The Rosetta Stone Hypothesis-Based Interaction of the Tumor Suppressor Proteins Nit1 and Fhit
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Antibodies
2.2. Plasmids
2.3. Co-Immunoprecipitation and Western Blot Analyses
2.4. Expression and Purification of Recombinant Proteins
2.5. Pull-Down Assays
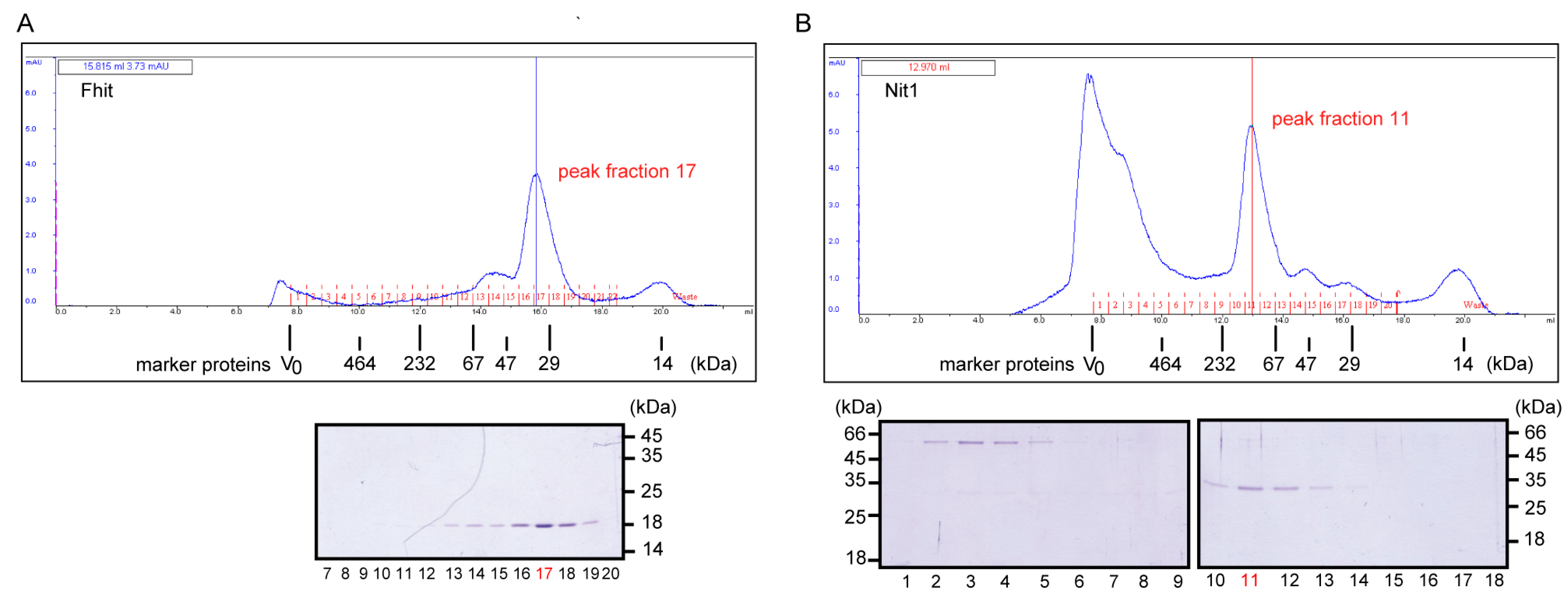
2.6. Size Exclusion Chromatography (SEC)
2.7. Proximity Ligation Assay
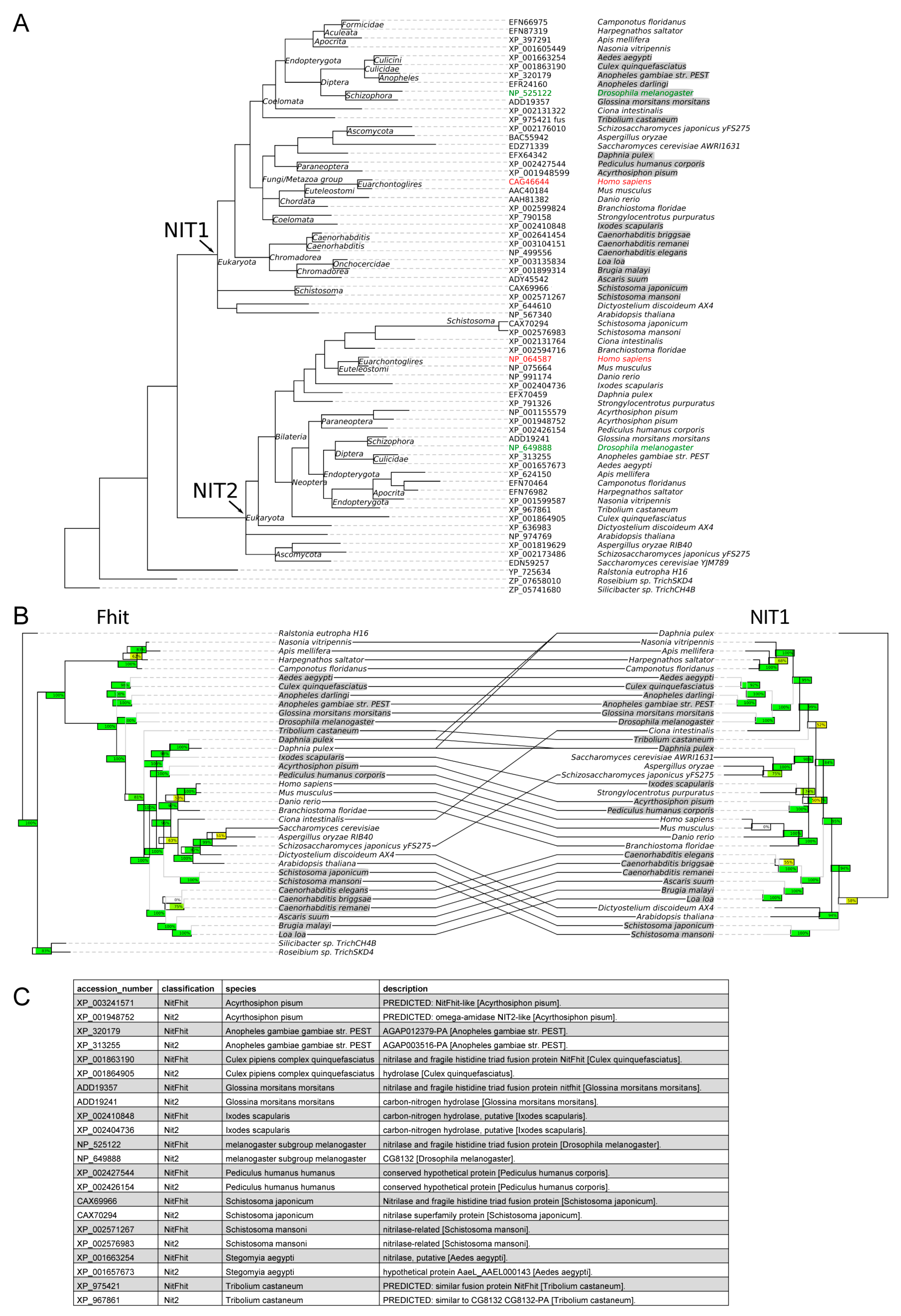
2.8. Phylogenetic Studies
3. Results
3.1. Co-Evolution of Nit1 and Fhit
3.2. Human Nit1 Forms Tetramers
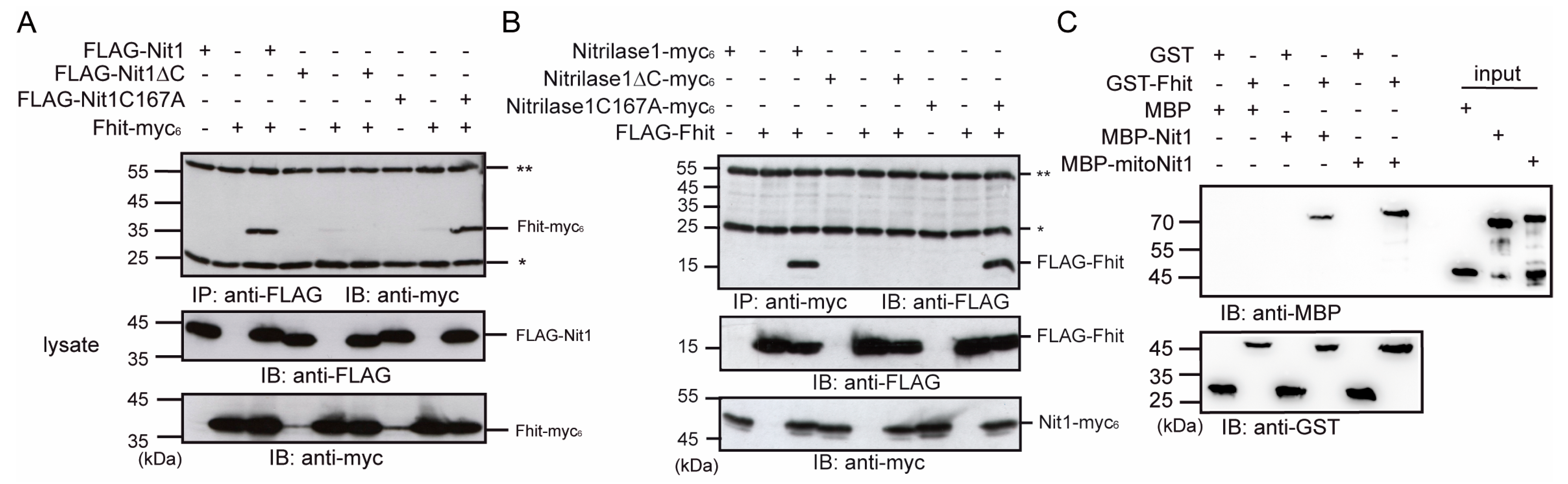
3.3. Human Nit1 Forms a Complex with Fhit
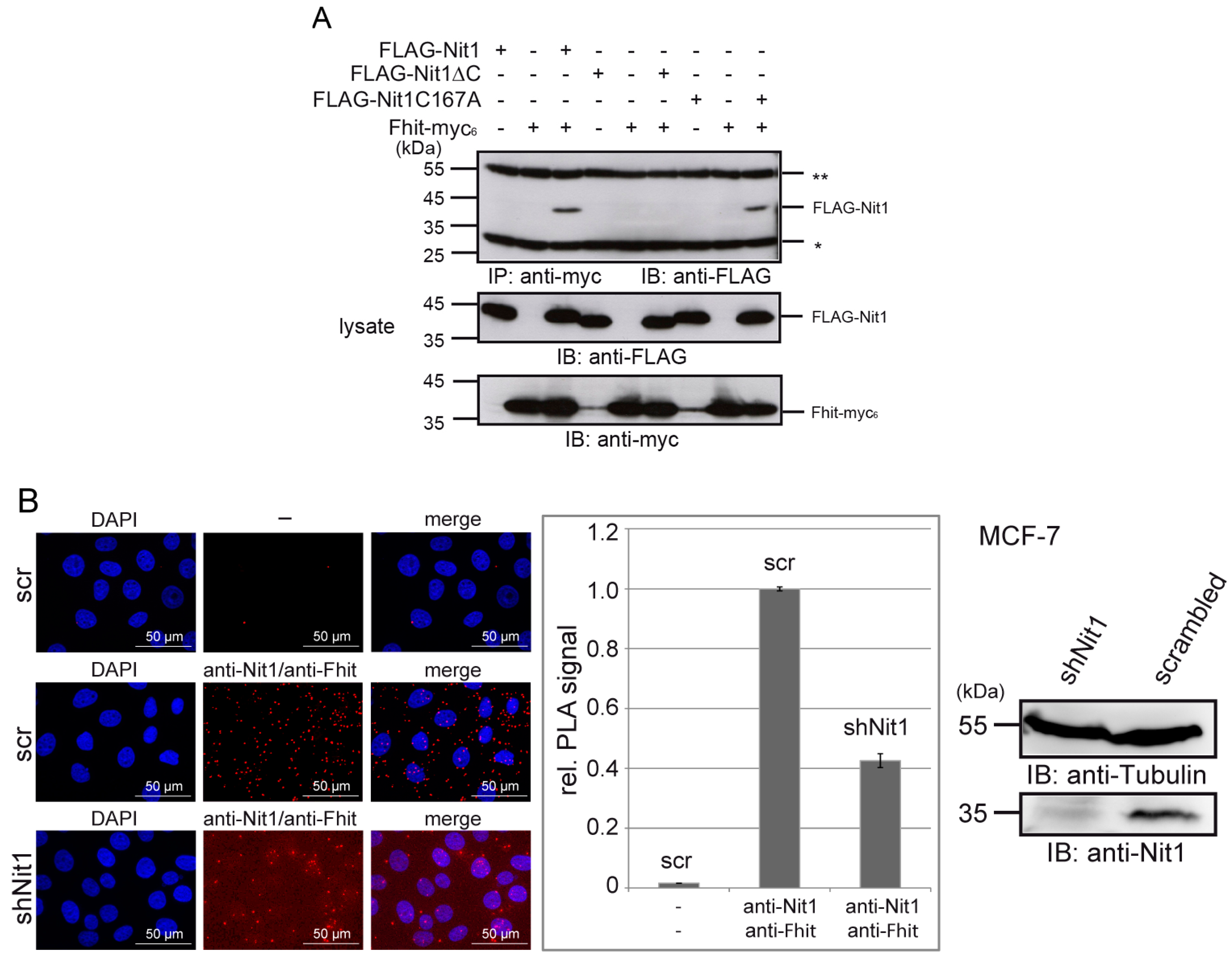
3.4. Human Nit1 Directly Interacts with Fhit
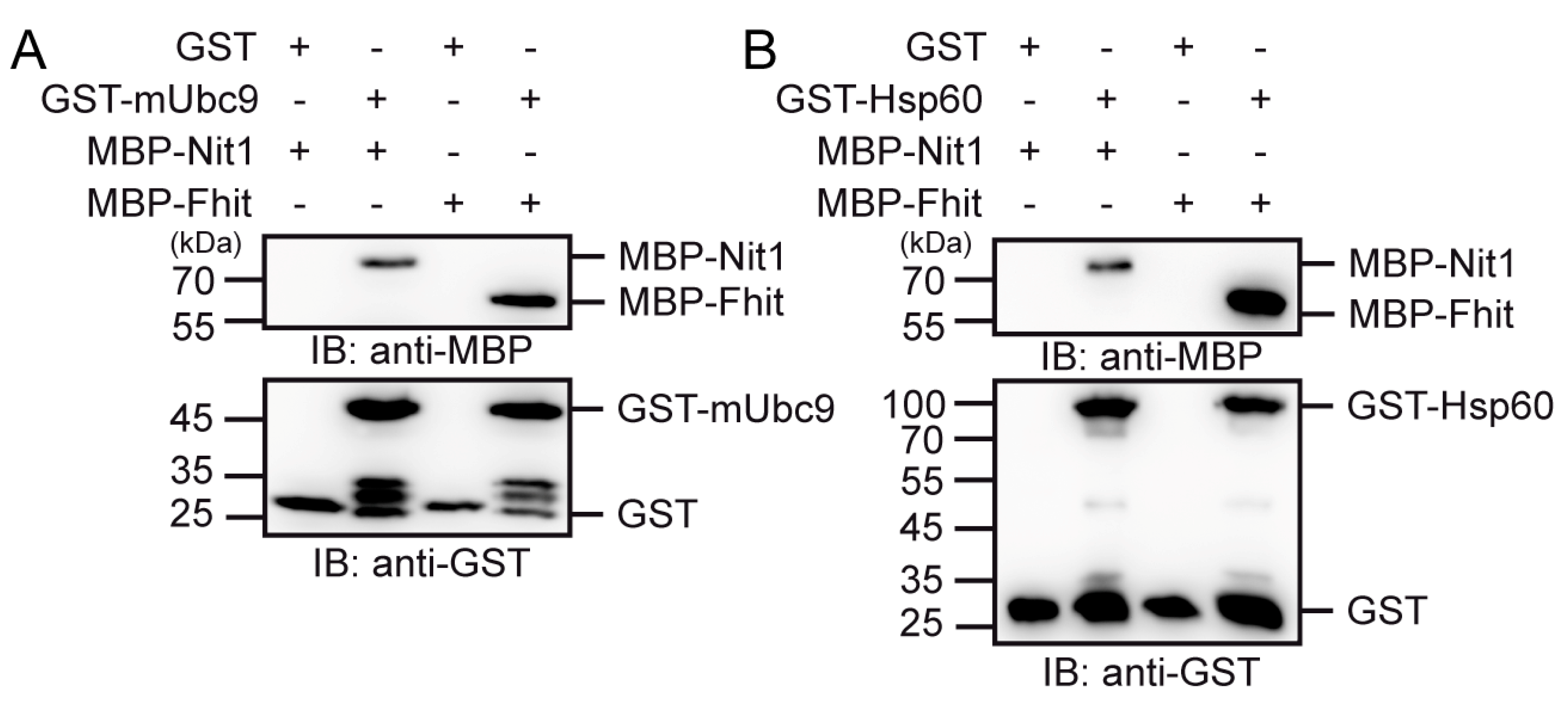
3.5. Nit1 Binds to Known Fhit Interaction Partners
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Marcotte, E.M.; Pellegrini, M.; Ng, H.L.; Rice, D.W.; Yeates, T.O.; Eisenberg, D. Detecting protein function and protein-protein interactions from genome sequences. Science 1999, 285, 751–753. [Google Scholar] [CrossRef] [PubMed]
- Marcotte, E.M.; Pellegrini, M.; Thompson, M.J.; Yeates, T.O.; Eisenberg, D. A combined algorithm for genome-wide prediction of protein function. Nature 1999, 402, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Veitia, R.A. Rosetta Stone proteins: “chance and necessity”? Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Campiglio, M.; Siprashvili, Z.; Druck, T.; Sedkov, Y.; Tillib, S.; Draganescu, A.; Wermuth, P.; Rothman, J.H.; Huebner, K.; et al. Nitrilase and Fhit homologs are encoded as fusion proteins in Drosophila melanogaster and Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 1998, 95, 8744–8749. [Google Scholar] [CrossRef] [PubMed]
