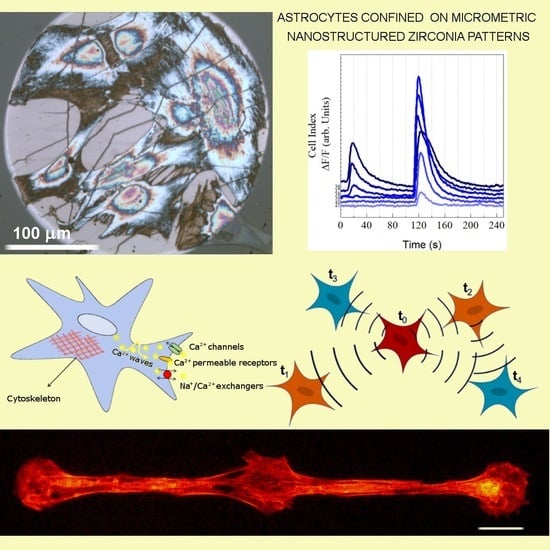
Nanotopography and Microconfinement Impact on Primary Hippocampal Astrocyte Morphology, Cytoskeleton and Spontaneous Calcium Wave Signalling
Abstract
1. Introduction
2. Materials and Methods
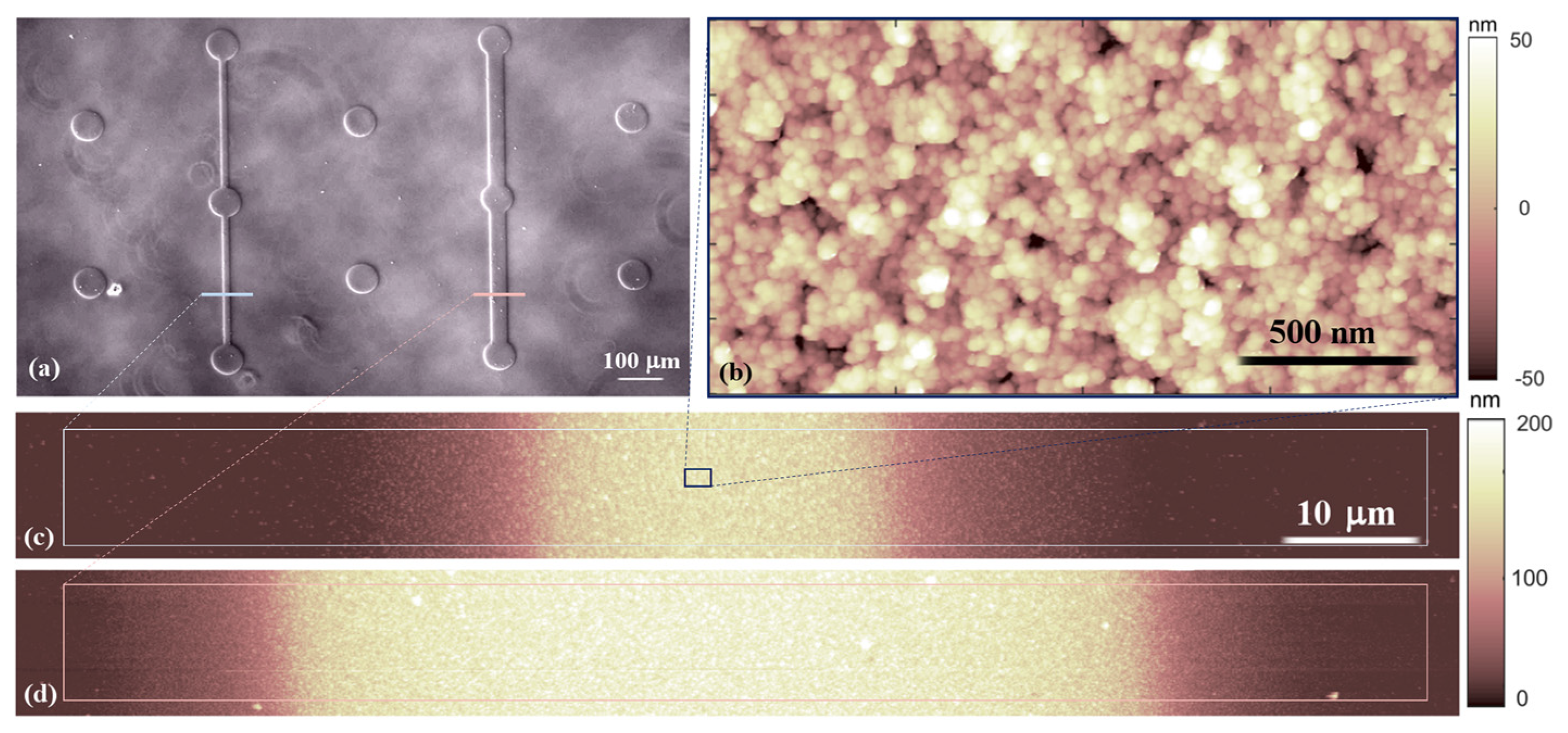
2.1. Fabrication of Micropatterned Nanostructured Substrates
2.2. Culture of Primary Astrocytes
2.3. Immunofluorescence Imaging
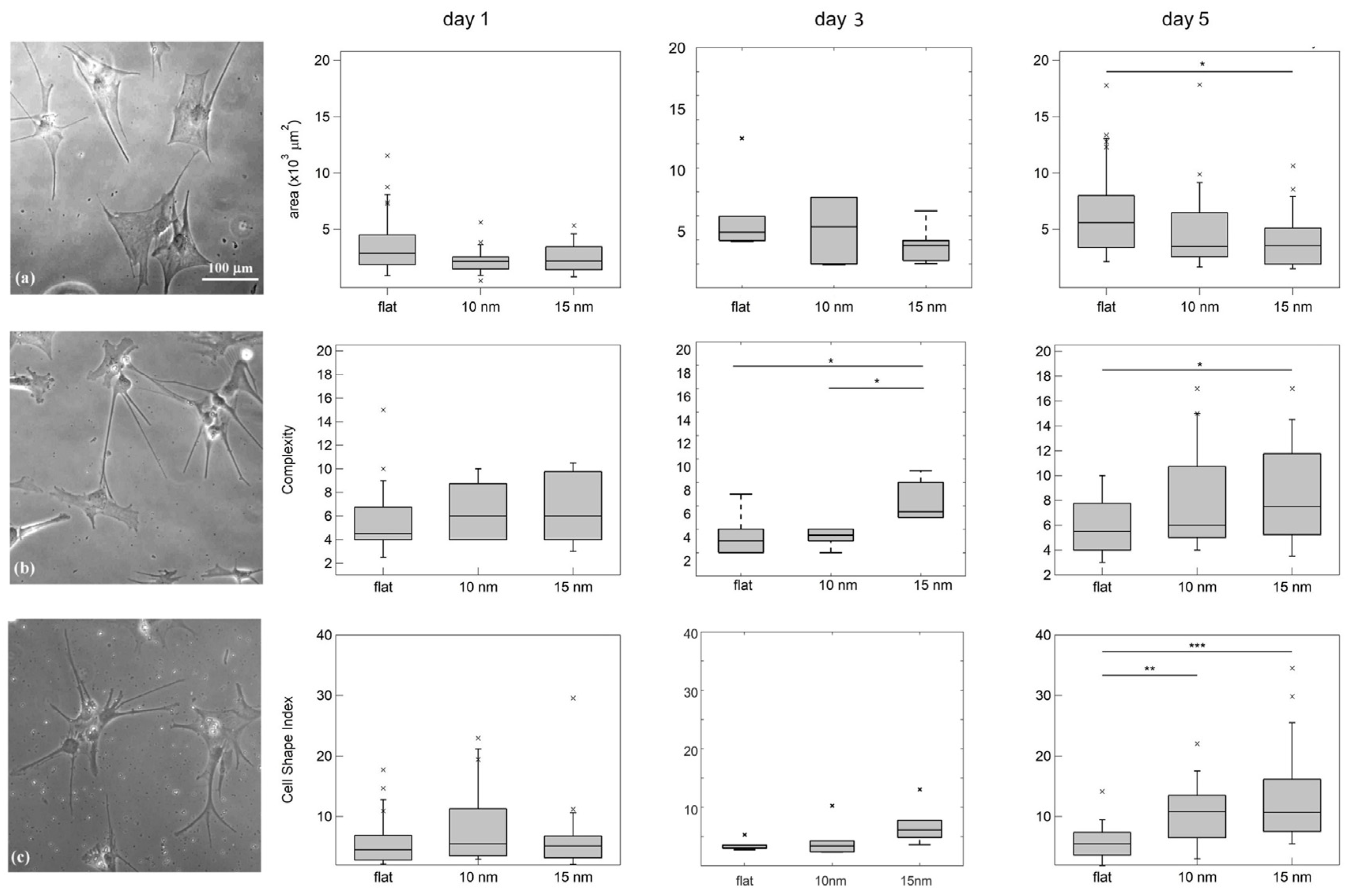
2.4. Characterisation of the Morphological Properties of the Astrocytes
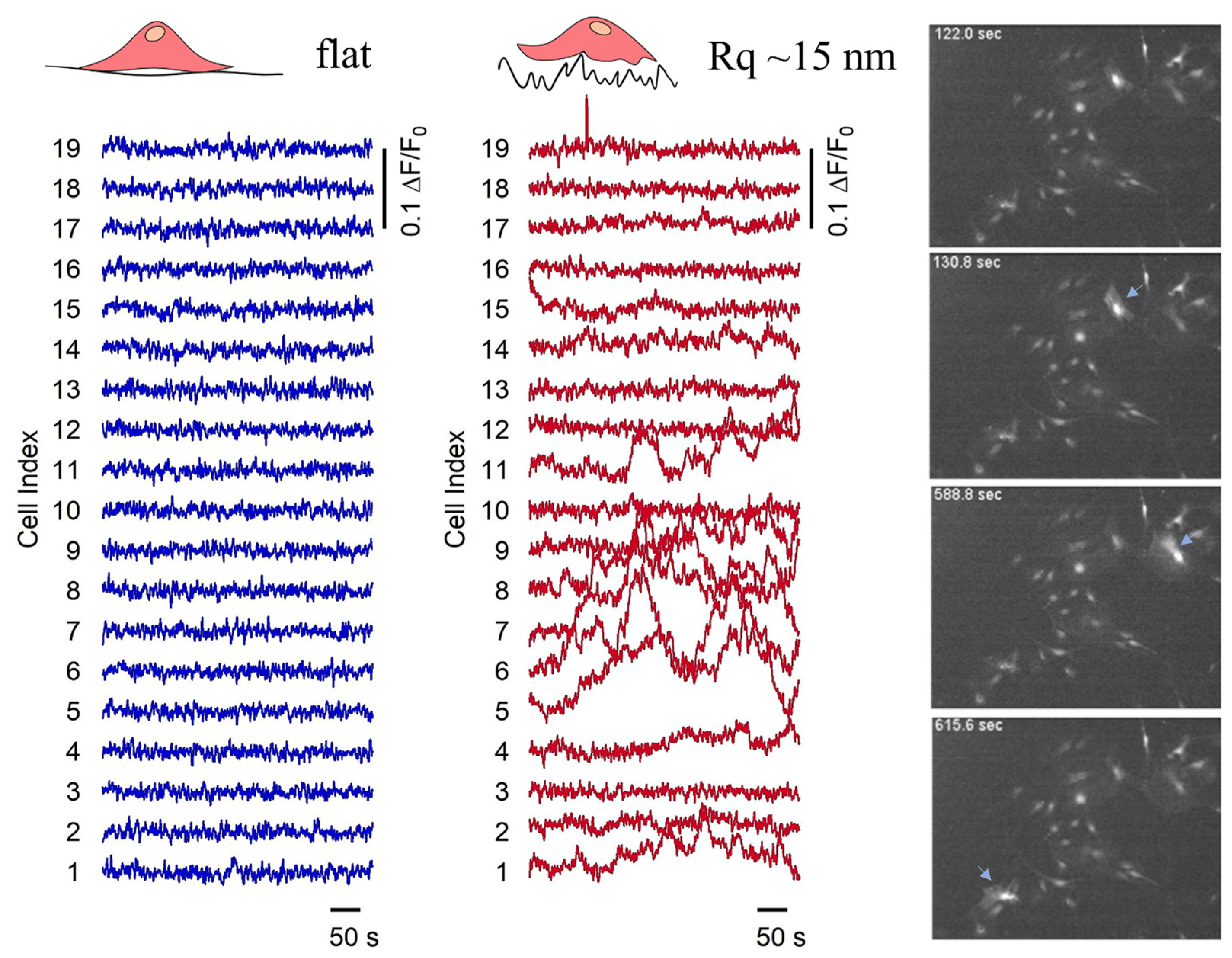
2.5. Calcium Imaging and Analysis of Calcium Wave Propagation
2.6. Statistical Analysis
3. Results and Discussion
3.1. Micrometric Patterns with Nanostructured Zirconia Films
3.2. Astrocyte/Nanotopography Interaction Induces Morphological, Cytoskeletal and Functional Differences
3.3. Pattern-Dependent Cell Network Properties
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jäkel, S.; Dimou, L. Glial Cells and Their Function in the Adult Brain: A Journey through the History of Their Ablation. Front. Cell. Neurosci. 