Genes Involved in DNA Repair and Mitophagy Protect Embryoid Bodies from the Toxic Effect of Methylmercury Chloride under Physioxia Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. MeHgCl Toxicity for hiPSCs Cultured in Atmospheric Oxygen Conditions
2.1.1. hiPSCs Culture and hiPSCs MeHgCl Treatment in Monolayer
2.1.2. Apoptosis Level Assay in hiPSC Monolayer Culture
2.1.3. Alamar Blue Cell Viability Assay in hiPSC Monolayer Culture
2.1.4. ROS Accumulation Assay in hiPSC Monolayer Culture
2.1.5. Mitochondrial Membrane Potential Assay in hiPSC Monolayer Culture
2.1.6. hiPSCs Ability to Form EBs after MeHgCl Treatment
2.2. MeHgCl Toxicity in hiPSC-Derived EBs Cultured under Atmospheric Conditions
2.2.1. EBs Generation and MeHgCl Treatment under 21% Oxygen Conditions
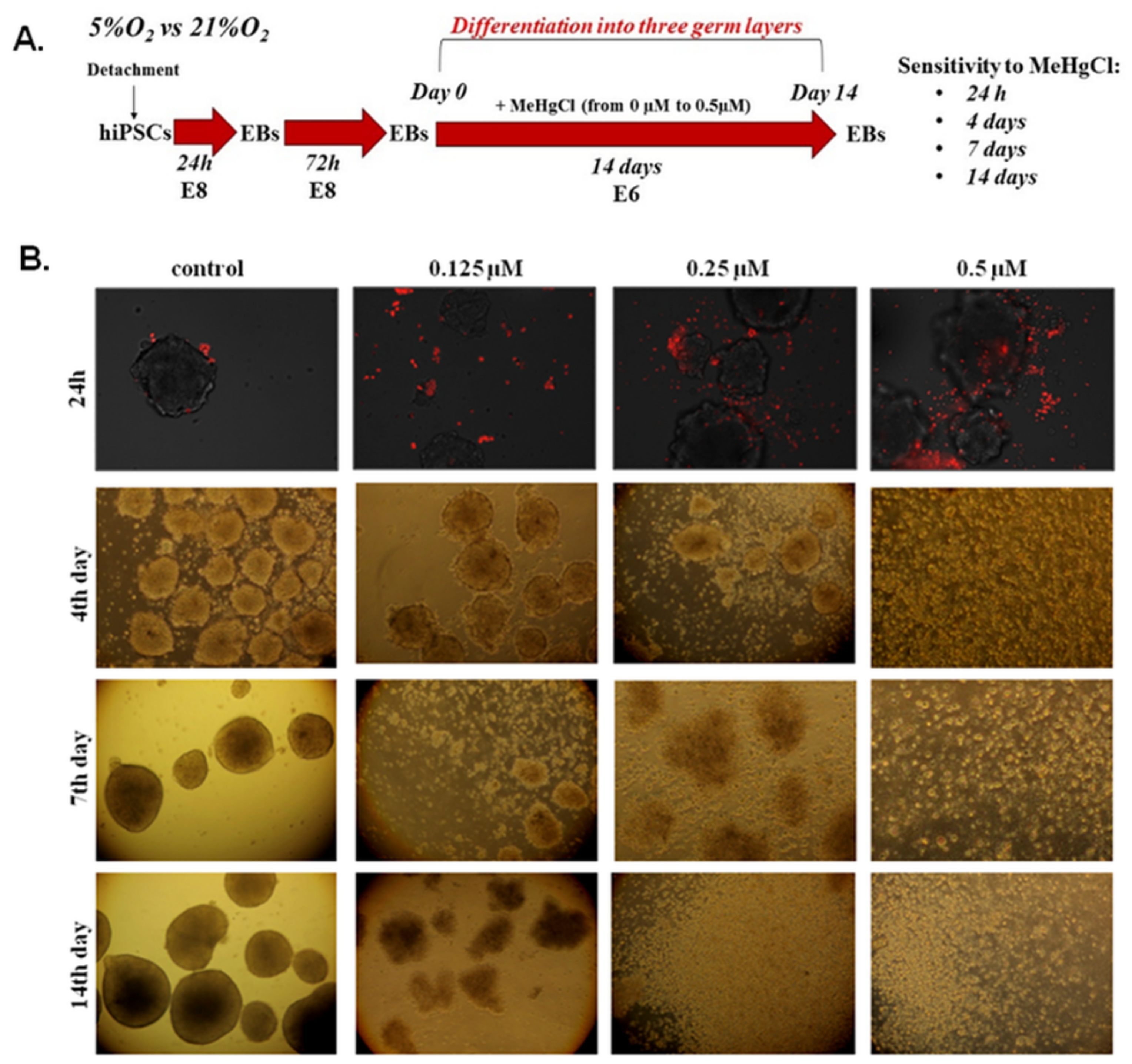
2.2.2. Analysis of EB Morphology Changes under MeHgCl Treatment
2.2.3. Visualization of Apoptosis in Suspension-Cultured EBs
2.3. Toxicity of a Chronic Low Dose of MeHgCl in hiPSC-Derived EBs Cultured under Atmospheric and Low Oxygen Conditions
2.3.1. EBs Generation and MeHgCl Treatment under 21% and 5% Oxygen Conditions
2.3.2. Total Genomic DNA Isolation
2.3.3. DNA Damage Assay
2.3.4. Total RNA Isolation
2.3.5. Reverse Transcription (RT)
2.3.6. Quantitative Polymerase Chain Reaction (qPCR)
2.3.7. qRT-PCR
2.3.8. Immunofluorescence Staining
2.3.9. Gene Function Prediction and Network Analysis
2.4. Statistical Analysis
3. Results
3.1. The Effect of MeHgCl on hiPSCs Grown in 21% O2
3.1.1. Apoptosis
3.1.2. Viability
3.1.3. ROS Accumulation
3.1.4. Mitochondrial Membrane Potential
3.1.5. The Effect of MeHgCl on hiPSC-Derived EB Formation under 21% O2 Conditions
3.2. The Effect of MeHgCl on EB Development under 21% O2 Conditions
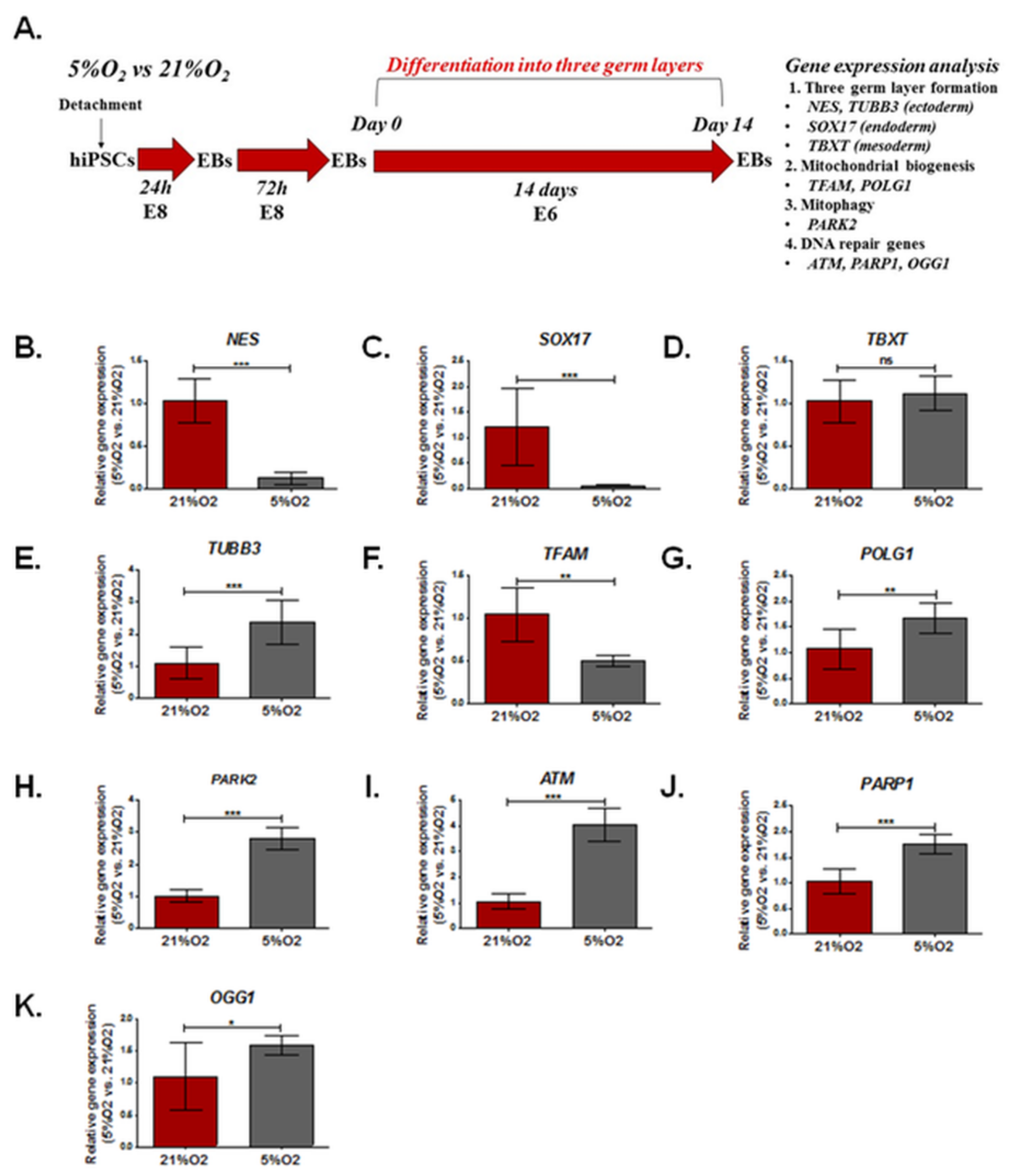
3.3. The Effect of Different Oxygen Concentrations (21%, 5%) on the Expression of Genes Involved in DNA Damage Repair, Mitochondrial Biogenesis, and Formation of the Three Germ Layers in EBs
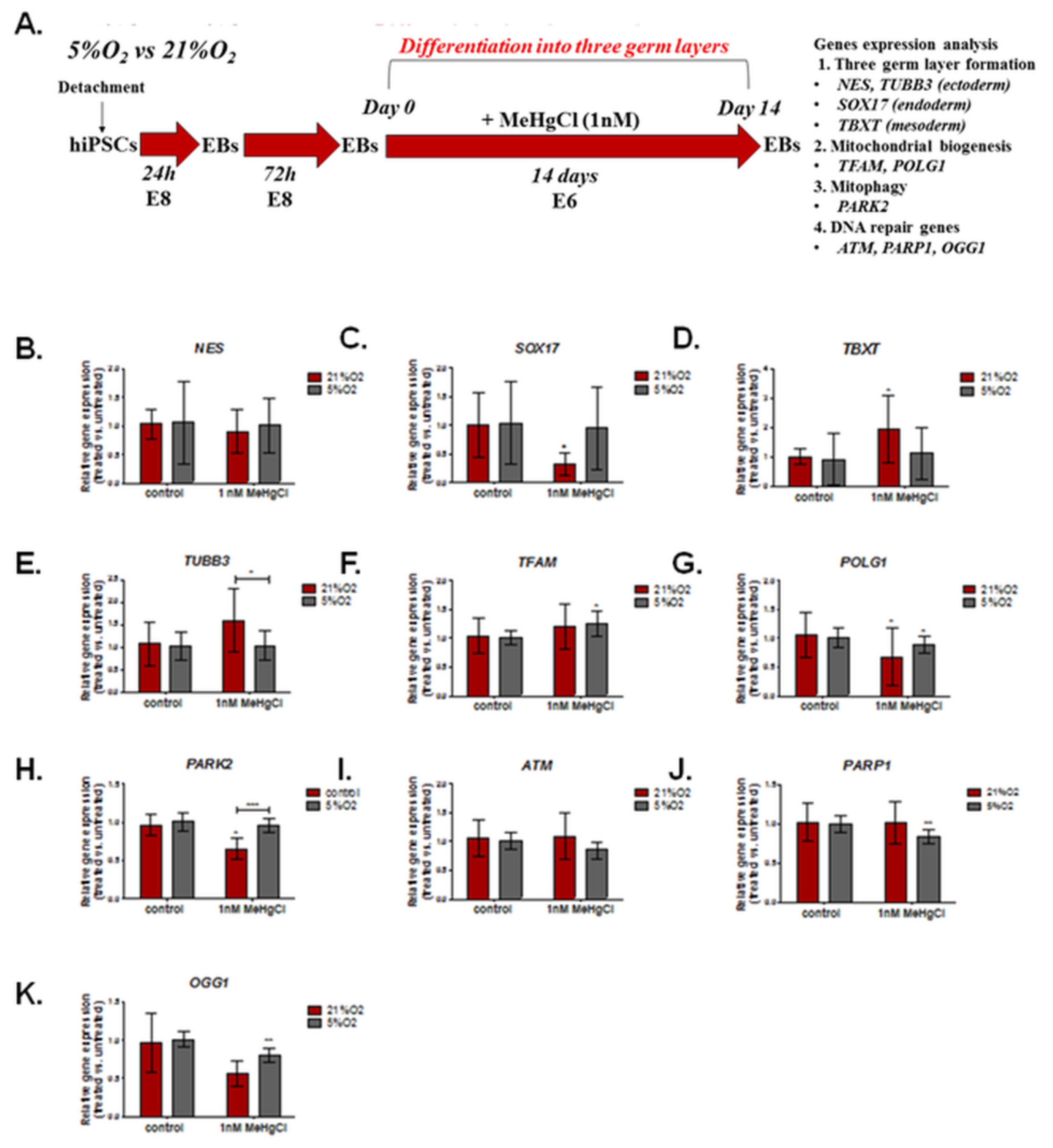
3.4. The Effect of Different Oxygen Concentrations and a Low Dose of MeHgCl (1 nM) on the Expression of Genes Involved in DNA Damage Repair, Mitochondrial Biogenesis, and Formation of the Three Germ Layers in EBs
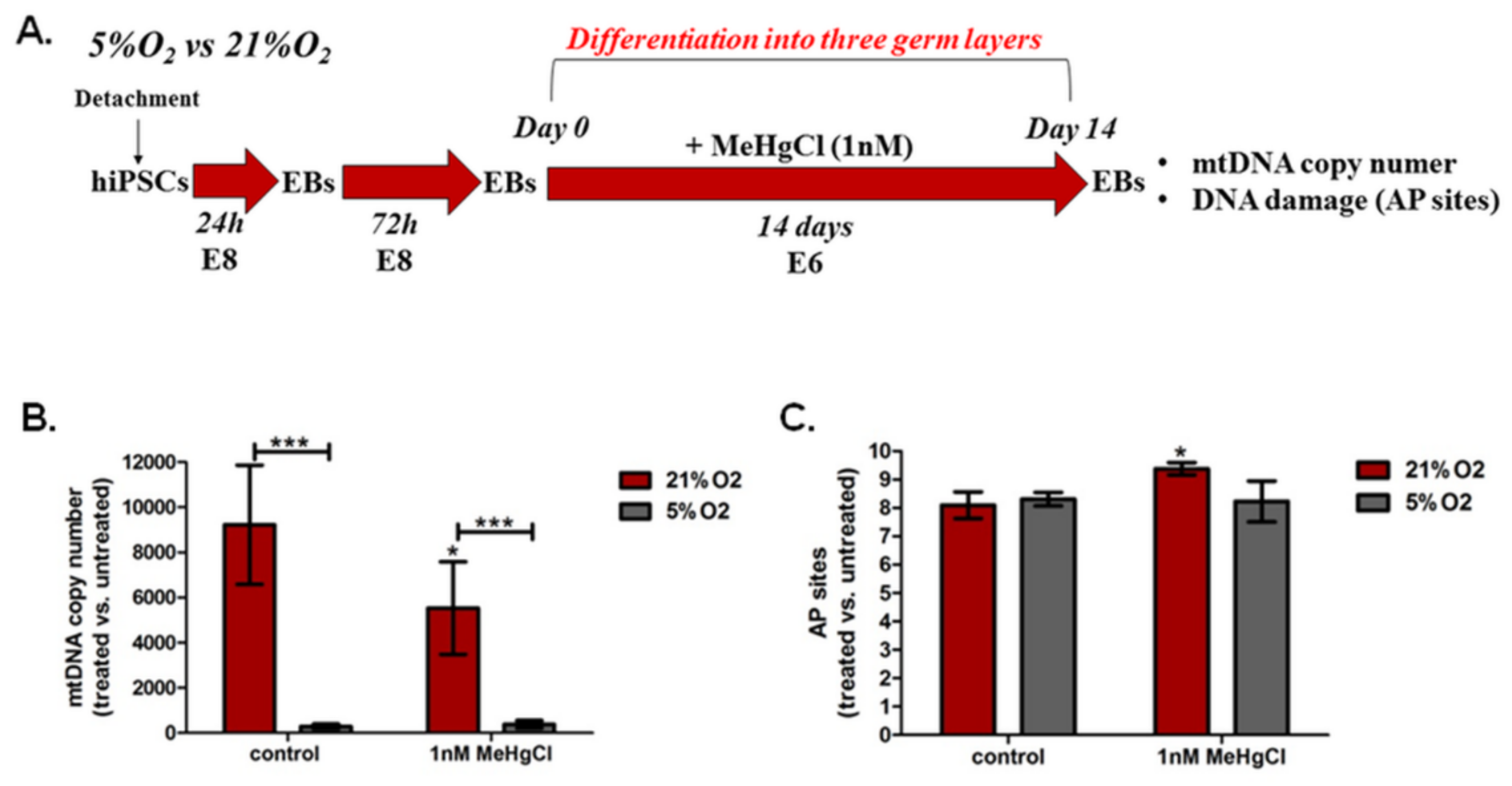
3.5. The Effect of Different Oxygen Concentrations and a Low Dose of MeHgCl (1 nM) on mtDNA Copy Number and DNA Damage in EBs
3.5.1. The Effect of Different Oxygen Concentrations and a Low Dose of MeHgCl (1 nM) on the mtDNA Copy Number in EBs
3.5.2. The Effect of Different Oxygen Concentrations and a Low Dose of MeHgCl (1 nM) on DNA Damage in EBs
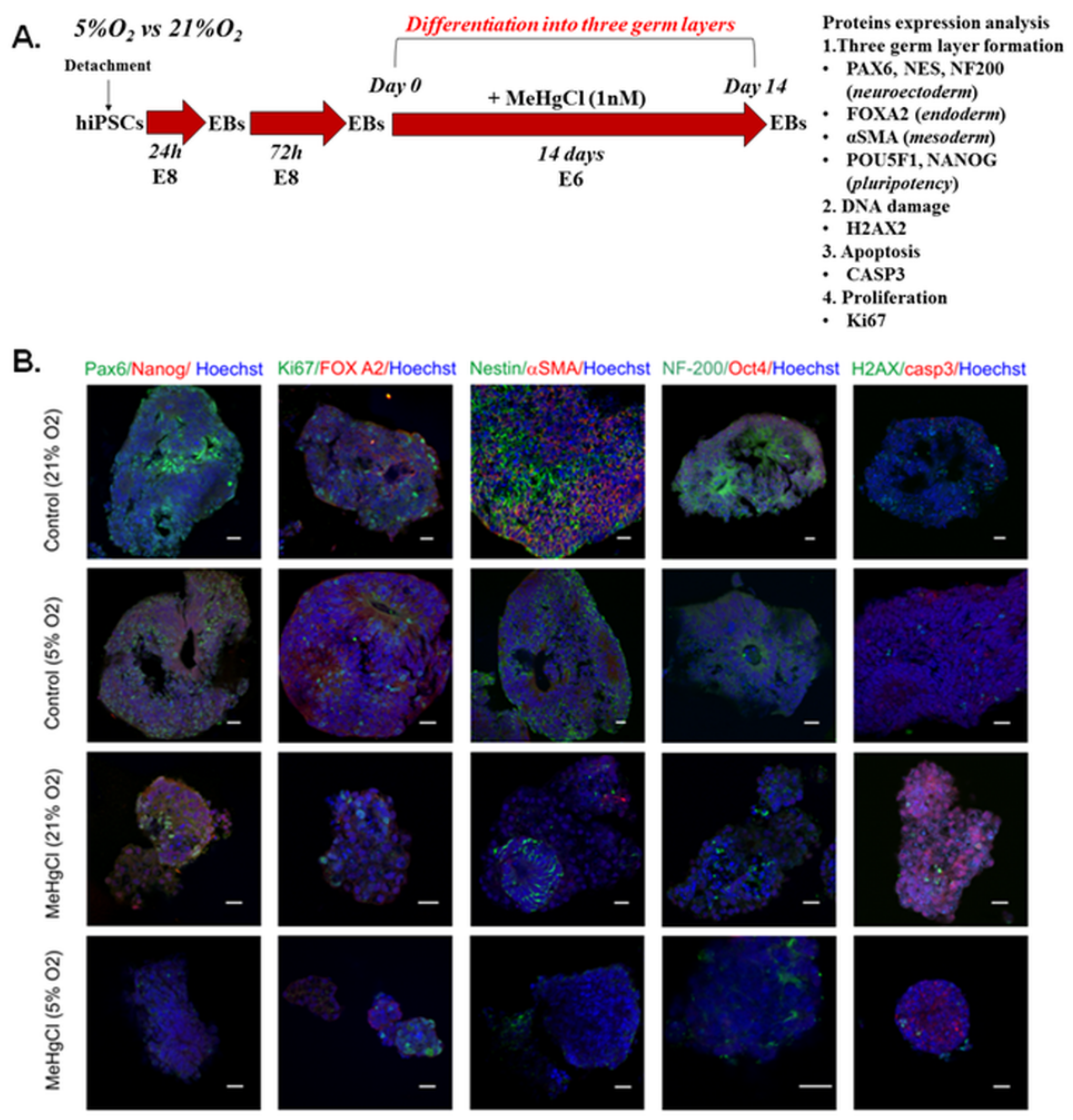
3.6. The Effect of Different Oxygen Concentrations and a Low Dose of MeHgCl (1 nM) on the Differentiation of EBs into Three Germ Layers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barbosa, R.V.; Point, D.; Médieu, A.; Allain, V.; Gillikin, D.P.; Couturier, L.I.E.; Munaron, J.M.; Roupsard, F.; Lorrain, A. Mercury Concentrations in Tuna Blood and Muscle Mirror Seawater Methylmercury in the Western and Central Pacific Ocean. Mar. Pollut. Bull. 2022, 180, 113801. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.; Mieiro, C.L.; Pereira, M.E.; Coelho, J.P. Mercury Bioaccessibility in Fish and Seafood: Effect of Method, Cooking and Trophic Level on Consumption Risk Assessment. Mar. Pollut. Bull. 2022, 179, 113736. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Kondo, M.; Akiyama, H.; Ogra, Y. Presence of Nano-Sized Mercury-Containing Particles in Seafoods, and an Estimate of Dietary Exposure. Environ. Pollut. 2022, 307, 119555. [Google Scholar] [CrossRef] [PubMed]