- Pace, H.C.; Hodawadekar, S.C.; Draganescu, A.; Huang, J.; Bieganowski, P.; Pekarsky, Y.; Croce, C.M.; Brenner, C. Crystal structure of the worm NitFhit Rosetta Stone protein reveals a Nit tetramer binding two Fhit dimers. Curr. Biol. 2000, 10, 907–917. [Google Scholar] [CrossRef]
- Lima, C.D.; DAmico, K.L.; Naday, I.; Rosenbaum, G.; Westbrook, E.M.; Hendrickson, W.A. MAD analysis of FHIT, a putative human tumor suppressor from the HIT protein family. Structure 1997, 5, 763–774. [Google Scholar] [CrossRef]
- Huebner, K.; Croce, C.M. Cancer and the FRA3B/FHIT fragile locus: It’s a HIT. Br. J. Cancer 2003, 88, 1501–1506. [Google Scholar] [CrossRef]
- Fu, Y.; Shan, X.J.; Song, W.Q.; Xu, K.; Jiao, C.Y.; Zhang, Q.B. Correlations of breast cancer FHIT gene with the incidence and prognosis of breast cancer. J. BUON 2019, 24, 40–47. [Google Scholar]
- Barnes, L.D.; Garrison, P.N.; Siprashvili, Z.; Guranowski, A.; Robinson, A.K.; Ingram, S.W.; Croce, C.M.; Ohta, M.; Huebner, F. Fhit, a putative tumor suppressor in humans, is a dinucleoside 5′,5‴-P-1,P-3-triphosphate hydrolase. Biochemistry 1996, 35, 11529–11535. [Google Scholar] [CrossRef]
- Siprashvili, Z.; Sozzi, G.; Barnes, L.D.; McCue, P.; Robinson, A.K.; Eryomin, V.; Sard, L.; Tagliabue, E.; Greco, A.; Fusetti, L.; et al. Replacement of Fhit in cancer cells suppresses tumorigenicity. Proc. Natl. Acad. Sci. USA 1997, 94, 13771–13776. [Google Scholar] [CrossRef]
- Garrison, P.N.; Robinson, A.K.; Pekarsky, Y.; Croce, C.M.; Barnes, L.D. Phosphorylation of the human fhit tumor suppressor on tyrosine 114 in Escherichia coli and unexpected steady state kinetics of the phosphorylated forms. Biochemistry 2005, 44, 6286–6292. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.; Garrison, P.N.; Palamarchuk, A.; Zanesi, N.; Aqeilan, R.I.; Huebner, K.; Barnes, L.D.; Croce, C.M. Fhit is a physiological target of the protein kinase Src. Proc. Natl. Acad. Sci. USA 2004, 101, 3775–3779. [Google Scholar] [CrossRef] [PubMed]
