HGCA2.0: An RNA-Seq Based Webtool for Gene Coexpression Analysis in Homo sapiens
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcriptomic Data Processing
2.2. Gene Coexpression Tree Construction
2.3. External Data Collection and Biological Term Enrichment Calculation
2.4. Webtool Implementation and Usage
2.5. API Access
2.6. STRING Analysis
3. Results
3.1. Use Cases
3.1.1. Ribosomal Proteins
3.1.2. Metallothioneins
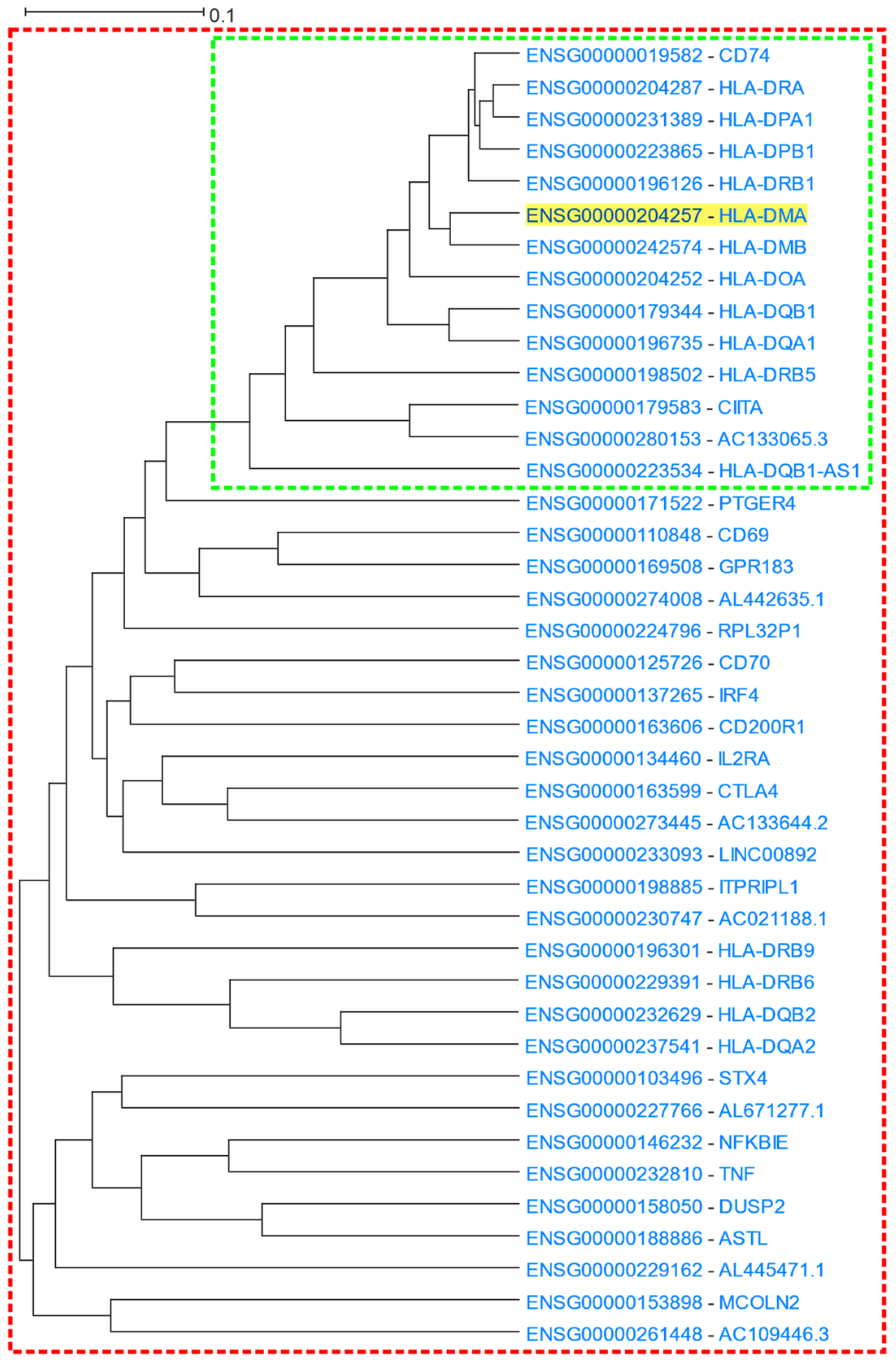
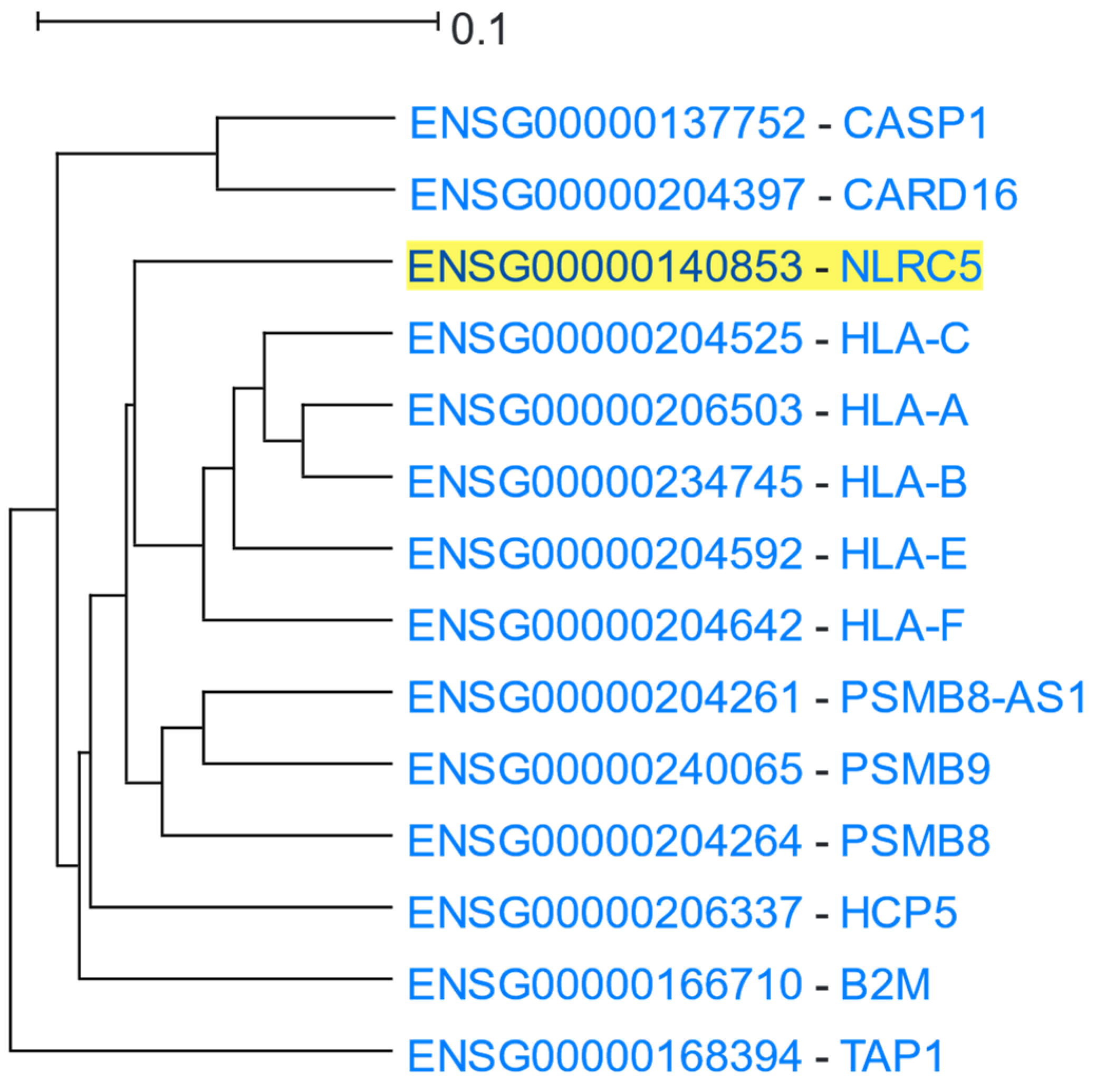
3.1.3. MHC Class I and Class II Protein Clusters
3.1.4. STAT1 Transcription Factor
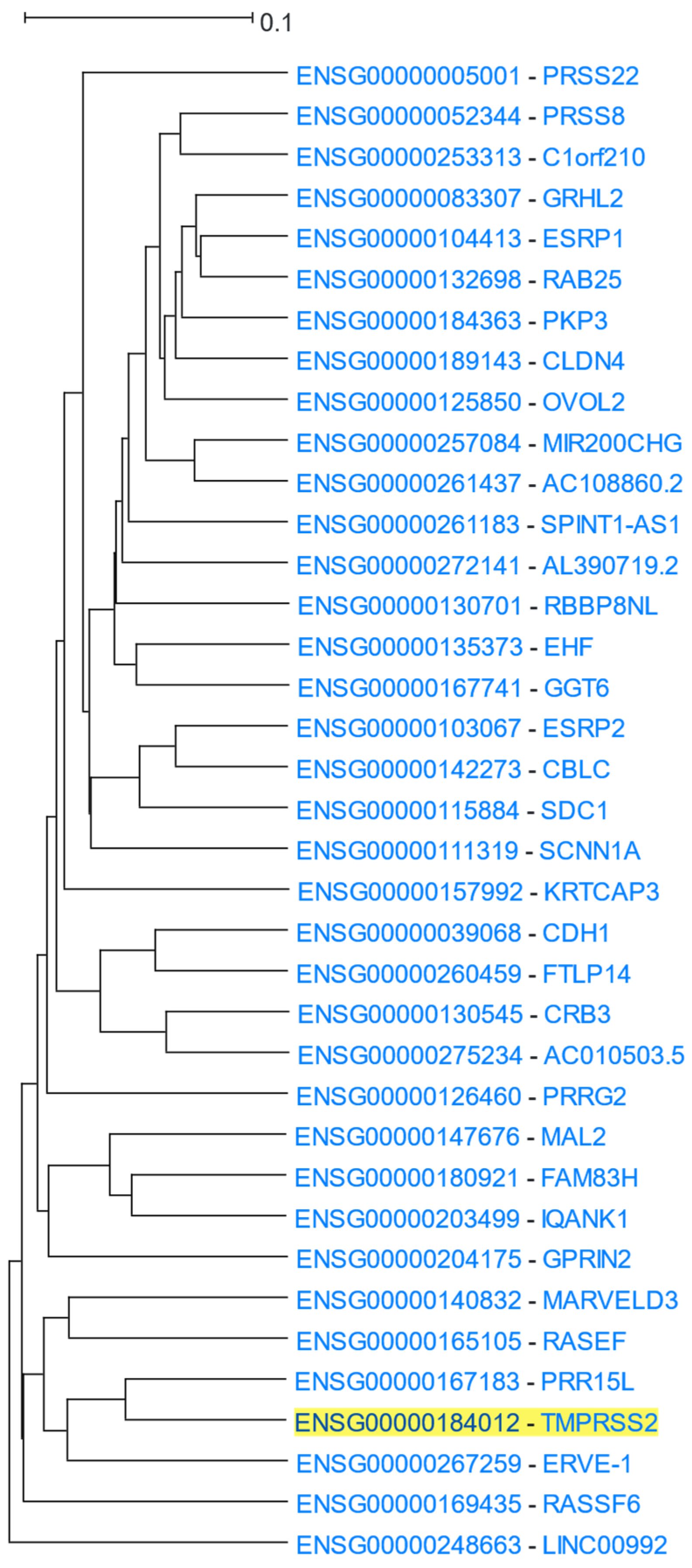
3.1.5. TMPRSS2 in Relation to COVID-19 Infection
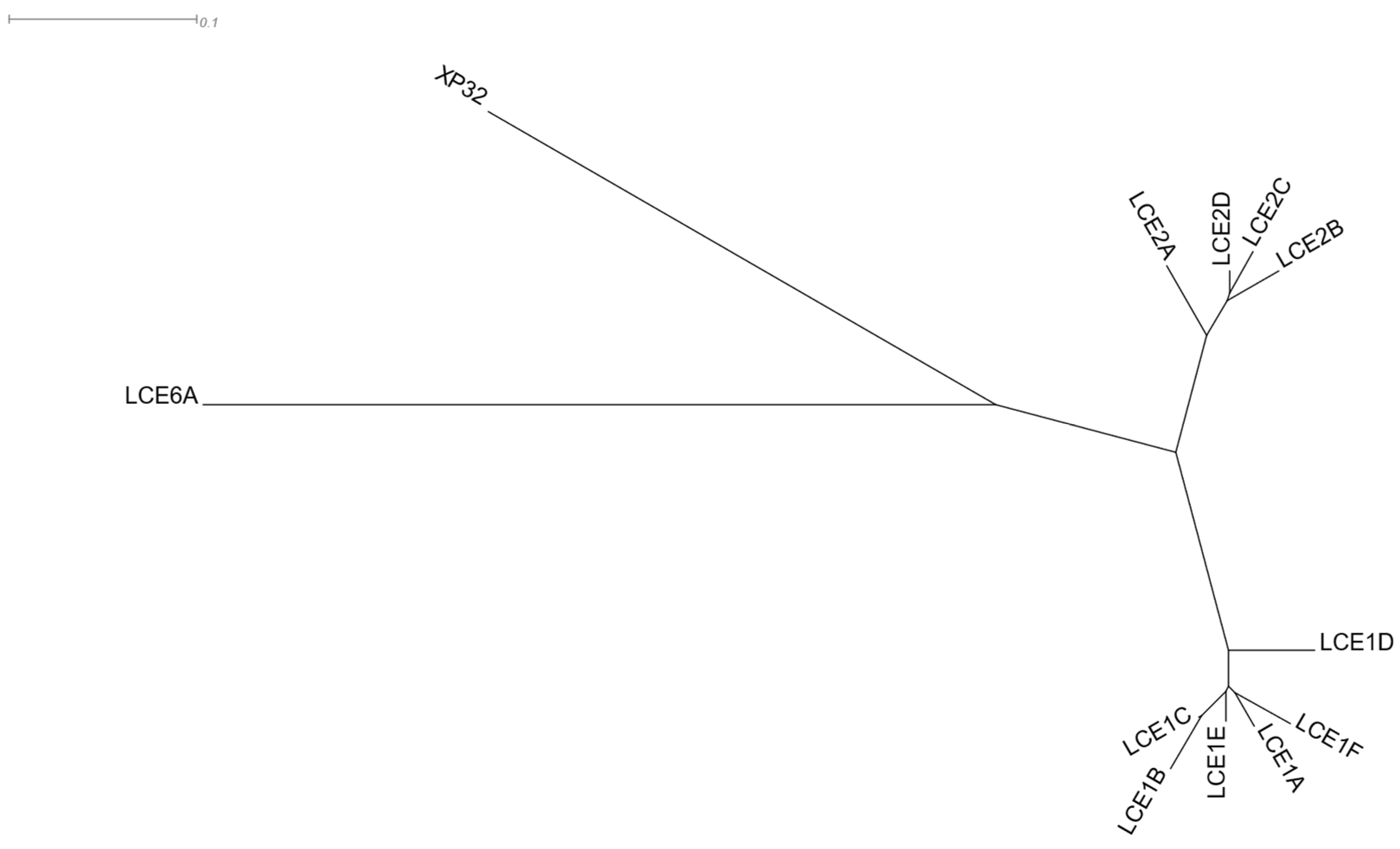
3.1.6. Late Cornified Envelope Genes
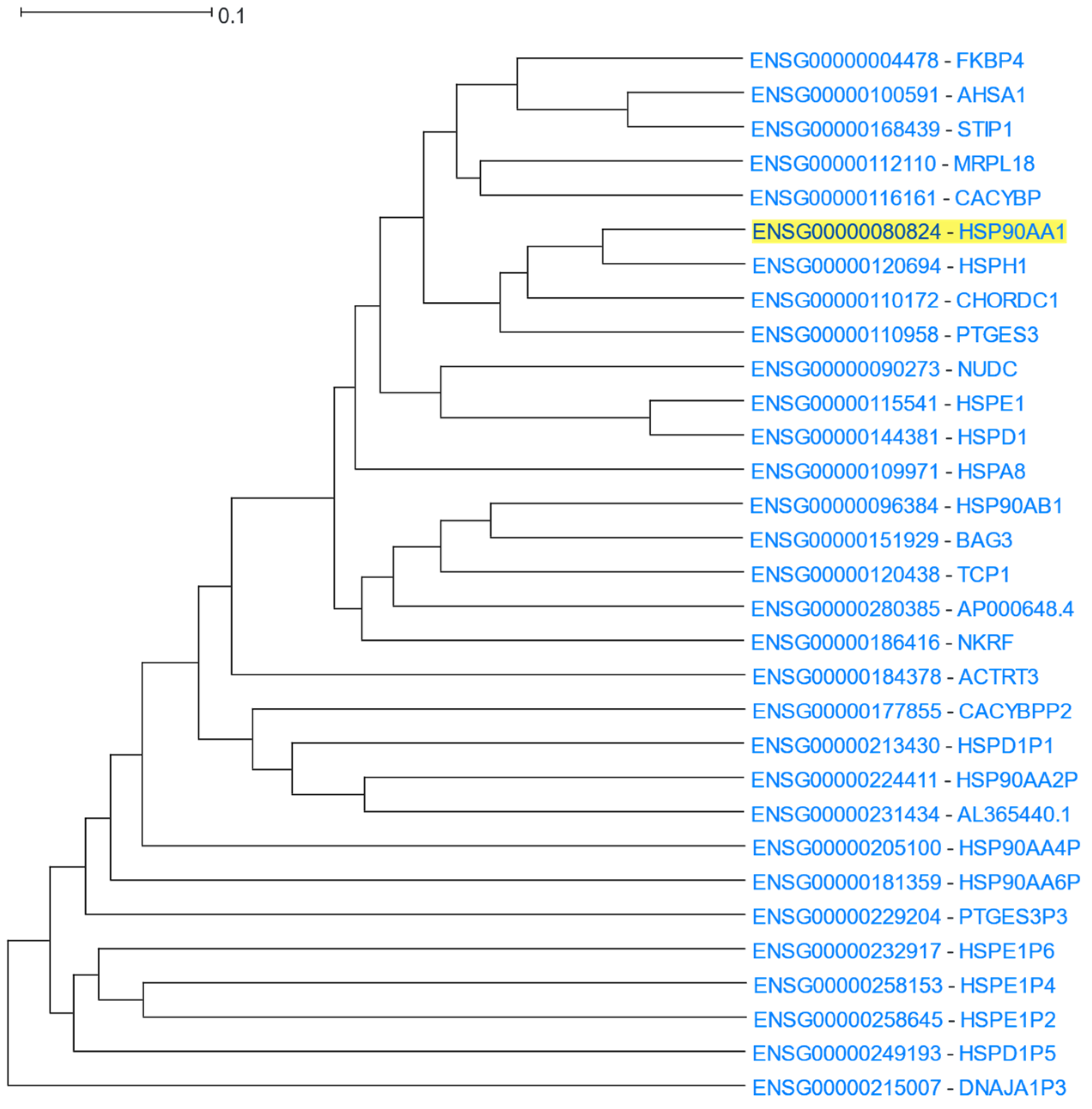
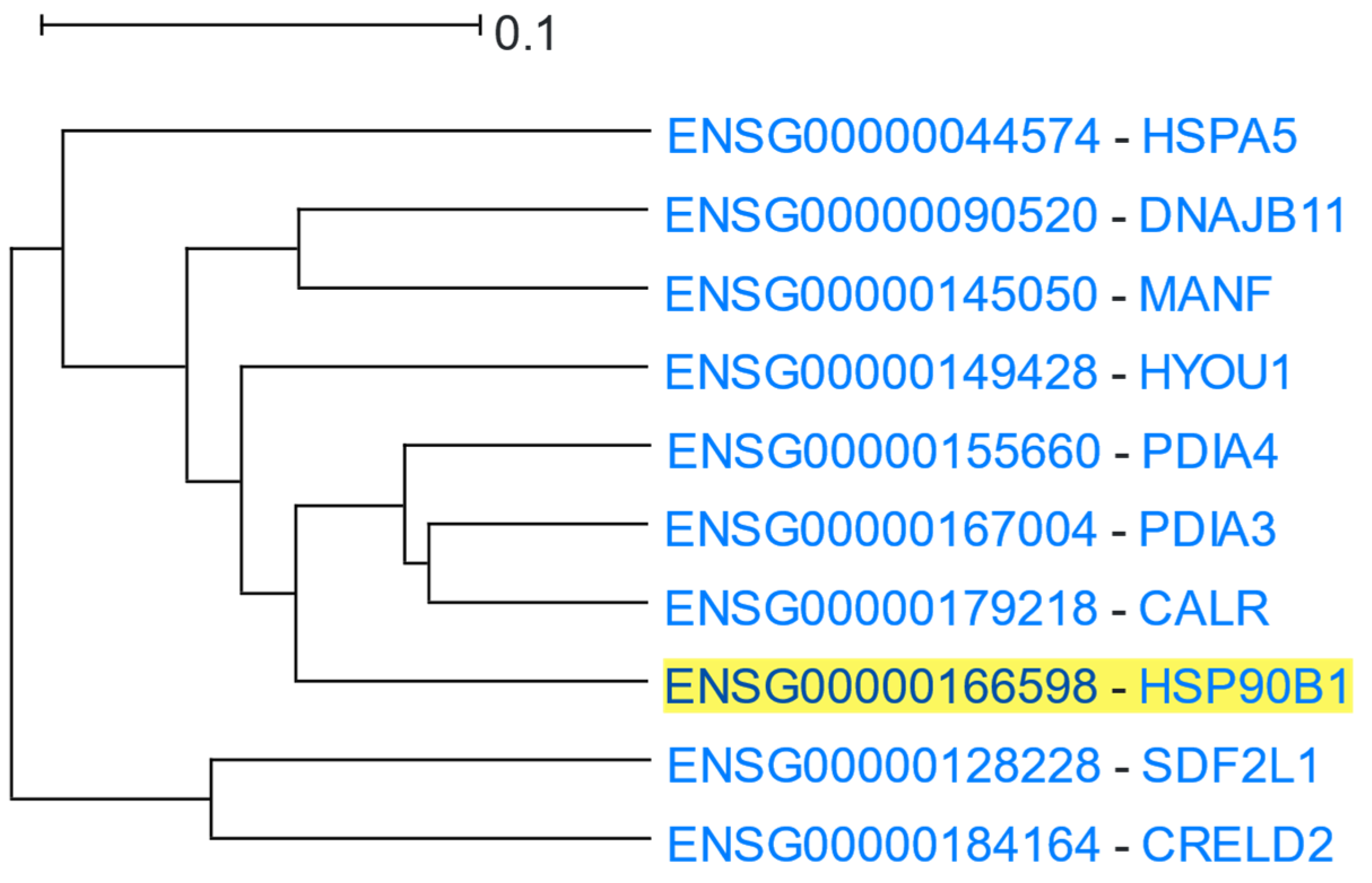
3.1.7. Heat Shock Protein 90
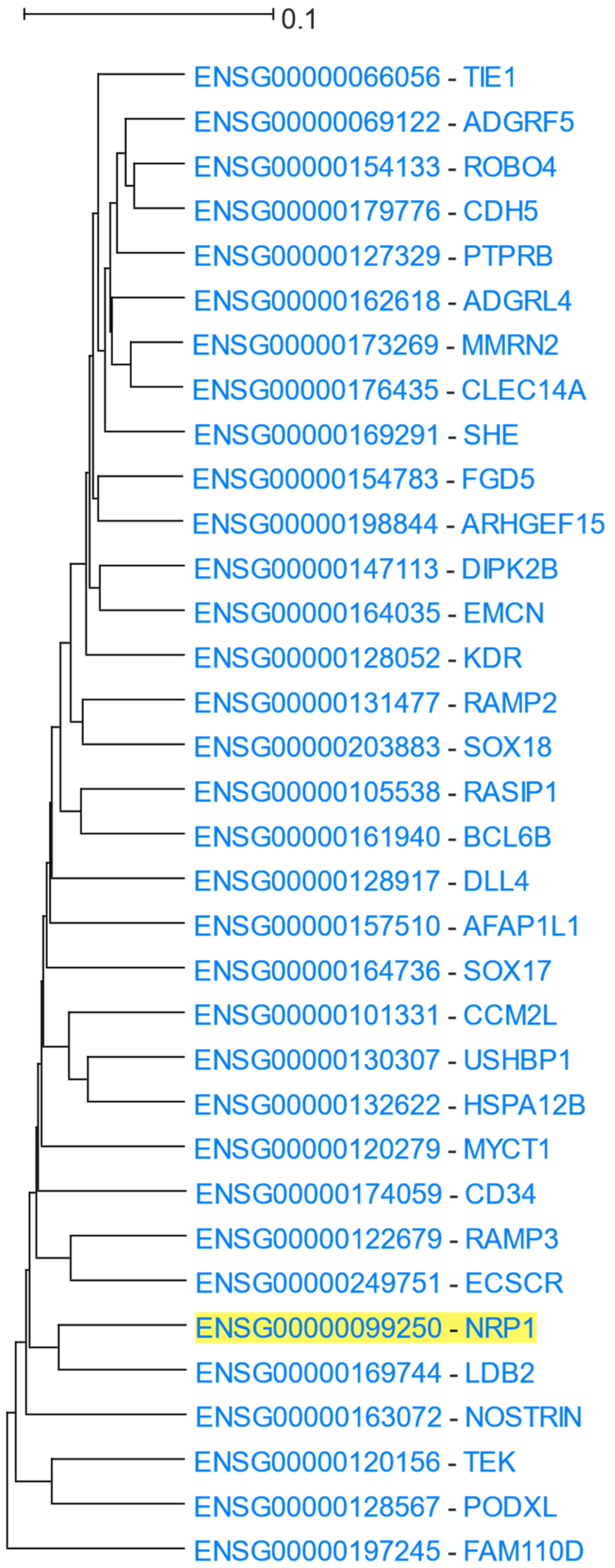
3.1.8. Neurovascular Genes
3.1.9. Olfactory Receptors
3.1.10. Glucocorticoid Receptor Signalling
3.1.11. ALS and LGMD Related Genes
3.1.12. Growth Hormones
3.1.13. Antisense Genes
3.2. Coexpression Tool Benchmarking
4. Discussion
4.1. Comparison with Previous HGCA Version
4.2. Comparison of Coexpression Webtools
4.3. Limitations
4.4. Interpretation of HGCA2.0 Predictions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zogopoulos, V.L.; Saxami, G.; Malatras, A.; Papadopoulos, K.; Tsotra, I.; Iconomidou, V.A.; Michalopoulos, I. Approaches in Gene Coexpression Analysis in Eukaryotes. Biology 2022, 11, 1019. [Google Scholar] [CrossRef]
- Zogopoulos, V.L.; Malatras, A.; Michalopoulos, I. Gene coexpression analysis in Arabidopsis thaliana based on public microarray data. STAR Protoc. 2022, 3, 101208. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Obayashi, T.; Mutwil, M.; Giorgi, F.M.; Bassel, G.W.; Tanimoto, M.; Chow, A.; Steinhauser, D.; Persson, S.; Provart, N.J. Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009, 32, 1633–1651. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Kodama, Y.; Shumway, M.; Leinonen, R.; International Nucleotide Sequence Database, C. The Sequence Read Archive: Explosive growth of sequencing data. Nucleic Acids Res. 2012, 40, D54–D56. [Google Scholar] [CrossRef]
- Kolesnikov, N.; Hastings, E.; Keays, M.; Melnichuk, O.; Tang, Y.A.; Williams, E.; Dylag, M.; Kurbatova, N.; Brandizi, M.; Burdett, T.; et al. ArrayExpress update--simplifying data submissions. Nucleic Acids Res. 2015, 43, D1113–D1116. [Google Scholar] [CrossRef]
- Papatheodorou, I.; Moreno, P.; Manning, J.; Fuentes, A.M.; George, N.; Fexova, S.; Fonseca, N.A.; Fullgrabe, A.; Green, M.; Huang, N.; et al. Expression Atlas update: From tissues to single cells. Nucleic Acids Res. 2020, 48, D77–D83. [Google Scholar] [CrossRef] [PubMed]
- Amid, C.; Alako, B.T.F.; Balavenkataraman Kadhirvelu, V.; Burdett, T.; Burgin, J.; Fan, J.; Harrison, P.W.; Holt, S.; Hussein, A.; Ivanov, E.; et al. The European Nucleotide Archive in 2019. Nucleic Acids Res. 2020, 48, D70–D76. [Google Scholar] [CrossRef]
- Hutter, C.; Zenklusen, J.C. The Cancer Genome Atlas: Creating Lasting Value beyond Its Data. Cell 2018, 173, 283–285. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulos, I.; Pavlopoulos, G.A.; Malatras, A.; Karelas, A.; Kostadima, M.A.; Schneider, R.; Kossida, S. Human gene correlation analysis (HGCA): A tool for the identification of transcriptionally co-expressed genes. BMC Res. Notes 2012, 5, 265. [Google Scholar] [CrossRef]
- Aoki, Y.; Okamura, Y.; Ohta, H.; Kinoshita, K.; Obayashi, T. ALCOdb: Gene Coexpression Database for Microalgae. Plant Cell Physiol. 2016, 57, e3. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Hirsch-Hoffmann, M.; Hennig, L.; Gruissem, W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004, 136, 2621–2632. [Google Scholar] [CrossRef]
- Tseng, K.C.; Li, G.Z.; Hung, Y.C.; Chow, C.N.; Wu, N.Y.; Chien, Y.Y.; Zheng, H.Q.; Lee, T.Y.; Kuo, P.L.; Chang, S.B.; et al. EXPath 2.0: An Updated Database for Integrating High-Throughput Gene Expression Data with Biological Pathways. Plant Cell Physiol. 2020, 61, 1818–1827. [Google Scholar] [CrossRef] [PubMed]
- Obayashi, T.; Hibara, H.; Kagaya, Y.; Aoki, Y.; Kinoshita, K. ATTED-II v11: A Plant Gene Coexpression Database Using a Sample Balancing Technique by Subagging of Principal Components. Plant Cell Physiol. 2022, 63, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Obayashi, T.; Kagaya, Y.; Aoki, Y.; Tadaka, S.; Kinoshita, K. COXPRESdb v7: A gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res. 2019, 47, D55–D62. [Google Scholar] [CrossRef] [PubMed]
- Raina, P.; Lopes, I.; Chatsirisupachai, K.; Farooq, Z.; de Magalhães, J.P. GeneFriends 2021: Updated co-expression databases and tools for human and mouse genes and transcripts. bioRxiv 2021, 2021:2021.2001.2010.426125. [Google Scholar] [CrossRef]
- Miller, H.E.; Bishop, A.J.R. Correlation AnalyzeR: Functional predictions from gene co-expression correlations. BMC Bioinform. 2021, 22, 206. [Google Scholar] [CrossRef] [PubMed]
- Fionda, V. Networks in Biology. In Encyclopedia of Bioinformatics and Computational Biology; Ranganathan, S., Gribskov, M., Nakai, K., Schönbach, C., Eds.; Academic Press: Oxford, UK, 2019; pp. 915–921. [Google Scholar]
