Hypoxia Induces Alterations in the Circadian Rhythm in Patients with Chronic Respiratory Diseases
Abstract
1. Introduction
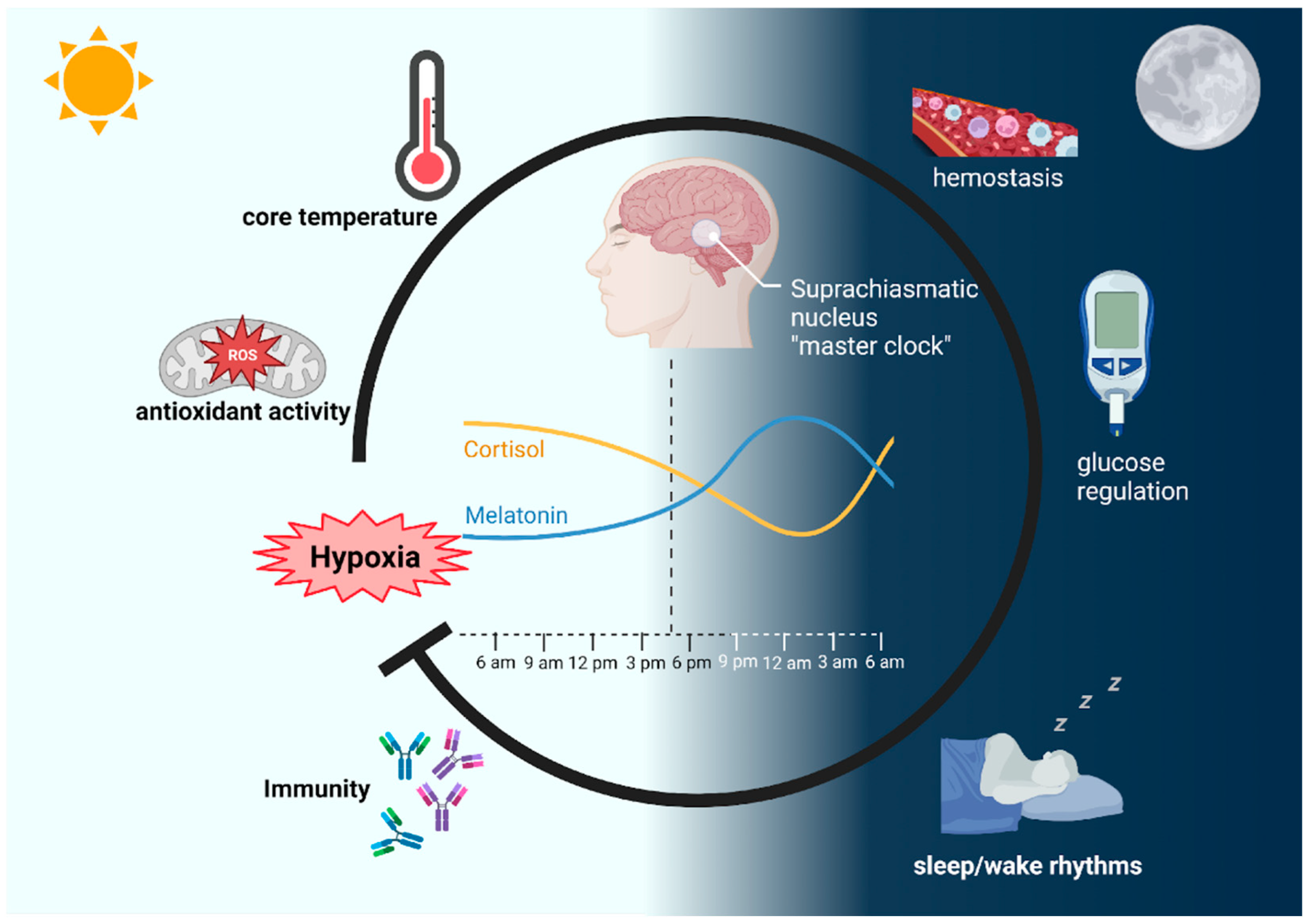
2. The Impact of Hypoxia on Physiological and Cellular Circadian Rhythms
3. Pulmonary Pathologies and Their Relationship with the Circadian Cycle
3.1. Asthma
3.2. Chronic Obstructive Pulmonary Disease
3.3. Lung Cancer
3.4. Idiopathic Pulmonary Fibrosis
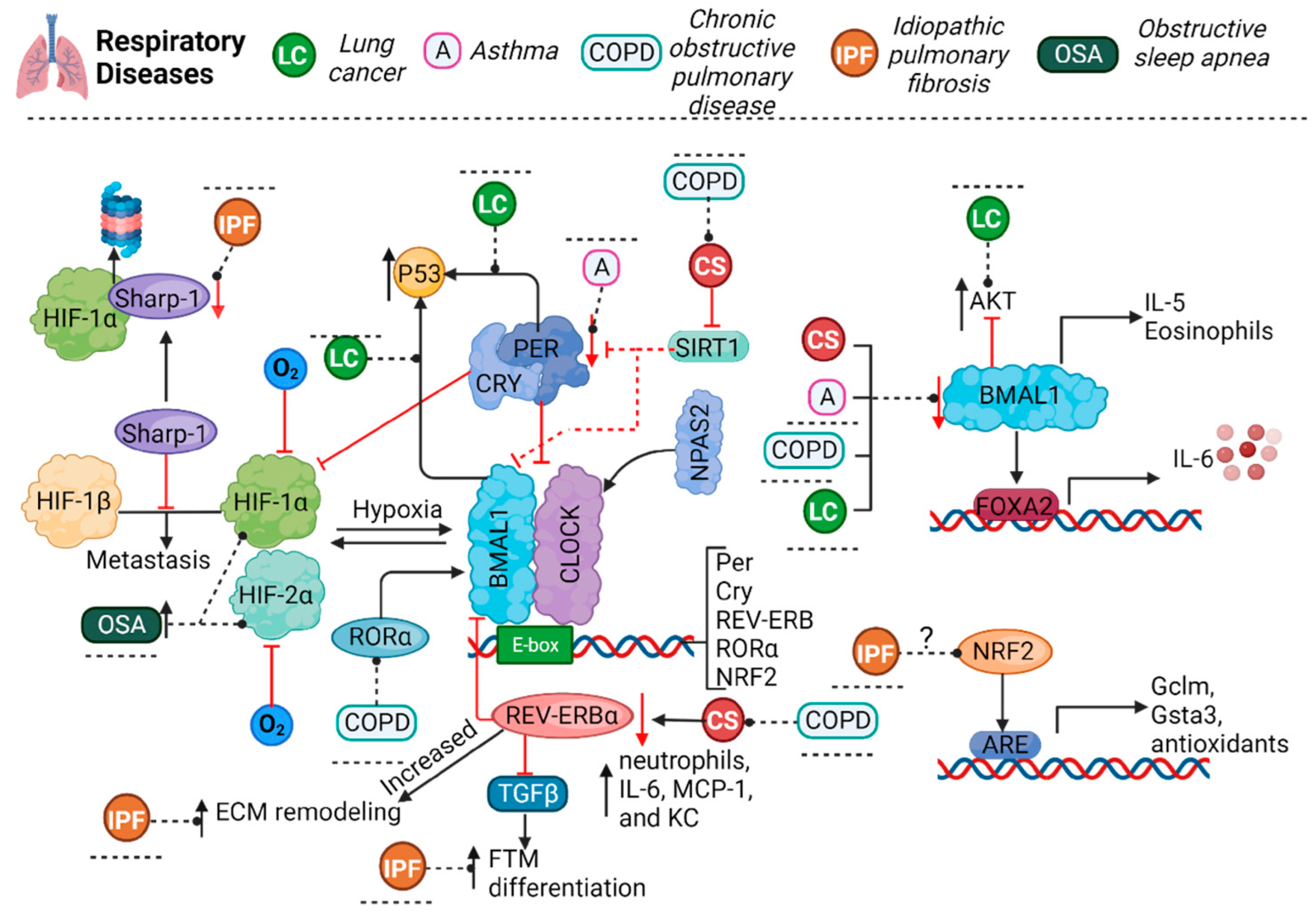
3.5. Obstructive Sleep Apnea
3.6. Influenza and COVID-19
4. Understanding Hypoxia and Circadian Rhythm Genes from a Systems Biology Perspective
5. Interactions between Aging, Hypoxia, and Circadian Rhythms
- (a)
- HIF can regulate the expression of clock genes, and vice versa.
- (b)
- The circadian clock regulates the expression of genes involved in hypoxia signaling.
- (c)
- Mitochondrial dysfunction, which is a common feature of aging, can disrupt both circadian clock function and hypoxia signaling.
