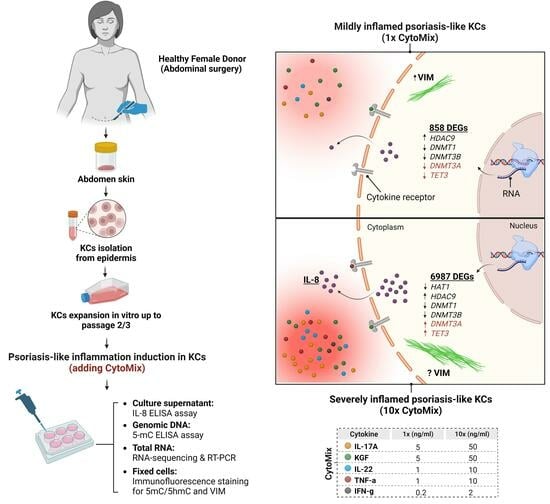
Unraveling Transcriptome Profile, Epigenetic Dynamics, and Morphological Changes in Psoriasis-like Keratinocytes: “Insights into Similarity with Psoriatic Lesional Epidermis”
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Culture of Skin Epidermal Keratinocyte Cells (KCs)
2.2. Cytokine Mixture (CytoMix) and Reagents
2.3. xCELLigence Real-Time Cell Analysis (RTCA)
2.4. Total RNA Isolation and Real-Time RT-PCR
2.5. IL-8 ELISA Assay
2.6. High-Throughput RNA Sequencing
2.7. Examining the Transcriptome Similarity between Mild and Severely Inflamed KCs and Psoriatic-Lesional Epidermis
2.8. Immunofluorescence Staining
2.9. Confocal Microscopy
2.10. DNA Isolation and Quantification of Global DNA Methylation Levels
2.11. Statistical Analysis and Graphical Abstract
3. Results
3.1. The Real-Time Cell Analysis (RTCA) Confirms the Significant Impact of Mild CytoMix Treatment on the Cellular Fate of KCs
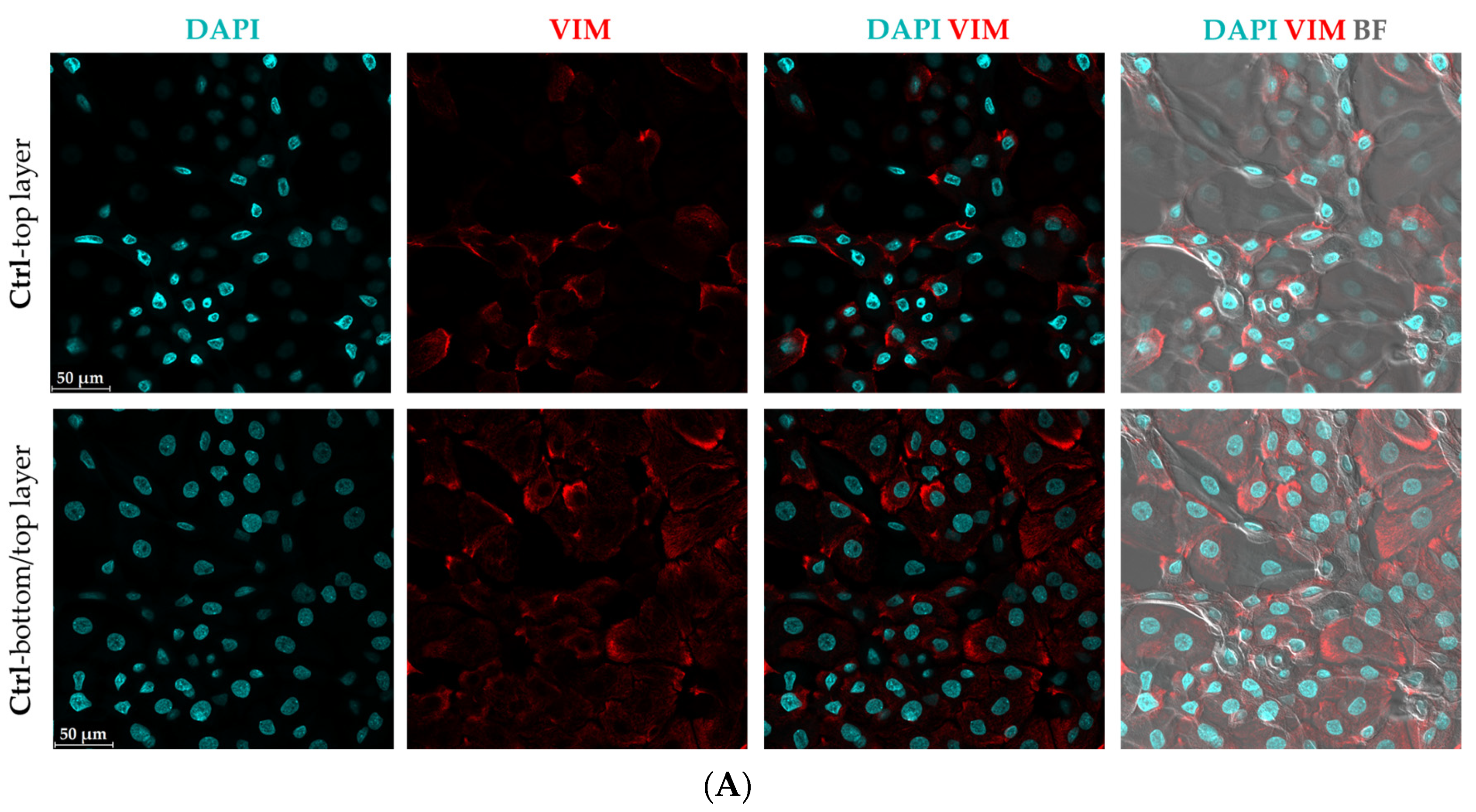
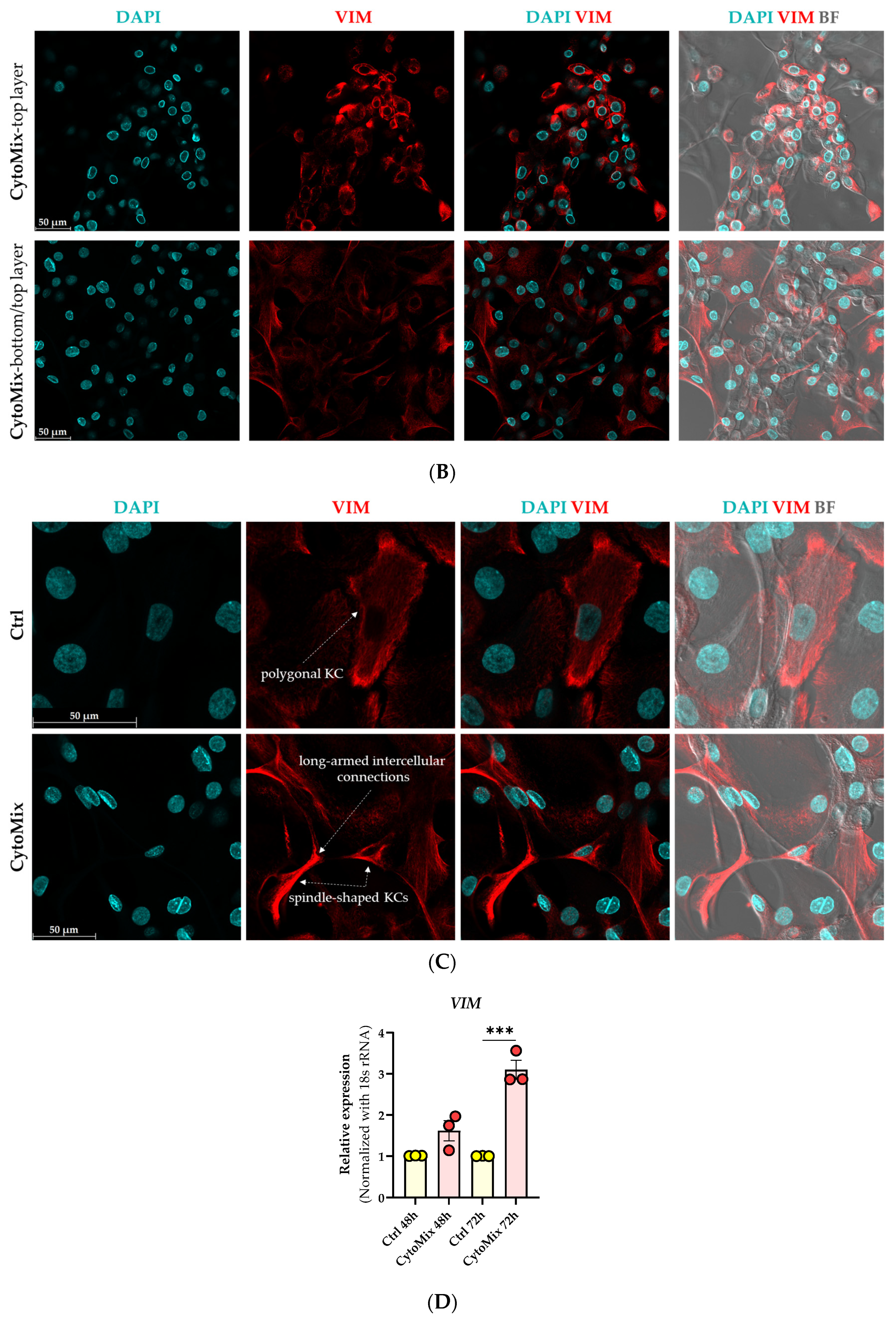
3.2. Morphological Changes and Increased Vimentin (VIM) Expression Suggest an Epithelial-Mesenchymal Transition (EMT)-like Phenomenon in Mild CytoMix-Treated KCs
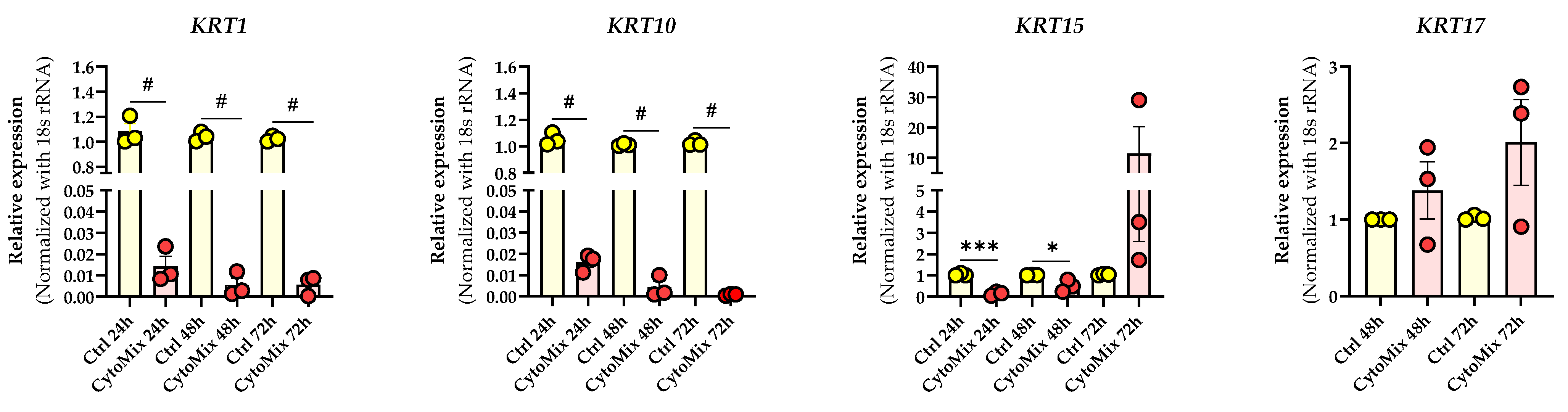
3.3. Mild CytoMix Treatment Induces Cytokeratin Gene Expression Changes
3.4. Severely Inflamed KCs Produce Significantly Higher Levels of Pro-Inflammatory Cytokines Than Mildly Inflamed Ones
3.5. 10× CytoMix-Treatment Results in a Higher Number of Differentially Expressed Genes (DEGs)
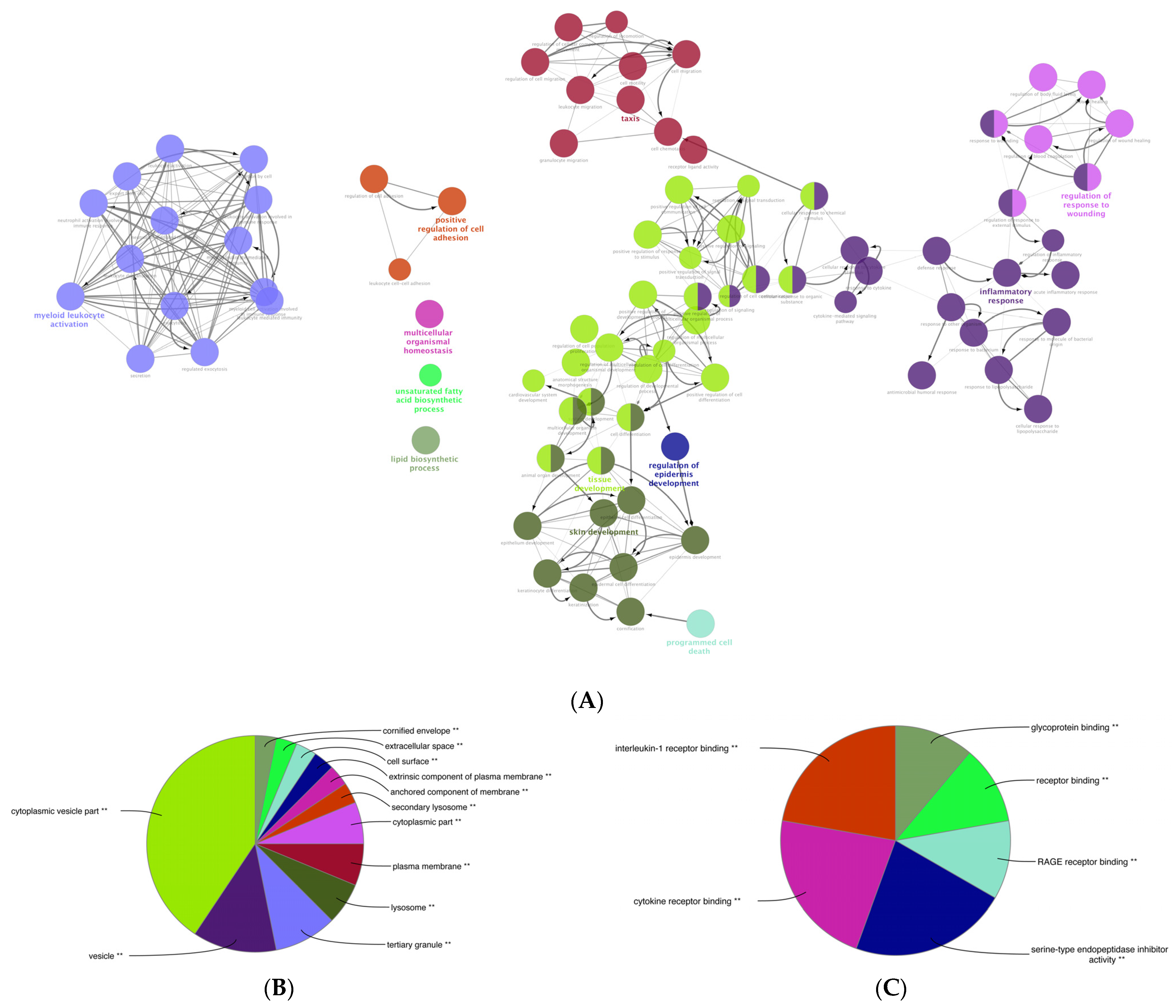
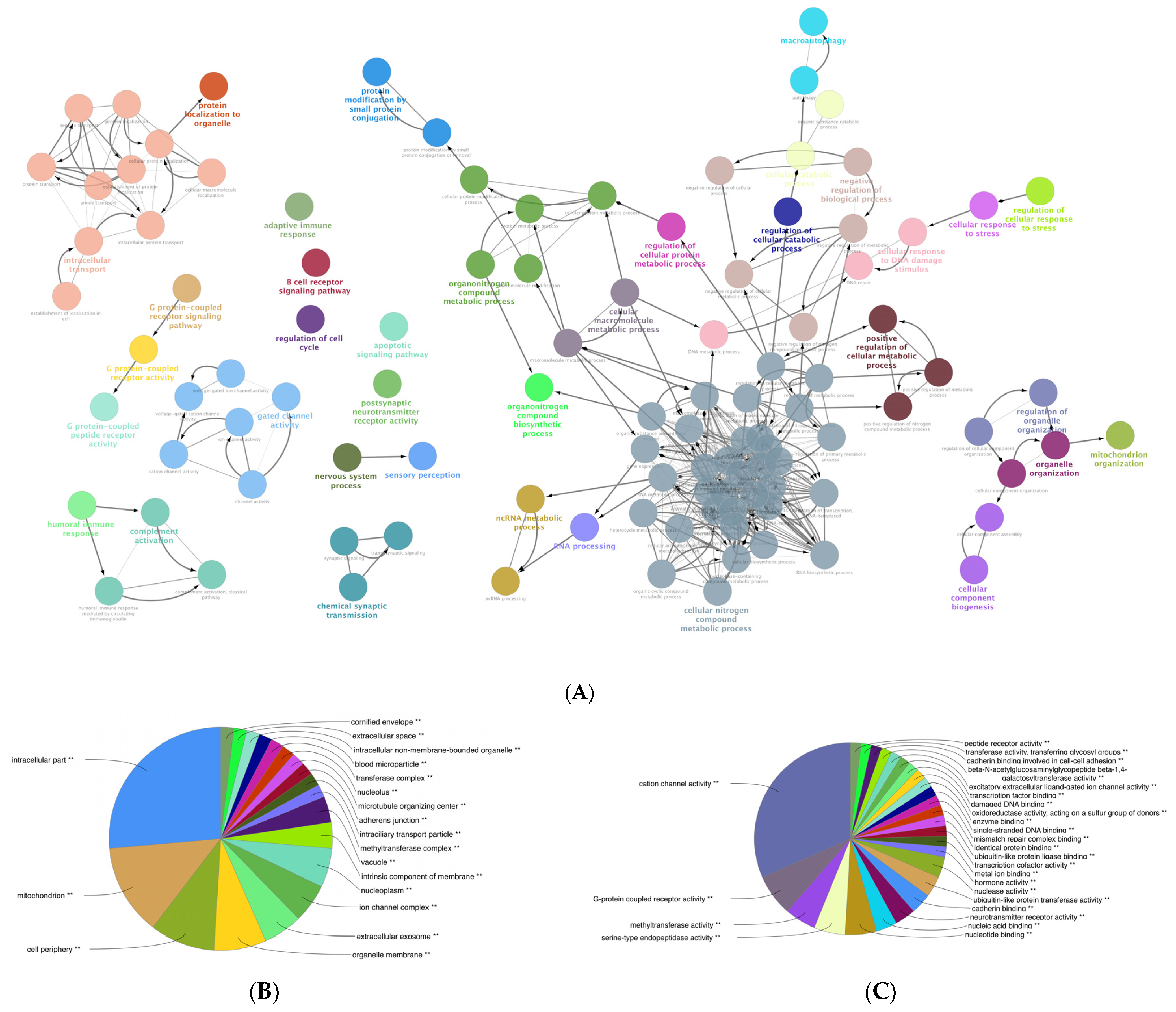
3.6. Gene Ontology (GO) Enrichment Analysis of the DEGs of Mildly and Severely Inflamed KCs
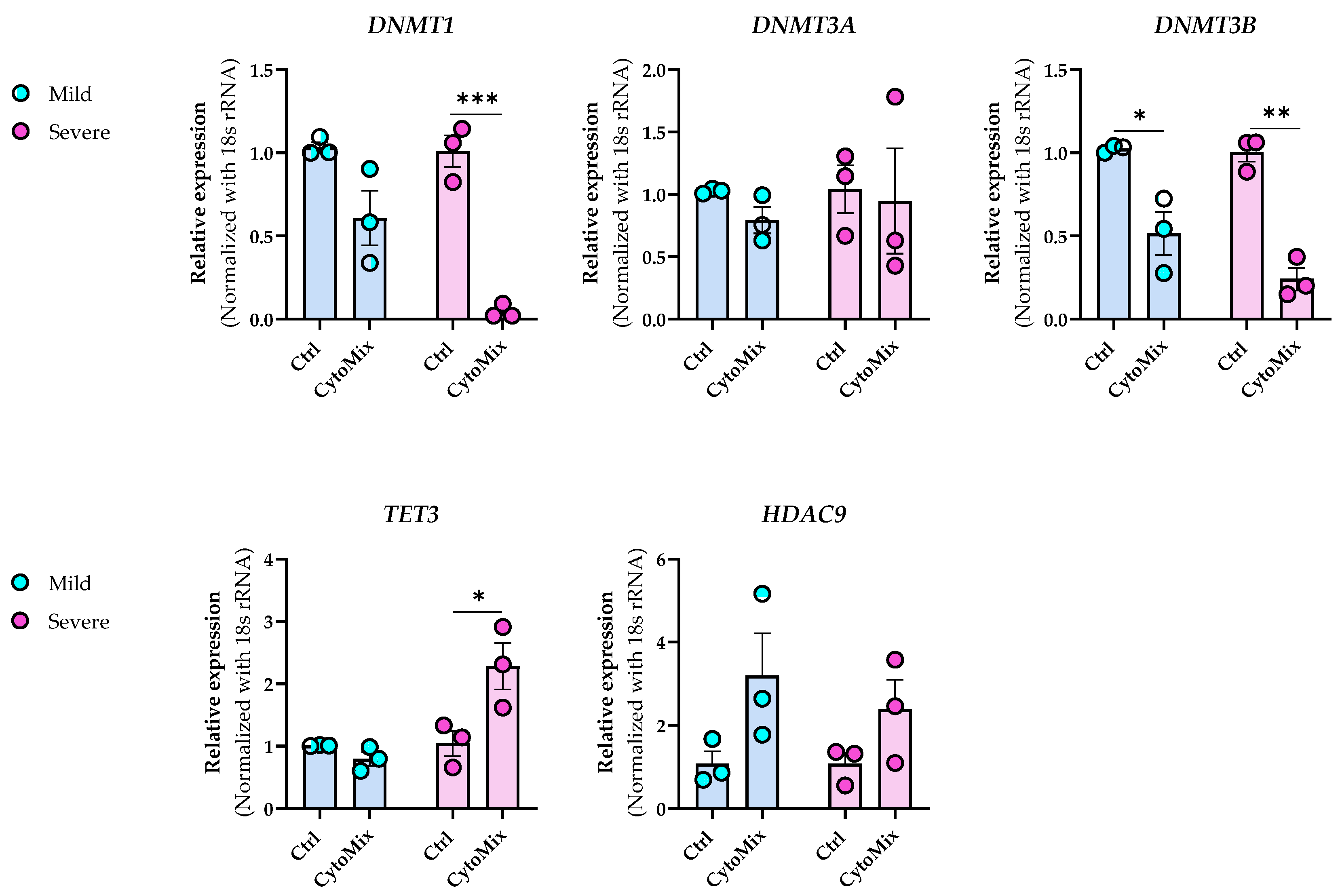
3.7. Dynamic Epigenetic Alterations in Psoriasis-like Mildly and Severely Inflamed Keratinocytes (KCs)
3.8. The Transcriptome of Mildly Inflamed Keratinocytes More Closely Mimicked That of the Psoriatic Epidermis Transcriptome Than the Severely Inflamed Keratinocytes
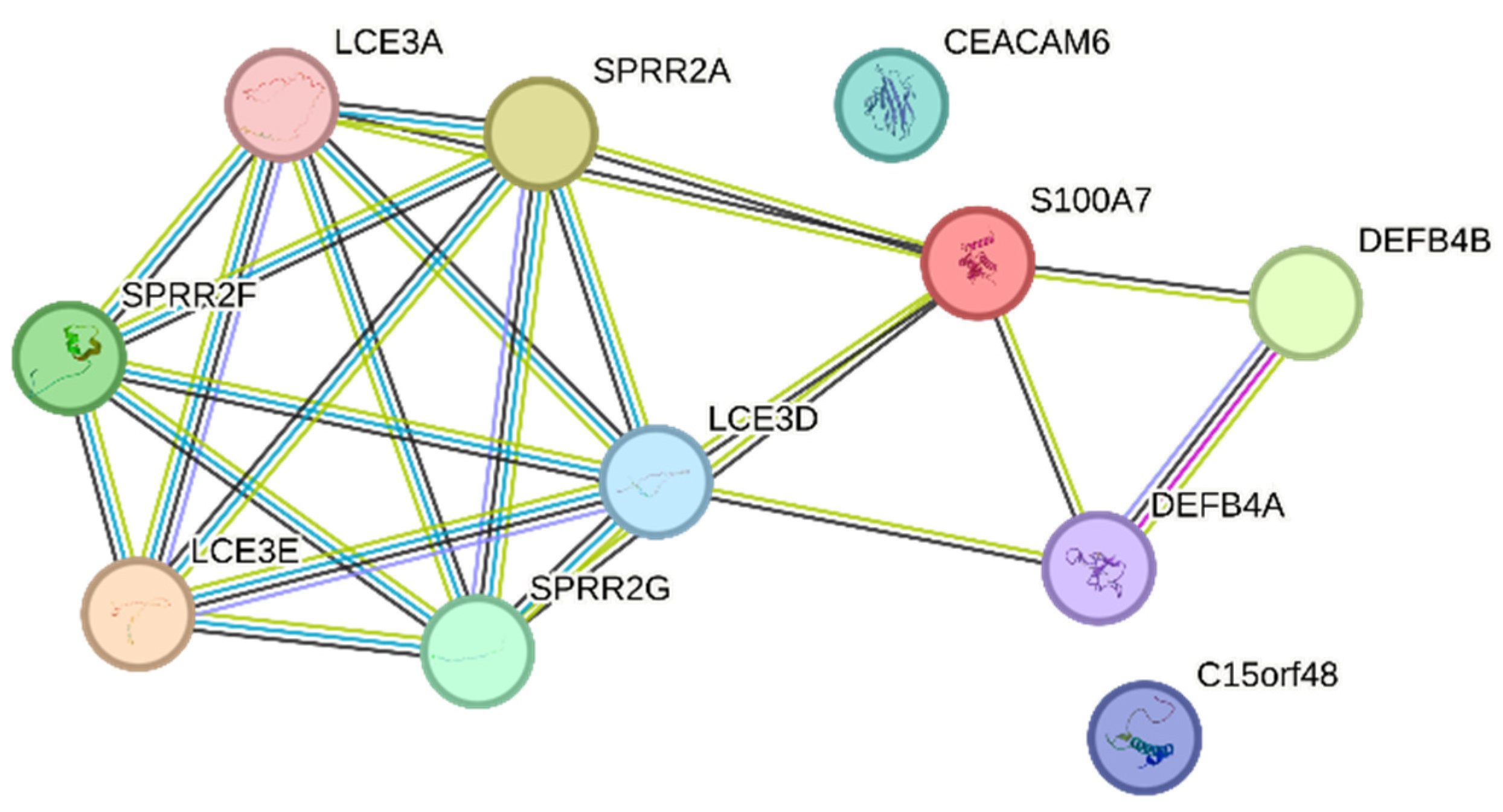
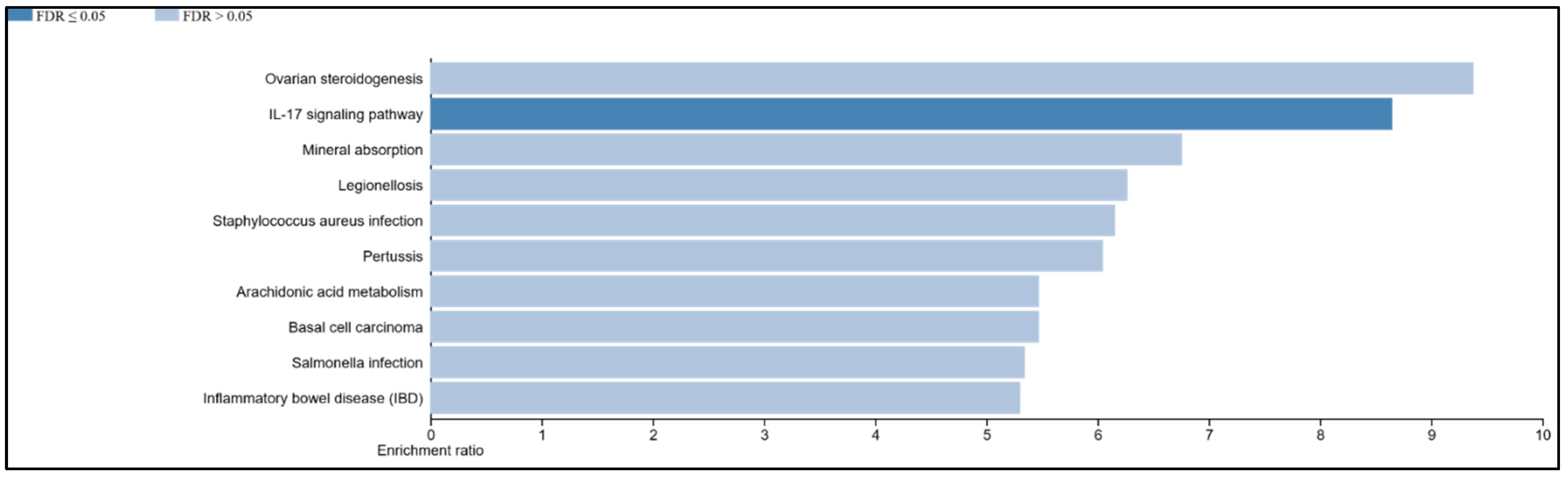
3.9. The IL-17 Signaling Pathway Is Enriched in the Overlapping Gene Set of the Mildly Inflamed Keratinocytes and Psoriatic Lesional Epidermis
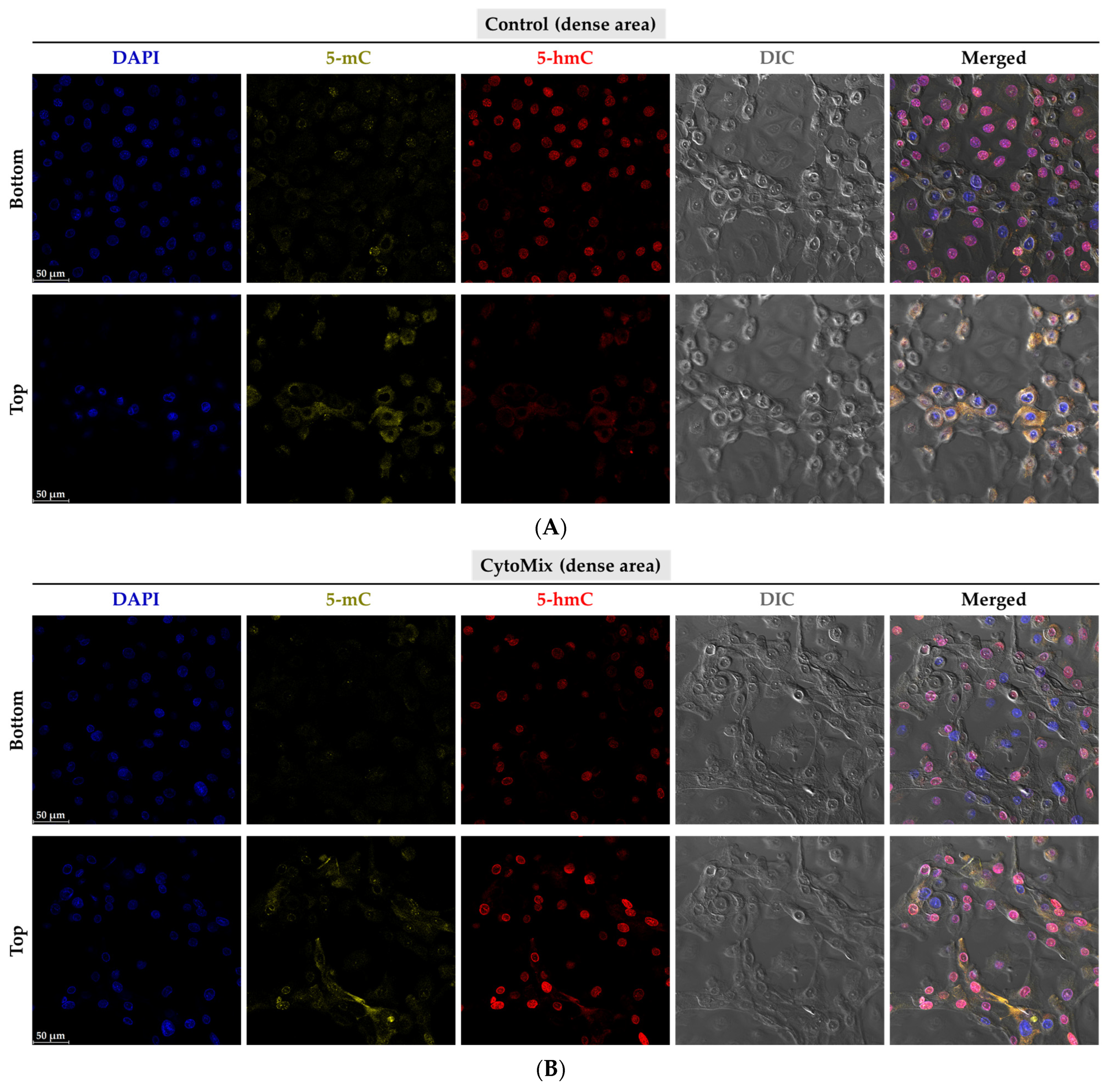
3.10. Exploring the Changes of 5-methylcytosine (5-mC) and 5-hydroxymethylcytosine (5-hmC) Epigenetic Marks in the Psoriasis-like Mild Inflammation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Griffiths, C.E.M.; van der Walt, J.M.; Ashcroft, D.M.; Flohr, C.; Naldi, L.; Nijsten, T.; Augustin, M. The global state of psoriasis disease epidemiology: A workshop report. Br. J. Dermatol. 2017, 177, e4–e7. [Google Scholar] [CrossRef] [PubMed]
- Lowes, M.A.; Suárez-Fariñas, M.; Krueger, J.G. Immunology of psoriasis. Annu. Rev. Immunol. 2014, 32, 227–255. [Google Scholar] [CrossRef] [PubMed]