- Serin, E.A.R.; Nijveen, H.; Hilhorst, H.W.M.; Ligterink, W. Learning from Co-expression Networks: Possibilities and Challenges. Front. Plant Sci. 2016, 7, 444. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Aguet, F.; Barbeira, A.N.; Bonazzola, R.; Brown, A.; Castel, S.E.; Jo, B.; Kasela, S.; Kim-Hellmuth, S.; Liang, Y.; Oliva, M.; et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. bioRxiv 2019, 2019:787903. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Kinsella, R.J.; Kahari, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database (Oxford) 2011, 2011, bar030. [Google Scholar] [CrossRef] [PubMed]
- Paulson, J.N.; Chen, C.Y.; Lopes-Ramos, C.M.; Kuijjer, M.L.; Platig, J.; Sonawane, A.R.; Fagny, M.; Glass, K.; Quackenbush, J. Tissue-aware RNA-Seq processing and normalization for heterogeneous and sparse data. BMC Bioinform. 2017, 18, 437. [Google Scholar] [CrossRef]
- Hicks, S.C.; Okrah, K.; Paulson, J.N.; Quackenbush, J.; Irizarry, R.A.; Bravo, H.C. Smooth quantile normalization. Biostatistics 2018, 19, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Pearson, K. VII. Note on regression and inheritance in the case of two parents. Proc. R. Soc. Lond. 1895, 58, 240–242. [Google Scholar] [CrossRef]
- D’Haeseleer, P. How does gene expression clustering work? Nat. Biotechnol. 2005, 23, 1499. [Google Scholar] [CrossRef] [PubMed]
- Schliep, K.; Potts, A.J.; Morrison, D.A.; Grimm, G.W. Intertwining phylogenetic trees and networks. Methods Ecol. Evol. 2017, 8, 1212–1220. [Google Scholar] [CrossRef]
- Sokal, R.R.; Michener, C.D. A statistical method for evaluating systematic relationships. Univ. Kansas Sci. Bull. 1958, 38, 1409–1438. [Google Scholar]
- Zogopoulos, V.L.; Saxami, G.; Malatras, A.; Angelopoulou, A.; Jen, C.H.; Duddy, W.J.; Daras, G.; Hatzopoulos, P.; Westhead, D.R.; Michalopoulos, I. Arabidopsis Coexpression Tool: A tool for gene coexpression analysis in Arabidopsis thaliana. iScience 2021, 24, 102848. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Distance Matrix Programs. Available online: http://evolution.genetics.washington.edu/phylip/doc/distance.html (accessed on 20 January 2023).
- Archie, J.; Day, H.E.W.; Felsenstein, J.; Maddison, W.; Meacham, C.; Rohlf, F.J.; Swofford, D. The Newick Tree Format. Available online: http://evolution.genetics.washington.edu/phylip/newicktree.html (accessed on 20 January 2023).
- Tweedie, S.; Braschi, B.; Gray, K.; Jones, T.E.M.; Seal, R.L.; Yates, B.; Bruford, E.A. Genenames.org: The HGNC and VGNC resources in 2021. Nucleic Acids Res. 2021, 49, D939–D946. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Martens, M.; Ammar, A.; Riutta, A.; Waagmeester, A.; Slenter, D.N.; Hanspers, K.; Ryan, A.M.; Digles, D.; Lopes, E.N.; Ehrhart, F.; et al. WikiPathways: Connecting communities. Nucleic Acids Res. 2021, 49, D613–D621. [Google Scholar] [CrossRef]
- Encode Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011, 9, e1001046. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016, 2016, baw100. [Google Scholar] [CrossRef] [PubMed]
- Cheneby, J.; Menetrier, Z.; Mestdagh, M.; Rosnet, T.; Douida, A.; Rhalloussi, W.; Bergon, A.; Lopez, F.; Ballester, B. ReMap 2020: A database of regulatory regions from an integrative analysis of Human and Arabidopsis DNA-binding sequencing experiments. Nucleic Acids Res. 2020, 48, D180–D188. [Google Scholar] [CrossRef] [PubMed]
- McKusick, V.A. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007, 80, 588–604. [Google Scholar] [CrossRef]
- Pinero, J.; Ramirez-Anguita, J.M.; Sauch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Forbes, C.; Evans, M.; Hastings, N.; Peacock, B. Statistical Distributions, 4th ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef] [PubMed]
- Rodchenkov, I.; Babur, O.; Luna, A.; Aksoy, B.A.; Wong, J.V.; Fong, D.; Franz, M.; Siper, M.C.; Cheung, M.; Wrana, M.; et al. Pathway Commons 2019 Update: Integration, analysis and exploration of pathway data. Nucleic Acids Res. 2020, 48, D489–D497. [Google Scholar] [CrossRef]
- Thanati, F.; Karatzas, E.; Baltoumas, F.A.; Stravopodis, D.J.; Eliopoulos, A.G.; Pavlopoulos, G.A. FLAME: A Web Tool for Functional and Literature Enrichment Analysis of Multiple Gene Lists. Biology 2021, 10, 665. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Obayashi, T.; Kodate, S.; Hibara, H.; Kagaya, Y.; Kinoshita, K. COXPRESdb v8: An animal gene coexpression database navigating from a global view to detailed investigations. Nucleic Acids Res. 2022, 51, D80–D87. [Google Scholar] [CrossRef]
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma’ayan, A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018, 9, 1366. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Wong, A.K.; Krishnan, A.; Aure, M.R.; Tadych, A.; Zhang, R.; Corney, D.C.; Greene, C.S.; Bongo, L.A.; Kristensen, V.N.; et al. Targeted exploration and analysis of large cross-platform human transcriptomic compendia. Nat. Methods 2015, 12, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ‘small-world’ networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Khatter, H.; Myasnikov, A.G.; Natchiar, S.K.; Klaholz, B.P. Structure of the human 80S ribosome. Nature 2015, 520, 640–645. [Google Scholar] [CrossRef]
