Scholar] [CrossRef]
- Vezzoli, E.; Caron, I.; Talpo, F.; Besusso, D.; Conforti, P.; Battaglia, E.; Sogne, E.; Falqui, A.; Petricca, L.; Verani, M.; et al. Inhibiting pathologically active ADAM10 rescues synaptic and cognitive decline in Huntington’s disease. J. Clin. Investig. 2019, 129, 2390–2403. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, R.W.; Elder, M.K.; Barrett, M.C.; Westlake, C.M.; Peppercorn, K.; Tate, W.P.; Abraham, W.C.; Williams, J.M. Secreted Amyloid Precursor Protein-Alpha Promotes Arc Protein Synthesis in Hippocampal Neurons. Front. Mol. Neurosci. 2019, 12, 198. [Google Scholar] [CrossRef] [PubMed]
- Mockett, B.G.; Guévremont, D.; Elder, M.K.; Parfitt, K.D.; Peppercorn, K.; Morrissey, J.; Singh, A.; Hintz, T.J.; Kochen, L.; Tom Dieck, S.; et al. Glutamate Receptor Trafficking and Protein Synthesis Mediate the Facilitation of LTP by Secreted Amyloid Precursor Protein-Alpha. J. Neurosci. 2019, 39, 3188–3203. [Google Scholar] [CrossRef]
- Ryan, M.M.; Morris, G.P.; Mockett, B.G.; Bourne, K.; Abraham, W.C.; Tate, W.P.; Williams, J.M. Time-dependent changes in gene expression induced by secreted amyloid precursor protein-alpha in the rat hippocampus. BMC Genom. 2013, 14, 376. [Google Scholar] [CrossRef]
- Claasen, A.M.; Guévremont, D.; Mason-Parker, S.E.; Bourne, K.; Tate, W.P.; Abraham, W.C.; Williams, J.M. Secreted amyloid precursor protein-alpha upregulates synaptic protein synthesis by a protein kinase G-dependent mechanism. Neurosci. Lett. 2009, 460, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Peppercorn, K.; Kleffmann, T.; Jones, O.; Hughes, S.; Tate, W. Secreted Amyloid Precursor Protein Alpha, a Neuroprotective Protein in the Brain Has Widespread Effects on the Transcriptome and Proteome of Human Inducible Pluripotent Stem Cell-Derived Glutamatergic Neurons Related to Memory Mechanisms. Front. Neurosci. 2022, 16, 858524. [Google Scholar] [CrossRef]
- Kumari, D.; Bhattacharya, A.; Nadel, J.; Moulton, K.; Zeak, N.M.; Glicksman, A.; Dobkin, C.; Brick, D.J.; Schwartz, P.H.; Smith, C.B.; et al. Identification of fragile X syndrome specific molecular markers in human fibroblasts: A useful model to test the efficacy of therapeutic drugs. Hum. Mutat. 2014, 35, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- Fleischer, J.G.; Schulte, R.; Tsai, H.H.; Tyagi, S.; Ibarra, A.; Shokhirev, M.N.; Huang, L.; Hetzer, M.W.; Navlakha, S. Predicting age from the transcriptome of human dermal fibroblasts. Genome Biol. 2018, 19, 221. [Google Scholar] [CrossRef]
- Rorteau, J.; Chevalier, F.P.; Bonnet, S.; Barthélemy, T.; Lopez-Gaydon, A.; Martin, L.S.; Bechetoille, N.; Lamartine, J. Maintenance of Chronological Aging Features in Culture of Normal Human Dermal Fibroblasts from Old Donors. Cells 2022, 11, 858. [Google Scholar] [CrossRef]
- Ivanov, N.A.; Tao, R.; Chenoweth, J.G.; Brandtjen, A.; Mighdoll, M.I.; Genova, J.D.; McKay, R.D.; Jia, Y.; Weinberger, D.R.; Kleinman, J.E.; et al. Strong Components of Epigenetic Memory in Cultured Human Fibroblasts Related to Site of Origin and Donor Age. PLoS Genet. 2016, 12, e1005819. [Google Scholar] [CrossRef] [PubMed]