2017, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Verkhratsky, A.; Kettenmann, H. Calcium signalling in glial cells. Trends Neurosci. 1996, 19, 346–352. [Google Scholar] [CrossRef]
- Verkhratsky, A.; Rodríguez, J.J.; Parpura, V. Calcium signalling in astroglia. Mol. Cell. Endocrinol. 2012, 353, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Saracino, E.; Cirillo, V.; Marrese, M.; Guarino, V.; Benfenati, V.; Zamboni, R.; Ambrosio, L. Structural and functional properties of astrocytes on PCL based electrospun fibres. Mater. Sci. Eng. C 2020, 118, 111363. [Google Scholar] [CrossRef]
- Verkhratsky, A.; Nedergaard, M. Physiology of Astroglia. Physiol. Rev. 2018, 98, 239–389. [Google Scholar] [CrossRef] [PubMed]
- Escartin, C.; Galea, E.; Lakatos, A.; O’Callaghan, J.P.; Petzold, G.C.; Serrano-Pozo, A.; Steinhäuser, C.; Volterra, A.; Carmignoto, G.; Agarwal, A.; et al. Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 2021, 24, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Marinval, N.; Chew, S.Y. Mechanotransduction assays for neural regeneration strategies: A focus on glial cells. APL Bioeng. 2021, 5, 021505. [Google Scholar] [CrossRef] [PubMed]
- Barnes, J.M.; Przybyla, L.; Weaver, V.M. Tissue mechanics regulate brain development, homeostasis and disease. J. Cell Sci. 2017, 130, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, J.D.; Dufresne, E.R.; Schwartz, M.A. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 2014, 15, 802–812. [Google Scholar] [CrossRef]
- Wang, H.; Tewari, A.; Einheber, S.; Salzer, J.; Melendez-Vasquez, C.V. Myosin II has distinct functions in PNS and CNS myelin sheath formation. J. Cell Biol. 2008, 182, 1171–1184. [Google Scholar] [CrossRef]
- Lourenço, T.; Paes De Faria, J.; Bippes, C.A.; Maia, J.; Lopes-Da-Silva, J.A.; Relvas, J.B.; Graõs, M. Modulation of oligodendrocyte differentiation and maturation by combined biochemical and mechanical cues. Sci. Rep. 2016, 6, 21563. [Google Scholar] [CrossRef] [PubMed]