- (d)
- Inflammation, which is another common feature of aging, can also disrupt both circadian clock function and hypoxia signaling.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vitaterna, M.H.; Takahashi, J.S.; Turek, F.W. Overview of circadian rhythms. Alcohol Res. Health J. Natl. Inst. Alcohol Abus. Alcohol. 2001, 25, 85–93. [Google Scholar]
- Huang, W.; Ramsey, K.M.; Marcheva, B.; Bass, J. Circadian rhythms, sleep, and metabolism. J. Clin. Investig. 2011, 121, 2133–2141. [Google Scholar] [CrossRef] [PubMed]
- Peek, C.B.; Ramsey, K.M.; Levine, D.C.; Marcheva, B.; Perelis, M.; Bass, J. Circadian Regulation of Cellular Physiology. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 165–184. [Google Scholar] [CrossRef]
- Yamazaki, S.; Numano, R.; Abe, M.; Hida, A.; Takahashi, R.; Ueda, M.; Block, G.D.; Sakaki, Y.; Menaker, M.; Tei, H. Resetting central and peripheral circadian oscillators in transgenic rats. Science 2000, 288, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Claustrat, B.; Leston, J. Melatonin: Physiological effects in humans. Neuro-Chirurgie 2015, 61, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Cipolla-Neto, J.; Amaral, F.G.D. Melatonin as a Hormone: New Physiological and Clinical Insights. Endocr. Rev. 2018, 39, 990–1028. [Google Scholar] [CrossRef] [PubMed]
- Kennaway, D.J.; Goble, F.C.; Stamp, G.E. Factors influencing the development of melatonin rhythmicity in humans. J. Clin. Endocrinol. Metab. 1996, 81, 1525–1532. [Google Scholar] [CrossRef]
- Price, D.A.; Close, G.C.; Fielding, B.A. Age of appearance of circadian rhythm in salivary cortisol values in infancy. Arch. Dis. Child. 1983, 58, 454–456. [Google Scholar] [CrossRef]
- Reddy, S.; Reddy, V.; Sharma, S. Physiology, Circadian Rhythm; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Quera-Salva, M.A.; Claustrat, B. Melatonin: Physiological and pharmacological aspects related to sleep: The interest of a prolonged-release formulation (Circadin ®) in insomnia. L’Encephale 2018, 44, 548–557. [Google Scholar] [CrossRef]
- Tavartkiladze, A.G.; Simoniia, G.V.; Kolbaia, D.T.; Shalashvili, A.G.; Petriashvili, T.G. Biochemical, pharmacological and clinical aspects of influencing methioninc, tryptophan, pyridoxine (vitamin B6), Ca2+ and high-calorie food on the synthesis and intensity of the secretion of melatonin. Georgian Med. News 2006, 132, 121–123. [Google Scholar]
- Ostrin, L.A. Ocular and systemic melatonin and the influence of light exposure. Clin. Exp. Optom. 2019, 102, 99–108. [Google Scholar] [CrossRef]
- Law, R.; Clow, A. Stress, the cortisol awakening response and cognitive function. Int. Rev. Neurobiol. Elsevier 2020, 150, 187–217. [Google Scholar] [CrossRef]
- Dinneen, S.; Alzaid, A.; Miles, J.; Rizza, R. Effects of the normal nocturnal rise in cortisol on carbohydrate and fat metabolism in IDDM. Am. J. Physiol. 1995, 268, E595–E603. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.; Korf, H.W.; Kuffer, L.; Groß, J.V.; Erren, T.C. Exercise Time Cues (zeitgebers) for Human Circadian Systems Can Foster Health and Improve Performance: A Systematic Review. BMJ Open Sport Exerc. Med. 2018, 4, e000443. [Google Scholar] [CrossRef] [PubMed]
- Hastings, M.H.; Maywood, E.S.; Brancaccio, M. Generation of Circadian Rhythms in the Suprachiasmatic Nucleus. Nat. Rev. Neurosci. 2018, 19, 453–469. [Google Scholar] [CrossRef] [PubMed]
- Giri, A.; Wang, Q.; Rahman, I.; Sundar, I.K. Circadian molecular clock disruption in chronic pulmonary diseases. Trends Mol. Med. 2022, 28, 513–527. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Zhou, Y.; Yin, D.; Lv, C.; Lin, J.; Bao, W.; Zhang, M. Age-related circadian rhythm and variability of large- and small-airway function in healthy non-smoking adults: Data from 7-day diurnal and nocturnal home monitoring using an electronic portable spirometer. Front. Public Health. 2022, 10, 946988. [Google Scholar] [CrossRef]
- Nosal, C.; Ehlers, A.; Haspel, J.A. Why Lungs Keep Time: Circadian Rhythms and Lung Immunity. Annu. Rev. Physiol. 2020, 82, 391–412. [Google Scholar] [CrossRef]
- Dodd, J.W.; Getov, S.V.; Jones, P.W. Cognitive Function in COPD. Eur. Respir. J. 2010, 35, 913–922. [Google Scholar] [CrossRef]
- Daulatzai, M.A. Evidence of Neurodegeneration in Obstructive Sleep Apnea: Relationship Between Obstructive Sleep Apnea and Cognitive Dysfunction in the Elderly. J. Neurosci. Res. 2015, 93, 1778–1794. [Google Scholar] [CrossRef]
- Kerner, N.A.; Roose, S.P. Obstructive Sleep Apnea Is Linked to Depression and Cognitive Impairment: Evidence and Potential Mechanisms. Am. J. Geriatr. Psychiatry 2016, 24, 496–508. [Google Scholar] [CrossRef]
- Mayfield, K.P.; Hong, E.J.; Carney, K.M.; D’Alecy, L.G. Potential adaptations to acute hypoxia: Hct, stress proteins, and set point for temperature regulation. Am. J. Physiol. 1994, 266, R1615–R1622. [Google Scholar] [CrossRef] [PubMed]
- Mesarwi, O.A.; Loomba, R.; Malhotra, A. Obstructive Sleep Apnea, Hypoxia, and Nonalcoholic Fatty Liver Disease. Am. J. Respir. Crit. Care Med. 2019, 199, 830–841. [Google Scholar] [CrossRef]
- Richalet, J.P.; Lhuissier, F.J. Aging, Tolerance to High Altitude, and Cardiorespiratory Response to Hypoxia. High Alt. Med. Biol. 2015, 16, 117–124. [Google Scholar] [CrossRef]
- O’Connell, E.J.; Martinez, C.A.; Liang, Y.G.; Cistulli, P.A.; Cook, K.M. Out of breath, out of time: Interactions between HIF and circadian rhythms. Am. J. Physiol. Cell Physiol. 2020, 319, C533–C540. [Google Scholar] [CrossRef] [PubMed]
- Adamovich, Y.; Ladeuix, B.; Sobel, J.; Manella, G.; Neufeld-Cohen, A.; Assadi, M.H.; Golik, M.; Kuperman, Y.; Tarasiuk, A.; Koeners, M.P. Oxygen and Carbon Dioxide Rhythms Are Circadian Clock Controlled and Differentially Directed by Behavioral Signals. Cell Metab. 2019, 29, 1092–1103.e3. [Google Scholar] [CrossRef]
- Adamovich, Y.; Ladeuix, B.; Golik, M.; Koeners, M.P.; Asher, G. Rhythmic Oxygen Levels Reset Circadian Clocks Through Hif1α. Cell Metab. 2017, 25, 93–101. [Google Scholar] [CrossRef]
- Prabhakar, N.R.; Peng, Y.-J.; Nanduri, J. Hypoxia-inducible Factors and Obstructive Sleep Apnea. J. Clin. Investig. 2020, 130, 5042–5051. [Google Scholar] [CrossRef] [PubMed]
- Hunyor, I.; Cook, K.M. Models of Intermittent Hypoxia and Obstructive Sleep Apnea: Molecular Pathways and Their Contribution to Cancer. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2018, 315, R669–R687. [Google Scholar] [CrossRef]
- Durgan, D.J.; Crossland, R.F.; Bryan, R.M. The Rat Cerebral Vasculature Exhibits Time-of-day-dependent Oscillations in Circadian Clock Genes and Vascular Function That Are Attenuated Following Obstructive Sleep Apnea. J. Cereb. Blood Flow Metab. 2017, 37, 2806–2819. [Google Scholar] [CrossRef]
