Figure 1.
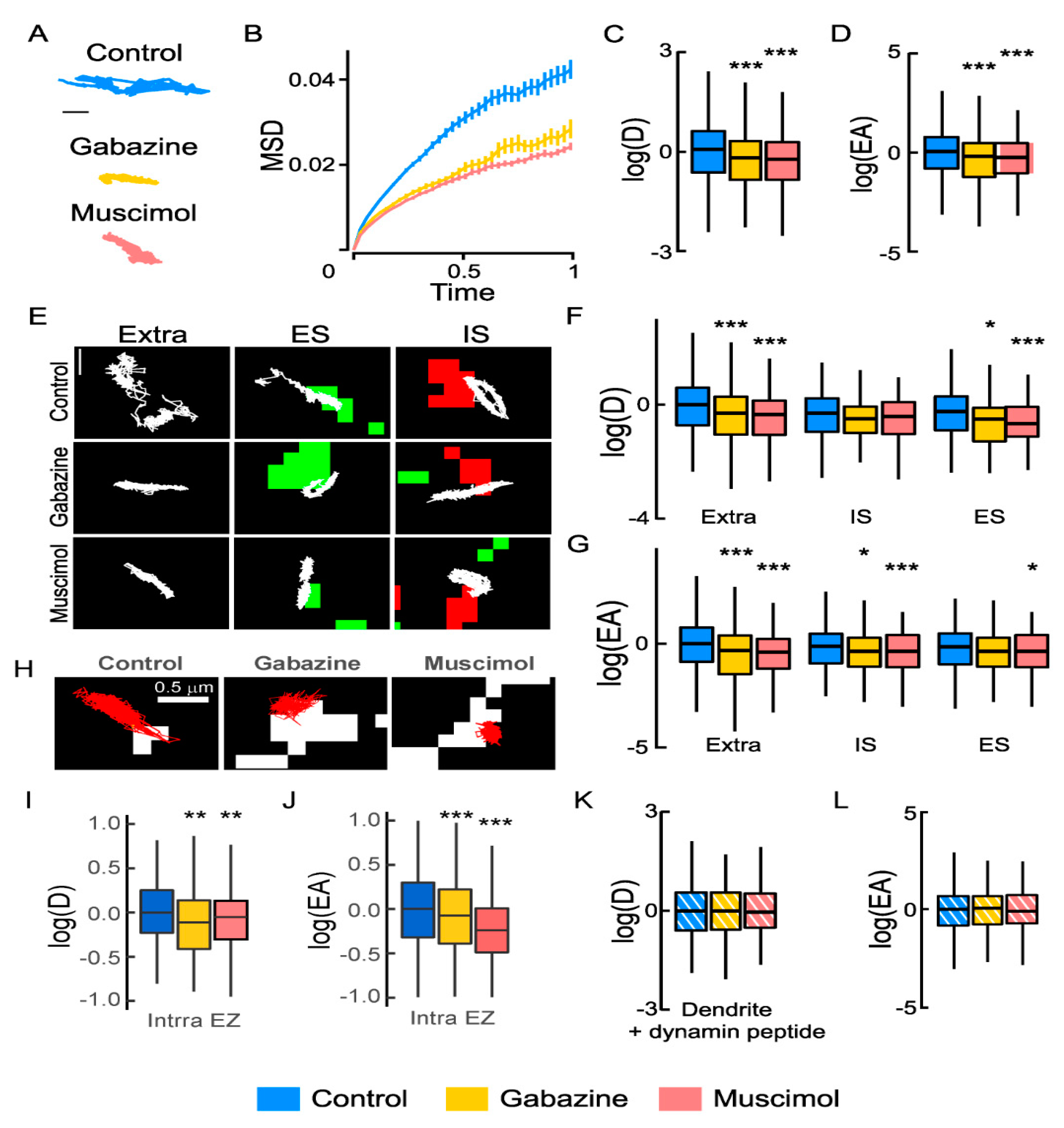
GABA type A receptor (GABAAR) activity regulates the Na+-K+-2Cl− cotransporter NKCC1 membrane dynamics: (A) Individual NKCC1 trajectories with reduced surface exploration upon gabazine or muscimol treatment. Scale bar, 0.5 µm. (B) Time-averaged mean square displacement (MSD) functions in control (blue) vs. gabazine (yellow) or muscimol (orange) conditions show increased confinement upon gabazine or muscimol application. (C,D) Boxplots of log diffusion coefficient (D) of NKCC1 in control condition (blue) or upon application of gabazine (yellow) or muscimol (orange) showing reduced diffusion upon drug treatments. N = 1558 quantum dots (QDs) (control, 41 cells), n = 387 QDs (gabazine, 18 cells), Welch t-test, p = 1.3 × 10−6, n = 545 QDs (muscimol, 27 cells), Welch t-test, p = 2.1 × 10−12, 5 cultures. (D) Median explored area EA in control vs. gabazine or muscimol conditions show increased confinement upon gabazine (Welch t-test, p = 2 × 10−8) or muscimol (Welch t-test, p = 4.2 × 10−11) treatment. (E) Trajectories (white) overlaid with clusters of homer1c-DsRed (green) or gephyrin-Finger-YFP (red) to identify extrasynaptic trajectories (extra), trajectories at excitatory (ES), and inhibitory synapses (IS). Scale bar, 0.4 µm. (F,G) Log(D) (F) and EA (G) of NKCC1 are decreased upon gabazine or muscimol application as compared with control condition. Note that the effect is more pronounced for extrasynaptic trajectories than for ES or IS trajectories. Diffusion coefficient (D): Extra, n = 899 QDs (control), 227 QDs (gabazine), p = 2.9 × 10−5, n = 268 QDs (muscimol) p = 2.2 × 10−9; IS, n = 244 QDs (control), 79 QDs (gabazine) p = 0.16; n = 142 QDs (muscimol) p = 0.19; ES, n = 415 QDs (control), n = 81 QDs (gabazine) p = 0.021, n = 135 QDs (muscimol) p = 0.00046. Explored area (EA): Extra, gabazine p = 2.9 × 10−5, muscimol p = 2.2 × 10−9; IS, gabazine p = 0.16, muscimol p = 0.19; ES, n = 415 QDs (control), n = 81 QDs (gabazine) p = 2.1 × 10−2, n = 135 QDs (muscimol) p = p = 4.6 × 10−4. (H) NKCC1 trajectories (red) in control vs. gabazine or muscimol conditions in relation to endocytic zones identified by clathrin-YFP clusters (white). Scale bar, 0.5 µm. (I,J) Reduced diffusion coefficient (I) and explored area (J) of NKCC1 within endocytic zones (Intra EZ) upon muscimol and gabazine treatment. Diffusion coefficient (D): Ctrl n = 117 QDs, 48 cells, Gbz n = 145 QDs, 64 cells, p = 1.4 × 10−3; Ctrl n = 273 QDs, 35 cells, Musc n = 247 QDs, 28 cells, p = 5.6 × 10−3, 3 cultures. Explored area (EA): Gbz, p = 5.46 × 10−5 and Musc, p = 2.2 × 10−16, 3 cultures. (K,L) No effect of gabazine or muscimol on log(D) (K) and EA (L) of dendritic NKCC1 in conditions of blockade of endocytosis (+ dynamin peptide). Diffusion coefficient (D): Bulk Ctrl n = 491 QDs, Gbz n = 238 QDs, p = 0.6 and Musc n = 243 QDs, p = p = 6.1 × 10−1, 5 cultures. Explored area (EA): Gbz, p = 0.84 and Musc, p = 8.1 × 10−1, 5 cultures. (B,F,I,K) D in µm2.s−1; C: MSD in µm2 vs. time(s); (D,G,J,L) EA in µm2. In all graphs, *, p < 5.0 × 10−2; **, p < 1.0 × 10−2; ***, p < 1.0 × 10−3.