- Semba, S.; Trapasso, F.; Fabbri, M.; McCorkell, K.A.; Volinia, S.; Druck, T.; Iliopoulos, D.; Pekarsky, Y.; Ishii, H.; Garrison, P.N.; et al. Fhit modulation of the Akt-survivin pathway in lung cancer cells: Fhit-tyrosine 114 (Y114) is essential. Oncogene 2006, 25, 2860–2872. [Google Scholar] [CrossRef] [PubMed]
- Trapasso, F.; Krakowiak, A.; Cesari, R.; Arkles, J.; Yendamuri, S.; Ishii, H.; Vecchione, A.; Kuroki, T.; Bieganowski, P.; Pace, H.C.; et al. Designed FHIT alleles establish that Fhit-induced apoptosis in cancer cells is limited by substrate binding. Proc. Natl. Acad. Sci. USA 2003, 100, 1592–1597. [Google Scholar] [CrossRef] [PubMed]
- Golebiowski, F.; Szulc, A.; Szutowicz, A.; Pawelczyk, T. Ubc9-induced inhibition of diadenosine triphosphate hydrolase activity of the putative tumor suppressor protein Fhit. Arch Biochem. Biophys 2004, 428, 160–164. [Google Scholar] [CrossRef]
- Shi, Y.F.; Zou, M.J.; Farid, N.R.; Paterson, M.C. Association of FHIT (fragile histidine triad), a candidate tumour suppressor gene, with the ubiquitin-conjugating enzyme hUBC9. Biochem. J. 2000, 352, 443–448. [Google Scholar] [CrossRef]
- Semba, S.; Han, S.Y.; Qin, H.R.; McCorkell, K.A.; Iliopoulos, D.; Pekarsky, Y.; Druck, T.; Trapasso, F.; Croce, C.M.; Huebner, K. Biological functions of mammalian NIT1, the counterpart of the invertebrate NitFhit Rosetta Stone protein, a possible tumor suppressor. J. Biol. Chem. 2006, 281, 28244–28253. [Google Scholar] [CrossRef]
- Pace, H.C.; Brenner, C. The nitrilase superfamily: Classification, structure and function. Genome Biol. 2001, 2, REVIEWS0001. [Google Scholar] [CrossRef]