- Chilov, D.; Hofer, T.; Bauer, C.; Wenger, R.H.; Gassmann, M. Hypoxia affects expression of circadian genes PER1 and CLOCK in mouse brain. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2001, 15, 2613–2622. [Google Scholar] [CrossRef]
- Hogenesch, J.B.; Gu, Y.-Z.; Jain, S.; Bradfield, C.A. The Basic-helix–loop–helix-pas Orphan MOP3 Forms Transcriptionally Active Complexes with Circadian and Hypoxia Factors. Proc. Natl. Acad. Sci. USA 1998, 95, 5474–5479. [Google Scholar] [CrossRef] [PubMed]
- Eckle, T.; Hartmann, K.; Bonney, S.; Reithel, S.; Mittelbronn, M.; Walker, L.A.; Lowes, B.D.; Han, J.; Borchers, C.H.; Buttrick, P.M. Adora2b-elicited Per2 Stabilization Promotes a Hif-dependent Metabolic Switch Crucial for Myocardial Adaptation to Ischemia. Nat. Med. 2012, 18, 774–782. [Google Scholar] [CrossRef] [PubMed]
- Gabryelska, A.; Turkiewicz, S.; Karuga, F.F.; Sochal, M.; Strzelecki, D.; Białasiewicz, P. Disruption of Circadian Rhythm Genes in Obstructive Sleep Apnea Patients—Possible Mechanisms Involved and Clinical Implication. Int. J. Mol. Sci. 2022, 23, 709. [Google Scholar] [CrossRef] [PubMed]
- Bartman, C.M.; Eckle, T. Circadian-hypoxia Link and Its Potential for Treatment of Cardiovascular Disease. Curr. Pharm. Des. 2019, 25, 1075–1090. [Google Scholar] [CrossRef] [PubMed]
- López-Cano, C.; Gutiérrez-Carrasquilla, L.; Barbé, F.; Sánchez, E.; Hernández, M.; Martí, R.; Ceperuelo-Mallafre, V.; Dalmases, M.; Fernández-Veledo, S.; Vendrell, J. Effect of Type 2 Diabetes Mellitus on the Hypoxia-inducible Factor 1-alpha Expression. Is There a Relationship with the Clock Genes? J. Clin. Med. 2020, 9, 2632. [Google Scholar] [CrossRef]
- Manella, G.; Aviram, R.; Bolshette, N.; Muvkadi, S.; Golik, M.; Smith, D.F.; Asher, G. Hypoxia Induces a Time- and Tissue-specific Response That Elicits Intertissue Circadian Clock Misalignment. Proc. Natl. Acad. Sci. USA 2020, 117, 779–786. [Google Scholar] [CrossRef]
- Dimova, E.Y.; Jakupovic, M.; Kubaichuk, K.; Mennerich, D.; Chi, T.F.; Tamanini, F.; Oklejewicz, M.; Hänig, J.; Byts, N.; Mäkelä, K.A. The Circadian Clock Protein CRY1 Is a Negative Regulator of Hif-1α. iScience 2019, 13, 284–304. [Google Scholar] [CrossRef]
- Sato, T.K.; Yamada, R.G.; Ukai, H.; Baggs, J.E.; Miraglia, L.J.; Kobayashi, T.J.; Welsh, D.K.; Kay, S.A.; Ueda, H.R.; Hogenesch, J.B. Feedback Repression Is Required for Mammalian Circadian Clock Function. Nat. Genet. 2006, 38, 312–319. [Google Scholar] [CrossRef]
- Wu, Y.; Tang, D.; Liu, N.; Xiong, W.; Huang, H.; Li, Y.; Ma, Z.; Zhao, H.; Chen, P.; Qi, X. Reciprocal Regulation Between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab. 2017, 25, 73–85. [Google Scholar] [CrossRef]
- Walton, Z.E.; Patel, C.H.; Brooks, R.C.; Yu, Y.; Ibrahim-Hashim, A.; Riddle, M.; Porcu, A.; Jiang, T.; Ecker, B.L.; Tameire, F. Acid Suspends the Circadian Clock in Hypoxia through Inhibition of Mtor. Cell 2018, 174, 72–87.e32. [Google Scholar] [CrossRef]
- Bando, H.; Nishio, T.; Van Der Horst, G.T.J.; Masubuchi, S.; Hisa, Y.; Okamura, H. Vagal Regulation of Respiratory Clocks in Mice. J. Neurosci. 2007, 27, 4359–4365. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J.E.; Beesley, S.; Plumb, J.; Singh, D.; Farrow, S.; Ray, D.W.; Loudon, A.S. Circadian timing in the lung; a specific role for bronchiolar epithelial cells. Endocrinology. 2009, 150, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Casale, R.; Natali, G.; Colantonio, D.; Pasqualetti, P. Circadian Rhythm of Peak Expiratory Flow in Children Passively Exposed and Not Exposed to Cigarette Smoke. Thorax 1992, 47, 801–803. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gebel, S.; Gerstmayer, B.; Kuhl, P.; Borlak, J.; Meurrens, K.; Müller, T. The kinetics of transcriptomic changes induced by cigarette smoke in rat lungs reveals a specific program of defense, inflammation, and circadian clock gene expression. Toxicol. Sci. Off. J. Soc. Toxicol. 2006, 93, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Janssens, J.P. Aging of the respiratory system: Impact on pulmonary function tests and adaptation to exertion. Clin. Chest Med. 2005, 26, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Yeo, E.-J. Hypoxia and Aging. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Holgate, S.T.; Wenzel, S.; Postma, D.S.; Weiss, S.T.; Renz, H.; Sly, P.D. Asthma. Nat. Rev. Dis. Primers 2015, 1, 15025. [Google Scholar] [CrossRef] [PubMed]
- Ballard, R.D.; Saathoff, M.C.; Patel, D.K.; Kelly, P.L.; Martin, R.J. Effect of sleep on nocturnal bronchoconstriction and ventilatory patterns in asthmatics. J. Appl. Physiol. 1989, 67, 243–249. [Google Scholar] [CrossRef]
- Scheer, F.A.J.L.; Hilton, M.F.; Evoniuk, H.L.; Shiels, S.A.; Malhotra, A.; Sugarbaker, R.; Ayers, R.T.; Israel, E.; Massaro, A.F.; Shea, S.A. The Endogenous Circadian System Worsens Asthma at Night Independent of Sleep and Other Daily Behavioral or Environmental Cycles. Proc. Natl. Acad. Sci. USA 2021, 118, e2018486118. [Google Scholar] [CrossRef]
- Sutherland, E.R.; Ellison, M.C.; Kraft, M.; Martin, R.J. Elevated serum melatonin is associated with the nocturnal worsening of asthma. J. Allergy Clin. Immunol. 2003, 112, 513–517. [Google Scholar] [CrossRef]
- Jönsson, E.; Mossberg, B. Impairment of ventilatory function by supine posture in asthma. Eur. J. Respir. Dis. 1984, 65, 496–503. [Google Scholar] [PubMed]
- Chen, W.Y.; Chai, H. Airway cooling and nocturnal asthma. Chest 1982, 81, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Maidstone, R.J.; Turner, J.; Vetter, C.; Dashti, H.S.; Saxena, R.; Scheer, F.A.J.L.; Shea, S.A.; Kyle, S.D.; Lawlor, D.A.; Loudon, A.S.I. Night Shift Work Is Associated with an Increased Risk of Asthma. Thorax 2021, 76, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Landstra, A.M.; Postma, D.S.; Boezen, H.M.; van Aalderen, W.M. R ole of serum cortisol levels in children with asthma. Am. J. Respir. Crit. Care Med. 2002, 165, 708–712. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, E.R.; Ellison, M.C.; Kraft, M.; Martin, R.J. Altered Pituitary-adrenal Interaction in Nocturnal Asthma. J. Allergy Clin. Immunol. 2003, 112, 52–57. [Google Scholar] [CrossRef]
- Ehlers, A.; Xie, W.; Agapov, E.; Brown, S.; Steinberg, D.; Tidwell, R.; Sajol, G.; Schutz, R.; Weaver, R.; Yu, H. BMAL1 Links the Circadian Clock to Viral Airway Pathology and Asthma Phenotypes. Mucosal Immunol. 2018, 11, 97–111. [Google Scholar] [CrossRef]
- Chen, H.-C.; Chen, Y.-C.; Wang, T.-N.; Fang, W.-F.; Chang, Y.-C.; Chen, Y.-M.; Chen, I.-Y.; Lin, M.-C.; Yang, M.-Y. Disrupted Expression of Circadian Clock Genes in Patients with Bronchial Asthma. J. Asthma Allergy 2021, 14, 371–380. [Google Scholar] [CrossRef]