Figure 1.
GABA type A receptor (GABAAR) activity regulates the Na+-K+-2Cl− cotransporter NKCC1 membrane dynamics: (A) Individual NKCC1 trajectories with reduced surface exploration upon gabazine or muscimol treatment. Scale bar, 0.5 µm. (B) Time-averaged mean square displacement (MSD) functions in control (blue) vs. gabazine (yellow) or muscimol (orange) conditions show increased confinement upon gabazine or muscimol application. (C,D) Boxplots of log diffusion coefficient (D) of NKCC1 in control condition (blue) or upon application of gabazine (yellow) or muscimol (orange) showing reduced diffusion upon drug treatments. N = 1558 quantum dots (QDs) (control, 41 cells), n = 387 QDs (gabazine, 18 cells), Welch t-test, p = 1.3 × 10−6, n = 545 QDs (muscimol, 27 cells), Welch t-test, p = 2.1 × 10−12, 5 cultures. (D) Median explored area EA in control vs. gabazine or muscimol conditions show increased confinement upon gabazine (Welch t-test, p = 2 × 10−8) or muscimol (Welch t-test, p = 4.2 × 10−11) treatment. (E) Trajectories (white) overlaid with clusters of homer1c-DsRed (green) or gephyrin-Finger-YFP (red) to identify extrasynaptic trajectories (extra), trajectories at excitatory (ES), and inhibitory synapses (IS). Scale bar, 0.4 µm. (F,G) Log(D) (F) and EA (G) of NKCC1 are decreased upon gabazine or muscimol application as compared with control condition. Note that the effect is more pronounced for extrasynaptic trajectories than for ES or IS trajectories. Diffusion coefficient (D): Extra, n = 899 QDs (control), 227 QDs (gabazine), p = 2.9 × 10−5, n = 268 QDs (muscimol) p = 2.2 × 10−9; IS, n = 244 QDs (control), 79 QDs (gabazine) p = 0.16; n = 142 QDs (muscimol) p = 0.19; ES, n = 415 QDs (control), n = 81 QDs (gabazine) p = 0.021, n = 135 QDs (muscimol) p = 0.00046. Explored area (EA): Extra, gabazine p = 2.9 × 10−5, muscimol p = 2.2 × 10−9; IS, gabazine p = 0.16, muscimol p = 0.19; ES, n = 415 QDs (control), n = 81 QDs (gabazine) p = 2.1 × 10−2, n = 135 QDs (muscimol) p = p = 4.6 × 10−4. (H) NKCC1 trajectories (red) in control vs. gabazine or muscimol conditions in relation to endocytic zones identified by clathrin-YFP clusters (white). Scale bar, 0.5 µm. (I,J) Reduced diffusion coefficient (I) and explored area (J) of NKCC1 within endocytic zones (Intra EZ) upon muscimol and gabazine treatment. Diffusion coefficient (D): Ctrl n = 117 QDs, 48 cells, Gbz n = 145 QDs, 64 cells, p = 1.4 × 10−3; Ctrl n = 273 QDs, 35 cells, Musc n = 247 QDs, 28 cells, p = 5.6 × 10−3, 3 cultures. Explored area (EA): Gbz, p = 5.46 × 10−5 and Musc, p = 2.2 × 10−16, 3 cultures. (K,L) No effect of gabazine or muscimol on log(D) (K) and EA (L) of dendritic NKCC1 in conditions of blockade of endocytosis (+ dynamin peptide). Diffusion coefficient (D): Bulk Ctrl n = 491 QDs, Gbz n = 238 QDs, p = 0.6 and Musc n = 243 QDs, p = p = 6.1 × 10−1, 5 cultures. Explored area (EA): Gbz, p = 0.84 and Musc, p = 8.1 × 10−1, 5 cultures. (B,F,I,K) D in µm2.s−1; C: MSD in µm2 vs. time(s); (D,G,J,L) EA in µm2. In all graphs, *, p < 5.0 × 10−2; **, p < 1.0 × 10−2; ***, p < 1.0 × 10−3.
![Cells 12 00464 g001 Cells 12 00464 g001]()
Figure 2.
Regulation of NKCC1 membrane clustering by GABAAR-mediated inhibition: (A) Conventional microscopy showing NKCC1 surface staining in hippocampal neurons at 21 days in vitro (DIV) in absence (Control) or presence of gabazine (Gabazine) or muscimol (Muscimol) for 30 min. Scale bar, 10 µm. (B) Quantification of the ratio of the surface/total pool of NKCC1 in control (blue), gabazine (yellow), and muscimol (orange) conditions showing no changes in the surface level of NKCC1 after gabazine or muscimol treatment. Ctrl n = 30 cells, Gbz, n = 41 cells, MW test p = 8.7 × 10−1, Musc n = 38 cells, p = 1.7 × 10−1, 8 cultures. (C–E), Quantification of NKCC1 cluster number (C), area (D), and intensity (E) showing reduced density and size of NKCC1 clusters upon muscimol treatment, while gabazine treatment reduced the size and intensity of NKCC1 clusters. Values were normalized to control values. MW test: Gabazine: cluster number (nb) p = 4.9 × 10−1, area p = 3.0 × 10−3, 0.003, intensity p = 1.7 × 10−2. Muscimol: cluster nb p = 2.7 × 10−2, area p = 2.9 × 10−2, intensity p = p = 9.2 × 10−1. (F–I) Stochastic Optical Reconstruction Microscopy (STORM) showing that gabazine and muscimol treatments alter NKCC1 nanoclusters at the surface of hippocampal neurons. (F) Representative STORM images of NKCC1 at the surface of neurons exposed 30 min to gabazine or muscimol. Scale bar, 0.1 µm. (G) Quantification of NKCC1 cluster area shows reduction in nanocluster size upon gabazine and muscimol treatment. Ctrl n = 550 nanoclusters, Gbz n = 192 nanoclusters, Monte Carlo simulations of the MW test p = 2.0 × 10−3, Musc n = 410 nanoclusters, p = 0.004, 4 cultures. (H) Quantification of the number of particles detected per nanocluster showing reduced number of detection upon gabazine (Monte Carlo simulations of MW test, p = 1.2 × 10−2) but not muscimol (p = 3.0 × 10−3) exposure. (I) Quantification of the density of NKCC1 molecules per square nanometer highlighting denser NKCC1 packing upon neuronal exposure to muscimol (Monte Carlo simulations of MW test, p = 1.6 × 10−2) but not gabazine (p = 9.7 × 10−1). (C) µm−1, (D) µm², (G) nm², (H) µm−1, (I) µm−2. In all graphs, *, p < 5.0 × 10−2; **, p < 1.0 × 10−2.
Figure 2.