- Razak, K.A.; Dominick, K.C.; Erickson, C.A. Developmental studies in fragile X syndrome. J. Neurodev. Disord. 2020, 12, 13. [Google Scholar] [CrossRef]
- Wen, T.H.; Lovelace, J.W.; Ethell, I.M.; Binder, D.K.; Razak, K.A. Developmental Changes in EEG Phenotypes in a Mouse Model of Fragile X Syndrome. Neuroscience 2019, 398, 126–143. [Google Scholar] [CrossRef]
- Hoeft, F.; Carter, J.C.; Lightbody, A.A.; Cody Hazlett, H.; Piven, J.; Reiss, A.L. Region-specific alterations in brain development in one- to three-year-old boys with fragile X syndrome. Proc. Natl. Acad. Sci. USA 2010, 107, 9335–9339. [Google Scholar] [CrossRef]
- Quintin, E.-M.; Jo, B.; Hall, S.S.; Bruno, J.L.; Chromik, L.C.; Raman, M.M.; Lightbody, A.A.; Martin, A.; Reiss, A.L. The cognitive developmental profile associated with fragile X syndrome: A longitudinal investigation of cognitive strengths and weaknesses through childhood and adolescence. Dev. Psychopathol. 2016, 28, 1457–1469. [Google Scholar] [CrossRef]
- Roberts, J.; Crawford, H.; Hogan, A.L.; Fairchild, A.; Tonnsen, B.; Brewe, A.; O’Connor, S.; Roberts, D.A.; Abbeduto, L. Social Avoidance Emerges in Infancy and Persists into Adulthood in Fragile X Syndrome. J. Autism Dev. Disord. 2019, 49, 3753–3766. [Google Scholar] [CrossRef] [PubMed]
- Ellis, K.; Oliver, C.; Stefanidou, C.; Apperly, I.; Moss, J. An Observational Study of Social Interaction Skills and Behaviors in Cornelia de Lange, Fragile X and Rubinstein-Taybi Syndromes. J. Autism Dev. Disord. 2020, 50, 4001–4010. [Google Scholar] [CrossRef]
- Usher, L.V.; DaWalt, L.S.; Hong, J.; Greenberg, J.S.; Mailick, M.R. Trajectories of Change in the Behavioral and Health Phenotype of Adolescents and Adults with Fragile X Syndrome and Intellectual Disability: Longitudinal Trends Over a Decade. J. Autism Dev. Disord. 2020, 50, 2779–2792. [Google Scholar] [CrossRef]
- Raspa, M.; Franco, V.; Bishop, E.; Wheeler, A.C.; Wylie, A.; Bailey, D.B.J. A comparison of functional academic and daily living skills in males with fragile X syndrome with and without autism. Res. Dev. Disabil. 2018, 78, 1–14. [Google Scholar] [CrossRef]
- Mockett, B.G.; Richter, M.; Abraham, W.C.; Müller, U.C. Therapeutic Potential of Secreted Amyloid Precursor Protein APPsα. Front. Mol. Neurosci. 2017, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Hick, M.; Herrmann, U.; Weyer, S.W.; Mallm, J.P.; Tschäpe, J.A.; Borgers, M.; Mercken, M.; Roth, F.C.; Draguhn, A.; Slomianka, L.; et al. Acute function of secreted amyloid precursor protein fragment APPsα in synaptic plasticity. Acta Neuropathol. 2015, 129, 21–37. [Google Scholar] [CrossRef] [PubMed]
- McLane, R.D.; Schmitt, L.M.; Pedapati, E.V.; Shaffer, R.C.; Dominick, K.C.; Horn, P.S.; Gross, C.; Erickson, C.A. Peripheral Amyloid Precursor Protein Derivative Expression in Fragile X Syndrome. Front. Integr. Neurosci. 2019, 13, 49. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.R.; Giunta, B.N.; Obregon, D.; Nikolic, W.V.; Tian, J.; Sanberg, C.D.; Sutton, D.T.; Tan, J. Peripheral biomarkers in Autism: Secreted amyloid precursor protein-alpha as a probable key player in early diagnosis. Int. J. Clin. Exp. Med. 2008, 1, 338–344. [Google Scholar]