- Tang, L.; Liu, L.; Sun, X.; Hu, P.; Zhang, H.; Wang, B.; Zhang, X.; Jiang, J.; Zhao, X.; Shi, X. BMAL1/FOXA2-induced rhythmic fluctuations in IL-6 contribute to nocturnal asthma attacks. Front. Immunol. 2022, 13, 947067. [Google Scholar] [CrossRef]
- Zasłona, Z.; Case, S.; Early, J.O.; Lalor, S.J.; McLoughlin, R.M.; Curtis, A.M.; O’Neill, L.A.J. The circadian protein BMAL1 in myeloid cells is a negative regulator of allergic asthma. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 312, L855–L860. [Google Scholar] [CrossRef]
- Partridge, M.R.; Karlsson, N.; Small, I.R. Patient insight into the impact of chronic obstructive pulmonary disease in the morning: An internet survey. Curr. Med. Res. Opin. 2009, 25, 2043–2048. [Google Scholar] [CrossRef]
- Vij, N.; Chandramani-Shivalingappa, P.; Van Westphal, C.; Hole, R.; Bodas, M. Cigarette Smoke-induced Autophagy Impairment Accelerates Lung Aging, Copd-emphysema Exacerbations and Pathogenesis. Am. J. Physiol. Cell Physiol. 2018, 314, C73–C87. [Google Scholar] [CrossRef] [PubMed]
- Casale, R.; Pasqualetti, P. Cosinor analysis of circadian peak expiratory flow variability in normal subjects, passive smokers, heavy smokers, patients with chronic obstructive pulmonary disease and patients with interstitial lung disease. Respir. Int. Rev. Thorac. Dis. 1997, 64, 251–256. [Google Scholar] [CrossRef]
- Dawkins, K.D.; Muers, M.F. Diurnal Variation in Airflow Obstruction in Chronic Bronchitis. Thorax 1981, 36, 618–621. [Google Scholar] [CrossRef]
- Calverley, P.M.A. Effect of Tiotropium Bromide on Circadian Variation in Airflow Limitation in Chronic Obstructive Pulmonary Disease. Thorax 2003, 58, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Sundar, I.K.; Yao, H.; Sellix, M.T.; Rahman, I. Circadian Molecular Clock in Lung Pathophysiology. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 309, L1056–L1075. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.W.; Sundar, I.K.; Yao, H.; Sellix, M.T.; Rahman, I. Circadian clock function is disrupted by environmental tobacco/cigarette smoke, leading to lung inflammation and injury via a SIRT1-BMAL1 pathway. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biology 2014, 28, 176–194. [Google Scholar] [CrossRef] [PubMed]
- Sundar, I.K.; Rashid, K.; Sellix, M.T.; Rahman, I. The Nuclear Receptor and Clock Gene Rev-erbα Regulates Cigarette Smoke-induced Lung Inflammation. Biochem. Biophys. Res. Commun. 2017, 493, 1390–1395. [Google Scholar] [CrossRef]
- Wang, Q.; Sundar, I.K.; Lucas, J.H.; Muthumalage, T.; Rahman, I. Molecular Clock Rev-erbα Regulates Cigarette Smoke–induced Pulmonary Inflammation and Epithelial-mesenchymal Transition. JCI Insight 2021, 6, e145200. [Google Scholar] [CrossRef]
- Shi, Y.; Cao, J.; Gao, J.; Zheng, L.; Goodwin, A.; An, C.H.; Patel, A.; Lee, J.S.; Duncan, S.R.; Kaminski, N. Retinoic Acid–related Orphan Receptor-α Is Induced in the Setting of DNA Damage and Promotes Pulmonary Emphysema. Am. J. Respir. Crit. Care Med. 2012, 186, 412–419. [Google Scholar] [CrossRef]
- Yao, H.; Sundar, I.K.; Huang, Y.; Gerloff, J.; Sellix, M.T.; Sime, P.J.; Rahman, I. Disruption of Sirtuin 1–mediated Control of Circadian Molecular Clock and Inflammation in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cell Mol. Biol. 2015, 53, 782–792. [Google Scholar] [CrossRef]
- Li, L.; Zhang, M.; Zhao, C.; Cheng, Y.; Liu, C.; Shi, M. Circadian Clock Gene Clock-bmal1 Regulates Cellular Senescence in Chronic Obstructive Pulmonary Disease. BMC Pulm. Med. 2022, 22, 435. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Larsson, S.C.; Carter, P.; Kar, S.; Vithayathil, M.; Mason, A.M.; Michaëlsson, K.; Burgess, S. Smoking, Alcohol Consumption, and Cancer: A Mendelian Randomisation Study in UK Biobank and International Genetic Consortia Participants. PLOS Med. 2020, 17, e1003178. [Google Scholar] [CrossRef] [PubMed]
- Gandini, S.; Botteri, E.; Iodice, S.; Boniol, M.; Lowenfels, A.B.; Maisonneuve, P.; Boyle, P. Tobacco Smoking and Cancer: A Meta-analysis. Int. J. Cancer 2008, 122, 155–164. [Google Scholar] [CrossRef]
- Aredo, J.V.; Luo, S.J.; Gardner, R.M.; Sanyal, N.; Choi, E.; Hickey, T.P.; Riley, T.L.; Huang, W.-Y.; Kurian, A.W.; Leung, A.N. Tobacco Smoking and Risk of Second Primary Lung Cancer. J. Thorac. Oncol. 2021, 16, 968–979. [Google Scholar] [CrossRef]
- Wang, J.; Tang, H.; Duan, Y.; Yang, S.; An, J. Association Between Sleep Traits and Lung Cancer: A Mendelian Randomization Study. J. Immunol. Res. 2021, 2021, 1893882. [Google Scholar] [CrossRef]
- Huang, B.-H.; Duncan, M.J.; Cistulli, P.A.; Nassar, N.; Hamer, M.; Stamatakis, E. Sleep and Physical Activity in Relation to All-cause, Cardiovascular Disease and Cancer Mortality Risk. Br. J. Sport. Med. 2022, 56, 718–724. [Google Scholar] [CrossRef]
- Stone, C.R.; Haig, T.R.; Fiest, K.M.; Mcneil, J.; Brenner, D.R.; Friedenreich, C.M. The Association between Sleep Duration and Cancer-specific Mortality: A Systematic Review and Meta-analysis. Cancer Causes Control 2019, 30, 501–525. [Google Scholar] [CrossRef]
- Xie, J.; Zhu, M.; Ji, M.; Fan, J.; Huang, Y.; Wei, X.; Jiang, X.; Xu, J.; Yin, R.; Wang, Y.; et al. Relatsh Between Sleep Trait Lung Cancer Risk: A Prospect. Cohort Study UK Biobank. Sleep 2021, 44, zsab089. [Google Scholar] [CrossRef]
- Huo, Z.; Ge, F.; Li, C.; Cheng, H.; Lu, Y.; Wang, R.; Wen, Y.; Yue, K.; Pan, Z.; Peng, H.; et al. Genetically predicted insomnia and lung cancer risk: A Mendelian randomization study. Sleep Med. 2021, 87, 183–190. [Google Scholar] [CrossRef]
- Cao, Q.; Zhang, Q.; Li, X.C.; Ren, C.F.; Qiang, Y. Impact of sleep status on lung adenocarcinoma risk: A prospective cohort study. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 7641–7648. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.L.; Wu, M.W.; Sun, J.; Sun, Y.L.; Cai, Y.C.; Huang, Y.J.; Xian, L.J. Effects of the biological clock gene Bmal1 on tumour growth and anti-cancer drug activity. J. Biochem. 2010, 148, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Sulli, G.; Lam, M.T.Y.; Panda, S. Interplay between Circadian Clock and Cancer: New Frontiers for Cancer Treatment. Trends Cancer 2019, 5, 475–494. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Chen, Y.B.; Jin, S.; Fang, X.F.; He, X.X.; Xiong, Z.F.; Yang, S.L. Research on circadian clock genes in non-small-cell lung carcinoma. Chronobiol. Int. 2019, 36, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Vasu, V.T.; Cross, C.E.; Gohil, K. Nr1d1, an important circadian pathway regulatory gene, is suppressed by cigarette smoke in murine lungs. Integr. Cancer Ther. 2009, 8, 321–328. [Google Scholar] [CrossRef]