Regulation of NKCC1 membrane clustering by GABAAR-mediated inhibition: (A) Conventional microscopy showing NKCC1 surface staining in hippocampal neurons at 21 days in vitro (DIV) in absence (Control) or presence of gabazine (Gabazine) or muscimol (Muscimol) for 30 min. Scale bar, 10 µm. (B) Quantification of the ratio of the surface/total pool of NKCC1 in control (blue), gabazine (yellow), and muscimol (orange) conditions showing no changes in the surface level of NKCC1 after gabazine or muscimol treatment. Ctrl n = 30 cells, Gbz, n = 41 cells, MW test p = 8.7 × 10−1, Musc n = 38 cells, p = 1.7 × 10−1, 8 cultures. (C–E), Quantification of NKCC1 cluster number (C), area (D), and intensity (E) showing reduced density and size of NKCC1 clusters upon muscimol treatment, while gabazine treatment reduced the size and intensity of NKCC1 clusters. Values were normalized to control values. MW test: Gabazine: cluster number (nb) p = 4.9 × 10−1, area p = 3.0 × 10−3, 0.003, intensity p = 1.7 × 10−2. Muscimol: cluster nb p = 2.7 × 10−2, area p = 2.9 × 10−2, intensity p = p = 9.2 × 10−1. (F–I) Stochastic Optical Reconstruction Microscopy (STORM) showing that gabazine and muscimol treatments alter NKCC1 nanoclusters at the surface of hippocampal neurons. (F) Representative STORM images of NKCC1 at the surface of neurons exposed 30 min to gabazine or muscimol. Scale bar, 0.1 µm. (G) Quantification of NKCC1 cluster area shows reduction in nanocluster size upon gabazine and muscimol treatment. Ctrl n = 550 nanoclusters, Gbz n = 192 nanoclusters, Monte Carlo simulations of the MW test p = 2.0 × 10−3, Musc n = 410 nanoclusters, p = 0.004, 4 cultures. (H) Quantification of the number of particles detected per nanocluster showing reduced number of detection upon gabazine (Monte Carlo simulations of MW test, p = 1.2 × 10−2) but not muscimol (p = 3.0 × 10−3) exposure. (I) Quantification of the density of NKCC1 molecules per square nanometer highlighting denser NKCC1 packing upon neuronal exposure to muscimol (Monte Carlo simulations of MW test, p = 1.6 × 10−2) but not gabazine (p = 9.7 × 10−1). (C) µm−1, (D) µm², (G) nm², (H) µm−1, (I) µm−2. In all graphs, *, p < 5.0 × 10−2; **, p < 1.0 × 10−2.
![Cells 12 00464 g002 Cells 12 00464 g002]()
Figure 3.
Lowering intracellular chloride increases the membrane diffusion, clustering, and stability of NKCC1: (A) Trajectories of NKCC1 (white) under low and high Cl− concentration in the extrasynaptic area (extra) and at inhibitory (IS) or excitatory (ES) synapses. Scale bar, 0.5 µm. (B) No major effect of a reduction in Cl− concentration on log(D) (B) of NKCC1. Note the increase in log(EA) for extrasynaptic trajectories (C) in conditions of low chloride. Diffusion coefficient (D): extra, low Cl− n = 128 QDs, high Cl− n = 93 QDs, Welch t-test p = 8.9 × 10−1; IS, low Cl− n = 64 QDs, high Cl− n = 56 QDs, Welch t-test p = 7.4 × 10−1; ES, low Cl− n = 62 QDs, high Cl− n = 68 QDs, Welch t-test p = 3.6 × 10−1, 2 cultures. Explored area (EA): extra, low Cl− Welch t-test p = 1.0 × 10−2; IS, Welch t-test p = 6.0 × 10−1; ES, Welch t-test p = 0.6. (D) Examples of NKCC1 trajectories (red) inside and outside clathrin-YFP fluorescent (white) endocytic zones in conditions of low (0 mM) vs. high (138 mM) Cl− concentration. Scale bar, 0.5 µm. (E,F) lowering intracellular chloride levels increases log(D) and log(EA) of NKCC1 located outside endocytic zones (Extra EZ) while the diffusion of NKCC1 inside endocytic zones (Intra EZ) is unchanged. Diffusion coefficient (D): extra EZ, low Cl− n = 209 QDs, high Cl− n = 238 QDs, Welch t-test p = 6.0 × 10−4; intra EZ, low Cl− n = 91 QDs, high Cl− n = 105 QDs, Welch t-test p = 2.3 × 10−1, 2 cultures. Explored area (EA): extra EZ, p = 7.12 × 10−8; intra EZ, p = 4.1 × 10−1. (G–K) Lowering intracellular chloride increases surface detection and clustering of NKCC1. G, HA surface staining in neurons expressing recombinant NKCC1-HA in conditions of high vs. low Cl− concentration for 30 min. Scale bar, 1 µm. (H) Quantification of the surface/total pool of NKCC1 in high (dark blue) and low (light blue) Cl− concentration showing increase in NKCC1 surface staining upon reduced Cl− level. Low Cl− n = 35 cells, high Cl− n = 40 cells, Welch t-test p = 1.0 × 10−1, 3 cultures. (I–K) Quantification of NKCC1-HA cluster number (I), area (J), and intensity (K) shows increased size and intensity of NKCC1 clusters upon reduction in Cl− levels. Values normalized to control values. Cluster number (Nb) MW test p = 2.0 × 10−1, area MW test p = 1.8 × 10−2, intensity MW test p = 5.6 × 10−3. (B,E) D in µm2.s−1; (C,F) EA in µm2; (I) µm−1; (J) µm². In all graphs, *, p < 5.0 × 10−2; **, p < 1.0 × 10−2; ***, p < 1.0 × 10−3.
Figure 3.
Lowering intracellular chloride increases the membrane diffusion, clustering, and stability of NKCC1: (A) Trajectories of NKCC1 (white) under low and high Cl− concentration in the extrasynaptic area (extra) and at inhibitory (IS) or excitatory (ES) synapses. Scale bar, 0.5 µm. (B) No major effect of a reduction in Cl− concentration on log(D) (B) of NKCC1. Note the increase in log(EA) for extrasynaptic trajectories (C) in conditions of low chloride. Diffusion coefficient (D): extra, low Cl− n = 128 QDs, high Cl− n = 93 QDs, Welch t-test p = 8.9 × 10−1; IS, low Cl− n = 64 QDs, high Cl− n = 56 QDs, Welch t-test p = 7.4 × 10−1; ES, low Cl− n = 62 QDs, high Cl− n = 68 QDs, Welch t-test p = 3.6 × 10−1, 2 cultures. Explored area (EA): extra, low Cl− Welch t-test p = 1.0 × 10−2; IS, Welch t-test p = 6.0 × 10−1; ES, Welch t-test p = 0.6. (D) Examples of NKCC1 trajectories (red) inside and outside clathrin-YFP fluorescent (white) endocytic zones in conditions of low (0 mM) vs. high (138 mM) Cl− concentration. Scale bar, 0.5 µm. (E,F) lowering intracellular chloride levels increases log(D) and log(EA) of NKCC1 located outside endocytic zones (Extra EZ) while the diffusion of NKCC1 inside endocytic zones (Intra EZ) is unchanged. Diffusion coefficient (D): extra EZ, low Cl− n = 209 QDs, high Cl− n = 238 QDs, Welch t-test p = 6.0 × 10−4; intra EZ, low Cl− n = 91 QDs, high Cl− n = 105 QDs, Welch t-test p = 2.3 × 10−1, 2 cultures. Explored area (EA): extra EZ, p = 7.12 × 10−8; intra EZ, p = 4.1 × 10−1. (G–K) Lowering intracellular chloride increases surface detection and clustering of NKCC1. G, HA surface staining in neurons expressing recombinant NKCC1-HA in conditions of high vs. low Cl− concentration for 30 min. Scale bar, 1 µm. (H) Quantification of the surface/total pool of NKCC1 in high (dark blue) and low (light blue) Cl− concentration showing increase in NKCC1 surface staining upon reduced Cl− level. Low Cl− n = 35 cells, high Cl− n = 40 cells, Welch t-test p = 1.0 × 10−1, 3 cultures. (I–K) Quantification of NKCC1-HA cluster number (I), area (J), and intensity (K) shows increased size and intensity of NKCC1 clusters upon reduction in Cl− levels. Values normalized to control values. Cluster number (Nb) MW test p = 2.0 × 10−1, area MW test p = 1.8 × 10−2, intensity MW test p = 5.6 × 10−3. (B,E) D in µm2.s−1; (C,F) EA in µm2; (I) µm−1; (J) µm². In all graphs, *, p < 5.0 × 10−2; **, p < 1.0 × 10−2; ***, p < 1.0 × 10−3.