- Counter, S.A.; Buchanan, L.H. Mercury Exposure in Children: A Review. Toxicol. Appl. Pharmacol. 2004, 198, 209–230. [Google Scholar] [CrossRef] [PubMed]
- Crespo-López, M.E.; Macêdo, G.L.; Pereira, S.I.D.; Arrifano, G.P.F.; Picanço-Diniz, D.L.W.; do Nascimento, J.L.M.; Herculano, A.M. Mercury and Human Genotoxicity: Critical Considerations and Possible Molecular Mechanisms. Pharmacol. Res. 2009, 60, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Zhu, Z.; Huangfu, D. Human Pluripotent Stem Cells: An Emerging Model in Developmental Biology. Development 2013, 140, 705–717. [Google Scholar] [CrossRef]
- Keller, G. Embryonic Stem Cell Differentiation: Emergence of a New Era in Biology and Medicine. Genes Dev. 2005, 19, 1129–1155. [Google Scholar] [CrossRef]
- Pera, M.F.; Trounson, A.O. Human Embryonic Stem Cells: Prospects for Development. Development 2004, 131, 5515–5525. [Google Scholar] [CrossRef]
- Fu, X.; Cui, K.; Yi, Q.; Yu, L.; Xu, Y. DNA Repair Mechanisms in Embryonic Stem Cells. Cell. Mol. Life Sci. 2017, 74, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Maynard, S.; Swistowska, A.M.; Lee, J.W.; Liu, Y.; Liu, S.-T.; da Cruz, A.B.; Rao, M.; de Souza-Pinto, N.C.; Zeng, X.; Bohr, V.A. Human Embryonic Stem Cells Have Enhanced Repair of Multiple Forms of DNA Damage. Stem Cells 2008, 26, 2266–2274. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.H.; Yoon, S.; Koh, Y.E.; Seo, Y.J.; Kim, K.P. Maintenance of Genome Integrity and Active Homologous Recombination in Embryonic Stem Cells. Exp. Mol. Med. 2020, 52, 1220–1229. [Google Scholar] [CrossRef]
- Guo, N.N.; Liu, L.P.; Zhang, Y.X.; Cai, Y.T.; Guo, Y.; Zheng, Y.W.; Li, Y.M. Early Prediction of the Differentiation Potential during the Formation of Human IPSC-Derived Embryoid Bodies. Biochem. Biophys. Res. Commun. 2019, 516, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Itskovitz-Eldor, J.; Schuldiner, M.; Karsenti, D.; Eden, A.; Yanuka, O.; Amit, M.; Soreq, H.; Benvenisty, N. Differentiation of Human Embryonic Stem Cells into Embryoid Bodies Compromising the Three Embryonic Germ Layers. Mol. Med. 2000, 6, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Fridley, K.M.; Nair, R.; McDevitt, T.C. Differential Expression of Extracellular Matrix and Growth Factors by Embryoid Bodies in Hydrodynamic and Static Cultures. Tissue Eng. Part C Methods 2014, 20, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Zeevaert, K.; Elsafi Mabrouk, M.H.; Wagner, W.; Goetzke, R. Cell Mechanics in Embryoid Bodies. Cells 2020, 9, 2270. [Google Scholar] [CrossRef] [PubMed]
- Isaja, L.; Ferriol-Laffouillere, S.L.; Mucci, S.; Rodríguez-Varela, M.S.; Romorini, L. Embryoid Bodies–Based Multilineage Differentiation of Human Embryonic Stem Cells Grown on Feeder-Free Conditions. Methods Mol. Biol. 2022, 2520, 189–198. [Google Scholar] [CrossRef]
- Bratt-Leal, A.M.; Carpenedo, R.L.; McDevitt, T.C. Engineering the Embryoid Body Microenvironment to Direct Embryonic Stem Cell Differentiation. Biotechnol. Prog. 2009, 25, 43–51. [Google Scholar] [CrossRef]
- Fuchs, C.; Scheinast, M.; Pasteiner, W.; Lagger, S.; Hofner, M.; Hoellrigl, A.; Schultheis, M.; Weitzer, G.; Univ, A.; Scheinast, F.; et al. Self-Organization Phenomena in Embryonic Stem Cell-Derived Embryoid Bodies: Axis Formation and Breaking of Symmetry during Cardiomyogenesis. Cells Tissues Organs 2012, 195, 377–391. [Google Scholar] [CrossRef]