- Barglow, K.T.; Cravatt, B.F. Substrate mimicry in an activity-based probe that targets the nitrilase family of enzymes. Angew. Chem. Int. Ed. 2006, 45, 7408–7411. [Google Scholar] [CrossRef]
- Jaisson, S.; Veiga-da-Cunha, M.; Van Schaftingen, E. Molecular identification of omega-amidase, the enzyme that is functionally coupled with glutamine transaminases, as the putative tumor suppressor Nit2. Biochimie 2009, 91, 1066–1071. [Google Scholar] [CrossRef]
- Peracchi, A.; Veiga-da-Cunha, M.; Kuhara, T.; Ellens, K.W.; Paczia, N.; Stroobant, V.; Seliga, A.K.; Marlaire, S.; Jaisson, S.; Bommer, G.T.; et al. Nit1 is a metabolite repair enzyme that hydrolyzes deaminated glutathione. Proc. Natl. Acad. Sci. USA 2017, 114, E3233–E3242. [Google Scholar] [CrossRef] [PubMed]
- Mittag, S.; Valenta, T.; Weiske, J.; Bloch, L.; Klingel, S.; Gradl, D.; Wetzel, F.; Chen, Y.; Petersen, I.; Basler, K.; et al. A novel role for the tumour suppressor Nitrilase1 modulating the Wnt/beta-catenin signalling pathway. Cell Discov. 2016, 2, 15039. [Google Scholar] [CrossRef] [PubMed]
- Pichiorri, F.; Okumura, H.; Nakamura, T.; Garrison, P.N.; Gasparini, P.; Suh, S.S.; Druck, T.; McCorkell, K.A.; Barnes, L.D.; Croce, C.M.; et al. Correlation of fragile histidine triad (Fhit) protein structural features with effector interactions and biological functions. J. Biol. Chem. 2009, 284, 1040–1049. [Google Scholar] [CrossRef] [PubMed]
- Weiske, J.; Albring, K.F.; Huber, O. The tumor suppressor Fhit acts as a repressor of beta-catenin transcriptional activity. Proc. Natl. Acad. Sci. USA 2007, 104, 20344–20349. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Okumura, H.; Yearsley, M.; Frankel, W.; Fong, L.Y.; Druck, T.; Huebner, K. Nit1 and Fhit tumor suppressor activities are additive. J. Cell. Biochem. 