- Albanesi, C.; Madonna, S.; Gisondi, P.; Girolomoni, G. The Interplay between Keratinocytes and Immune Cells in the Pathogenesis of Psoriasis. Front. Immunol. 2018, 9, 1549. [Google Scholar] [CrossRef] [PubMed]
- Di Meglio, P.; Villanova, F.; Nestle, F.O. Psoriasis. Cold Spring Harb. Perspect. Med. 2014, 4, a015354. [Google Scholar] [CrossRef] [PubMed]
- Guilloteau, K.; Paris, I.; Pedretti, N.; Boniface, K.; Juchaux, F.; Huguier, V.; Guillet, G.; Bernard, F.-X.; Lecron, J.-C.; Morel, F. Skin Inflammation Induced by the Synergistic Action of IL-17A, IL-22, Oncostatin M, IL-1α, and TNF-α Recapitulates Some Features of Psoriasis. J. Immunol. 2010, 184, 5263–5270. [Google Scholar] [CrossRef] [PubMed]
- Szabó, K.; Bata-Csörgő, Z.; Dallos, A.; Bebes, A.; Francziszti, L.; Dobozy, A.; Kemény, L.; Széll, M. Regulatory networks contributing to psoriasis susceptibility. Acta Derm. Venereol. 2014, 94, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Rabeony, H.; Petit-Paris, I.; Garnier, J.; Barrault, C.; Pedretti, N.; Guilloteau, K.; Jegou, J.-F.; Guillet, G.; Huguier, V.; Lecron, J.-C.; et al. Inhibition of keratinocyte differentiation by the synergistic effect of IL-17A, IL-22, IL-1α, TNFα and oncostatin M. PLoS ONE 2014, 9, e101937. [Google Scholar] [CrossRef]
- Bocheńska, K.; Smolińska, E.; Moskot, M.; Jakóbkiewicz-Banecka, J.; Gabig-Cimińska, M. Models in the Research Process of Psoriasis. Int. J. Mol. Sci. 2017, 18, 2514. [Google Scholar] [CrossRef]
- Nograles, K.E.; Zaba, L.C.; Guttman-Yassky, E.; Fuentes-Duculan, J.; Suárez-Fariñas, M.; Cardinale, I.; Khatcherian, A.; Gonzalez, J.; Pierson, K.C.; White, T.R.; et al. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br. J. Dermatol. 2008, 159, 1092–1102. [Google Scholar] [CrossRef]
- Mee, J.B.; Johnson, C.M.; Morar, N.; Burslem, F.; Groves, R.W. The psoriatic transcriptome closely resembles that induced by interleukin-1 in cultured keratinocytes: Dominance of innate immune responses in psoriasis. Am. J. Pathol. 2007, 171, 32–42. [Google Scholar] [CrossRef]
- Banno, T.; Gazel, A.; Blumenberg, M. Effects of tumor necrosis factor-α (TNF α) in epidermal keratinocytes revealed using global transcriptional profiling. J. Biol. Chem. 2004, 279, 32633–32642. [Google Scholar] [CrossRef]
- Finelt, N.; Gazel, A.; Gorelick, S.; Blumenberg, M. Transcriptional responses of human epidermal keratinocytes to Oncostatin-M. Cytokine 2005, 31, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Gudjonsson, J.E.; Ding, J.; Johnston, A.; Tejasvi, T.; Guzman, A.M.; Nair, R.P.; Voorhees, J.J.; Abecasis, G.R.; Elder, J.T. Assessment of the psoriatic transcriptome in a large sample: Additional regulated genes and comparisons with in vitro models. J. Investig. Dermatol. 2010, 130, 1829–1840. [Google Scholar] [CrossRef]
- Shanmugam, M.K.; Sethi, G. Role of epigenetics in inflammation-associated diseases. Subcell. Biochem. 2013, 61, 627–657. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Su, Y.; Chen, H.; Zhao, M.; Lu, Q. Abnormal DNA methylation in skin lesions and PBMCs of patients with psoriasis vulgaris. J. Dermatol. Sci. 2010, 60, 40–42. [Google Scholar] [CrossRef] [PubMed]