- Narla, A.; Ebert, B.L. Ribosomopathies: Human disorders of ribosome dysfunction. Blood 2010, 115, 3196–3205. [Google Scholar] [CrossRef] [PubMed]
- Gazda, H.T.; Sheen, M.R.; Vlachos, A.; Choesmel, V.; O’Donohue, M.F.; Schneider, H.; Darras, N.; Hasman, C.; Sieff, C.A.; Newburger, P.E.; et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am. J. Hum. Genet. 2008, 83, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.J.; Kimura, T.; Sato, B.G.; Shi, Y.; Andrews, G.K. Metallothionein induction by hypoxia involves cooperative interactions between metal-responsive transcription factor-1 and hypoxia-inducible transcription factor-1alpha. Mol. Cancer Res. 2008, 6, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Holling, T.M.; Schooten, E.; van Den Elsen, P.J. Function and regulation of MHC class II molecules in T-lymphocytes: Of mice and men. Hum. Immunol. 2004, 65, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Roche, P.A. HLA-DM: An in vivo facilitator of MHC class II peptide loading. Immunity 1995, 3, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Kropshofer, H.; Hammerling, G.J.; Vogt, A.B. The impact of the non-classical MHC proteins HLA-DM and HLA-DO on loading of MHC class II molecules. Immunol. Rev. 1999, 172, 267–278. [Google Scholar] [CrossRef]
- Mellins, E.D.; Stern, L.J. HLA-DM and HLA-DO, key regulators of MHC-II processing and presentation. Curr. Opin. Immunol. 2014, 26, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.L.; Wei, X.S.; Zhang, M.; Niu, Y.R.; Zhou, Q. The Significance of Tumor Necrosis Factor Receptor Type II in CD8(+) Regulatory T Cells and CD8(+) Effector T Cells. Front. Immunol. 2018, 9, 583. [Google Scholar] [CrossRef]
- Faustman, D.L.; Davis, M. TNF Receptor 2 and Disease: Autoimmunity and Regenerative Medicine. Front. Immunol. 2013, 4, 478. [Google Scholar] [CrossRef]
- Zhang, T.; Ma, C.; Zhang, Z.; Zhang, H.; Hu, H. NF-kappaB signaling in inflammation and cancer. MedComm (2020) 2021, 2, 618–653. [Google Scholar] [CrossRef]
- Kearns, J.D.; Basak, S.; Werner, S.L.; Huang, C.S.; Hoffmann, A. IkappaBepsilon provides negative feedback to control NF-kappaB oscillations, signaling dynamics, and inflammatory gene expression. J. Cell Biol. 2006, 173, 659–664. [Google Scholar] [CrossRef]
- Alves, B.N.; Tsui, R.; Almaden, J.; Shokhirev, M.N.; Davis-Turak, J.; Fujimoto, J.; Birnbaum, H.; Ponomarenko, J.; Hoffmann, A. IkappaBepsilon is a key regulator of B cell expansion by providing negative feedback on cRel and RelA in a stimulus-specific manner. J. Immunol. 2014, 192, 3121–3132. [Google Scholar] [CrossRef]
- Nam, S.; Lim, J.S. Essential role of interferon regulatory factor 4 (IRF4) in immune cell development. Arch. Pharm. Res. 2016, 39, 1548–1555. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhai, S.; Zhang, J.; Zhang, D.; Wang, S.; Wang, L.; Yu, J. Interferon Regulatory Factor 4 Correlated With Immune Cells Infiltration Could Predict Prognosis for Patients With Lung Adenocarcinoma. Front Oncol. 2021, 11, 698465. [Google Scholar] [CrossRef] [PubMed]
- Meissner, T.B.; Li, A.; Biswas, A.; Lee, K.H.; Liu, Y.J.; Bayir, E.; Iliopoulos, D.; van den Elsen, P.J.; Kobayashi, K.S. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc. Natl. Acad. Sci. USA 2010, 107, 13794–13799. [Google Scholar] [CrossRef]
- Kobayashi, K.S.; van den Elsen, P.J. NLRC5: A key regulator of MHC class I-dependent immune responses. Nat. Rev. Immunol. 2012, 12, 813–820. [Google Scholar] [CrossRef]
- Davis, B.K.; Roberts, R.A.; Huang, M.T.; Willingham, S.B.; Conti, B.J.; Brickey, W.J.; Barker, B.R.; Kwan, M.; Taxman, D.J.; Accavitti-Loper, M.A.; et al. Cutting edge: NLRC5-dependent activation of the inflammasome. J. Immunol. 2011, 186, 1333–1337. [Google Scholar] [CrossRef] [PubMed]
- Najjar, I.; Fagard, R. STAT1 and pathogens, not a friendly relationship. Biochimie 2010, 92, 425–444. [Google Scholar] [CrossRef]
- Fraser, B.J.; Beldar, S.; Seitova, A.; Hutchinson, A.; Mannar, D.; Li, Y.; Kwon, D.; Tan, R.; Wilson, R.P.; Leopold, K.; et al. Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation. Nat. Chem. Biol. 2022, 18, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Gkogkou, E.; Barnasas, G.; Vougas, K.; Trougakos, I.P. Expression profiling meta-analysis of ACE2 and TMPRSS2, the putative anti-inflammatory receptor and priming protease of SARS-CoV-2 in human cells, and identification of putative modulators. Redox Biol. 2020, 36, 101615. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.M.; Bai, P.; He, W.; Wu, F.; Liu, X.F.; Han, D.M.; Liu, S.; Yang, J.K. Gender Differences in Patients With COVID-19: Focus on Severity and Mortality. Front. Public Health 2020, 8, 152. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Boon, A.C.M.; Michelson, A.P.; Foraker, R.E.; Zhan, M.; Payne, P.R.O. Estrogen hormone is an essential sex factor inhibiting inflammation and immune response in COVID-19. Sci. Rep. 2022, 12, 9462. [Google Scholar] [CrossRef] [PubMed]
- Henry, J.; Toulza, E.; Hsu, C.Y.; Pellerin, L.; Balica, S.; Mazereeuw-Hautier, J.; Paul, C.; Serre, G.; Jonca, N.; Simon, M. Update on the epidermal differentiation complex. Front. Biosci. (Landmark Ed.) 2012, 17, 1517–1532. [Google Scholar] [CrossRef]