![Cells 12 00464 g003 Cells 12 00464 g003]()
Figure 4.
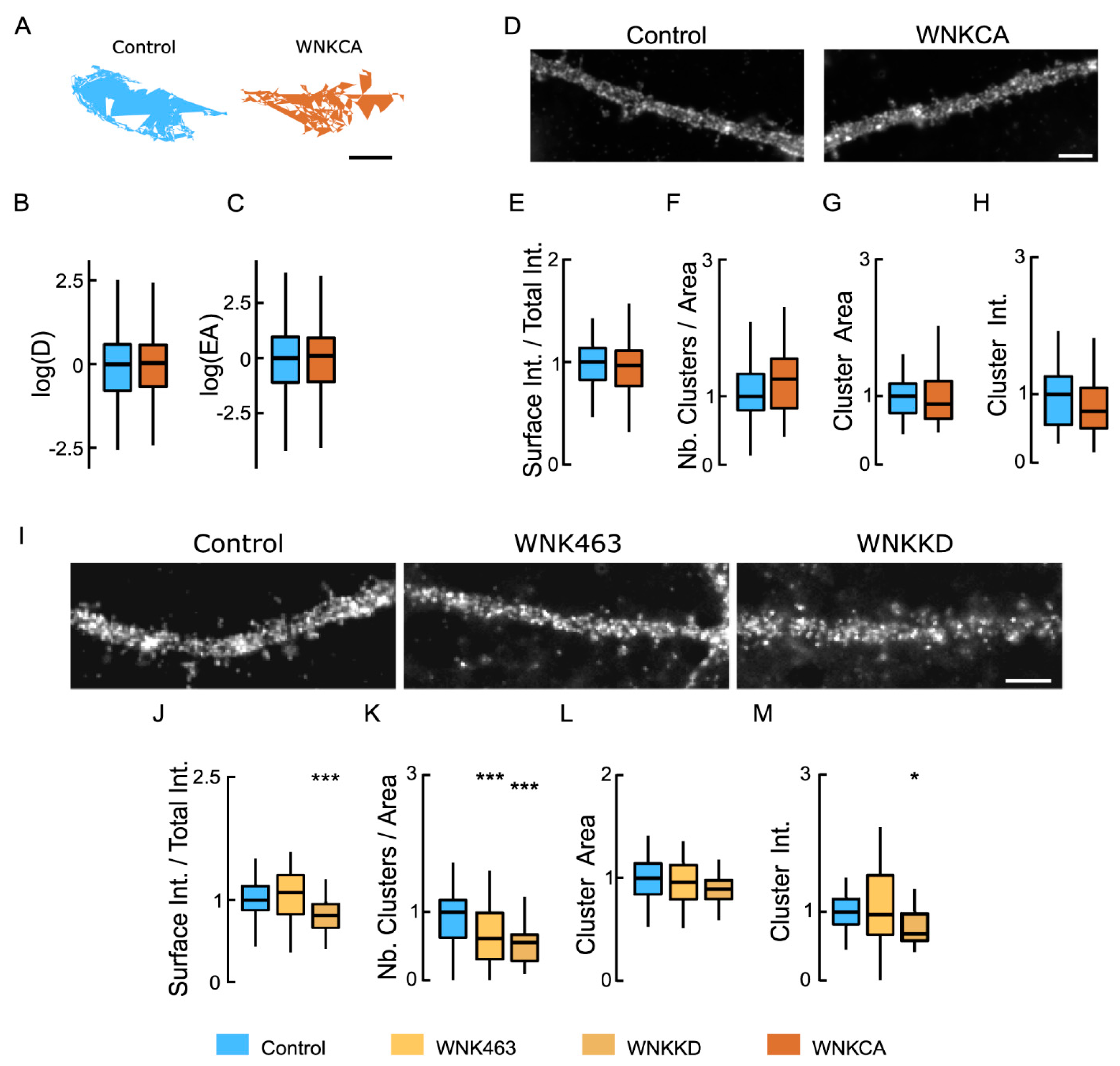
Inhibiting WNK1 reduces the clustering of NKCC1 at the neuronal surface: (A) Representative trajectories of NKCC1 in neurons expressing constitutively active WNK1 (WNK-CA, brown) vs. control (blue). Bar: 0.4 µm. (B,C) WNK-CA over-expression does not change NKCC1 diffusion at the plasma membrane, with no effect on D (B) and EA (C). Diffusion coefficient (D): Bulk, Ctrl n = 305 QDs, WNK-CA n = 358 QDs, Welch t-test p = 8.1 × 10−1, 3 cultures; Explored area (EA): Welch t-test p = 4.9 × 10−1. (D) HA surface staining in hippocampal neurons expressing recombinant NKCC1-HA together with “constitutively active” WNK1 (WNK1-CA) or a control plasmid. Scale bar, 4 µm. (E) Overexpressing WNK-CA does not modify the level of NKCC1 expressed at the cell surface. Ctrl n = 45 cells, WNK-CA: n = 43 cells, Welch t-test p = 2.6 × 10−1, 3 cultures. (F–H) No effect of WNK-CA expression on NKCC1 cluster number (F), area (G), and intensity (H). All values were normalized to the control values. Cluster number (Nb) MW test p = 1.4 × 10−1, area MW test p = 3.2 × 10−1, intensity MW test p = 0.25. (I) Impact of inhibition of WNK1 on HA surface staining of neurons transfected with NKCC1-HA. WNK1 was inhibited either by incubating the neurons for 30 min with a specific inhibitor, WNK-463, or by co-expressing kinase-dead WNK1 (WNK1-KD). Scale bar, 4 µm. (J) Incubating the neurons with WNK-463 (orange) does not modify the surface level of NKCC1 compared to controls (blue); however, over-expression of WNK-KD (brown) sharply reduced NKCC1 surface levels. WNK-463 experiment: Ctrl n = 24 cells, WNK-463 n = 25 cells, Welch t-test p = 5.8 × 10−1. WNK-KD experiment: Ctrl n = 34 cells, WNK-KD n = 24 cells, Welch t-test p = 9.6 × 10−3, p = 0.0096, 4 cultures. (K–M) Loss of NKCC1 clusters (K) and reduced cluster size (L) but not cluster intensity (M) upon WNK1 activity suppression with either WNK-KD or WNK-463, as compared to control conditions. Values were normalized to the corresponding control values. MW test: WNK-KD experiment: Cluster number (Nb) p = 4.5 × 10−7, area p = 1.2 × 10−2, intensity p = 3.1 × 10−3. WNK-463 experiment: Cluster number (Nb) p = 8.0 × 10−4, area p = 8.6 × 10−1, intensity p = 8.3 × 10−1. In all graphs, *, p < 5.0 × 10−2; ***, p < 1.0 × 10−3.