2009, 107, 1097–1106. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Zhang, J.M.; Lu, Y.X.; Li, X.M.; Zhang, W.J.; Zhang, W.; Lin, W.H.; Zheng, L.; Li, X.N. NIT1 suppresses tumour proliferation by activating the TGF beta 1-Smad2/3 signalling pathway in colorectal cancer. Cell Death Dis. 2018, 9, 263. [Google Scholar] [CrossRef]
- Weiske, J.; Huber, O. The histidine triad protein Hint1 triggers apoptosis independent of its enzymatic activity. J. Biol. Chem. 2006, 281, 27356–27366. [Google Scholar] [CrossRef]
- Wetzel, F.; Mittag, S.; Cano-Cortina, M.; Wagner, T.; Kramer, O.; Niedenthal, R.; Gonzalez-Mariscal, L.; Huber, O. SUMOylation regulates the intracellular fate of ZO-2. Cell. Mol. Life Sci. 2017, 74, 373–392. [Google Scholar] [CrossRef]
- Weiske, J.; Huber, O. The histidine triad protein Hint1 interacts with Pontin and Reptin and inhibits TCF-beta-catenin-mediated transcription. J. Cell Sci. 2005, 118, 3117–3129. [Google Scholar] [CrossRef]
- Scheich, C.; Kummel, D.; Soumailakakis, D.; Heinemann, U.; Bussow, K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007, 35, e43. [Google Scholar] [CrossRef]
- Söderberg, O.; Gullberg, M.; Jarvius, M.; Ridderstrale, K.; Leuchowius, K.J.; Jarvius, J.; Wester, K.; Hydbring, P.; Bahram, F.; Larsson, L.G.; et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 2006, 3, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Griebel, T.; Brinkmeyer, M.; Bocker, S. EPoS: A modular software framework for phylogenetic analysis. Bioinformatics 2008, 24, 2399–2400. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef]
- Böcker, S.; Hüffner, F.; Truss, A.; Wahlström, M. A faster fixed-parameter approach to drawing binary tanglegrams. In Parameterized and Exact Computation; Chen, J., Fomin, F.V., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 38–49. [Google Scholar]