- Deng, Z.; Matsuda, K.; Tanikawa, C.; Lin, J.; Furukawa, Y.; Hamamoto, R.; Nakamura, Y. Late Cornified Envelope Group I, a novel target of p53, regulates PRMT5 activity. Neoplasia 2014, 16, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gislason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Sillitoe, I.; Bordin, N.; Dawson, N.; Waman, V.P.; Ashford, P.; Scholes, H.M.; Pang, C.S.M.; Woodridge, L.; Rauer, C.; Sen, N.; et al. CATH: Increased structural coverage of functional space. Nucleic Acids Res. 2021, 49, D266–D273. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kubler, J.; Lozajic, M.; Gabler, F.; Soding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Huson, D.H.; Scornavacca, C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012, 61, 1061–1067. [Google Scholar] [CrossRef]
- Ritossa, F. Discovery of the heat shock response. Cell Stress Chaperones 1996, 1, 97–98. [Google Scholar] [CrossRef] [PubMed]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef]
- Wandinger, S.K.; Richter, K.; Buchner, J. The Hsp90 chaperone machinery. J. Biol. Chem. 2008, 283, 18473–18477. [Google Scholar] [CrossRef]
- Chen, B.; Zhong, D.; Monteiro, A. Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genom. 2006, 7, 156. [Google Scholar] [CrossRef] [PubMed]
- Zuehlke, A.D.; Beebe, K.; Neckers, L.; Prince, T. Regulation and function of the human HSP90AA1 gene. Gene 2015, 570, 8–16. [Google Scholar] [CrossRef]
- Fan, G.; Tu, Y.; Wu, N.; Xiao, H. The expression profiles and prognostic values of HSPs family members in Head and neck cancer. Cancer Cell Int. 2020, 20, 220. [Google Scholar] [CrossRef]
- Gano, J.J.; Simon, J.A. A proteomic investigation of ligand-dependent HSP90 complexes reveals CHORDC1 as a novel ADP-dependent HSP90-interacting protein. Mol. Cell. Proteom. MCP 2010, 9, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Shelton, L.B.; Koren, J., 3rd; Blair, L.J. Imbalances in the Hsp90 Chaperone Machinery: Implications for Tauopathies. Front. Neurosci. 2017, 11, 724. [Google Scholar] [CrossRef] [PubMed]
- Pelham, H.R.B. Speculations on the functions of the major heat shock and glucose-regulated proteins. Cell 1986, 46, 959–961. [Google Scholar] [CrossRef]
- Domingues, A.; Fantin, A. Neuropilin 1 Regulation of Vascular Permeability Signaling. Biomolecules 2021, 11, 666. [Google Scholar] [CrossRef] [PubMed]
- Trimmer, C.; Keller, A.; Murphy, N.R.; Snyder, L.L.; Willer, J.R.; Nagai, M.H.; Katsanis, N.; Vosshall, L.B.; Matsunami, H.; Mainland, J.D. Genetic variation across the human olfactory receptor repertoire alters odor perception. Proc. Natl. Acad. Sci. USA 2019, 116, 9475–9480. [Google Scholar] [CrossRef] [PubMed]
- Gilad, Y.; Lancet, D. Population differences in the human functional olfactory repertoire. Mol. Biol. Evol. 2003, 20, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Chess, A.; Simon, I.; Cedar, H.; Axel, R. Allelic inactivation regulates olfactory receptor gene expression. Cell 1994, 78, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Sokal, R.; Rohlf, F. The comparison of dendrograms by objective methods. Taxon 1962, 11, 33–40. [Google Scholar] [CrossRef]
- Nicolaides, N.C.; Charmandari, E.; Chrousos, G.P.; Kino, T. Recent advances in the molecular mechanisms determining tissue sensitivity to glucocorticoids: Novel mutations, circadian rhythm and ligand-induced repression of the human glucocorticoid receptor. BMC Endocr. Disord. 2014, 14, 71. [Google Scholar] [CrossRef] [PubMed]
- Grad, I.; Picard, D. The glucocorticoid responses are shaped by molecular chaperones. Mol. Cell Endocrinol. 2007, 275, 2–12. [Google Scholar] [CrossRef]
- Oakley, R.H.; Cidlowski, J.A. The biology of the glucocorticoid receptor: New signaling mechanisms in health and disease. J. Allergy Clin. Immunol. 2013, 132, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Cole, T.J.; Blendy, J.A.; Monaghan, A.P.; Krieglstein, K.; Schmid, W.; Aguzzi, A.; Fantuzzi, G.; Hummler, E.; Unsicker, K.; Schutz, G. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 1995, 9, 1608–1621. [Google Scholar] [CrossRef] [PubMed]
- Harbour, J.W.; Luo, R.X.; Santi, A.D.; Postigo, A.A.; Dean, D.C. Cdk Phosphorylation Triggers Sequential Intramolecular Interactions that Progressively Block Rb Functions as Cells Move through G1. Cell 1999, 98, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Rogatsky, I.; Trowbridge, J.M.; Garabedian, M.J. Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol. Cell Biol. 1997, 17, 3181–3193. [Google Scholar] [CrossRef]
- Zhang, Z.; Turer, E.; Li, X.; Zhan, X.; Choi, M.; Tang, M.; Press, A.; Smith, S.R.; Divoux, A.; Moresco, E.M.; et al. Insulin resistance and diabetes caused by genetic or diet-induced KBTBD2 deficiency in mice. Proc. Natl. Acad. Sci. USA 2016, 113, E6418–E6426. [Google Scholar] [CrossRef]
- Geering, B.; Cutillas, P.R.; Nock, G.; Gharbi, S.I.; Vanhaesebroeck, B. Class IA phosphoinositide 3-kinases are obligate p85-p110 heterodimers. Proc. Natl. Acad. Sci. USA 2007, 104, 7809–7814. [Google Scholar] [CrossRef]