- Pekkanen-Mattila, M.; Pelto-Huikko, M.; Kujala, V.; Suuronen, R.; Skottman, H.; Aalto-Setälä, K.; Kerkelä, E. Spatial and Temporal Expression Pattern of Germ Layer Markers during Human Embryonic Stem Cell Differentiation in Embryoid Bodies. Histochem. Cell Biol. 2010, 133, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Dvash, T.; Mayshar, Y.; Darr, H.; McElhaney, M.; Barker, D.; Yanuka, O.; Kotkow, K.J.; Rubin, L.L.; Benvenisty, N.; Eiges, R. Temporal Gene Expression during Differentiation of Human Embryonic Stem Cells and Embryoid Bodies. Hum. Reprod. 2004, 19, 2875–2883. [Google Scholar] [CrossRef]
- Brickman, J.M.; Serup, P. Properties of Embryoid Bodies. Wiley Interdiscip. Rev. Dev. Biol. 2017, 6, e259. [Google Scholar] [CrossRef]
- Weatherbee, B.A.T.; Cui, T.; Zernicka-Goetz, M. Modeling Human Embryo Development with Embryonic and Extra-Embryonic Stem Cells. Dev. Biol. 2021, 474, 91–99. [Google Scholar] [CrossRef]
- Molè, M.A.; Weberling, A.; Zernicka-Goetz, M. Comparative Analysis of Human and Mouse Development: From Zygote to Pre-Gastrulation. Curr. Top. Dev. Biol. 2020, 136, 113–138. [Google Scholar] [CrossRef]
- Wamaitha, S.E.; Niakan, K.K. Human Pre-Gastrulation Development. Curr. Top. Dev. Biol. 2018, 128, 295–338. [Google Scholar] [CrossRef] [PubMed]
- Augustyniak, J.; Lenart, J.; Gaj, P.; Kolanowska, M.; Jazdzewski, K.; Stepien, P.P.; Buzanska, L. Bezafibrate Upregulates Mitochondrial Biogenesis and Influence Neural Differentiation of Human-Induced Pluripotent Stem Cells. Mol. Neurobiol. 2019, 56, 4346–4363. [Google Scholar] [CrossRef]
- Augustyniak, J.; Lenart, J.; Zychowicz, M.; Lipka, G.; Gaj, P.; Kolanowska, M.; Stepien, P.P.; Buzanska, L. Sensitivity of HiPSC-Derived Neural Stem Cells (NSC) to Pyrroloquinoline Quinone Depends on Their Developmental Stage. Toxicol. Vitr. 2017, 45, 434–444. [Google Scholar] [CrossRef]
- Augustyniak, J.; Lipka, G.; Kozlowska, H.; Caloni, F.; Buzanska, L. Oxygen as an Important Factor Modulating in Vitro MeHgCl Toxicity Associated with Mitochondrial Genes in HiPSCs. Ecotoxicol. Environ. Saf. 2022, 241, 113737. [Google Scholar] [CrossRef] [PubMed]
- Augustyniak, J.; Lenart, J.; Zychowicz, M.; Stepien, P.P.; Buzanska, L. Mitochondrial Biogenesis and Neural Differentiation of Human IPSC Is Modulated by Idebenone in a Developmental Stage-Dependent Manner. Biogerontology 2017, 18, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Konala, V.B.R.; Nandakumar, S.; Surendran, H.; Datar, S.; Bhonde, R.; Pal, R. Neuronal and Cardiac Toxicity of Pharmacological Compounds Identified through Transcriptomic Analysis of Human Pluripotent Stem Cell-Derived Embryoid Bodies. Toxicol. Appl. Pharmacol. 2021, 433, 115792. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, S.Y.; Ahn, C.; Yoo, Y.M.; Kim, C.W.; Kim, J.E.; Jo, N.R.; Kang, H.Y.; Jung, E.M.; Kim, K.S.; et al. Second-Phase Validation Study of an Alternative Developmental Toxicity Test Using Mouse Embryonic Stem Cell-Derived Embryoid Bodies. J. Physiol. Pharmacol. 2020, 71, 1–11. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, S.Y.; Ahn, C.; Kim, C.W.; Kim, J.; Kim, J.E.; Jo, N.R.; Kang, H.Y.; Yoo, Y.M.; Jung, E.M.; et al. Pre-Validation Study of Alternative Developmental Toxicity Test Using Mouse Embryonic Stem Cell-Derived Embryoid Bodies. Food Chem. Toxicol. 2019, 123, 50–56. [Google Scholar] [CrossRef]