- Ray, B.; Long, J.M.; Sokol, D.K.; Lahiri, D.K. Increased secreted amyloid precursor protein-α (sAPPα) in severe autism: Proposal of a specific, anabolic pathway and putative biomarker. PLoS ONE 2011, 6, e20405. [Google Scholar] [CrossRef]
- Sokol, D.K.; Chen, D.; Farlow, M.R.; Dunn, D.W.; Maloney, B.; Zimmer, J.A.; Lahiri, D.K. High levels of Alzheimer beta-amyloid precursor protein (APP) in children with severely autistic behavior and aggression. J. Child Neurol. 2006, 21, 444–449. [Google Scholar] [CrossRef]
- Li, X.; Zhou, P.; Li, Q.; Peng, B.; Cun, Y.; Dai, Y.; Wei, H.; Liu, X.; Yu, Y.; Jiang, Z.; et al. Regressive Autism Spectrum Disorder: High Levels of Total Secreted Amyloid Precursor Protein and Secreted Amyloid Precursor Protein-α in Plasma. Front. Psychiatry 2022, 13, 809543. [Google Scholar] [CrossRef]
- Sokol, D.K.; Maloney, B.; Westmark, C.J.; Lahiri, D.K. Novel Contribution of Secreted Amyloid-β Precursor Protein to White Matter Brain Enlargement in Autism Spectrum Disorder. Front. Psychiatry 2019, 10, 165. [Google Scholar] [CrossRef]
- Erickson, C.A.; Wink, L.K.; Baindu, B.; Ray, B.; Schaefer, T.L.; Pedapati, E.V.; Lahiri, D.K. Analysis of peripheral amyloid precursor protein in Angelman Syndrome. Am. J. Med. Genet. A 2016, 170, 2334–2337. [Google Scholar] [CrossRef]
- Ardhanareeswaran, K.; Mariani, J.; Coppola, G.; Abyzov, A.; Vaccarino, F.M. Human induced pluripotent stem cells for modelling neurodevelopmental disorders. Nat. Rev. Neurol. 2017, 13, 265–278. [Google Scholar] [CrossRef]
- Linda, K.; Fiuza, C.; Nadif Kasri, N. The promise of induced pluripotent stem cells for neurodevelopmental disorders. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 84, 382–391. [Google Scholar] [CrossRef]
- Sharma, A.; Sances, S.; Workman, M.J.; Svendsen, C.N. Multi-lineage Human iPSC-Derived Platforms for Disease Modeling and Drug Discovery. Cell Stem Cell 2020, 26, 309–329. [Google Scholar] [CrossRef]
- Utami, K.H.; Skotte, N.H.; Colaço, A.R.; Yusof, N.A.B.M.; Sim, B.; Yeo, X.Y.; Bae, H.-G.; Garcia-Miralles, M.; Radulescu, C.I.; Chen, Q.; et al. Integrative Analysis Identifies Key Molecular Signatures Underlying Neurodevelopmental Deficits in Fragile X Syndrome. Biol. Psychiatry 2020, 88, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Brighi, C.; Salaris, F.; Soloperto, A.; Cordella, F.; Ghirga, S.; de Turris, V.; Rosito, M.; Porceddu, P.F.; D’Antoni, C.; Reggiani, A.; et al. Novel fragile X syndrome 2D and 3D brain models based on human isogenic FMRP-KO iPSCs. Cell Death Dis. 2021, 12, 498. [Google Scholar] [CrossRef]
- Zhang, A.; Sokolova, I.; Domissy, A.; Davis, J.; Rao, L.; Hana Utami, K.; Wang, Y.; Hagerman, R.J.; Pouladi, M.A.; Sanna, P.; et al. Maturation Delay of Human GABAergic Neurogenesis in Fragile X Syndrome Pluripotent Stem Cells. Stem Cells Transl. Med. 2022, 11, 613–629. [Google Scholar] [CrossRef] [PubMed]