- Barglow, K.T.; Saikatendu, K.S.; Bracey, M.H.; Huey, R.; Morris, G.M.; Olson, A.J.; Stevens, R.C.; Cravatt, B.F. Functional proteomic and structural insights into molecular recognition in the nitrilase family enzymes. Biochemistry 2008, 47, 13514–13523. [Google Scholar] [CrossRef]
- Chien, C.H.; Gao, Q.Z.; Cooper, A.J.; Lyu, J.H.; Sheu, S.Y. Structural insights into the catalytic active site and activity of human Nit2/omega-amidase: Kinetic assay and molecular dynamics simulation. J. Biol. Chem. 2012, 287, 25715–25726. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Zanesi, N.; Palamarchuk, A.; Huebner, K.; Croce, C.M. FHIT: From gene discovery to cancer treatment and prevention. Lancet Oncol. 2002, 3, 748–754. [Google Scholar] [CrossRef]
- Krasnikov, B.F.; Chien, C.H.; Nostramo, R.; Pinto, J.T.; Nieves, E.; Callaway, M.; Sun, J.; Huebner, K.; Cooper, A.J. Identification of the putative tumor suppressor Nit2 as omega-amidase, an enzyme metabolically linked to glutamine and asparagine transamination. Biochimie 2009, 91, 1072–1080. [Google Scholar] [CrossRef]
- Doskocilova, A.; Kohoutova, L.; Volc, J.; Kourova, H.; Benada, O.; Chumova, J.; Plihal, O.; Petrovska, B.; Halada, P.; Bogre, L.; et al. NITRILASE1 regulates the exit from proliferation, genome stability and plant development. N. Phytol. 2013, 198, 685–698. [Google Scholar] [CrossRef]
- Kowara, R.; Karaczyn, A.A.; Fivash, M.J., Jr.; Kasprzak, K.S. In vitro inhibition of the enzymatic activity of tumor suppressor FHIT gene product by carcinogenic transition metals. Chem. Res. Toxicol 2002, 15, 319–325. [Google Scholar] [CrossRef]