- Worley, K.E.; Rico-Varela, J.; Ho, D.; Wan, L.Q. Teratogen Screening with Human Pluripotent Stem Cells. Integr. Biol. 2018, 10, 491–501. [Google Scholar] [CrossRef]
- Ivanovic, Z. Respect the Anaerobic Nature of Stem Cells to Exploit Their Potential in Regenerative Medicine. Regen. Med. 2013, 8, 677–680. [Google Scholar] [CrossRef]
- Mohyeldin, A.; Garzón-Muvdi, T.; Quiñones-Hinojosa, A. Oxygen in Stem Cell Biology: A Critical Component of the Stem Cell Niche. Cell Stem Cell 2010, 7, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Fathollahipour, S.; Patil, P.S.; Leipzig, N.D. Oxygen Regulation in Development: Lessons from Embryogenesis towards Tissue Engineering. Cells Tissues Organs 2018, 205, 350. [Google Scholar] [CrossRef]
- Kirkegaard, K.; Hindkjaer, J.J.; Ingerslev, H.J. Effect of Oxygen Concentration on Human Embryo Development Evaluated by Time-Lapse Monitoring. Fertil. Steril. 2013, 99, 738–744.e4. [Google Scholar] [CrossRef] [PubMed]
- Herbemont, C.; Labrosse, J.; Bennani-Smires, B.; Cedrin-Durnerin, I.; Peigne, M.; Sermondade, N.; Sarandi, S.; Vivot, A.; Vicaut, E.; Talib, Z.; et al. Impact of Oxygen Tension According to Embryo Stage of Development: A Prospective Randomized Study. Sci. Rep. 2021, 11, 22313. [Google Scholar] [CrossRef]
- Szablowska-Gadomska, I.; Zayat, V.; Winiarska, H.; Buzanska, L. Influence of Low Oxygen Tensions on Expression of Pluripotency Genes in HUCB-NSC. Acta Neurobiol. Exp. 2011, 71, 86–93. [Google Scholar]
- Szablowska-Gadomska, I.; Sypecka, J.; Zayat, V.; Podobinska, M.; Pastwinska, A.; Pienkowska-Grela, B.; Buzanska, L. Treatment with Small Molecules Is an Important Milestone towards the Induction of Pluripotency in Neural Stem Cells Derived from Human Cord Blood. Acta Neurobiol. Exp. 2012, 72, 337–350. [Google Scholar]
- Augustyniak, J.; Zychowicz, M.; Podobinska, M.; Barta, T.; Buzanska, L. Reprogramming of Somatic Cells: Possible Methods to Derive Safe, Clinical-Grade Human Induced Pluripotent Stem Cells. Acta Neurobiol. Exp. 2014, 74, 373–382. [Google Scholar]
- Nit, K.; Tyszka-Czochara, M.; Bobis-Wozowicz, S. Oxygen as a Master Regulator of Human Pluripotent Stem Cell Function and Metabolism. J. Pers. Med. 2021, 11, 905. [Google Scholar] [CrossRef]
- Zhu, S.; Li, W.; Zhou, H.; Wei, W.; Ambasudhan, R.; Lin, T.; Kim, J.; Zhang, K.; Ding, S. Reprogramming of Human Primary Somatic Cells by OCT4 and Chemical Compounds. Cell Stem Cell 2010, 7, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Prigione, A.; Fauler, B.; Lurz, R.; Lehrach, H.; Adjaye, J. The Senescence-Related Mitochondrial/Oxidative Stress Pathway Is Repressed in Human Induced Pluripotent Stem Cells. Stem Cells 2010, 28, 721–733. [Google Scholar] [CrossRef]
- Kaplan, A.R.; Glazer, P.M. Impact of Hypoxia on DNA Repair and Genome Integrity. Mutagenesis 2020, 35, 61–67. [Google Scholar] [CrossRef]
- Scanlon, S.E.; Glazer, P.M. Multifaceted Control of DNA Repair Pathways by the Hypoxic Tumor Microenvironment. DNA Repair 2015, 32, 180–189. [Google Scholar] [CrossRef]
- Bindra, R.S.; Crosby, M.E.; Glazer, P.M. Regulation of DNA Repair in Hypoxic Cancer Cells. Cancer Metastasis Rev. 2007, 26, 249–260. [Google Scholar] [CrossRef]