- Jiang, W.; Zhao, S.; Jiang, X.; Zhang, E.; Hu, G.; Hu, B.; Zheng, P.; Xiao, J.; Lu, Z.; Lu, Y.; et al. The circadian clock gene Bmal1 acts as a potential anti-oncogene in pancreatic cancer by activating the p53 tumor suppressor pathway. Cancer Lett. 2016, 371, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, T.; Vila-Caballer, M.; Santos, C.S.; Liu, J.; Yang, J.; Finkielstein, C.V. The Circadian Factor Period 2 Modulates P53 Stability and Transcriptional Activity in Unstressed Cells. Mol. Biol. Cell 2014, 25, 3081–3093. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, T.; Kim, J.K.; Liu, J.; Vila-Caballer, M.; Stauffer, P.E.; Tyson, J.J.; Finkielstein, C.V. Model-driven Experimental Approach Reveals the Complex Regulatory Distribution of P53 by the Circadian Factor Period 2. Proc. Natl. Acad. Sci. USA 2016, 113, 13516–13521. [Google Scholar] [CrossRef]
- Gotoh, T.; Vila-Caballer, M.; Liu, J.; Schiffhauer, S.; Finkielstein, C.V. Association of the Circadian Factor Period 2 to P53 Influences P53′s Function in Dna-damage Signaling. Mol. Biol. Cell 2015, 26, 359–372. [Google Scholar] [CrossRef]
- Zhanfeng, N.; Chengquan, W.; Hechun, X.; Jun, W.; Lijian, Z.; Dede, M.; Wenbin, L.; Lei, Y. Period2 Downregulation Inhibits Glioma Cell Apoptosis by Activating the MDM2-TP53 Pathway. Oncotarget 2016, 7, 27350–27362. [Google Scholar] [CrossRef]
- Fu, L.; Pelicano, H.; Liu, J.; Huang, P.; Lee, C.C. The Circadian Gene Period2 Plays an Important Role in Tumor Suppression and DNA Damage Response in Vivo. Cell 2002, 111, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.-H.; Kim, E.M.; Park, J.K.; Hwang, S.-G.; Moon, S.-K.; Kim, W.-J.; Um, H.-D. Bmal1 Suppresses Cancer Cell Invasion by Blocking the Phosphoinositide 3-kinase-akt-mmp-2 Signaling Pathway. Oncol. Rep. 2013, 29, 2109–2113. [Google Scholar] [CrossRef] [PubMed]
- Iksen; Pothongsrisit, S.; Pongrakhananon, V. Targeting the Pi3k/akt/mtor Signaling Pathway in Lung Cancer: An Update Regarding Potential Drugs and Natural Products. Molecules 2021, 26, 4100. [Google Scholar] [CrossRef] [PubMed]
- Bastani, S.; Akbarzadeh, M.; Rastgar Rezaei, Y.; Farzane, A.; Nouri, M.; Mollapour Sisakht, M.; Fattahi, A.; Akbarzadeh, M.; Reiter, R.J. Melatonin as a Therapeutic Agent for the Inhibition of Hypoxia-induced Tumor Progression: A Description of Possible Mechanisms Involved. Int. J. Mol. Sci. 2021, 22, 10874. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wu, Q.-Q.; Cao, L.-F.; Qing, H.-Y.; Zhang, C.; Chen, Y.-H.; Wang, H.; Liu, R.-R.; Xu, D.-X. Melatonin Inhibits Endoplasmic Reticulum Stress and Epithelial-mesenchymal Transition During Bleomycin-induced Pulmonary Fibrosis in Mice. PLoS ONE 2014, 9, e97266. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Cook, K.; Gee, H.E.; Hau, E. Hypoxia, Metabolism, and the Circadian Clock: New Links to Overcome Radiation Resistance in High-grade Gliomas. J. Exp. Clin. Cancer Res. 2020, 39, 129. [Google Scholar] [CrossRef]
- Balsara, B.R.; Pei, J.; Mitsuuchi, Y.; Page, R.; Klein-Szanto, A.; Wang, H.; Unger, M.; Testa, J.R. Frequent activation of AKT in non-small cell lung carcinomas and preneoplastic bronchial lesions. Carcinogenesis 2004, 25, 2053–2059. [Google Scholar] [CrossRef]
- Scheffler, M.; Bos, M.; Gardizi, M.; König, K.; Michels, S.; Fassunke, J.; Heydt, C.; Künstlinger, H.; Ihle, M.; Ueckeroth, F. PIK3CA Mutations in Non-small Cell Lung Cancer (NSCLC): Genetic Heterogeneity, Prognostic Impact and Incidence of Prior Malignancies. Oncotarget 2015, 6, 1315–1326. [Google Scholar] [CrossRef]
- Zhao, D.; Dong, Y.; Duan, M.; He, D.; Xie, Q.; Peng, W.; Cui, W.; Jiang, J.; Cheng, Y.; Zhang, H. Circadian Gene ARNTL Initiates Circgucy1a2 Transcription to Suppress Non-small Cell Lung Cancer Progression via Mir-200c-3p/pten Signaling. J. Exp. Clin. Cancer Res. 2023, 42, 229. [Google Scholar] [CrossRef]
- Rossner, M.J.; Oster, H.; Wichert, S.P.; Reinecke, L.; Wehr, M.C.; Reinecke, J.; Eichele, G.; Taneja, R.; Nave, K.-A. Disturbed Clockwork Resetting in Sharp-1 and Sharp-2 Single and Double Mutant Mice. PLoS ONE 2008, 3, e2762. [Google Scholar] [CrossRef]
- Liu, Q.; Wu, Y.; Seino, H.; Haga, T.; Yoshizawa, T.; Morohashi, S.; Kijima, H. Correlation between DEC1/DEC2 and Epithelial-mesenchymal Transition in Human Prostate Cancer PC-3 Cells. Mol. Med. Rep. 2018, 18, 3859–3865. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sato, H.; Suzuki, T.; Yoshizawa, T.; Morohashi, S.; Seino, H.; Kawamoto, T.; Fujimoto, K.; Kato, Y.; Kijima, H. Involvement of C-myc in the Proliferation of MCF-7 Human Breast Cancer Cells Induced by Bhlh Transcription Factor DEC2. Int. J. Mol. Med. 2015, 35, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Amelio, I.; Melino, G. The “sharp” blade against Hif-mediated Metastasis. Cell Cycle 2012, 11, 4530–4535. [Google Scholar] [CrossRef]
- Montagner, M.; Enzo, E.; Forcato, M.; Zanconato, F.; Parenti, A.; Rampazzo, E.; Basso, G.; Leo, G.; Rosato, A.; Bicciato, S. SHARP1 Suppresses Breast Cancer Metastasis by Promoting Degradation of Hypoxia-inducible Factors. Nature 2012, 487, 380–384. [Google Scholar] [CrossRef]
- Liao, Y.; Lu, W.; Che, Q.; Yang, T.; Qiu, H.; Zhang, H.; He, X.; Wang, J.; Qiu, M.; Zou, Y. SHARP1 Suppresses Angiogenesis of Endometrial Cancer by Decreasing Hypoxia-inducible Factor-1α Level. PLoS ONE 2014, 9, e99907. [Google Scholar] [CrossRef] [PubMed]
- Piccolo, S.; Enzo, E.; Montagner, M. p63, Sharp1, and HIFs: Master regulators of metastasis in triple-negative breast cancer. Cancer Res. 2013, 73, 4978–4981. [Google Scholar] [CrossRef]
- Falvella, F.S.; Colombo, F.; Spinola, M.; Campiglio, M.; Pastorino, U.; Dragani, T.A. BHLHB3: A Candidate Tumor Suppressor in Lung Cancer. Oncogene 2008, 27, 3761–3764. [Google Scholar] [CrossRef]
- Gallo, C.; Fragliasso, V.; Donati, B.; Torricelli, F.; Tameni, A.; Piana, S.; Ciarrocchi, A. The Bhlh Transcription Factor DEC1 Promotes Thyroid Cancer Aggressiveness by the Interplay with NOTCH1. Cell Death Dis. 2018, 9, 871. [Google Scholar] [CrossRef]
- Liao, Y.; He, X.; Qiu, H.; Che, Q.; Wang, F.; Lu, W.; Chen, Z.; Qiu, M.; Wang, J.; Wang, H. Suppression of the Epithelial-mesenchymal Transition by SHARP1 Is Linked to the NOTCH1 Signaling Pathway in Metastasis of Endometrial Cancer. BMC Cancer 2014, 14, 487. [Google Scholar] [CrossRef]
- Liu, J.-J.; Chung, T.-K.; Li, J.; Taneja, R. Sharp-1 Modulates the Cellular Response to DNA Damage. FEBS Lett. 2010, 584, 619–624. [Google Scholar] [CrossRef]