- Zhou, X.; Pan, Y.; Wang, Y.; Wang, B.; Yan, Y.; Qu, Y.; Ke, X. Tanshinones induce tumor cell apoptosis via directly targeting FHIT. Sci. Rep. 2021, 11, 12217. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.; Hacker, S.M.; Schmid, P.; Scheffner, M.; Marx, A. Small-molecule inhibitors of the tumor suppressor Fhit. Chembiochem 2017, 18, 1707–1711. [Google Scholar] [CrossRef] [PubMed]
- Herzog, D.; Jansen, J.; Missun, M.; Diederichs, K.; Stengel, F.; Marx, A. Chemical proteomics of the tumor suppressor Fhit covalently bound to the cofactor Ap3A elucidates its inhibitory action on translation. J. Am. Chem. Soc. 2022, 144, 8613–8623. [Google Scholar] [CrossRef] [PubMed]
- Despotovic, D.; Brandis, A.; Savidor, A.; Levin, Y.; Fumagalli, L.; Tawfik, D.S. Diadenosine tetraphosphate (Ap4A)—An E. coli alarmone or a damage metabolite? FEBS J. 2017, 284, 2194–2215. [Google Scholar] [CrossRef]
- Fisher, D.I.; McLennan, A.G. Correlation of intracellular diadenosine triphosphate (Ap3A) with apoptosis in Fhit-positive HEK293 cells. Cancer Lett. 2008, 259, 186–191. [Google Scholar] [CrossRef]
- Pietrowska-Borek, M.; Nuc, K.; Zielezinska, M.; Guranowski, A. Diadenosine polyphosphates (Ap3A and Ap4A) behave as alarmones triggering the synthesis of enzymes of the phenylpropanoid pathway in Arabidopsis thaliana. FEBS Open Bio. 2011, 1, 1–6. [Google Scholar] [CrossRef]
- Yu, J.; Liu, Z.; Liang, Y.; Luo, F.; Zhang, J.; Tian, C.; Motzik, A.; Zheng, M.; Kang, J.; Zhong, G.; et al. Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in FcepsilonRI-activated mast cells. Nat. Commun. 2019, 10, 4664. [Google Scholar] [CrossRef]
- Huebner, K.; Saldivar, J.C.; Sun, J.; Shibata, H.; Druck, T. Hits, Fhits and Nits: Beyond enzymatic function. Adv. Enzym. Regul. 2011, 51, 208–217. [Google Scholar] [CrossRef]