- Bindra, R.S.; Schaffer, P.J.; Meng, A.; Woo, J.; Måseide, K.; Roth, M.E.; Lizardi, P.; Hedley, D.W.; Bristow, R.G.; Glazer, P.M. Alterations in DNA Repair Gene Expression under Hypoxia: Elucidating the Mechanisms of Hypoxia-Induced Genetic Instability. Ann. N. Y. Acad. Sci. 2005, 1059, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Wells, P.G.; McCallum, G.P.; Lam, K.C.H.; Henderson, J.T.; Ondovcik, S.L. Oxidative DNA Damage and Repair in Teratogenesis and Neurodevelopmental Deficits. Birth Defects Res. C Embryo Today 2010, 90, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Khokhlova, E.V.; Fesenko, Z.S.; Sopova, J.V.; Leonova, E.I. Features of DNA Repair in the Early Stages of Mammalian Embryonic Development. Genes 2020, 11, 1138. [Google Scholar] [CrossRef] [PubMed]
- Juan, H.C.; Lin, Y.; Chen, H.R.; Fann, M.J. Cdk12 Is Essential for Embryonic Development and the Maintenance of Genomic Stability. Cell Death Differ. 2016, 23, 1038. [Google Scholar] [CrossRef]
- Hamdoun, A.; Epel, D. Embryo Stability and Vulnerability in an Always Changing World. Proc. Natl. Acad. Sci. USA 2007, 104, 1745–1750. [Google Scholar] [CrossRef]
- Ganapathy, S.; Farrell, E.R.; Vaghela, S.; Joshee, L.; Ford, E.G.; Uchakina, O.; McKallip, R.J.; Barkin, J.L.; Bridges, C.C. Transport and Toxicity of Methylmercury-Cysteine in Cultured BeWo Cells. Int. J. Mol. Sci. 2022, 23, 394. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Chen, G.; Li, W.; Kepp, O.; Zhu, Y.; Chen, Q. Mitophagy, Mitochondrial Homeostasis, and Cell Fate. Front. Cell Dev. Biol. 2020, 8, 467. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.L.; Fornace, A.J. Mammalian DNA Damage-Inducible Genes Associated with Growth Arrest and Apoptosis. Mutat. Res. 1996, 340, 109–124. [Google Scholar] [CrossRef]
- Zychowicz, M.; Pietrucha, K.; Podobinska, M.; Kowalska-Wlodarczyk, M.; Lenart, J.; Augustyniak, J.; Buzanska, L. The Collagen Scaffold Supports HiPSC-Derived NSC Growth and Restricts HiPSC. Front. Biosci. Sch. 2019, 11, 105–121. [Google Scholar] [CrossRef]
- Mesnage, R.; Teixeira, M.; Mandrioli, D.; Falcioni, L.; Ibragim, M.; Ducarmon, Q.R.; Zwittink, R.D.; Amiel, C.; Panoff, J.M.; Bourne, E.; et al. Multi-Omics Phenotyping of the Gut-Liver Axis Reveals Metabolic Perturbations from a Low-Dose Pesticide Mixture in Rats. Commun. Biol. 2021, 4, 471. [Google Scholar] [CrossRef]
- Rooney, J.P.; Ryde, I.T.; Sanders, L.H.; Howlett, E.v.; Colton, M.D.; Germ, K.E.; Mayer, G.D.; Timothy Greenamyre, J.; Meyer, J.N. PCR Based Determination of Mitochondrial DNA Copy Number in Multiple Species. Methods Mol. Biol. 2015, 1241, 23–28. [Google Scholar] [CrossRef]
- Augustyniak, J.; Lenart, J.; Lipka, G.; Stepien, P.P.; Buzanska, L. Reference Gene Validation via RT–QPCR for Human IPSC-Derived Neural Stem Cells and Neural Progenitors. Mol. Neurobiol. 2019, 56, 6820. [Google Scholar] [CrossRef]
- Akutsu, H.; Machida, M.; Kanzaki, S.; Sugawara, T.; Ohkura, T.; Nakamura, N.; Yamazaki-Inoue, M.; Miura, T.; Vemuri, M.C.; Rao, M.S.; et al. Xenogeneic-Free Defined Conditions for Derivation and Expansion of Human Embryonic Stem Cells with Mesenchymal Stem Cells. Regen. Ther. 2015, 1, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Beltcheva, M.; Gontarz, P.; Zhang, B.; Popli, P.; Fischer, L.A.; Khan, S.A.; Park, K.M.; Yoon, E.J.; Xing, X.; et al. Derivation of Trophoblast Stem Cells from Naïve Human Pluripotent Stem Cells. eLife 2020, 9, e52504. [Google Scholar] [CrossRef]
- Flamier, A.; Singh, S.; Rasmussen, T.P. A Standardized Human Embryoid Body Platform for the Detection and Analysis of Teratogens. PLoS ONE 2017, 12, e0171101. [Google Scholar] [CrossRef] [PubMed]
- Ungrin, M.D.; Joshi, C.; Nica, A.; Bauwens, C.; Zandstra, P.W. Reproducible, Ultra High-Throughput Formation of Multicellular Organization from Single Cell Suspension-Derived Human Embryonic Stem Cell Aggregates. PLoS ONE 2008, 3, e1565. [Google Scholar] [CrossRef]