- Roberson, E.D.O.; Liu, Y.; Ryan, C.; Joyce, C.E.; Duan, S.; Cao, L.; Martin, A.; Liao, W.; Menter, A.; Bowcock, A.M. A subset of methylated CpG sites differentiate psoriatic from normal skin. J. Investig. Dermatol. 2012, 132, 583–592. [Google Scholar] [CrossRef]
- Zhou, F.; Wang, W.; Shen, C.; Li, H.; Zuo, X.; Zheng, X.; Yue, M.; Zhang, C.; Yu, L.; Chen, M.; et al. Epigenome-Wide Association Analysis Identified Nine Skin DNA Methylation Loci for Psoriasis. J. Investig. Dermatol. 2016, 136, 779–787. [Google Scholar] [CrossRef]
- Li, F.; Yuan, C.W.; Xu, S.; Zu, T.; Woappi, Y.; Lee, C.A.A.; Abarzua, P.; Wells, M.; Ramsey, M.R.; Frank, N.Y.; et al. Loss of the Epigenetic Mark 5-hmC in Psoriasis: Implications for Epidermal Stem Cell Dysregulation. J. Investig. Dermatol. 2020, 140, 1266–1275.e3. [Google Scholar] [CrossRef]
- Kovacs, D.; Falchi, M.; Cardinali, G.; Raffa, S.; Carducci, M.; Cota, C.; Amantea, A.; Torrisi, M.R.; Picardo, M. Immunohistochemical analysis of keratinocyte growth factor and fibroblast growth factor 10 expression in psoriasis. Exp. Dermatol. 2005, 14, 130–137. [Google Scholar] [CrossRef]
- Bolla, B.S.; Erdei, L.; Urbán, E.; Burián, K.; Kemény, L.; Szabó, K. Cutibacterium acnes regulates the epidermal barrier properties of HPV-KER human immortalized keratinocyte cultures. Sci. Rep. 2020, 10, 12815. [Google Scholar] [CrossRef]
- Bozó, R.; Szél, E.; Danis, J.; Gubán, B.; Bata-Csörgő, Z.; Szabó, K.; Kemény, L.; Groma, G. Cartilage Oligomeric Matrix Protein Negatively Influences Keratinocyte Proliferation via α5β1-Integrin: Potential Relevance of Altered Cartilage Oligomeric Matrix Protein Expression in Psoriasis. J. Investig. Dermatol. 2020, 140, 1733–1742.e7. [Google Scholar] [CrossRef]
- Stefanowicz-Hajduk, J.; Ochocka, J.R. Real-time cell analysis system in cytotoxicity applications: Usefulness and comparison with tetrazolium salt assays. Toxicol. Rep. 2020, 7, 335–344. [Google Scholar] [CrossRef]
- Watson, R.E.B.; Poddar, R.; Walker, J.M.; McGuill, I.; Hoare, L.M.; Griffiths, C.E.M.; O’neill, C.A. Altered claudin expression is a feature of chronic plaque psoriasis. J. Pathol. 2007, 212, 450–458. [Google Scholar] [CrossRef]
- Furuse, M.; Hata, M.; Furuse, K.; Yoshida, Y.; Haratake, A.; Sugitani, Y.; Noda, T.; Kubo, A.; Tsukita, S. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: A lesson from claudin-1-deficient mice. J. Cell Biol. 2002, 156, 1099–1111. [Google Scholar] [CrossRef]
- Kitano, Y.; Okada, N. Separation of the epidermal sheet by dispase. Br. J. Dermatol. 1983, 108, 555–560. [Google Scholar] [CrossRef]