- Nakamura, H.; Tanimoto, K.; Hiyama, K.; Yunokawa, M.; Kawamoto, T.; Kato, Y.; Yoshiga, K.; Poellinger, L.; Hiyama, E.; Nishiyama, M. Human Mismatch Repair Gene, MLH1, Is Transcriptionally Repressed by the Hypoxia-inducible Transcription Factors, DEC1 and DEC2. Oncogene 2008, 27, 4200–4209. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.; Lin, X.-Y.; Wang, J.; Yu, J.-H.; Miao, Y.; Wang, E.-H. The Transcription Factor DEC1 (BHLHE40/STRA13/SHARP-2) Is Negatively Associated with TNM Stage in Non-small-cell Lung Cancer and Inhibits the Proliferation through Cyclin D1 in A549 and BE1 Cells. Tumor Biol. 2013, 34, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Raghu, G.; Collard, H.R.; Egan, J.J.; Martinez, F.J.; Behr, J.; Brown, K.K.; Colby, T.V.; Cordier, J.-F.; Flaherty, K.R.; Lasky, J.A. An Official ATS/ERS/JRS/ALAT Statement: Idiopathic Pulmonary Fibrosis: Evidence-based Guidelines for Diagnosis and Management. Am. J. Respir. Crit. Care Med. 2011, 183, 788–824. [Google Scholar] [CrossRef] [PubMed]
- Noble, P.W.; Barkauskas, C.E.; Jiang, D. Pulmonary Fibrosis: Patterns and Perpetrators. J. Clin. Investig. 2012, 122, 2756–2762. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.; Meyer, K.C. Therapies for interstitial lung disease: Past, present and future. Ther. Adv. Respir. Dis. 2008, 2, 319–338. [Google Scholar] [CrossRef]
- Yue, X.; Shan, B.; Lasky, J.A. TGF-β: Titan of Lung Fibrogenesis. Curr. Enzym. Inhib. 2010, 6, 67–77. [Google Scholar] [CrossRef]
- Gharaee-Kermani, M.; Hu, B.; Phan, S.H.; Gyetko, M.R. Recent advances in molecular targets and treatment of idiopathic pulmonary fibrosis: Focus on TGFbeta signaling and the myofibroblast. Curr. Med. Chem. 2009, 16, 1400–1417. [Google Scholar] [CrossRef]
- Khalil, N.; Xu, Y.D.; O’Connor, R.; Duronio, V. Proliferation of Pulmonary Interstitial Fibroblasts Is Mediated by Transforming Growth Factor-β1-induced Release of Extracellular Fibroblast Growth Factor-2 and Phosphorylation of P38 MAPK and JNK. J. Biol. Chem. 2005, 280, 43000–43009. [Google Scholar] [CrossRef]
- King, T.E., Jr.; Pardo, A.; Selman, M. Idiopathic pulmonary fibrosis. Lancet 2011, 378, 1949–1961. [Google Scholar] [CrossRef]
- Zhang, K.; Flanders, K.C.; Phan, S.H. Cellular localization of transforming growth factor-beta expression in bleomycin-induced pulmonary fibrosis. Am. J. Pathol. 1995, 147, 352–361. [Google Scholar]
- Sime, P.J.; Xing, Z.; Graham, F.L.; Csaky, K.G.; Gauldie, J. Adenovector-mediated Gene Transfer of Active Transforming Growth Factor-beta1 Induces Prolonged Severe Fibrosis in Rat Lung. J. Clin. Investig. 1997, 100, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Warshamana, G.S.; Pociask, D.A.; Fisher, K.J.; Liu, J.-Y.; Sime, P.J.; Brody, A.R. Titration of Non-replicating Adenovirus as a Vector for Transducing Active Tgf-β1 Gene Expression Causing Inflammation and Fibrogenesis in the Lungs of C57BL/6 Mice. Int. J. Exp. Pathol. 2002, 83, 183–202. [Google Scholar] [CrossRef]
- Gibbs, J.; Ince, L.; Matthews, L.; Mei, J.; Bell, T.; Yang, N.; Saer, B.; Begley, N.; Poolman, T.; Pariollaud, M. An Epithelial Circadian Clock Controls Pulmonary Inflammation and Glucocorticoid Action. Nat. Med. 2014, 20, 919–926. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, P.S.; Meijer, P.; Nazgiewicz, A.; Anderson, S.G.; Borthwick, L.A.; Bagnall, J.; Kitchen, G.B.; Lodyga, M.; Begley, N.; Venkateswaran, R.V. The Circadian Clock Protein Reverbα Inhibits Pulmonary Fibrosis Development. Proc. Natl. Acad. Sci. USA 2020, 117, 1139–1147. [Google Scholar] [CrossRef] [PubMed]
- Prasad, C.; Hahn, K.; Duraisamy, S.K.; Salathe, M.A.; Huang, S.K.; Burris, T.P.; Sundar, I.K. Rev-erbα agonists suppresses TGFβ1-induced fibroblast-to-myofibroblast transition and pro-fibrotic phenotype in human lung fibroblasts. Biochem. Biophys. Res. Commun. 2023, 669, 120–127. [Google Scholar] [CrossRef]
- Wang, Q.; Sundar, I.K.; Lucas, J.H.; Park, J.-G.; Nogales, A.; Martinez-Sobrido, L.; Rahman, I. Circadian Clock Molecule Rev-erbα Regulates Lung Fibrotic Progression Through Collagen Stabilization. Nat. Commun. 2023, 14, 1295. [Google Scholar] [CrossRef]
- Hatanaka, F.; Matsubara, C.; Myung, J.; Yoritaka, T.; Kamimura, N.; Tsutsumi, S.; Kanai, A.; Suzuki, Y.; Sassone-Corsi, P.; Aburatani, H. Genome-wide Profiling of the Core Clock Protein BMAL1 Targets Reveals a Strict Relationship with Metabolism. Mol. Cell. Biol. 2010, 30, 5636–5648. [Google Scholar] [CrossRef]
- Ingle, K.A.; Kain, V.; Goel, M.; Prabhu, S.D.; Young, M.E.; Halade, G.V. Cardiomyocyte-specific Bmal1 Deletion in Mice Triggers Diastolic Dysfunction, Extracellular Matrix Response, and Impaired Resolution of Inflammation. Am. J. Physiol.-Heart Circ. Physiol. 2015, 309, H1827–H1836. [Google Scholar] [CrossRef]
- Dong, C.; Gongora, R.; Sosulski, M.L.; Luo, F.; Sanchez, C.G. Regulation of Transforming Growth Factor-beta1 (tgf-β1)-induced Pro-fibrotic Activities by Circadian Clock Gene BMAL1. Respir. Res. 2016, 17, 4. [Google Scholar] [CrossRef]
- Bunger, M.K.; Wilsbacher, L.D.; Moran, S.M.; Clendenin, C.; Radcliffe, L.A.; Hogenesch, J.B.; Simon, M.C.; Takahashi, J.S.; Bradfield, C.A. Mop3 Is an Essential Component of the Master Circadian Pacemaker in Mammals. Cell 2000, 103, 1009–1017. [Google Scholar] [CrossRef]
- Pekovic-Vaughan, V.; Gibbs, J.; Yoshitane, H.; Yang, N.; Pathiranage, D.; Guo, B.; Sagami, A.; Taguchi, K.; Bechtold, D.; Loudon, A. The Circadian Clock Regulates Rhythmic Activation of the Nrf2/glutathione-mediated Antioxidant Defense Pathway to Modulate Pulmonary Fibrosis. Genes Dev. 2014, 28, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Romero, Y.; Balderas-Martínez, Y.I.; Vargas-Morales, M.A.; Castillejos-López, M.; Vázquez-Pérez, J.A.; Calyeca, J.; Torres-Espíndola, L.M.; Patiño, N.; Camarena, A.; Carlos-Reyes, Á. Effect of Hypoxia in the Transcriptomic Profile of Lung Fibroblasts from Idiopathic Pulmonary Fibrosis. Cells 2022, 11, 3014. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.R. Obstructive Sleep Apnea. Ann. Intern. Med. 2019, 171, ITC81–ITC96. [Google Scholar] [CrossRef] [PubMed]
- Rundo, J.V. Obstructive Sleep Apnea Basics. Clevel. Clin. J. Med. 2019, 86, 2–9. [Google Scholar] [CrossRef]
- Dempsey, J.A. Central Sleep Apnea: Misunderstood and Mistreated! F1000Research 2019, 8, 981. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, O.; Oks, M. Central Sleep Apnea. Clin. Geriatr. Med. 2021, 37, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Şenel, G.B.; Aliş, C.; Karadeniz, D. Are We Underestimating the Central Components of the Mixed Apneas?-A Hypothesis for Revised Scoring. J. Clin. Neurophysiol. Off. Publ. Am. Electroencephalogr. Soc. 2023, 40, 165–172. [Google Scholar] [CrossRef]
- Semelka, M.; Wilson, J.; Floyd, R. Diagnosis and Treatment of Obstructive Sleep Apnea in Adults. Am. Fam. Physician 2016, 94, 355–360. [Google Scholar]