- Zhou, D.; Shao, L.; Spitz, D.R. Reactive Oxygen Species in Normal and Tumor Stem Cells. Adv. Cancer Res. 2014, 122, 1–67. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, Y.; Zhang, Y. Mitochondrial Reactive Oxygen Species Cause Major Oxidative Mitochondrial DNA Damages and Repair Pathways. J. Biosci. 2020, 45, 84. [Google Scholar] [CrossRef]
- Chen, H.; Chan, D.C. Mitochondrial Dynamics in Regulating the Unique Phenotypes of Cancer and Stem Cells. Cell Metab. 2017, 26, 39–48. [Google Scholar] [CrossRef]
- Hu, C.; Fan, L.; Cen, P.; Chen, E.; Jiang, Z.; Li, L. Energy Metabolism Plays a Critical Role in Stem Cell Maintenance and Differentiation. Int. J. Mol. Sci. 2016, 17, 253. [Google Scholar] [CrossRef] [PubMed]
- Klungland, A.; Bjelland, S. Oxidative Damage to Purines in DNA: Role of Mammalian Ogg1. DNA Repair 2007, 6, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Pietrzak, J.; Spickett, C.M.; Płoszaj, T.; Virág, L.; Robaszkiewicz, A. PARP1 Promoter Links Cell Cycle Progression with Adaptation to Oxidative Environment. Redox Biol. 2018, 18, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Eustermann, S.; Wu, W.F.; Langelier, M.F.; Yang, J.C.; Easton, L.E.; Riccio, A.A.; Pascal, J.M.; Neuhaus, D. Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1. Mol. Cell 2015, 60, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, L.H.; Luz, A.L.; Cao, X.; Maurer, L.L.; Blawas, A.M.; Aballay, A.; Pan, W.K.Y.; Meyer, J.N. Effects of Methyl and Inorganic Mercury Exposure on Genome Homeostasis and Mitochondrial Function in Caenorhabditis Elegans. DNA Repair 2017, 52, 31–48. [Google Scholar] [CrossRef]
- Pieper, I.; Wehe, C.A.; Bornhorst, J.; Ebert, F.; Leffers, L.; Holtkamp, M.; Höseler, P.; Weber, T.; Mangerich, A.; Bürkle, A.; et al. Mechanisms of Hg Species Induced Toxicity in Cultured Human Astrocytes: Genotoxicity and DNA-Damage Response. Metallomics 2014, 6, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Kaguni, L.S. DNA Polymerase Gamma, the Mitochondrial Replicase. Annu. Rev. Biochem. 2004, 73, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Bogenhagen, D.F.; Pinz, K.G.; Perez-Jannotti, R.M. Enzymology of Mitochondrial Base Excision Repair. Prog. Nucleic Acid Res. Mol. Biol. 2001, 68, 257–271. [Google Scholar] [CrossRef]
- Hance, N.; Ekstrand, M.I.; Trifunovic, A. Mitochondrial DNA Polymerase Gamma Is Essential for Mammalian Embryogenesis. Hum. Mol. Genet. 2005, 14, 1775–1783. [Google Scholar] [CrossRef]
- Sunderland, P.; Augustyniak, J.; Lenart, J.; Bużańska, L.; Carlessi, L.; Delia, D.; Sikora, E. ATM-Deficient Neural Precursors Develop Senescence Phenotype with Disturbances in Autophagy. Mech. Ageing Dev. 2020, 190, 111296. [Google Scholar] [CrossRef]
- Shiloh, Y. ATM and Related Protein Kinases: Safeguarding Genome Integrity. Nat. Rev. Cancer 2003, 3, 155–168. [Google Scholar] [CrossRef]
- Lu, J.; Li, H.; Baccei, A.; Sasaki, T.; Gilbert, D.M.; Lerou, P.H. Influence of ATM-Mediated DNA Damage Response on Genomic Variation in Human Induced Pluripotent Stem Cells. Stem Cells Dev. 2016, 25, 740–747. [Google Scholar] [CrossRef]
- Bhuller, Y.; Wells, P.G. A Developmental Role for Ataxia-Telangiectasia Mutated in Protecting the Embryo from Spontaneous and Phenytoin-Enhanced Embryopathies in Culture. Toxicol. Sci. 2006, 93, 156–163. [Google Scholar] [CrossRef]