- Szabad, G.; Kormos, B.; Pivarcsi, A.; Széll, M.; Kis, K.; Kenderessy Szabó, A.; Dobozy, A.; Kemény, L.; Bata-Csörgo, Z. Human adult epidermal melanocytes cultured without chemical mitogens express the EGF receptor and respond to EGF. Arch. Dermatol. Res. 2007, 299, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Kho, D.; MacDonald, C.; Johnson, R.; Unsworth, C.P.; O’Carroll, S.J.; du Mez, E.; Angel, C.E.; Graham, E.S. Application of xCELLigence RTCA Biosensor Technology for Revealing the Profile and Window of Drug Responsiveness in Real Time. Biosensors 2015, 5, 199–222. [Google Scholar] [CrossRef] [PubMed]
- Andrianne, M.; Assabban, A.; La, C.; Mogilenko, D.; Salle, D.S.; Fleury, S.; Doumont, G.; Van Simaeys, G.; Nedospasov, S.A.; Blackshear, P.J.; et al. Tristetraprolin expression by keratinocytes controls local and systemic inflammation. JCI Insight 2017, 2, 92979. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Eden, E.; Lipson, D.; Yogev, S.; Yakhini, Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 2007, 3, e39. [Google Scholar] [CrossRef]
- Velez-delValle, C.; Marsch-Moreno, M.; Castro-Muñozledo, F.; Galván-Mendoza, I.J.; Kuri-Harcuch, W. Epithelial cell migration requires the interaction between the vimentin and keratin intermediate filaments. Sci. Rep. 2016, 6, 24389. [Google Scholar] [CrossRef]
- Yan, C.; Grimm, W.A.; Garner, W.L.; Qin, L.; Travis, T.; Tan, N.; Han, Y.-P. Epithelial to mesenchymal transition in human skin wound healing is induced by tumor necrosis factor-α through bone morphogenic protein-2. Am. J. Pathol. 2010, 176, 2247–2258. [Google Scholar] [CrossRef] [PubMed]
- Man, X.-Y.; Chen, X.-B.; Li, W.; Landeck, L.; Dou, T.-T.; Chen, J.-Q.; Zhou, J.; Cai, S.-Q.; Zheng, M. Analysis of epithelial-mesenchymal transition markers in psoriatic epidermal keratinocytes. Open Biol. 2015, 5, 150032. [Google Scholar] [CrossRef] [PubMed]
- Bernerd, F.; Magnaldo, T.; Darmon, M. Delayed onset of epidermal differentiation in psoriasis. J. Investig. Dermatol. 1992, 98, 902–910. [Google Scholar] [CrossRef] [PubMed]
- Waseem, A.; Dogan, B.; Tidman, N.; Alam, Y.; Purkis, P.; Jackson, S.; Lalli, A.; Machesney, M.; Leigh, I.M. Keratin 15 expression in stratified epithelia: Downregulation in activated keratinocytes. J. Investig. Dermatol. 1999, 112, 362–369. [Google Scholar] [CrossRef]
- Zhang, X.; Yin, M.; Zhang, L.-J. Keratin 6, 16 and 17-Critical Barrier Alarmin Molecules in Skin Wounds and Psoriasis. Cells 2019, 8, 807. [Google Scholar] [CrossRef]
- Tuschil, A.; Lam, C.; Haslberger, A.; Lindley, I. Interleukin-8 stimulates calcium transients and promotes epidermal cell proliferation. J. Investig. Dermatol. 1992, 99, 294–298. [Google Scholar] [CrossRef] [PubMed]
- Piskin, G.; Sylva-Steenland, R.M.R.; Bos, J.D.; Teunissen, M.B.M. In vitro and in situ expression of IL-23 by keratinocytes in healthy skin and psoriasis lesions: Enhanced expression in psoriatic skin. J. Immunol. 2006, 176, 1908–1915. [Google Scholar] [CrossRef] [PubMed]
- Thewes, M.; Stadler, R.; Korge, B.; Mischke, D. Normal psoriatic epidermis expression of hyperproliferation-associated keratins. Arch. Dermatol. Res. 1991, 283, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Gordon, K.; Kochkodan, J.J.; Blatt, H.; Lin, S.Y.; Kaplan, N.; Johnston, A.; Swindell, W.R.; Hoover, P.; Schlosser, B.J.; Elder, J.T.; et al. Alteration of the EphA2/Ephrin-A signaling axis in psoriatic epidermis. J. Investig. Dermatol. 2013, 133, 712–722. [Google Scholar] [CrossRef]