- Utami, K.H.; Yusof, N.A.B.M.; Kwa, J.E.; Peteri, U.-K.; Castrén, M.L.; Pouladi, M.A. Elevated de novo protein synthesis in FMRP-deficient human neurons and its correction by metformin treatment. Mol. Autism 2020, 11, 41. [Google Scholar] [CrossRef]
- Lee, A.; Xu, J.; Wen, Z.; Jin, P. Across Dimensions: Developing 2D and 3D Human iPSC-Based Models of Fragile X Syndrome. Cells 2022, 11, 1725. [Google Scholar] [CrossRef] [PubMed]
- Telias, M.; Mayshar, Y.; Amit, A.; Ben-Yosef, D. Molecular mechanisms regulating impaired neurogenesis of Fragile X syndrome human embryonic stem cells. Stem Cells Dev. 2015, 24, 2353–2365. [Google Scholar] [CrossRef] [PubMed]
- Graef, J.D.; Wu, H.; Ng, C.; Sun, C.; Villegas, V.; Qadir, D.; Jesseman, K.; Warren, S.T.; Jaenisch, R.; Cacace, A.; et al. Partial FMRP expression is sufficient to normalize neuronal hyperactivity in Fragile X neurons. Eur. J. Neurosci. 2020, 51, 2143–2157. [Google Scholar] [CrossRef] [PubMed]
- Das Sharma, S.; Pal, R.; Reddy, B.K.; Selvaraj, B.T.; Raj, N.; Samaga, K.K.; Srinivasan, D.J.; Ornelas, L.; Sareen, D.; Livesey, M.R.; et al. Cortical neurons derived from human pluripotent stem cells lacking FMRP display altered spontaneous firing patterns. Mol. Autism 2020, 11, 52. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, S.D.; Theriault, K.M.; Reis, S.A.; Zhou, F.; Madison, J.M.; Daheron, L.; Loring, J.F.; Haggarty, S.J. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS ONE 2011, 6, e26203. [Google Scholar] [CrossRef]
- Doers, M.E.; Musser, M.T.; Nichol, R.; Berndt, E.R.; Baker, M.; Gomez, T.M.; Zhang, S.C.; Abbeduto, L.; Bhattacharyya, A. IPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev. 2014, 23, 1777–1787. [Google Scholar] [CrossRef]
- Raj, N.; McEachin, Z.T.; Harousseau, W.; Zhou, Y.; Zhang, F.; Merritt-Garza, M.E.; Taliaferro, J.M.; Kalinowska, M.; Marro, S.G.; Hales, C.M.; et al. Cell-type-specific profiling of human cellular models of fragile X syndrome reveal PI3K-dependent defects in translation and neurogenesis. Cell Rep. 2021, 35, 108991. [Google Scholar] [CrossRef]
- Baratchi, S.; Evans, J.; Tate, W.P.; Abraham, W.C.; Connor, B. Secreted amyloid precursor proteins promote proliferation and glial differentiation of adult hippocampal neural progenitor cells. Hippocampus 2012, 22, 1517–1527. [Google Scholar] [CrossRef]
- Caillé, I.; Allinquant, B.; Dupont, E.; Bouillot, C.; Langer, A.; Müller, U.; Prochiantz, A. Soluble form of amyloid precursor protein regulates proliferation of progenitors in the adult subventricular zone. Development 2004, 131, 2173–2181. [Google Scholar] [CrossRef] [PubMed]
- Demars, M.P.; Bartholomew, A.; Strakova, Z.; Lazarov, O. Soluble amyloid precursor protein: A novel proliferation factor of adult progenitor cells of ectodermal and mesodermal origin. Stem Cell Res. Ther. 2011, 2, 36. [Google Scholar] [CrossRef]