Figure 4.
Inhibiting WNK1 reduces the clustering of NKCC1 at the neuronal surface: (A) Representative trajectories of NKCC1 in neurons expressing constitutively active WNK1 (WNK-CA, brown) vs. control (blue). Bar: 0.4 µm. (B,C) WNK-CA over-expression does not change NKCC1 diffusion at the plasma membrane, with no effect on D (B) and EA (C). Diffusion coefficient (D): Bulk, Ctrl n = 305 QDs, WNK-CA n = 358 QDs, Welch t-test p = 8.1 × 10−1, 3 cultures; Explored area (EA): Welch t-test p = 4.9 × 10−1. (D) HA surface staining in hippocampal neurons expressing recombinant NKCC1-HA together with “constitutively active” WNK1 (WNK1-CA) or a control plasmid. Scale bar, 4 µm. (E) Overexpressing WNK-CA does not modify the level of NKCC1 expressed at the cell surface. Ctrl n = 45 cells, WNK-CA: n = 43 cells, Welch t-test p = 2.6 × 10−1, 3 cultures. (F–H) No effect of WNK-CA expression on NKCC1 cluster number (F), area (G), and intensity (H). All values were normalized to the control values. Cluster number (Nb) MW test p = 1.4 × 10−1, area MW test p = 3.2 × 10−1, intensity MW test p = 0.25. (I) Impact of inhibition of WNK1 on HA surface staining of neurons transfected with NKCC1-HA. WNK1 was inhibited either by incubating the neurons for 30 min with a specific inhibitor, WNK-463, or by co-expressing kinase-dead WNK1 (WNK1-KD). Scale bar, 4 µm. (J) Incubating the neurons with WNK-463 (orange) does not modify the surface level of NKCC1 compared to controls (blue); however, over-expression of WNK-KD (brown) sharply reduced NKCC1 surface levels. WNK-463 experiment: Ctrl n = 24 cells, WNK-463 n = 25 cells, Welch t-test p = 5.8 × 10−1. WNK-KD experiment: Ctrl n = 34 cells, WNK-KD n = 24 cells, Welch t-test p = 9.6 × 10−3, p = 0.0096, 4 cultures. (K–M) Loss of NKCC1 clusters (K) and reduced cluster size (L) but not cluster intensity (M) upon WNK1 activity suppression with either WNK-KD or WNK-463, as compared to control conditions. Values were normalized to the corresponding control values. MW test: WNK-KD experiment: Cluster number (Nb) p = 4.5 × 10−7, area p = 1.2 × 10−2, intensity p = 3.1 × 10−3. WNK-463 experiment: Cluster number (Nb) p = 8.0 × 10−4, area p = 8.6 × 10−1, intensity p = 8.3 × 10−1. In all graphs, *, p < 5.0 × 10−2; ***, p < 1.0 × 10−3.
![Cells 12 00464 g004 Cells 12 00464 g004]()
Figure 5.
Inhibiting SPAK-OSR1 activity tunes NKCC1 membrane diffusion and clustering: (A) NKCC1 trajectories showing decreased exploration in the presence of closantel (pink) compared to connrol (blue). Scale bar, 0.5 µm. (B,C), Log (D) (B) and EA (C) of NKCC1 are decreased upon closantel application (pink) as compared with control condition (blue), indicating reduced mobility and increased confinement. Diffusion coefficient (D): Bulk, Ctrl n = 433 QDs, closantel n = 371 QDs, Welch t-test p = 5.5 × 10−5, 2 cultures. Explored area (EA): Welch t-test p = 4.8 × 10−11. (D–H), Standard epifluorescence microscopy showing no effect of closantel on NKCC1 membrane immunoreactivity. (D) HA surface staining in neurons expressing NKCC1-HA and treated or not with closantel. Scale bar, 4 µm. (E) Closantel treatment does not modify the surface level of NKCC1. Ctrl n = 51 cells, closantel n = 61 cells, Welch t-test p = 4.9 × 10−1, 7 cultures. (F–H) Closantel has no impact on NKCC1 cluster number (F), area (G), and intensity (H). Cluster Number (Nb) MW test p = 6.0 × 10−1, area MW test p = 9.0 × 10−2, intensity MW test p = 8.1 × 10−1. (I–L) STORM showing that closantel affects NKCC1 organization at the nanoscale. (I) Representative images of NKCC1 at the surface of neurons exposed 30 min to closantel vs. control condition. Scale bar, 0.1 µm. Quantification of NKCC1 cluster area (J), number of cluster (K) and density of detections in the cluster (L) reveal reduction in cluster size upon closantel treatment. Ctrl n = 218 clusters, closantel n = 147 clusters, 2 cultures. Cluster area: Monte Carlo simulations of the MW test p < 0.001; Nb detection: MW test p = 5.8 × 10−2, density: MW test p < 0.001. In all graphs, ***, p < 1.0 × 10−3.
Figure 5.
Inhibiting SPAK-OSR1 activity tunes NKCC1 membrane diffusion and clustering: (A) NKCC1 trajectories showing decreased exploration in the presence of closantel (pink) compared to connrol (blue). Scale bar, 0.5 µm. (B,C), Log (D) (B) and EA (C) of NKCC1 are decreased upon closantel application (pink) as compared with control condition (blue), indicating reduced mobility and increased confinement. Diffusion coefficient (D): Bulk, Ctrl n = 433 QDs, closantel n = 371 QDs, Welch t-test p = 5.5 × 10−5, 2 cultures. Explored area (EA): Welch t-test p = 4.8 × 10−11. (D–H), Standard epifluorescence microscopy showing no effect of closantel on NKCC1 membrane immunoreactivity. (D) HA surface staining in neurons expressing NKCC1-HA and treated or not with closantel. Scale bar, 4 µm. (E) Closantel treatment does not modify the surface level of NKCC1. Ctrl n = 51 cells, closantel n = 61 cells, Welch t-test p = 4.9 × 10−1, 7 cultures. (F–H) Closantel has no impact on NKCC1 cluster number (F), area (G), and intensity (H). Cluster Number (Nb) MW test p = 6.0 × 10−1, area MW test p = 9.0 × 10−2, intensity MW test p = 8.1 × 10−1. (I–L) STORM showing that closantel affects NKCC1 organization at the nanoscale. (I) Representative images of NKCC1 at the surface of neurons exposed 30 min to closantel vs. control condition. Scale bar, 0.1 µm. Quantification of NKCC1 cluster area (J), number of cluster (K) and density of detections in the cluster (L) reveal reduction in cluster size upon closantel treatment. Ctrl n = 218 clusters, closantel n = 147 clusters, 2 cultures. Cluster area: Monte Carlo simulations of the MW test p < 0.001; Nb detection: MW test p = 5.8 × 10−2, density: MW test p < 0.001. In all graphs, ***, p < 1.0 × 10−3.
![Cells 12 00464 g005 Cells 12 00464 g005]()
Figure 6.