- Arancibia, S.; Benitez, D.; Nunez, L.E.; Jewell, C.M.; Langjahr, P.; Candia, E.; Zapata-Torres, G.; Cidlowski, J.A.; Gonzalez, M.J.; Hermoso, M.A. Phosphatidylinositol 3-kinase interacts with the glucocorticoid receptor upon TLR2 activation. J. Cell Mol. Med. 2011, 15, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Heiska, L.; Carpen, O. Src phosphorylates ezrin at tyrosine 477 and induces a phosphospecific association between ezrin and a kelch-repeat protein family member. J. Biol. Chem. 2005, 280, 10244–10252. [Google Scholar] [CrossRef]
- Gautreau, A.; Poullet, P.; Louvard, D.; Arpin, M. Ezrin, a plasma membrane-microfilament linker, signals cell survival through the phosphatidylinositol 3-kinase/Akt pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 7300–7305. [Google Scholar] [CrossRef]
- van Dam, S.; Vosa, U.; van der Graaf, A.; Franke, L.; de Magalhaes, J.P. Gene co-expression analysis for functional classification and gene-disease predictions. Brief. Bioinform. 2018, 19, 575–592. [Google Scholar] [CrossRef]
- Kino, T.; De Martino, M.U.; Charmandari, E.; Mirani, M.; Chrousos, G.P. Tissue glucocorticoid resistance/hypersensitivity syndromes. J. Steroid Biochem. Mol. Biol. 2003, 85, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Stortz, M.; Pecci, A.; Presman, D.M.; Levi, V. Unraveling the molecular interactions involved in phase separation of glucocorticoid receptor. BMC Biol. 2020, 18, 59. [Google Scholar] [CrossRef]
- Rowland, L.P.; Shneider, N.A. Amyotrophic lateral sclerosis. N. Engl. J. Med. 2001, 344, 1688–1700. [Google Scholar] [CrossRef]
- Chio, A.; Logroscino, G.; Traynor, B.J.; Collins, J.; Simeone, J.C.; Goldstein, L.A.; White, L.A. Global epidemiology of amyotrophic lateral sclerosis: A systematic review of the published literature. Neuroepidemiology 2013, 41, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Connolly, O.; Le Gall, L.; McCluskey, G.; Donaghy, C.G.; Duddy, W.J.; Duguez, S. A Systematic Review of Genotype-Phenotype Correlation across Cohorts Having Causal Mutations of Different Genes in ALS. J. Pers. Med. 2020, 10, 58. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, A.; Kenna, K.P.; Renton, A.E.; Ticozzi, N.; Faghri, F.; Chia, R.; Dominov, J.A.; Kenna, B.J.; Nalls, M.A.; Keagle, P.; et al. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron 2018, 97, 1268–1283. [Google Scholar] [CrossRef] [PubMed]
- Volk, A.E.; Weishaupt, J.H.; Andersen, P.M.; Ludolph, A.C.; Kubisch, C. Current knowledge and recent insights into the genetic basis of amyotrophic lateral sclerosis. Med. Genet. 2018, 30, 252–258. [Google Scholar] [CrossRef]
- Van Damme, P. How much of the missing heritability of ALS is hidden in known ALS genes? J. Neurol. Neurosurg. Psychiatry 2018, 89, 794. [Google Scholar] [CrossRef]
- Vijayakumar, U.G.; Milla, V.; Cynthia Stafford, M.Y.; Bjourson, A.J.; Duddy, W.; Duguez, S.M. A Systematic Review of Suggested Molecular Strata, Biomarkers and Their Tissue Sources in ALS. Front. Neurol. 2019, 10, 400. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, L.; Anakor, E.; Connolly, O.; Vijayakumar, U.G.; Duddy, W.J.; Duguez, S. Molecular and Cellular Mechanisms Affected in ALS. J. Pers. Med. 2020, 10, 101. [Google Scholar] [CrossRef] [PubMed]
- Vissing, J. Limb girdle muscular dystrophies: Classification, clinical spectrum and emerging therapies. Curr. Opin. Neurol. 2016, 29, 635–641. [Google Scholar] [CrossRef]
- Liewluck, T.; Milone, M. Untangling the complexity of limb-girdle muscular dystrophies. Muscle Nerve 2018, 58, 167–177. [Google Scholar] [CrossRef]
- Straub, V.; Murphy, A.; Udd, B.; group, L.W.S. 229th ENMC international workshop: Limb girdle muscular dystrophies—Nomenclature and reformed classification Naarden, the Netherlands, 17–19 March 2017. Neuromuscul. Disord. 2018, 28, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Barton, E.R.; Pacak, C.A.; Stoppel, W.L.; Kang, P.B. The ties that bind: Functional clusters in limb-girdle muscular dystrophy. Skelet Muscle 2020, 10, 22. [Google Scholar] [CrossRef]
- Nguengang Wakap, S.; Lambert, D.M.; Olry, A.; Rodwell, C.; Gueydan, C.; Lanneau, V.; Murphy, D.; Le Cam, Y.; Rath, A. Estimating cumulative point prevalence of rare diseases: Analysis of the Orphanet database. Eur. J. Hum. Genet. 2020, 28, 165–173. [Google Scholar] [CrossRef]
- Fifita, J.A.; Williams, K.L.; Sundaramoorthy, V.; McCann, E.P.; Nicholson, G.A.; Atkin, J.D.; Blair, I.P. A novel amyotrophic lateral sclerosis mutation in OPTN induces ER stress and Golgi fragmentation in vitro. Amyotroph Lateral Scler. Front. Degener. 2017, 18, 126–133. [Google Scholar] [CrossRef]
- Barsh, G.S.; Seeburg, P.H.; Gelinas, R.E. The human growth hormone gene family: Structure and evolution of the chromosomal locus. Nucleic Acids Res. 1983, 11, 3939–3958. [Google Scholar] [CrossRef]
- Dehkhoda, F.; Lee, C.M.M.; Medina, J.; Brooks, A.J. The Growth Hormone Receptor: Mechanism of Receptor Activation, Cell Signaling, and Physiological Aspects. Front. Endocrinol. (Lausanne) 2018, 9, 35. [Google Scholar] [CrossRef]
- Strous, G.J.; Almeida, A.D.S.; Putters, J.; Schantl, J.; Sedek, M.; Slotman, J.A.; Nespital, T.; Hassink, G.C.; Mol, J.A. Growth Hormone Receptor Regulation in Cancer and Chronic Diseases. Front. Endocrinol. (Lausanne) 2020, 11, 597573. [Google Scholar] [CrossRef] [PubMed]