- Ohline, S.M.; Chan, C.; Schoderboeck, L.; Wicky, H.E.; Tate, W.P.; Hughes, S.M.; Abraham, W.C. Effect of soluble amyloid precursor protein-alpha on adult hippocampal neurogenesis in a mouse model of Alzheimer’s disease. Mol. Brain 2022, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Bergström, P.; Agholme, L.; Nazir, F.H.; Satir, T.M.; Toombs, J.; Wellington, H.; Strandberg, J.; Bontell, T.O.; Kvartsberg, H.; Holmström, M.; et al. Amyloid precursor protein expression and processing are differentially regulated during cortical neuron differentiation. Sci. Rep. 2016, 6, 29200. [Google Scholar] [CrossRef]
- Berry-Kravis, E.; Filipink, R.A.; Frye, R.E.; Golla, S.; Morris, S.M.; Andrews, H.; Choo, T.-H.; Kaufmann, W.E. Seizures in Fragile X Syndrome: Associations and Longitudinal Analysis of a Large Clinic-Based Cohort. Front. Pediatr. 2021, 9, 736255. [Google Scholar] [CrossRef] [PubMed]
- Buckley, A.W.; Holmes, G.L. Epilepsy and Autism. Cold Spring Harb. Perspect. Med. 2016, 6, a022749. [Google Scholar] [CrossRef]
- Kwon, C.-S.; Wirrell, E.C.; Jetté, N. Autism Spectrum Disorder and Epilepsy. Neurol. Clin. 2022, 40, 831–847. [Google Scholar] [CrossRef] [PubMed]
- Leung, H.T.T.; Ring, H. Epilepsy in four genetically determined syndromes of intellectual disability. J. Intellect. Disabil. Res. 2013, 57, 3–20. [Google Scholar] [CrossRef]
- Samanta, D. Epilepsy in Angelman syndrome: A scoping review. Brain Dev. 2021, 43, 32–44. [Google Scholar] [CrossRef]
- Richter, M.C.; Ludewig, S.; Winschel, A.; Abel, T.; Bold, C.; Salzburger, L.R.; Klein, S.; Han, K.; Weyer, S.W.; Fritz, A.; et al. Distinct in vivo roles of secreted APP ectodomain variants APP sα and APP sβ in regulation of spine density, synaptic plasticity, and cognition. EMBO J. 2018, 37, e98335. [Google Scholar] [CrossRef] [PubMed]
- Rice, H.C.; De Malmazet, D.; Schreurs, A.; Frere, S.; Van Molle, I.; Volkov, A.N.; Creemers, E.; Vertkin, I.; Nys, J.; Ranaivoson, F.M.; et al. Secreted amyloid-b precursor protein functions as a GABA B R1a ligand to modulate synaptic transmission. Science 2019, 363, 6423. [Google Scholar] [CrossRef]
- Yang, H.; Mei, J.; Xu, W.; Ma, X.; Sun, B.; Ai, H. Identification of the probable structure of the sAPPα-GABA(B)R1a complex and theoretical solutions for such cases. Phys. Chem. Chem. Phys. 2022, 24, 12267–12280. [Google Scholar] [CrossRef]
- Haass, C.; Willem, M. Secreted APP Modulates Synaptic Activity: A Novel Target for Therapeutic Intervention? Neuron 2019, 101, 557–559. [Google Scholar] [CrossRef]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, G.; Brambilla, L.; Rossi, D. Peptides in clinical development for the treatment of brain tumors. Curr. Opin. Pharmacol. 2019, 47, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Ukidve, A.; Kim, J.; Mitragotri, S. Targeting Strategies for Tissue-Specific Drug Delivery. Cell 2020, 181, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Smith, Q.R.; Liu, X. Brain penetrating peptides and peptide-drug conjugates to overcome the blood-brain barrier and target CNS diseases. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2021, 13, e1695. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, P.; Ma, Z.; Lu, P.; Kebebe, D.; Liu, Z. Combination of cell-penetrating peptides with nanomaterials for the potential therapeutics of central nervous system disorders: A review. J. Nanobiotechnol. 2021, 19, 255. [Google Scholar] [CrossRef]