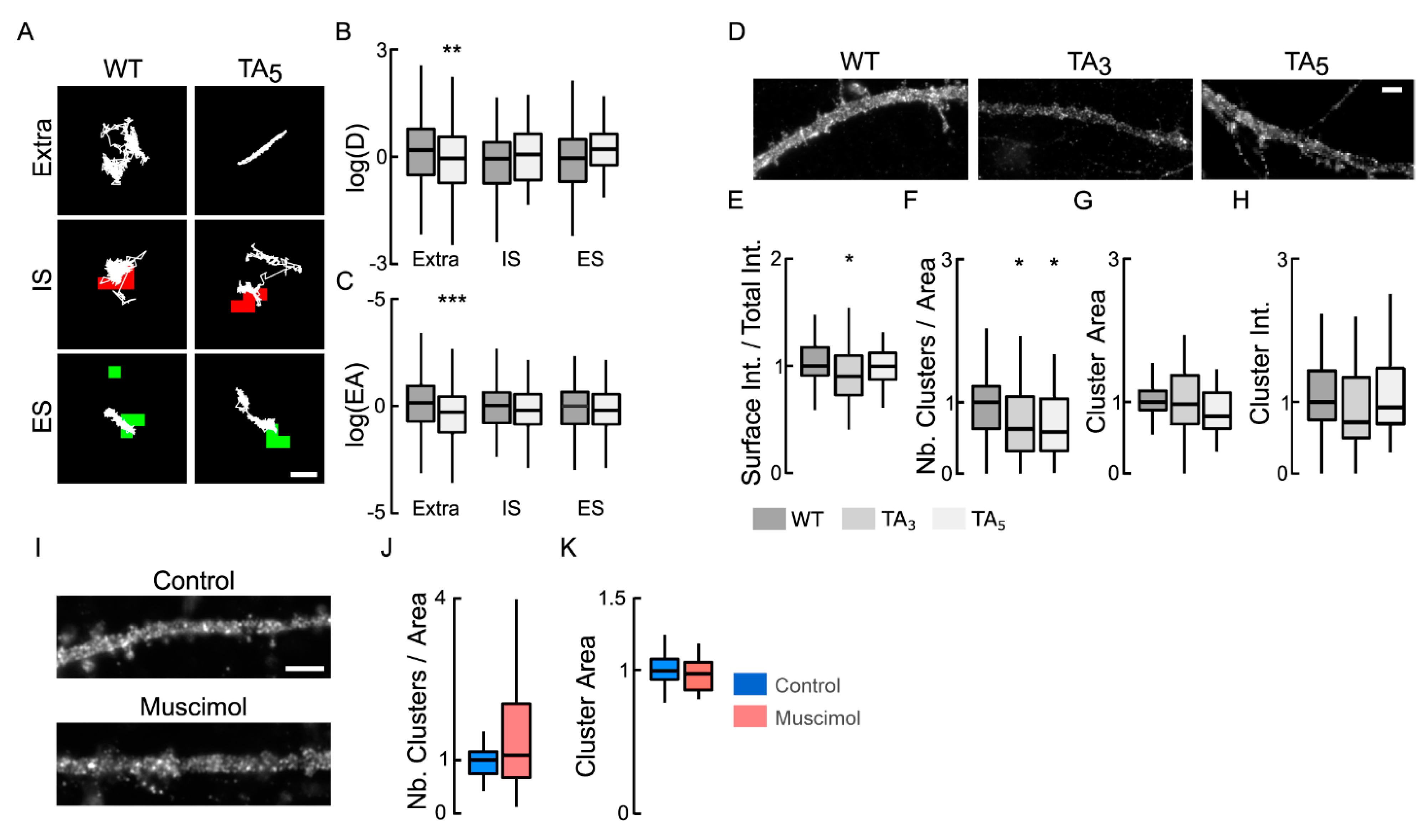
NKCC1 membrane dynamics, stability, and clustering are dependent on NKCC1 phosphorylation of T203/207/212/217/230. (A) Examples of NKCC1-T203/207/212/217/230 (WT) and NKCC1-T203/207/212/217/230A (TA5) trajectories (white) in resting condition at extrasynaptic site (extra), at inhibitory (IS) and excitatory glutamatergic (ES) synapses. Scale bar, 0.5 µm. (B,C) Log(D) (B) and EA (C) show that the dephospho-mimetic NKCC1-TA5 (light gray) is slower and more confined than NKCC1-WT (gray) at extrasynaptic sites but not near synapses. Diffusion coefficient (D): WT: extra, n = 189 QDs, IS, n = 42 QDs, ES n = 30 QDs; TA5: extra, n = 166 QDs, IS, n = 33 QDs, ES n = 34 QDs; from 67 cells and 2 cultures. Welch t-test: extra p = 8.8 × 10−3, IS p = 2.8 × 10−1, ES p = 7.3 × 10−1. Explored area (EA): extra p = 4.5 × 10−4, IS p = 7.1 × 10−1, ES p = 4.5 × 10−1. (D) HA surface staining in hippocampal neurons expressing recombinant NKCC1-WT (WT) vs. NKCC1-TA3 (TA3) or NKCC1-TA5 (TA5) in resting conditions. Scale bar, 4 µm. (E) Expression of NKCC1-TA3 is slightly reduced at the plasma membrane as compared to NKCC1-WT. WT (dark gray) n = 68 cells, TA3 (gray) n = 43 cells, TA5 (light gray) n = 36 cells (9 cultures); WT vs. TA3 Welch t-test p = 4.1 × 10−2, WT vs. TA5 p = 8.7 × 10−1, 9 cultures. (F–H) Quantification of cluster number (F), area (G), and intensity (H) for NKCC1-TA3, NKCC1-TA5 vs. NKCC1-WT. Note the reduced density of cluster (F) for NKCC1-TA3 and NKCC1-TA5 as compared to NKCC1-WT. Cluster number (Nb) WT vs. TA3 MW test p = 4.2 × 10−2, WT vs. TA5 MW test p = 1.4 × 10−2; area: WT vs. TA3 MW test p = 4.6 × 10−1, WT vs. TA5 MW test p = 2.2 × 10−1; intensity WT vs. TA3 MW test p = 3.0 × 10−1, WT vs. TA5 MW test p = 8.6 × 10−1. (I) HA surface staining in hippocampal neurons expressing recombinant NKCC1-TA5 exposed or not to muscimol. Scale bar, 4 µm. (J–K) Muscimol application has no effect on NKCC1-TA5 cluster number: MW test, p = 4.5 × 10−1, (J) and area: MW test, p = 3.8 × 10−1, (K) Control: 26 cells, muscimol: 22 cells, 3 cultures. In all graphs, *, p < 5.0 × 10−2; **, p < 1.0 × 10−2; ***, p < 1.0 × 10−3.
Figure 6.