- Lai, Y.; Gallo, R.L. AMPed up immunity: How antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009, 30, 131–141. [Google Scholar] [CrossRef]
- Zhang, C.; Hu, Z.; Lone, A.G.; Artami, M.; Edwards, M.; Zouboulis, C.C.; Stein, M.; Harris-Tryon, T.A. Small proline-rich proteins (SPRRs) are epidermally produced antimicrobial proteins that defend the cutaneous barrier by direct bacterial membrane disruption. eLife 2022, 11, e76729. [Google Scholar] [CrossRef]
- D’Erme, A.M.; Wilsmann-Theis, D.; Wagenpfeil, J.; Hölzel, M.; Ferring-Schmitt, S.; Sternberg, S.; Wittmann, M.; Peters, B.; Bosio, A.; Bieber, T.; et al. IL-36γ (IL-1F9) Is a Biomarker for Psoriasis Skin Lesions. J. Investig. Dermatol. 2015, 135, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Niehues, H.; Tsoi, L.C.; van der Krieken, D.A.; Jansen, P.A.M.; Oortveld, M.A.W.; Rodijk-Olthuis, D.; van Vlijmen, I.M.J.J.; Hendriks, W.J.A.J.; Helder, R.W.; Bouwstra, J.A.; et al. Psoriasis-Associated Late Cornified Envelope (LCE) Proteins Have Antibacterial Activity. J. Investig. Dermatol. 2017, 137, 2380–2388. [Google Scholar] [CrossRef] [PubMed]
- Borchers, N.S.; Santos-Valente, E.; Toncheva, A.A.; Wehkamp, J.; Franke, A.; Gaertner, V.D.; Nordkild, P.; Genuneit, J.; Jensen, B.A.H.; Kabesch, M. Human β-Defensin 2 Mutations Are Associated With Asthma and Atopy in Children and Its Application Prevents Atopic Asthma in a Mouse Model. Front. Immunol. 2021, 12, 636061. [Google Scholar] [CrossRef]
- De Jongh, G.J.; Zeeuwen, P.L.J.M.; Kucharekova, M.; Pfundt, R.; van der Valk, P.G.; Blokx, W.; Dogan, A.; Hiemstra, P.S.; van de Kerkhof, P.C.; Schalkwijk, J. High expression levels of keratinocyte antimicrobial proteins in psoriasis compared with atopic dermatitis. J. Investig. Dermatol. 2005, 125, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Clayton, S.A.; Daley, K.K.; MacDonald, L.; Fernandez-Vizarra, E.; Bottegoni, G.; O’Neil, J.D.; Major, T.; Griffin, D.; Zhuang, Q.; Adewoye, A.B.; et al. Inflammation causes remodeling of mitochondrial cytochrome c oxidase mediated by the bifunctional gene C15orf48. Sci. Adv. 2021, 7, eabl5182. [Google Scholar] [CrossRef]
- Fujisawa, A. CEA (Carcinoembryonic Antigen) and CEACAM6 (CEA-Related Cell Adhesion Molecul 6) are Expressed in Psoriasis Vulgaris. Open Dermatol. J. 2013, 7, 47–52. [Google Scholar] [CrossRef]
- Parthun, M.R. Hat1: The emerging cellular roles of a type B histone acetyltransferase. Oncogene 2007, 26, 5319–5328. [Google Scholar] [CrossRef]
- Koeneke, E.; Witt, O.; Oehme, I. HDAC Family Members Intertwined in the Regulation of Autophagy: A Druggable Vulnerability in Aggressive Tumor Entities. Cells 2015, 4, 135–168. [Google Scholar] [CrossRef]