- Bole-Feysot, C.; Goffin, V.; Edery, M.; Binart, N.; Kelly, P.A. Prolactin (PRL) and its receptor: Actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocr. Rev. 1998, 19, 225–268. [Google Scholar] [CrossRef] [PubMed]
- Angulo, M.A.; Butler, M.G.; Cataletto, M.E. Prader-Willi syndrome: A review of clinical, genetic, and endocrine findings. J. Endocrinol. Invest. 2015, 38, 1249–1263. [Google Scholar] [CrossRef]
- Ehrhart, F.; Janssen, K.J.M.; Coort, S.L.; Evelo, C.T.; Curfs, L.M.G. Prader-Willi syndrome and Angelman syndrome: Visualisation of the molecular pathways for two chromosomal disorders. World J. Biol. Psychiatry 2019, 20, 670–682. [Google Scholar] [CrossRef]
- Chen, E.Y.; Liao, Y.C.; Smith, D.H.; Barrera-Saldana, H.A.; Gelinas, R.E.; Seeburg, P.H. The human growth hormone locus: Nucleotide sequence, biology, and evolution. Genomics 1989, 4, 479–497. [Google Scholar] [CrossRef]
- Pelechano, V.; Steinmetz, L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013, 14, 880–893. [Google Scholar] [CrossRef]
- Lan, T.; Li, H.; Zhang, D.; Xu, L.; Liu, H.; Hao, X.; Yan, X.; Liao, H.; Chen, X.; Xie, K.; et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 2019, 18, 186. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, S.; Cai, K.; Zheng, D.; Zhu, C.; Li, L.; Wang, F.; He, Z.; Yu, C.; Sun, C. Hypoxia-induced long noncoding RNA NR2F1-AS1 maintains pancreatic cancer proliferation, migration, and invasion by activating the NR2F1/AKT/mTOR axis. Cell Death Dis. 2022, 13, 232. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Piccolo, S.R.; Sun, Y.; Campbell, J.D.; Lenburg, M.E.; Bild, A.H.; Johnson, W.E. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics 2012, 100, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Kel, A.E.; Gossling, E.; Reuter, I.; Cheremushkin, E.; Kel-Margoulis, O.V.; Wingender, E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003, 31, 3576–3579. [Google Scholar] [CrossRef] [PubMed]
- Matys, V.; Fricke, E.; Geffers, R.; Gossling, E.; Haubrock, M.; Hehl, R.; Hornischer, K.; Karas, D.; Kel, A.E.; Kel-Margoulis, O.V.; et al. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003, 31, 374–378. [Google Scholar] [CrossRef] [PubMed]
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv 1802, 2018:1802.03426. [Google Scholar] [CrossRef]
- Riley, B.; Williamson, M.; Collier, D.; Wilkie, H.; Makoff, A. A 3-Mb map of a large Segmental duplication overlapping the alpha7-nicotinic acetylcholine receptor gene (CHRNA7) at human 15q13-q14. Genomics 2002, 79, 197–209. [Google Scholar] [CrossRef]
- Florio, M.; Albert, M.; Taverna, E.; Namba, T.; Brandl, H.; Lewitus, E.; Haffner, C.; Sykes, A.; Wong, F.K.; Peters, J.; et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 2015, 347, 1465–1470. [Google Scholar] [CrossRef]
- Heide, M.; Huttner, W.B. Human-Specific Genes, Cortical Progenitor Cells, and Microcephaly. Cells 2021, 10, 1209. [Google Scholar] [CrossRef] [PubMed]
- Gable, A.L.; Szklarczyk, D.; Lyon, D.; Matias Rodrigues, J.F.; von Mering, C. Systematic assessment of pathway databases, based on a diverse collection of user-submitted experiments. Brief. Bioinform. 2022, 23, bbac355. [Google Scholar] [CrossRef] [PubMed]
- Gutteck, N.; Savov, P.; Panian, M.; Wohlrab, D.; Zeh, A.; Delank, K.S. Preliminary results of a plantar plate for Lapidus arthrodesis. Foot Ankle Surg. 2018, 24, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.; Gupta, T.; Bal, A.; Sahni, D.; Singh, G. Role of Melatonin in Breast Carcinoma: Correlation of Expression Patterns of Melatonin-1 Receptor With Estrogen, Progesterone, and HER2 Receptors. Appl. Immunohistochem. Mol. Morphol. 2020, 28, 518–523. [Google Scholar] [CrossRef] [PubMed]








| Category | p-Value | Term ID | Description |
|---|---|---|---|
| GO: Biological Process | 7.9 × 10−174 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane |
| GO: Molecular Function | 1.7 × 10−149 | GO:0003735 | Structural constituent of ribosome |
| GO: Cellular Component | 2.0 × 10−175 | GO:0022626 | Cytosolic ribosome |
| 4.8 × 10−148 | GO:0044391 | Ribosomal subunit | |
| KEGG | 4.4 × 10−133 | hsa03010 | Ribosome—Homo sapiens (human) |
| WikiPathways | 1.7 × 10−154 | WP477_r108309 | Cytoplasmic Ribosomal Proteins |
| DisGeNet | 4.2 × 10−47 | C1260899 | Anemia, Diamond-Blackfan |
| Pfam | 3.9 × 10−5 | Ribosomal_L7Ae | Ribosomal protein L7Ae/L30e/S12e/Gadd45 family |
| Category | p-Value | Term ID | Description |
|---|---|---|---|
| GO: Biological Process | 2.6 × 10−14 | GO:1990169 | Stress response to copper ion |
| 2.6 × 10−14 | GO:0010273 | Detoxification of copper ion | |
| 2.9 × 10−14 | GO:0097501 | Stress response to metal ion | |
| 2.9 × 10−14 | GO:0061687 | Detoxification of inorganic compound | |
| GO: Molecular Function | 8.8 × 10−6 | GO:0008270 | Zinc ion binding |
| 1.8 × 10−5 | GO:0046914 | Transition metal ion binding | |
| KEGG | 1.8 × 10−11 | hsa04978 | Mineral absorption—Homo sapiens (human) |
| WikiPathways | 7.9 × 10−13 | WP3529_r106738 | Zinc homeostasis |
| 3.9 × 10−12 | WP3286_r106367 | Copper homeostasis | |
| Pfam | 3.0 × 10−16 | Metallothio | Metallothionein |