- Druck, T.; Cheung, D.G.; Park, D.; Trapasso, F.; Pichiorri, F.; Gaspari, M.; Palumbo, T.; Aqeilan, R.I.; Gaudio, E.; Okumura, H.; et al. Fhit-Fdxr interaction in the mitochondria: Modulation of reactive oxygen species generation and apoptosis in cancer cells. Cell Death Dis. 2019, 10, 147. [Google Scholar] [CrossRef]
- Caruso Bavisotto, C.; Alberti, G.; Vitale, A.M.; Paladino, L.; Campanella, C.; Rappa, F.; Gorska, M.; Conway de Macario, E.; Cappello, F.; Macario, A.J.L.; et al. Hsp60 post-translational modifications: Functional and pathological consequences. Front. Mol. Biosci. 2020, 7, 95. [Google Scholar] [CrossRef]
- Chatterjee, S.; Burns, T.F. Targeting heat shock proteins in cancer: A promising therapeutic approach. Int. J. Mol. Sci. 2017, 18, 1978. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Li, G.; Yu, Q.; Liu, D.; Tang, X. HSP60 in cancer: A promising biomarker for diagnosis and a potentially useful target for treatment. J. Drug Target 2022, 30, 31–45. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mittag, S.; Wetzel, F.; Müller, S.Y.; Huber, O. The Rosetta Stone Hypothesis-Based Interaction of the Tumor Suppressor Proteins Nit1 and Fhit. Cells 2023, 12, 353. https://doi.org/10.3390/cells12030353
Mittag S, Wetzel F, Müller SY, Huber O. The Rosetta Stone Hypothesis-Based Interaction of the Tumor Suppressor Proteins Nit1 and Fhit. Cells. 2023; 12(3):353. https://doi.org/10.3390/cells12030353
Chicago/Turabian StyleMittag, Sonnhild, Franziska Wetzel, Sebastian Y. Müller, and Otmar Huber. 2023. "The Rosetta Stone Hypothesis-Based Interaction of the Tumor Suppressor Proteins Nit1 and Fhit" Cells 12, no. 3: 353. https://doi.org/10.3390/cells12030353
APA StyleMittag, S., Wetzel, F., Müller, S. Y., & Huber, O. (2023). The Rosetta Stone Hypothesis-Based Interaction of the Tumor Suppressor Proteins Nit1 and Fhit. Cells, 12(3), 353. https://doi.org/10.3390/cells12030353