- Bhuller, Y.; Jeng, W.; Wells, P.G. Variable in Vivo Embryoprotective Role for Ataxia-Telangiectasia-Mutated against Constitutive and Phenytoin-Enhanced Oxidative Stress in Atm Knockout Mice. Toxicol. Sci. 2006, 93, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Laposa, R.R.; Henderson, J.T.; Xu, E.; Wells, P.G. Atm-Null Mice Exhibit Enhanced Radiation-Induced Birth Defects and a Hybrid Form of Embryonic Programmed Cell Death Indicating a Teratological Suppressor Function for ATM. FASEB J. 2004, 18, 896–898. [Google Scholar] [CrossRef] [PubMed]
- Lezza, A.M.S. Mitochondrial Transcription Factor A (TFAM): One Actor for Different Roles. Front. Biol. 2012, 7, 30–39. [Google Scholar] [CrossRef]
- Kang, D.; Kim, S.H.; Hamasaki, N. Mitochondrial Transcription Factor A (TFAM): Roles in Maintenance of MtDNA and Cellular Functions. Mitochondrion 2007, 7, 39–44. [Google Scholar] [CrossRef]
- Larsson, N.-G.; Wang, J.; Wilhelmsson, H.; Oldfors, A.; Rustin, P.; Lewandoski, M.; Barsh, G.S.; Clayton, D.A. Mitochondrial transcription factor A is necessary for mtDNA maintance and embryogenesis in mice. Nat. Genet. 1998, 18, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L. Mitochondrial DNA Degradation: A Quality Control Measure for Mitochondrial Genome Maintenance and Stress Response. Enzymes 2019, 45, 311–341. [Google Scholar] [CrossRef]
- Canugovi, C.; Maynard, S.; Bayne, A.C.V.; Sykora, P.; Tian, J.; de Souza-Pinto, N.C.; Croteau, D.L.; Bohr, V.A. The Mitochondrial Transcription Factor A Functions in Mitochondrial Base Excision Repair. DNA Repair 2010, 9, 1080–1089. [Google Scholar] [CrossRef]
- Xu, W.; Boyd, R.M.; Tree, M.O.; Samkari, F.; Zhao, L. Mitochondrial Transcription Factor A Promotes DNA Strand Cleavage at Abasic Sites. Proc. Natl. Acad. Sci. USA 2019, 116, 17792–17799. [Google Scholar] [CrossRef]
- Onishi, M.; Yamano, K.; Sato, M.; Matsuda, N.; Okamoto, K. Molecular Mechanisms and Physiological Functions of Mitophagy. EMBO J. 2021, 40, e104705. [Google Scholar] [CrossRef]
- Binarová, P.; Tuszynski, J. Tubulin: Structure, Functions and Roles in Disease. Cells 2019, 8, 1294. [Google Scholar] [CrossRef]
- Vicente, J.J.; Wordeman, L. The Quantification and Regulation of Microtubule Dynamics in the Mitotic Spindle. Curr. Opin. Cell Biol. 2019, 60, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]

| Gene Symbol | GenBank Number | Primer Sequence | Amplicon Length (bp) |
|---|---|---|---|
| HBB F | NG_059281.1 | GCTCGGTGCCTTTAGTGATG | 136 |
| HBB R | ACATCAAGCGTCCCATAGAC | ||
| ND1 F | DQ092356.1 | TACGGGCTACTACAACCCTTC | 77 |
| ND1 R | ATGGTAGATGTGGCGGGTTT |
| Primers | GenBank Number | Primer Sequence | Amplicon Length |
|---|---|---|---|
| ATMF | NM_001351834.2 | AGGAATCACTGGATCGCTGTC | 100 |
| AMTR | CGTGAACACCGGACAAGAGT | ||
| OGG1F | NM_002542.6 | AGACCAACAAGGAACTGGGAAAC | 81 |
| OGG1R | CACTGAACAGCACCGCTTGG | ||
| PARP1F | NM_001618.4 | CAGCTTCTGGAGGACGACAA | 108 |
| PARP1R | CTCCTTGGACGGCATCTGTT | ||
| TFAM F | NM_005011.4 | TGAAAGATTCCAAGAAGCTAAGGGT | 132 |
| TFAM R | TAACGAGTTTCGTCCTCTTTAGCAT | ||
| POLG1 F | NM_002693.3 | GGCATTGTTGCTTGTTGGGT | 144 |
| POLG1 R | TTTCCCCTTCTAGGGCACTG | ||
| PARK2 F | NM_004562.3 | TCCCAGTGGAGGTCGATTCT | 105 |
| PARK2 R | CCTGCGAAAATCACACGCAA | ||
| SOX17F | NM_022454.4 | TGGACCGCACGGAATTTGAA | 101 |
| SOX17R | GCTGTCGGGGAGATTCACAC | ||
| NES F | NM_006617.2 | CCCCGTCGGTCTCTTTTCTC | 96 |
| NES R | TCGTCTGACCCACTGAGGAT | ||
| TBXT F | NM_003181.4 | CGATCCTGGGTGTGCGTAA | 100 |
| TBXT R | CCGATGCCTCAACTCTCCAG |
| Primers | GenBank Number | Primer Sequence | Amplicon Length |