- Orsmond, A.; Bereza-Malcolm, L.; Lynch, T.; March, L.; Xue, M. Skin Barrier Dysregulation in Psoriasis. Int. J. Mol. Sci. 2021, 22, 10841. [Google Scholar] [CrossRef]
- Mosca, M.; Hong, J.; Hadeler, E.; Hakimi, M.; Liao, W.; Bhutani, T. The Role of IL-17 Cytokines in Psoriasis. Immunotargets Ther. 2021, 10, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Peserico, A.; Simone, C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 2011, 2011, 371832. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Rao, A.; Ko, M. TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp. Mol. Med. 2017, 49, e323. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, M.K.; Uppala, R.; Zeng, C.; Billi, A.C.; Tsoi, L.C.; Kidder, A.; Xing, X.; Perez White, B.E.; Shao, S.; Plazyo, O.; et al. Keratinocytes sense and eliminate CRISPR DNA through STING/IFN-κ activation and APOBEC3G induction. J. Clin. Investig. 2023, 133, e159393. [Google Scholar] [CrossRef]
- Budunova, I.; Trubetskoy, D.; Klopot, A.; Shi, B.; Tsoi, L.C.; White, B.E.P. 1315 Differential response of 3D African American and White Non-Hispanic skin organoids to major pro-inflammatory cytokines. J. Investig. Dermatol. 2023, 143, S225. [Google Scholar] [CrossRef]





| Samples | DEGs | Down-Regulated | Up-Regulated |
|---|---|---|---|
| Mildly inflamed KCs | 858 | 285 (33.21%) | 573 (66.79%) |
| Severely inflamed KCs (10× CytoMix) | 6987 | 3655 (52.31%) | 3332 (47.69%) |
| Mildly-Inflamed KCs DEGs (CytoMix-Treated vs. Control) | |||||||||
| Down-regulated DEGs | Up-regulated DEGs | ||||||||
| Number | Gene Symbol | Description | FC | Log FC | Number | Gene Symbol | Description | FC | Log FC |
| 1 | KRT1 | keratin 1 | −151.88 | −7.25 | 1 | S100A7A | S100 calcium binding protein A7A | 4453.61 | 12.12 |
| 2 | KRT125P | keratin 125 pseudogene | −82.84 | −6.37 | 2 | S100A7 | S100 calcium binding protein A7 | 1299.46 | 10.34 |
| 3 | KRT10 | keratin 10 | −62.23 | −5.96 | 3 | DEFB4A | defensin beta 4A | 963.52 | 9.91 |
| 4 | CXCL14 | C-X-C motif chemokine ligand 14 | −52.00 | −5.70 | 4 | DEFB4B | defensin beta 4B | 782.22 | 9.61 |
| 5 | KRT77 | keratin 77 | −38.38 | −5.26 | 5 | CEACAM6 | carcinoembryonic antigen related cell adhesion molecule 6 | 337.34 | 8.40 |
| 6 | DSC1 | desmocollin 1 | −26.53 | −4.73 | 6 | SPRR2C | small proline rich protein 2C (pseudogene) | 294.24 | 8.20 |
| 7 | CHP2 | calcineurin like EF-hand protein 2 | −21.70 | −4.44 | 7 | C15orf48 | chromosome 15 open reading frame 48 | 293.36 | 8.20 |
| 8 | CALML3 | calmodulin like 3 | −21.13 | −4.40 | 8 | SPRR2A | small proline rich protein 2A | 287.57 | 8.17 |
| 9 | ZBTB16 | zinc finger and BTB domain containing 16 | −19.90 | −4.31 | 9 | SPRR2F | small proline rich protein 2F | 287.26 | 8.17 |
| 10 | BBOX1 | gamma-butyrobetaine hydroxylase 1 | −16.34 | −4.03 | 10 | CLDN17 | claudin 17 | 246.05 | 7.94 |
| 11 | AADACL2 | arylacetamide deacetylase like 2 | −15.30 | −3.94 | 11 | LCE3A | late cornified envelope 3A | 233.99 | 7.87 |