NKCC1 membrane dynamics, stability, and clustering are dependent on NKCC1 phosphorylation of T203/207/212/217/230. (A) Examples of NKCC1-T203/207/212/217/230 (WT) and NKCC1-T203/207/212/217/230A (TA5) trajectories (white) in resting condition at extrasynaptic site (extra), at inhibitory (IS) and excitatory glutamatergic (ES) synapses. Scale bar, 0.5 µm. (B,C) Log(D) (B) and EA (C) show that the dephospho-mimetic NKCC1-TA5 (light gray) is slower and more confined than NKCC1-WT (gray) at extrasynaptic sites but not near synapses. Diffusion coefficient (D): WT: extra, n = 189 QDs, IS, n = 42 QDs, ES n = 30 QDs; TA5: extra, n = 166 QDs, IS, n = 33 QDs, ES n = 34 QDs; from 67 cells and 2 cultures. Welch t-test: extra p = 8.8 × 10−3, IS p = 2.8 × 10−1, ES p = 7.3 × 10−1. Explored area (EA): extra p = 4.5 × 10−4, IS p = 7.1 × 10−1, ES p = 4.5 × 10−1. (D) HA surface staining in hippocampal neurons expressing recombinant NKCC1-WT (WT) vs. NKCC1-TA3 (TA3) or NKCC1-TA5 (TA5) in resting conditions. Scale bar, 4 µm. (E) Expression of NKCC1-TA3 is slightly reduced at the plasma membrane as compared to NKCC1-WT. WT (dark gray) n = 68 cells, TA3 (gray) n = 43 cells, TA5 (light gray) n = 36 cells (9 cultures); WT vs. TA3 Welch t-test p = 4.1 × 10−2, WT vs. TA5 p = 8.7 × 10−1, 9 cultures. (F–H) Quantification of cluster number (F), area (G), and intensity (H) for NKCC1-TA3, NKCC1-TA5 vs. NKCC1-WT. Note the reduced density of cluster (F) for NKCC1-TA3 and NKCC1-TA5 as compared to NKCC1-WT. Cluster number (Nb) WT vs. TA3 MW test p = 4.2 × 10−2, WT vs. TA5 MW test p = 1.4 × 10−2; area: WT vs. TA3 MW test p = 4.6 × 10−1, WT vs. TA5 MW test p = 2.2 × 10−1; intensity WT vs. TA3 MW test p = 3.0 × 10−1, WT vs. TA5 MW test p = 8.6 × 10−1. (I) HA surface staining in hippocampal neurons expressing recombinant NKCC1-TA5 exposed or not to muscimol. Scale bar, 4 µm. (J–K) Muscimol application has no effect on NKCC1-TA5 cluster number: MW test, p = 4.5 × 10−1, (J) and area: MW test, p = 3.8 × 10−1, (K) Control: 26 cells, muscimol: 22 cells, 3 cultures. In all graphs, *, p < 5.0 × 10−2; **, p < 1.0 × 10−2; ***, p < 1.0 × 10−3.
![Cells 12 00464 g006 Cells 12 00464 g006]()
Figure 7.
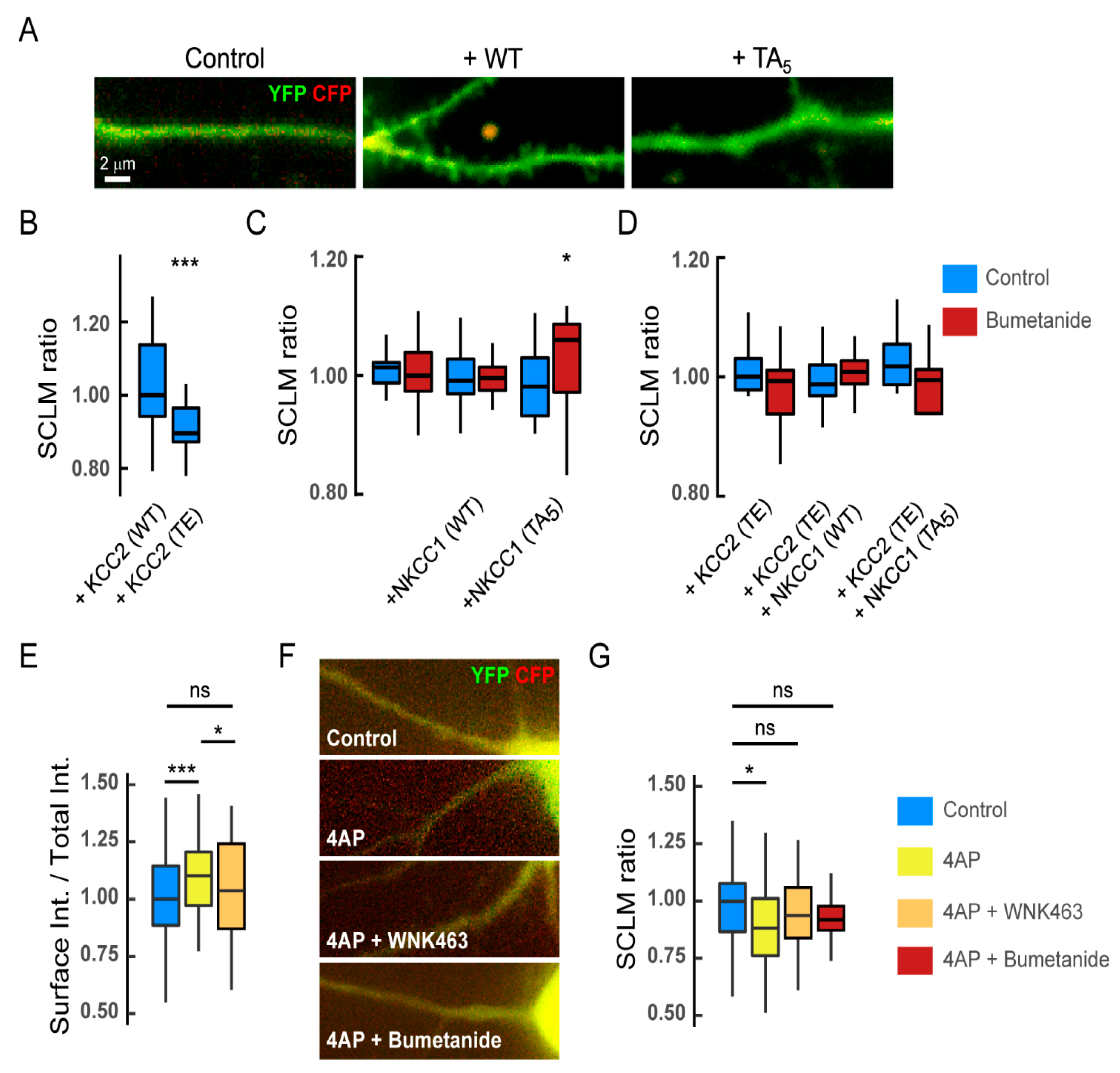
Functional impact of NKCC1 regulation by the WNK signaling on chloride homeostasis. (A–D) In basal activity conditions, expression of recombinant WT or dephospho-mimetic NKCC1 has no major impact on [Cl−]i in mature hippocampal neurons: (A) composite images of CFP (red) and YFP (green) in neurons expressing SuperClomeleon (SCLM) alone (control) or in combination with NKCC1-T203/207/212/217/230 (WT) or NKCC1-T203/207/212/217/230A (TA5) in resting conditions. Scale bar, 2 µm. (B) CFP/YFP fluorescence ratio in hippocampal neurons expressing SCLM together with KCC2-T906/T1007 (WT) or phospho-mimetic KCC2-T906/T1007E (TE). KCC2-WT n = 36 cells, KCC2-TE n = 39 cells, MW test p = 7.1 × 10−6, 3 cultures. (C) CFP/YFP fluorescence ratio in neurons expressing SCLM alone or in combination with NKCC1-WT or NKCC1-TA5 before (blue) and after (red) 10–30 min application of bumetanide. SCLM Ctrl n = 20 cells, Bumet n = 17 cells; SCLM + NKCC1-WT Ctrl n = 18 cells, Bumet n = 20 cells; SCLM + NKCC1-TA5 Ctrl n = 21 cells, Bumet n = 16 cells; 3 cultures. Ctrl vs. Bumet: SCLM MW test p = 9.6 × 10−1, SCLM + NKCC1-WT MW test p = 5.3 × 10−1, SCLM + NKCC1-TA5 MW test p = 2.9 × 10−2. SCLM vs. SCLM + NKCC1-WT, MW test p = 7.1 × 10−1, SCLM vs. SCLM + NKCC1-TA5, MW test p = 6.9 × 10−1; SCLM + NKCC1-WT vs. SCLM + NKCC1-TA5, MW test p = 8.9 × 10−1. (D) CFP/YFP fluorescence ratio in neurons expressing the same recombinant proteins as in B but in the presence of KCC2-TE before (blue) and after (red) bumetanide treatment. SCLM + KCC2-TE Ctrl n = 15 cells, Bumet n = 15 cells, MW test p = 1.5 × 10−1; SCLM + KCC2-TE + NKCC1-WT Ctrl n = 13 cells, Bumet n = 16 cells, MW test p = 5.6 × 10−1; SCLM + KCC2-TE + NKCC1-TA5 Ctrl n = 21 cells, Bumet n = 8 cells, MW test p = 9.2 × 10−2; SCLM + KCC2-TE vs. SCLM + KCC2-TE + NKCC1-WT, MW test p = 6.5 × 10−1, SCLM vs. SCLM + KCC2-TE + NKCC1-TA5, MW test p = 2.0 × 10−1, SCLM + KCC2-TE + NKCC1-WT vs. SuperClomeleon + KCC2-TE + NKCC1-TA5, MW test