|---|---|---|---|
| ACTB F | NM_001101.3 | GCTCACCATGGATGATGATATCGC | 169 |
| ACTB R | CACATAGGAATCCTTCTGACCCAT | ||
| EEF1A1 F | NM_001402.5 | TGTTCCTTTGGTCAACACCGA | 122 |
| EEF1A1 R | ACAACCCTATTCTCCACCCA | ||
| EID2 F | NM_153232.3 | GGCATCGCTCTGTCCAGTTA | 74 |
| EID2 R | GCTTGGACATCTCAGACCGT | ||
| GAPDH F | NM_002046.5 | GTTCGACAGTCAGCCGCATC | 90 |
| GAPDH R | TCCGTTGACTCCGACCTTCA | ||
| RPLP0 F | NM_001002.3 | CCTCGTGGAAGTGACATCGT | 76 |
| RPLP0 R | CTGTCTTCCCTGGGCATCAC | ||
| TBP F | NM_003194.4 | GCAAGGGTTTCTGGTTTGCC | 80 |
| TBP R | CAAGCCCTGAGCGTAAGGTG |
| Antibodies | Catalog Number | Company | Dilution |
|---|---|---|---|
| anti-OCT4, (rabbit/IgG) | PA5-27438 | Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA | 1:1000 |
| anti-NANOG, (rabbit/IgG) | PA1-097 | Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA | 1:200 |
| anti-PAX6, (mouse/IgG1) | Ma1-109 | Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA | 1:200 |
| anti-NESTIN, (mouse/IgG1) | Mab5326 | Sigma-Aldrich, Merck KGaA, Darmstadt, Germany | 1:500 |
| anti-NF200, (mouse/IgG1) | N0142 | Sigma-Aldrich, Merck KGaA, Darmstadt, Germany | 1:200 |
| anti-αSMA, (mouse/IgG2a) | A2547 | Sigma-Aldrich, Merck KGaA, Darmstadt, Germany | 1:600 |
| anti-FOXA2, (mouse/IgG2aκ) | WH0003170M1 | Sigma-Aldrich, Merck KGaA, Darmstadt, Germany | 1:300 |
| anti-Histone H2A.X phosphorylated at Ser139, (mouse/IgG1) | 05-636 | Sigma-Aldrich, Merck KGaA, Darmstadt, Germany | 1:100 |
| anti-CASP3, (rabbit/IgG) | 9664 | Cell Signaling Technologies, Danvers, MA, USA | 1:1000 |
| anti-Ki67, (rabbit/IgG) | AB9260 | Sigma-Aldrich, Merck KGaA, Darmstadt, Germany | 1:1000 |
| Antibodies | Catalog Number | Company | Dilution |
|---|---|---|---|
| Alexa Fluor 488-conjugated goat anti-rabbit IgG (H+L) cross-adsorbed secondary antibody | A11008 | Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA | 1:1000 |
| Alexa Fluor 546-conjugated goat anti-rabbit IgG (H+L) highly cross-adsorbed secondary antibody | A11035 | Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA | 1:1000 |
| Alexa Fluor 488-conjugated goat anti-mouse IgG1 cross-adsorbed secondary antibody | A-21121 | Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA | 1:1000 |
| Alexa Fluor 546-conjugated goat anti-mouse IgG2a cross-adsorbed secondary antibody | A21133 | Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA | 1:1000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Augustyniak, J.; Kozlowska, H.; Buzanska, L. Genes Involved in DNA Repair and Mitophagy Protect Embryoid Bodies from the Toxic Effect of Methylmercury Chloride under Physioxia Conditions. Cells 2023, 12, 390. https://doi.org/10.3390/cells12030390
Augustyniak J, Kozlowska H, Buzanska L. Genes Involved in DNA Repair and Mitophagy Protect Embryoid Bodies from the Toxic Effect of Methylmercury Chloride under Physioxia Conditions. Cells. 2023; 12(3):390. https://doi.org/10.3390/cells12030390
Chicago/Turabian StyleAugustyniak, Justyna, Hanna Kozlowska, and Leonora Buzanska. 2023. "Genes Involved in DNA Repair and Mitophagy Protect Embryoid Bodies from the Toxic Effect of Methylmercury Chloride under Physioxia Conditions" Cells 12, no. 3: 390. https://doi.org/10.3390/cells12030390
APA StyleAugustyniak, J., Kozlowska, H., & Buzanska, L. (2023). Genes Involved in DNA Repair and Mitophagy Protect Embryoid Bodies from the Toxic Effect of Methylmercury Chloride under Physioxia Conditions. Cells, 12(3), 390. https://doi.org/10.3390/cells12030390