| 12 | DSG1 | desmoglein 1 | −15.22 | −3.93 | 12 | SPRR2G | small proline rich protein 2G | 231.50 | 7.85 |
| 13 | POU3F1 | POU class 3 homeobox 1 | −15.16 | −3.92 | 13 | S100A12 | S100 calcium binding protein A12 | 216.71 | 7.76 |
| 14 | MYO3B | myosin IIIB | −14.94 | −3.90 | 14 | LCE3D | late cornified envelope 3D | 215.56 | 7.75 |
| 15 | THEM5 | thioesterase superfamily member 5 | −14.30 | −3.84 | 15 | SPRR2B | small proline rich protein 2B | 206.64 | 7.69 |
| 16 | EPHB6 | EPH receptor B6 | −13.49 | −3.75 | 16 | SLC6A14 | solute carrier family 6 member 14 | 177.68 | 7.47 |
| 17 | CCDC3 | coiled-coil domain containing 3 | −13.43 | −3.75 | 17 | IL36G | interleukin 36, gamma | 171.82 | 7.42 |
| 18 | DNASE1L3 | deoxyribonuclease 1 like 3 | −13.34 | −3.74 | 18 | S100A9 | S100 calcium binding protein A9 | 153.24 | 7.26 |
| 19 | FRG2HP | FSHD region gene 2 family member H, pseudogene | −13.21 | −3.72 | 19 | RHCG | Rh family C glycoprotein | 152.64 | 7.25 |
| 20 | PALMD | palmdelphin | −12.44 | −3.64 | 20 | LCE3E | late cornified envelope 3E | 133.70 | 7.06 |
| Severely-Inflamed KCs DEGs (CytoMix-Treated vs. Control) | |||||||||
| Down-regulated DEGs | Up-regulated DEGs | ||||||||
| Number | Gene Symbol | Description | FC | Log FC | Number | Gene Symbol | Description | FC | Log FC |
| 1 | ID3 | inhibitor of DNA binding 3, HLH protein | −571.72 | −9.16 | 1 | DEFB103B | defensin beta 103B | 8680.30 | 13.08 |
| 2 | DSC1 | desmocollin 1 | −487.55 | −8.93 | 2 | LCE3A | late cornified envelope 3A | 6831.53 | 12.74 |
| 3 | EPHB6 | EPH receptor B6 | −429.75 | −8.75 | 3 | DEFB103A | defensin beta 103A | 6802.54 | 12.73 |
| 4 | APCDD1 | APC down-regulated 1 | −335.49 | −8.39 | 4 | S100A7 | S100 calcium binding protein A7 | 4901.32 | 12.26 |
| 5 | ECM2 | extracellular matrix protein 2 | −333.54 | −8.38 | 5 | SPRR2C | small proline rich protein 2C (pseudogene) | 4883.04 | 12.25 |
| 6 | CCDC3 | coiled-coil domain containing 3 | −318.71 | −8.32 | 6 | DEFB4A | defensin beta 4A | 4175.80 | 12.03 |
| 7 | PALMD | palmdelphin | −277.46 | −8.12 | 7 | CXCL9 | C-X-C motif chemokine ligand 9 | 3962.80 | 11.95 |
| 8 | KRT1 | keratin 1 | −277.14 | −8.11 | 8 | DEFB4B | defensin beta 4B | 3775.58 | 11.88 |
| 9 | TXNIP | thioredoxin interacting protein | −264.49 | −8.05 | 9 | CEACAM6 | carcinoembryonic antigen related cell adhesion molecule 6 | 3679.34 | 11.85 |
| 10 | EPHX2 | epoxide hydrolase 2 | −264.19 | −8.05 | 10 | SPRR2A | small proline rich protein 2A | 3117.66 | 11.61 |
| 11 | KLRG2 | killer cell lectin like receptor G2 | −264.06 | −8.04 | 11 | ACP7 | acid phosphatase 7, tartrate resistant (putative) | 2982.39 | 11.54 |
| 12 | LYRM7 | LYR motif containing 7 | −262.17 | −8.03 | 12 | LCE3D | late cornified envelope 3D | 2980.27 | 11.54 |
| 13 | IGFL2 | IGF like family member 2 | −250.56 | −7.97 | 13 | SPRR2G | small proline rich protein 2G | 2869.05 | 11.49 |
| 14 | PIR | pirin | −240.98 | −7.91 | 14 | CRCT1 | cysteine rich C-terminal 1 | 2807.03 | 11.45 |