| ReMap | 7.6 × 10−5 | ZNF879 | Zinc finger protein 879 |
| 1.7 × 10−2 | ZNF26 | Zinc finger protein 26 |
| Category | p-Value | Term ID | Description |
|---|---|---|---|
| GO: Biological Process | 1.9 × 10−19 | GO:0019886 | Antigen processing and presentation of exogenous peptide antigen via MHC class II |
| 1.9 × 10−19 | GO:0002495 | Antigen processing and presentation of peptide antigen via MHC class II | |
| 1.9 × 10−19 | GO:0002504 | Antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | |
| GO: Molecular Function | 1.8 × 10−15 | GO:0023026 | MHC class II protein complex binding |
| 1.2 × 10−14 | GO:0032395 | MHC class II receptor activity | |
| GO: Cellular Component | 1.9 × 10−34 | GO:0042613 | MHC class II protein complex |
| KEGG | 2.1 × 10−24 | hsa04612 | Antigen processing and presentation—Homo sapiens (human) |
| Pfam | 1.6 × 10−25 | C1-set | Immunoglobulin C1-set domain |
| 4.4 × 10−16 | MHC_II_alpha | Class II histocompatibility antigen, alpha domain | |
| 1.0 × 10−15 | MHC_II_beta | Class II histocompatibility antigen, beta domain | |
| ReMap | 2.8 × 10−4 | SMAD5 | SMAD family member 5 |
| 2.8 × 10−4 | ZBED1 | Zinc finger BED-type containing 1 |
| Category | FDR | Term ID | Description |
|---|---|---|---|
| GO: Biological Process | 3.1 × 10−17 | GO:0042590 | Antigen processing and presentation of exogenous peptide antigen via MHC class I |
| 1.0 × 10−12 | GO:0019221 | Cytokine-mediated signaling pathway | |
| GO: Molecular Function | 3.3 × 10−4 | GO:0042288 | MHC class I protein binding |
| GO: Cellular Component | 4.7 × 10−17 | GO:0042612 | MHC class I protein complex |
| KEGG Pathways | 2.2 × 10−11 | hsa04612 | Antigen processing and presentation—Homo sapiens (human) |
| Pfam | 2.8 × 10−13 | MHC_I_C | MHC_I C-terminus |
| 1.4 × 10−4 | CARD | Caspase recruitment domain |
| Category | p-Value | Term ID | Description |
|---|---|---|---|
| GO: Biological Process | 2.2 × 10−33 | GO:0051607 | Defense response to virus |
| KEGG | 7.7 × 10−11 | hsa05160 | Hepatitis C—Homo sapiens (human) |
| 8.4 × 10−11 | hsa05164 | Influenza A—Homo sapiens (human) | |
| WikiPathways | 2.1 × 10−11 | WP4880_r109979 | Host-pathogen interaction of human corona viruses—Interferon induction |
| 3.4 × 10−8 | WP4868_r109974 | Type I Interferon Induction and Signaling During SARS-CoV-2 Infection | |
| Pfam | 6.0 × 10−11 | OAS1_C | 2’-5’-oligoadenylate synthetase 1, domain 2, C-terminus |
| DisGeNet | 4.7 × 10−28 | C0021400 | Influenza |
| 6.3 × 10−13 | C0042769 | Virus Diseases | |
| 1.8 × 10−10 | C0019196 | Hepatitis C | |
| ENCODE | 2.6 × 10−40 | STAT2 | Signal transducer and activator of transcription 2 |
| ReMap | 5.9 × 10−16 | STAT2 | Signal transducer and activator of transcription 2 |
| Category | p-Value | Term ID | Description |
|---|---|---|---|
| GO: Biological Process | 1.7 × 10−4 | GO:0007043 | Cell-cell junction assembly |
| 3.2 × 10−4 | GO:0030855 | Epithelial cell differentiation | |
| 3.2 × 10−4 | GO:0045216 | Cell-cell junction organization | |
| 9.6 × 10−4 | GO:0060429 | Epithelium development | |
| GO: Cellular Component | 1.4 × 10−3 | GO:0043296 | Apical junction complex |
| 1.4 × 10−3 | GO:0005911 | Cell-cell junction | |
| KEGG | 1.5 × 10−3 | hsa04514 | Cell adhesion molecules (CAMs)—Homo sapiens (human) |
| 1.5 × 10−3 | hsa04530 | Tight junction—Homo sapiens (human) | |
| Encode | 8.9 × 10−5 | ZNF217 | Zinc finger protein 217 |
| 4.2 × 10−4 | ESR1 | Estrogen receptor 1 | |
| 4.3 × 10−3 | ZBTB7A | Zinc finger and BTB domain containing 7A |
| Category | p-Value | Term ID | Description |
|---|---|---|---|
| GO: Biological Process | 2.8 × 10−19 | GO:0031424 | Keratinization |
| 1.6 × 10−18 | GO:0008544 | Epidermis development | |
| WikiPathways | 6.9 × 10−5 | WP2877_r105854 | Vitamin D Receptor Pathway |
| Pfam | 2.1 × 10−31 | LCE | Late cornified envelope |
| Chromosome Band | 7.4 × 10−29 | 1q21.3 |
| Category | p-Value | Term ID | Description |
|---|---|---|---|
| GO: Biological Process | 2.9 × 10−17 | GO:0006457 | Protein folding |
| 7.7 × 10−13 | GO:1900034 | Regulation of cellular response to heat | |
| 1.1 × 10−9 | GO:0061077 | Chaperone-mediated protein folding | |
| GO: Molecular Function | 7.8 × 10−12 | GO:0051082 | Unfolded protein binding |
| 1.5 × 10−10 | GO:0051087 | Chaperone binding | |
| 3.4 × 10−10 | GO:0031072 | Heat shock protein binding | |
| GO: Cellular Component | 3.1 × 10−12 | GO:0101031 | Chaperone complex |
| ENCODE | 1.1 × 10−21 | HSF1 | Heat shock transcription factor 1 |
| 2.6 × 10−20 | PPARGC1A | PPARG coactivator 1 alpha (PPARGC1A) | |
| DisGeNET | 6.4 × 10−4 | C0949664 | Tauopathies |
| Pfam | 3.4 × 10−9 | CS | CS domain |
| 2.0 × 10−5 | HSP90 | Hsp90 protein | |
| 1.1 × 10−4 | HSP70 | Hsp70 protein |
| Category | p-Value | Term ID | Description |
|---|---|---|---|
| GO: Biological Process | 2.8 × 10−14 | GO:0034976 | Response to endoplasmic reticulum stress |
| 1.3 × 10−10 | GO:0035966 | Response to topologically incorrect protein | |
| 6.7 × 10−10 | GO:0034975 | Protein folding in endoplasmic reticulum | |
| GO: Molecular Function | 2.4 × 10−6 | GO:0003756 | Protein disulfide isomerase activity |
| 2.4 × 10−6 | GO:0016864 | Intramolecular oxidoreductase activity, transposing S-S bonds | |