- Corti, S.; Faravelli, I.; Cardano, M.; Conti, L. Human pluripotent stem cells as tools for neurodegenerative and neurodevelopmental disease modeling and drug discovery. Expert Opin. Drug Discov. 2015, 10, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Engle, S.J.; Blaha, L.; Kleiman, R.J. Best Practices for Translational Disease Modeling Using Human iPSC-Derived Neurons. Neuron 2018, 100, 783–797. [Google Scholar] [CrossRef]
- Ernst, C. A roadmap for neurodevelopmental disease modeling for non-stem cell biologists. Stem Cells Transl. Med. 2020, 9, 567–574. [Google Scholar] [CrossRef]
- Berry-Kravis, E. Disease-Targeted Treatment Translation in Fragile X Syndrome as a Model for Neurodevelopmental Disorders. J. Child Neurol. 2022, 37, 797–812. [Google Scholar] [CrossRef]
- Richter, J.D.; Bassell, G.J.; Klann, E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat. Rev. Neurosci. 2015, 16, 595–605. [Google Scholar] [CrossRef]
- Brubaker, D.K.; Lauffenburger, D.A. Translating preclinical models to humans. Science 2020, 367, 742–743. [Google Scholar] [CrossRef] [PubMed]
- Prox, J.; Bernreuther, C.; Altmeppen, H.; Grendel, J.; Glatzel, M.; D’Hooge, R.; Stroobants, S.; Ahmed, T.; Balschun, D.; Willem, M.; et al. Postnatal disruption of the disintegrin/metalloproteinase ADAM10 in brain causes epileptic seizures, learning deficits, altered spine morphology, and defective synaptic functions. J. Neurosci. 2013, 33, 12915–12928. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cencelli, G.; Pacini, L.; De Luca, A.; Messia, I.; Gentile, A.; Kang, Y.; Nobile, V.; Tabolacci, E.; Jin, P.; Farace, M.G.; et al. Age-Dependent Dysregulation of APP in Neuronal and Skin Cells from Fragile X Individuals. Cells 2023, 12, 758. https://doi.org/10.3390/cells12050758
Cencelli G, Pacini L, De Luca A, Messia I, Gentile A, Kang Y, Nobile V, Tabolacci E, Jin P, Farace MG, et al. Age-Dependent Dysregulation of APP in Neuronal and Skin Cells from Fragile X Individuals. Cells. 2023; 12(5):758. https://doi.org/10.3390/cells12050758
Chicago/Turabian StyleCencelli, Giulia, Laura Pacini, Anastasia De Luca, Ilenia Messia, Antonietta Gentile, Yunhee Kang, Veronica Nobile, Elisabetta Tabolacci, Peng Jin, Maria Giulia Farace, and et al. 2023. "Age-Dependent Dysregulation of APP in Neuronal and Skin Cells from Fragile X Individuals" Cells 12, no. 5: 758. https://doi.org/10.3390/cells12050758
APA StyleCencelli, G., Pacini, L., De Luca, A., Messia, I., Gentile, A., Kang, Y., Nobile, V., Tabolacci, E., Jin, P., Farace, M. G., & Bagni, C. (2023). Age-Dependent Dysregulation of APP in Neuronal and Skin Cells from Fragile X Individuals. Cells, 12(5), 758. https://doi.org/10.3390/cells12050758