- Qaseem, A.; Dallas, P.; Owens, D.K.; Starkey, M.; Holty, J.E.; Shekelle, P.; Clinical Guidelines Committee of the American College of Physicians. Diagnosis of obstructive sleep apnea in adults: A clinical practice guideline from the American College of Physicians. Ann. Intern. Med. 2014, 161, 210–220. [Google Scholar] [CrossRef]
- Walia, R.; Achilefu, A.; Crawford, S.; Jain, V.; Wigley, S.D.; McCarthy, L.H. Are at-home sleep studies performed using portable monitors (PMs) as effective at diagnosing obstructive sleep apnea (OSA) in adults as sleep laboratory-based polysomnography (PSG)? J. Okla. State Med. Assoc. 2014, 107, 642–644. [Google Scholar]
- Epstein, L.J.; Kristo, D.; Strollo, P.J., Jr.; Friedman, N.; Malhotra, A.; Patil, S.P.; Ramar, K.; Rogers, R.; Schwab, R.J.; Weaver, E.M.; et al. Adult Obstructive Sleep Apnea Task Force of the American Academy of Sleep Medicine. Clinical guideline for the evaluation, management and long-term care of obstructive sleep apnea in adults. J. Clin. Sleep Med. 2009, 5, 263–276. [Google Scholar] [PubMed]
- Bonsignore, M.R.; Baiamonte, P.; Mazzuca, E.; Castrogiovanni, A.; Marrone, O. Obstructive Sleep Apnea and Comorbidities: A Dangerous Liaison. Multidiscip. Respir. Med. 2019, 14, 8. [Google Scholar] [CrossRef] [PubMed]
- Marin-Oto, M.; Vicente, E.E.; Marin, J.M. Long Term Management of Obstructive Sleep Apnea and Its Comorbidities. Multidiscip. Respir. Med. 2019, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Coste, O.; Beaumont, M.; Batéjat, D.; Beers, P.V.; Touitou, Y. Prolonged mild hypoxia modifies human circadian core body temperature and may be associated with sleep disturbances. Chronobiol. Int. 2004, 21, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Coste, O.; Beaumont, M.; Batéjat, D.; Beers, P.V.; Charbuy, H.; Touitou, Y. Hypoxic Depression of Melatonin Secretion After Simulated Long Duration Flights in Man. J. Pineal Res. 2004, 37, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Coste, O.; Beers, P.V.; Bogdan, A.; Charbuy, H.; Touitou, Y. Hypoxic alterations of cortisol circadian rhythm in man after simulation of a long duration flight. Steroids 2005, 70, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Yang, S.-L.; Fang, X.; Jiang, J.-X.; Sun, C.-Y.; Huang, T. Hypoxia Disrupts the Expression Levels of Circadian Rhythm Genes in Hepatocellular Carcinoma. Mol. Med. Rep. 2015, 11, 4002–4008. [Google Scholar] [CrossRef]
- Yang, M.Y.; Lin, P.W.; Lin, H.C.; Lin, P.M.; Chen, I.Y.; Friedman, M.; Hung, C.F.; Salapatas, A.M.; Lin, M.C.; Lin, S.F. Alternations of Circadian Clock Genes Expression and Oscillation in Obstructive Sleep Apnea. J. Clin. Med. 2019, 8, 1634. [Google Scholar] [CrossRef]
- Pellegrino, R.; Kavakli, I.H.; Goel, N.; Cardinale, C.J.; Dinges, D.F.; Kuna, S.T.; Maislin, G.; Van Dongen, H.P.; Tufik, S.; Hogenesch, J.B.; et al. A novel BHLHE41 variant is associated with short sleep and resistance to sleep deprivation in humans. Sleep 2014, 37, 1327–1336. [Google Scholar] [CrossRef]
- Clayville, L.R. Influenza update: A review of currently available vaccines. P T A Peer-Rev. J. Formul. Manag. 2011, 36, 659–684. [Google Scholar]
- Gaitonde, D.Y.; Moore, F.C.; Morgan, M.K. Influenza: Diagnosis and Treatment. Am. Fam. Physician. 2019, 100, 751–758. [Google Scholar] [PubMed]
- Javanian, M.; Barary, M.; Ghebrehewet, S.; Koppolu, V.; Vasigala, V.; Ebrahimpour, S. A brief review of influenza virus infection. J. Med. Virol. 2021, 93, 4638–4646. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Noma, H.; Kotake, O.; Motohashi, R.; Yasuda, K.; Shimura, M. Optic Neuritis and Acute Anterior Uveitis Associated with Influenza A Infection: A Case Report. Int. Med. Case Rep. J. 2017, 10, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Dharmapalan, D. Influenza. Indian J. Pediatr. 2020, 87, 828–832. [Google Scholar] [CrossRef] [PubMed]
- Sundar, I.K.; Ahmad, T.; Yao, H.; Hwang, J.-W.; Gerloff, J.; Lawrence, B.P.; Sellix, M.T.; Rahman, I. Influenza A Virus-dependent Remodeling of Pulmonary Clock Function in a Mouse Model of COPD. Sci. Rep. 2015, 5, 9927. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hunter, L.; Wu, G.; Maidstone, R.; Mizoro, Y.; Vonslow, R.; Fife, M.; Hopwood, T.; Begley, N.; Saer, B. Genome-wide Effect of Pulmonary Airway Epithelial Cell–specific Bmal1 Deletion. FASEB J. 2019, 33, 6226–6238. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Magri, A.; Hill, M.; Lai, A.G.; Kumar, A.; Rambhatla, S.B.; Donald, C.L.; Lopez-Clavijo, A.F.; Rudge, S.; Pinnick, K. The Circadian Clock Components BMAL1 and Rev-erbα Regulate Flavivirus Replication. Nat. Commun. 2019, 10, 377. [Google Scholar] [CrossRef]
- Borrmann, H.; Davies, R.; Dickinson, M.; Pedroza-Pacheco, I.; Schilling, M.; Vaughan-Jackson, A.; Magri, A.; James, W.; Balfe, P.; Borrow, P. Pharmacological Activation of the Circadian Component REV-ERB Inhibits HIV-1 Replication. Sci. Rep. 2020, 10, 13271. [Google Scholar] [CrossRef]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.C. Clinical Characteristics of Coronavirus Disease 2019 in China. New Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R. A Novel Coronavirus from Patients with Pneumonia in China, 2019. New Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Morin, C.M.; Carrier, J.; Bastien, C.; Godbout, R. Sleep and Circadian Rhythm in Response to the COVID-19 Pandemic. Can. J. Public Health 2020, 111, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Ma, S.; Wang, Y.; Cai, Z.; Hu, J.; Wei, N.; Wu, J.; Du, H.; Chen, T.; Li, R. Factors Associated with Mental Health Outcomes Among Health Care Workers Exposed to Coronavirus Disease 2019. JAMA Netw. Open 2020, 3, e203976. [Google Scholar] [CrossRef] [PubMed]
- Irwin, M.R. Sleep and Inflammation: Partners in Sickness and in Health. Nat. Rev. Immunol. 2019, 19, 702–715. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.; Reiter, R.J. Melatonin: Roles in Influenza, COVID-19, and Other Viral Infections. Rev. Med. Virol. 2020, 30, e2109. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J.E.; Blaikley, J.; Beesley, S.; Matthews, L.; Simpson, K.D.; Boyce, S.H.; Farrow, S.N.; Else, K.J.; Singh, D.; Ray, D.W. The Nuclear Receptor Rev-erbα Mediates Circadian Regulation of Innate Immunity Through Selective Regulation of Inflammatory Cytokines. Proc. Natl. Acad. Sci. USA 2012, 109, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Haspel, J.A.; Anafi, R.; Brown, M.K.; Cermakian, N.; Depner, C.; Desplats, P.; Gelman, A.E.; Haack, M.; Jelic, S.; Kim, B.S. Perfect Timing: Circadian Rhythms, Sleep, and Immunity—An NIH Workshop Summary. JCI Insight 2020, 5, e131487. [Google Scholar] [CrossRef] [PubMed]