| 3.2 × 10−6 | GO:0051087 | Chaperone binding | |
| 4.6 × 10−6 | GO:0051082 | Unfolded protein binding | |
| GO: Cellular Component | 5.8 × 10−14 | GO:0005788 | Endoplasmic reticulum lumen |
| 2.9 × 10−10 | GO:0034663 | Endoplasmic reticulum chaperone complex | |
| ENCODE | 3.1 × 10−6 | SP2 | Sp2 transcription factor |
| DisGeNET | 1.4 × 10−7 | C1846707 | Spinocerebellar Ataxia 17 |
| Category | p-Value | Term ID | Description |
|---|---|---|---|
| GO: Biological Process | 4.6 × 10−19 | GO:0001944 | Vasculature development |
| 4.6 × 10−19 | GO:0072358 | Cardiovascular system development | |
| 5.5 × 10−19 | GO:0048514 | Blood vessel morphogenesis | |
| GO: Molecular Function | 3.1 × 10−4 | GO:0004714 | Transmembrane receptor protein tyrosine kinase activity |
| 3.1 × 10−4 | GO:0001605 | Adrenomedullin receptor activity | |
| GO: Cellular Component | 3.4 × 10−4 | GO:1903143 | Adrenomedullin receptor complex |
| KEGG Pathways | 3.8 × 10−2 | hsa04514 | Cell adhesion molecules (CAMs)—Homo sapiens (human) |
| 3.8 × 10−2 | hsa05418 | Fluid shear stress and atherosclerosis—Homo sapiens (human) | |
| 3.8 × 10−2 | hsa04270 | Vascular smooth muscle contraction—Homo sapiens (human) | |
| WikiPathways | 6.5 × 10−4 | WP3943_r106492 | Robo4 and VEGF Signaling Pathways Crosstalk |
| 6.5 × 10−4 | WP3888_r108912 | VEGFA-VEGFR2 Signaling Pathway | |
| DisGeNET | 2.7 × 10−8 | C1519670 | Tumour Angiogenesis |
| 1.2 × 10−7 | C1658953 | Tumour vasculature | |
| Pfam | 3.9 × 10−5 | RAMP | Receptor activity modifying family |
| 3.9 × 10−5 | CD34_antigen | CD34/Podocalyxin family | |
| 3.9 × 10−5 | Sox17_18_mid | Sox 17/18 central domain |
| Category | p-Value | Term ID | Description |
|---|---|---|---|
| GO: Biological Process | 7.2 × 10−269 | GO:0050911 | Detection of chemical stimulus involved in sensory perception of smell |
| GO: Molecular Function | 1.5 × 10−285 | GO:0004984 | Olfactory receptor activity |
| 9.1 × 10−236 | GO:0004930 | G protein-coupled receptor activity | |
| GO: Cellular Component | 9.3 × 10−65 | GO:0016021 | Integral component of membrane |
| KEGG | 5.9 × 10−225 | hsa04740 | Olfactory transduction—Homo sapiens (human) |
| WikiPathways | 1.2 × 10−18 | WP455_r106426 | GPCRs, Class A Rhodopsin-like |
| 3.8 × 10−8 | WP4558_r107928 | Overview of interferons-mediated signaling pathway | |
| Pfam | 1.2 × 10−278 | 7tm_4 | Olfactory receptor |
| Category | p-Value | Term ID | Description |
|---|---|---|---|
| GO: Biological Process | 9.7 × 10−4 | GO:0030522 | Intracellular steroid hormone receptor signaling pathway |
| GO: Molecular Function | 1.2 × 10−2 | GO:0003713 | Transcription coactivator activity |
| 1.3 × 10−2 | GO:0140030 | Modification-dependent protein binding | |
| 1.3 × 10−2 | GO:0035257 | Nuclear hormone receptor binding | |
| GO: Cellular Component | 1.6 × 10−2 | GO:0005654 | Nucleoplasm |
| Chromosome Band | 4.5 × 10−4 | 5q31.3 |
| Category | p-Value | Term ID | Description |
|---|---|---|---|
| GO: Biological Process | 1.5 × 10−12 | GO:0060416 | Response to growth hormone |
| 7.5 × 10−12 | GO:0060396 | Growth hormone receptor signaling pathway | |
| 7.3 × 10−9 | GO:0040008 | Regulation of growth | |
| GO: Molecular Function | 6.6 × 10−19 | GO:0005179 | Hormone activity |
| 7.8 × 10−13 | GO:0005131 | Growth hormone receptor binding | |
| KEGG Pathway | 2.3 × 10−16 | hsa04080 | Neuroactive ligand-receptor interaction—Homo sapiens (human) |
| 3.1 × 10−9 | hsa04935 | Growth hormone synthesis, secretion and action—Homo sapiens (human) | |
| DisGeNET | 2.1 × 10−15 | C0013338 | Pituitary dwarfism |
| 2.7 × 10−15 | C0032002 | Pituitary Diseases | |
| WikiPathways | 3.9 × 10−5 | WP3998_r106536 | Prader-Willi and Angelman Syndrome |
| Chromosome Band | 3.4 × 10−10 | 17q23.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zogopoulos, V.L.; Malatras, A.; Kyriakidis, K.; Charalampous, C.; Makrygianni, E.A.; Duguez, S.; Koutsi, M.A.; Pouliou, M.; Vasileiou, C.; Duddy, W.J.; et al. HGCA2.0: An RNA-Seq Based Webtool for Gene Coexpression Analysis in Homo sapiens. Cells 2023, 12, 388. https://doi.org/10.3390/cells12030388
Zogopoulos VL, Malatras A, Kyriakidis K, Charalampous C, Makrygianni EA, Duguez S, Koutsi MA, Pouliou M, Vasileiou C, Duddy WJ, et al. HGCA2.0: An RNA-Seq Based Webtool for Gene Coexpression Analysis in Homo sapiens. Cells. 2023; 12(3):388. https://doi.org/10.3390/cells12030388
Chicago/Turabian StyleZogopoulos, Vasileios L., Apostolos Malatras, Konstantinos Kyriakidis, Chrysanthi Charalampous, Evanthia A. Makrygianni, Stéphanie Duguez, Marianna A. Koutsi, Marialena Pouliou, Christos Vasileiou, William J. Duddy, and et al. 2023. "HGCA2.0: An RNA-Seq Based Webtool for Gene Coexpression Analysis in Homo sapiens" Cells 12, no. 3: 388. https://doi.org/10.3390/cells12030388
APA StyleZogopoulos, V. L., Malatras, A., Kyriakidis, K., Charalampous, C., Makrygianni, E. A., Duguez, S., Koutsi, M. A., Pouliou, M., Vasileiou, C., Duddy, W. J., Agelopoulos, M., Chrousos, G. P., Iconomidou, V. A., & Michalopoulos, I. (2023). HGCA2.0: An RNA-Seq Based Webtool for Gene Coexpression Analysis in Homo sapiens. Cells, 12(3), 388. https://doi.org/10.3390/cells12030388