p = 1.2 × 10−1. Two cultures. (E) Quantification of the surface/total pool of NKCC1 in control (blue), 4-AP (yellow), and 4-AP + WNK463 (orange) conditions showing a significant increase in surface NKCC1 upon 4AP treatment, an effect prevented by pre-incubation of neurons with WNK463 (orange). Ctrl n = 63 cells, 4-AP, n = 64 cells, MW test p = 4.6 × 10−4, 4-AP + WNK463 n = 57 cells, 4-AP vs. 4-AP + WNK463 p = 3.0 × 10−2, Ctrl vs. 4-AP + WNK463 p = 1.9 × 10−1, 4 cultures. (F) Overlay images of CFP (red) and YFP (green) in neurons expressing SuperClomeleon (SCLM) and NKCC1 in control vs. 4AP, 4AP + WNK463 or 4AP + bumetanide conditions. Scale bar, 2 µm. (G) CFP/YFP fluorescence ratio in neurons expressing SCLM and NKCC1 in control (blue) vs. 4-AP (yellow), 4-AP + WNK463 (orange), or 4-AP + bumetanide (red) conditions. Ctrl n = 53 cells, 4-AP n = 48 cells, MW test p = 1.0 × 10−2; 4-AP + WNK463 n = 41 cells, MW test p = 2.0 × 10−1; 4-AP + bumetanide n = 29 cells, MW test p = 2.1 × 10−1, 3–6 cultures. In all graphs, n, not significant; *, p < 5.0 × 10−2; ***, p < 1.0 × 10−3.
Figure 7.
Functional impact of NKCC1 regulation by the WNK signaling on chloride homeostasis. (A–D) In basal activity conditions, expression of recombinant WT or dephospho-mimetic NKCC1 has no major impact on [Cl−]i in mature hippocampal neurons: (A) composite images of CFP (red) and YFP (green) in neurons expressing SuperClomeleon (SCLM) alone (control) or in combination with NKCC1-T203/207/212/217/230 (WT) or NKCC1-T203/207/212/217/230A (TA5) in resting conditions. Scale bar, 2 µm. (B) CFP/YFP fluorescence ratio in hippocampal neurons expressing SCLM together with KCC2-T906/T1007 (WT) or phospho-mimetic KCC2-T906/T1007E (TE). KCC2-WT n = 36 cells, KCC2-TE n = 39 cells, MW test p = 7.1 × 10−6, 3 cultures. (C) CFP/YFP fluorescence ratio in neurons expressing SCLM alone or in combination with NKCC1-WT or NKCC1-TA5 before (blue) and after (red) 10–30 min application of bumetanide. SCLM Ctrl n = 20 cells, Bumet n = 17 cells; SCLM + NKCC1-WT Ctrl n = 18 cells, Bumet n = 20 cells; SCLM + NKCC1-TA5 Ctrl n = 21 cells, Bumet n = 16 cells; 3 cultures. Ctrl vs. Bumet: SCLM MW test p = 9.6 × 10−1, SCLM + NKCC1-WT MW test p = 5.3 × 10−1, SCLM + NKCC1-TA5 MW test p = 2.9 × 10−2. SCLM vs. SCLM + NKCC1-WT, MW test p = 7.1 × 10−1, SCLM vs. SCLM + NKCC1-TA5, MW test p = 6.9 × 10−1; SCLM + NKCC1-WT vs. SCLM + NKCC1-TA5, MW test p = 8.9 × 10−1. (D) CFP/YFP fluorescence ratio in neurons expressing the same recombinant proteins as in B but in the presence of KCC2-TE before (blue) and after (red) bumetanide treatment. SCLM + KCC2-TE Ctrl n = 15 cells, Bumet n = 15 cells, MW test p = 1.5 × 10−1; SCLM + KCC2-TE + NKCC1-WT Ctrl n = 13 cells, Bumet n = 16 cells, MW test p = 5.6 × 10−1; SCLM + KCC2-TE + NKCC1-TA5 Ctrl n = 21 cells, Bumet n = 8 cells, MW test p = 9.2 × 10−2; SCLM + KCC2-TE vs. SCLM + KCC2-TE + NKCC1-WT, MW test p = 6.5 × 10−1, SCLM vs. SCLM + KCC2-TE + NKCC1-TA5, MW test p = 2.0 × 10−1, SCLM + KCC2-TE + NKCC1-WT vs. SuperClomeleon + KCC2-TE + NKCC1-TA5, MW test p = 1.2 × 10−1. Two cultures. (E) Quantification of the surface/total pool of NKCC1 in control (blue), 4-AP (yellow), and 4-AP + WNK463 (orange) conditions showing a significant increase in surface NKCC1 upon 4AP treatment, an effect prevented by pre-incubation of neurons with WNK463 (orange). Ctrl n = 63 cells, 4-AP, n = 64 cells, MW test p = 4.6 × 10−4, 4-AP + WNK463 n = 57 cells, 4-AP vs. 4-AP + WNK463 p = 3.0 × 10−2, Ctrl vs. 4-AP + WNK463 p = 1.9 × 10−1, 4 cultures. (F) Overlay images of CFP (red) and YFP (green) in neurons expressing SuperClomeleon (SCLM) and NKCC1 in control vs. 4AP, 4AP + WNK463 or 4AP + bumetanide conditions. Scale bar, 2 µm. (G) CFP/YFP fluorescence ratio in neurons expressing SCLM and NKCC1 in control (blue) vs. 4-AP (yellow), 4-AP + WNK463 (orange), or 4-AP + bumetanide (red) conditions. Ctrl n = 53 cells, 4-AP n = 48 cells, MW test p = 1.0 × 10−2; 4-AP + WNK463 n = 41 cells, MW test p = 2.0 × 10−1; 4-AP + bumetanide n = 29 cells, MW test p = 2.1 × 10−1, 3–6 cultures. In all graphs, n, not significant; *, p < 5.0 × 10−2; ***, p < 1.0 × 10−3.
![Cells 12 00464 g007 Cells 12 00464 g007]()