| 15 | RAB7B | RAB7B, member RAS oncogene family | −240.34 | −7.91 | 15 | C15orf48 | chromosome 15 open reading frame 48 | 2718.28 | 11.41 |
| 16 | IL20RA | interleukin 20 receptor subunit alpha | −212.41 | −7.73 | 16 | SPRR2F | small proline rich protein 2F | 2608.19 | 11.35 |
| 17 | CYP39A1 | cytochrome P450 family 39 subfamily A member 1 | −198.56 | −7.63 | 17 | IGFN1 | immunoglobulin-like and fibronectin type III domain containing 1 | 2348.78 | 11.20 |
| 18 | CCDC152 | coiled-coil domain containing 152 | −181.65 | −7.50 | 18 | LCE3E | late cornified envelope 3E | 2325.20 | 11.18 |
| 19 | FLG | filaggrin | −181.30 | −7.50 | 19 | PRSS22 | protease, serine 22 | 2291.30 | 11.16 |
| 20 | S1PR5 | sphingosine-1-phosphate receptor 5 | −180.17 | −7.49 | 20 | MMP10 | matrix metallopeptidase 10 | 2194.50 | 11.10 |
| Gene Symbol | Mildly Inflamed KCs vs. Controls | Severely Inflamed KCs vs. Controls | ||
|---|---|---|---|---|
| Fold of Change (FC) | p-Value | Fold of Change (FC) | p-Value | |
| HAT1 | −1.10 | 0.6981141 | −2.45 | 2.708402 × 10−4 |
| HDAC9 | +4.60 | 0.035447594 | +7.40 | 1.0198189 × 10−4 |
| DNMT1 | −1.31 | 0.017961252 | −7.30 | 0.0012681335 |
| DNMT3A | −1.06 | 0.52116096 | +2.24 | 3.4366465 × 10−5 |
| DNMT3B | −2.20 | 0.22995375 | −1.38 | 0.04189076 |
| TET3 | −1.25 | 0.5099057 | +3.97 | 3.0937628 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghaffarinia, A.; Póliska, S.; Ayaydin, F.; Goblos, A.; Parvaneh, S.; Manczinger, M.; Balogh, F.; Erdei, L.; Veréb, Z.; Szabó, K.; et al. Unraveling Transcriptome Profile, Epigenetic Dynamics, and Morphological Changes in Psoriasis-like Keratinocytes: “Insights into Similarity with Psoriatic Lesional Epidermis”. Cells 2023, 12, 2825. https://doi.org/10.3390/cells12242825
Ghaffarinia A, Póliska S, Ayaydin F, Goblos A, Parvaneh S, Manczinger M, Balogh F, Erdei L, Veréb Z, Szabó K, et al. Unraveling Transcriptome Profile, Epigenetic Dynamics, and Morphological Changes in Psoriasis-like Keratinocytes: “Insights into Similarity with Psoriatic Lesional Epidermis”. Cells. 2023; 12(24):2825. https://doi.org/10.3390/cells12242825
Chicago/Turabian StyleGhaffarinia, Ameneh, Szilárd Póliska, Ferhan Ayaydin, Aniko Goblos, Shahram Parvaneh, Máté Manczinger, Fanni Balogh, Lilla Erdei, Zoltán Veréb, Kornélia Szabó, and et al. 2023. "Unraveling Transcriptome Profile, Epigenetic Dynamics, and Morphological Changes in Psoriasis-like Keratinocytes: “Insights into Similarity with Psoriatic Lesional Epidermis”" Cells 12, no. 24: 2825. https://doi.org/10.3390/cells12242825
APA StyleGhaffarinia, A., Póliska, S., Ayaydin, F., Goblos, A., Parvaneh, S., Manczinger, M., Balogh, F., Erdei, L., Veréb, Z., Szabó, K., Bata-Csörgő, Z., & Kemény, L. (2023). Unraveling Transcriptome Profile, Epigenetic Dynamics, and Morphological Changes in Psoriasis-like Keratinocytes: “Insights into Similarity with Psoriatic Lesional Epidermis”. Cells, 12(24), 2825. https://doi.org/10.3390/cells12242825