- Costantini, C.; Renga, G.; Sellitto, F.; Borghi, M.; Stincardini, C.; Pariano, M.; Zelante, T.; Chiarotti, F.; Bartoli, A.; Mosci, P.; et al. Microbes Era Circadian Medicine. Front. Cell. Infect. Microbiol. 2020, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Rambhatla, S.B.; Lai, A.G.; Mckeating, J.A. Interplay between Circadian Clock and Viral Infection. J. Mol. Med. 2017, 95, 1283–1289. [Google Scholar] [CrossRef]
- Mazzoccoli, G.; Vinciguerra, M.; Carbone, A.; Relógio, A. The Circadian Clock, the Immune System, and Viral Infections: The Intricate Relationship between Biological Time and Host-virus Interaction. Pathogens 2020, 9, 83. [Google Scholar] [CrossRef]
- Pariollaud, M.; Gibbs, J.E.; Hopwood, T.W.; Brown, S.; Begley, N.; Vonslow, R.; Poolman, T.; Guo, B.; Saer, B.; Jones, D.H. Circadian Clock Component Rev-erbα Controls Homeostatic Regulation of Pulmonary Inflammation. J. Clin. Investig. 2018, 128, 2281–2296. [Google Scholar] [CrossRef]
- Perry, M.G.; Kirwan, J.R.; Jessop, D.S.; Hunt, L.P. Overnight Variations in Cortisol, Interleukin 6, Tumour Necrosis Factor A and Other Cytokines in People with Rheumatoid Arthritis. Ann. Rheum. Dis. 2009, 68, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.; Choi, Y.-H.; Seo, S.W.; Lee, J.-M. Scale-integrated Network Hubs of the White Matter Structural Network. Sci. Rep. 2017, 7, 2449. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Inestrosa, N.C.; Tapia-Rojas, C.; Lindsay, C.B.; Zolezzi, J.M. Wnt Signaling Pathway Dysregulation in the Aging Brain: Lessons From the Octodon degus. Front. Cell Dev. Biol. 2020, 8, 734. [Google Scholar] [CrossRef]
- Raslan, A.A.; Yoon, J.K. WNT Signaling in Lung Repair and Regeneration. Mol. Cells 2020, 43, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, Q.; Xu, H. Wnt/β-catenin Signal Transduction Pathway in Prostate Cancer and Associated Drug Resistance. Discov. Oncol. 2021, 12, 40. [Google Scholar] [CrossRef] [PubMed]
- Gajos-Michniewicz, A.; Czyz, M. WNT Signaling in Melanoma. Int. J. Mol. Sci. 2020, 21, 4852. [Google Scholar] [CrossRef]
- Morrisey, E.E. Wnt signaling and pulmonary fibrosis. Am. J. Pathol. 2003, 162, 1393–1397. [Google Scholar] [CrossRef]
- Villanueva, M.T. Selective activation of Wnt ameliorates idiopathic pulmonary fibrosis. Nat. Rev. Drug Discov. 2023, 22, 619. [Google Scholar] [CrossRef]
- Rojas, M.; Mora, A.L.; Kapetanaki, M.; Weathington, N.; Gladwin, M.; Eickelberg, O. Aging and Lung Disease. Clin. Impact Cell. Mol. Pathways. Ann. Am. Thorac. Soc. 2015, 12, S222–S227. [Google Scholar] [CrossRef]
- Faner, R.; Rojas, M.; Macnee, W.; Agustí, A. Abnormal lung aging in chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2012, 186, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.L.; Rowe, J.H.; Garcia-De-Alba, C.; Kim, C.F.; Sharpe, A.H.; Haigis, M.C. The Aging Lung: Physiology, Disease, and Immunity. Cell 2021, 184, 1990–2019. [Google Scholar] [CrossRef] [PubMed]
- Angelidis, I.; Simon, L.M.; Fernandez, I.E.; Strunz, M.; Mayr, C.H.; Greiffo, F.R.; Tsitsiridis, G.; Ansari, M.; Graf, E.; Strom, T.-M. An Atlas of the Aging Lung Mapped by Single Cell Transcriptomics and Deep Tissue Proteomics. Nat. Commun. 2019, 10, 963. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, Y.; Zhao, C.; Liu, L.; He, Q.; Li, Y.; Zhou, D.; Jin, F. Effects of Biological Clock Gene BMAL1 and Hypoxia-inducible Factor Hif-1α on Proliferation, Migration and Radiotherapy Sensitivity of Nasopharyngeal Carcinoma Cells HONE1. Holist. Integr. Oncol. 2023, 2, 26. [Google Scholar] [CrossRef]
- Yuan, Y.; Cruzat, V.F.; Newsholme, P.; Cheng, J.; Chen, Y.; Lu, Y. Regulation of SIRT1 in aging: Roles in mitochondrial function and biogenesis. Mech. Ageing Dev. 2016, 155, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Sack, M.N. The Role of SIRT3 in Mitochondrial Homeostasis and Cardiac Adaptation to Hypertrophy and Aging. J. Mol. Cell. Cardiol. 2012, 52, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Hurd, M.W.; Ralph, M.R. The significance of circadian organization for longevity in the golden hamster. J. Biol. Rhythm. 1998, 13, 430–436. [Google Scholar] [CrossRef]
- Takeda, N.; Maemura, K. Circadian clock and vascular disease. Hypertens. Res. Off. J. Jpn. Soc. Hypertens. 2010, 33, 645–651. [Google Scholar] [CrossRef]
- Scheer, F.A.J.L.; Hilton, M.F.; Mantzoros, C.S.; Shea, S.A. Adverse Metabolic and Cardiovascular Consequences of Circadian Misalignment. Proc. Natl. Acad. Sci. USA 2009, 106, 4453–4458. [Google Scholar] [CrossRef]
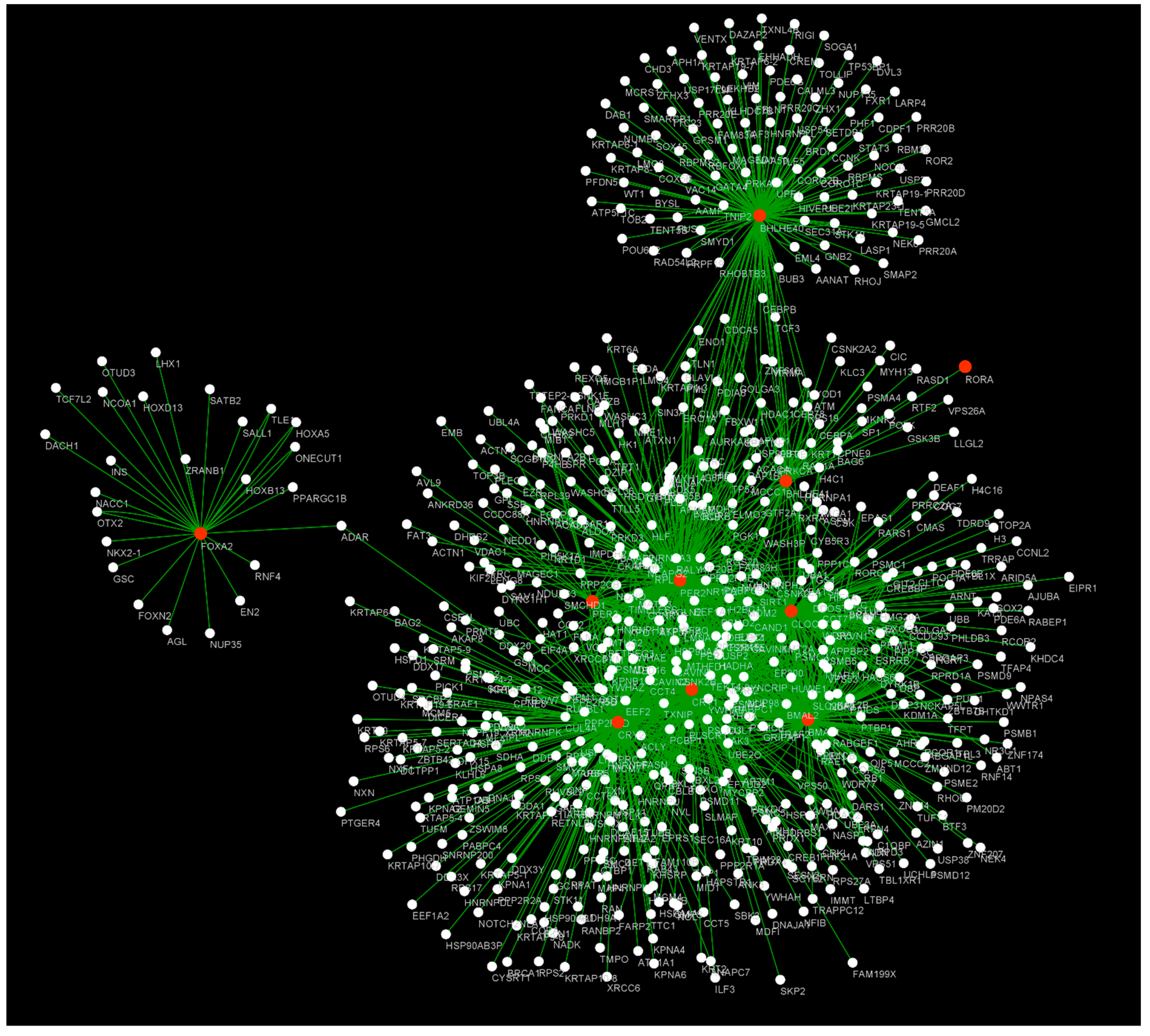
| Pathway | p-Value | Database |
|---|---|---|
| Regulation of circadian rhythm | 1.1 × 10−9 | DAVID/GO—CTD |
| Positive regulation of transcription | 6 × 10−21 | DAVID/GO |
| Proteasome | 3.8 × 10−8 | DAVID/KEEG—CTD |
| Regulation of telomerase RNA localization to Cajal bodies | 1.7 × 10−7 | DAVID/GO |
| Regulation of splicing | 9.1 × 10−5 | DAVID/GO |
| Regulation of protein localization to telomere | 1.7 × 10−7 | DAVID/GO |
| Viral carcinogenesis | 6.0 × 10−10 | DAVID/KEEG |
| Cell cycle | 4.5 × 10−25 | DAVID/KEEG—CTD |
| Ubiquitin-mediated proteolysis | 2 × 10−5 | CTD |
| Wnt signaling pathways | 6 × 10−5 | CTD |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castillejos-López, M.; Romero, Y.; Varela-Ordoñez, A.; Flores-Soto, E.; Romero-Martinez, B.S.; Velázquez-Cruz, R.; Vázquez-Pérez, J.A.; Ruiz, V.; Gomez-Verjan, J.C.; Rivero-Segura, N.A.; et al. Hypoxia Induces Alterations in the Circadian Rhythm in Patients with Chronic Respiratory Diseases. Cells 2023, 12, 2724. https://doi.org/10.3390/cells12232724
Castillejos-López M, Romero Y, Varela-Ordoñez A, Flores-Soto E, Romero-Martinez BS, Velázquez-Cruz R, Vázquez-Pérez JA, Ruiz V, Gomez-Verjan JC, Rivero-Segura NA, et al. Hypoxia Induces Alterations in the Circadian Rhythm in Patients with Chronic Respiratory Diseases. Cells. 2023; 12(23):2724. https://doi.org/10.3390/cells12232724
Chicago/Turabian StyleCastillejos-López, Manuel, Yair Romero, Angelica Varela-Ordoñez, Edgar Flores-Soto, Bianca S. Romero-Martinez, Rafael Velázquez-Cruz, Joel Armando Vázquez-Pérez, Víctor Ruiz, Juan C. Gomez-Verjan, Nadia A. Rivero-Segura, and et al. 2023. "Hypoxia Induces Alterations in the Circadian Rhythm in Patients with Chronic Respiratory Diseases" Cells 12, no. 23: 2724. https://doi.org/10.3390/cells12232724
APA StyleCastillejos-López, M., Romero, Y., Varela-Ordoñez, A., Flores-Soto, E., Romero-Martinez, B. S., Velázquez-Cruz, R., Vázquez-Pérez, J. A., Ruiz, V., Gomez-Verjan, J. C., Rivero-Segura, N. A., Camarena, Á., Torres-Soria, A. K., Gonzalez-Avila, G., Sommer, B., Solís-Chagoyán, H., Jaimez, R., Torres-Espíndola, L. M., & Aquino-Gálvez, A. (2023). Hypoxia Induces Alterations in the Circadian Rhythm in Patients with Chronic Respiratory Diseases. Cells, 12(23), 2724. https://doi.org/10.3390/cells12232724