- Pathak, M.M.; Nourse, J.L.; Tran, T.; Hwe, J.; Arulmoli, J.; Dai Trang, T.L.; Bernardis, E.; Flanagan, L.A.; Tombola, F. Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, 16148–16153. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xia, J.; Li, J.; Hagemann, T.L.; Jones, J.R.; Fraenkel, E.; Weitz, D.A.; Zhang, S.-C.; Messing, A.; Feany, M.B. Tissue and cellular rigidity and mechanosensitive signaling activation in Alexander disease. Nat. Commun. 2018, 9, 1899. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.A.; Stys, P.K.; Lusardi, T.; Meaney, D.; Smith, D.H. Traumatic Axonal Injury Induces Calcium Influx Modulated by Tetrodotoxin-Sensitive Sodium Channels. J. Neurosci. 2001, 21, 1923–1930. [Google Scholar] [CrossRef]
- Smith, D.H.; Johnson, V.E.; Stewart, W. Chronic neuropathologies of single and repetitive TBI: Substrates of dementia? Nat. Rev. Neurol. 2013, 9, 211–221. [Google Scholar] [CrossRef]
- Nogueira, M.L.; Lafitte, O.; Steyaert, J.; Bakardjian, H.; Dubois, B.; Hampel, H.; Schwartz, L. Mechanical stress related to brain atrophy in Alzheimer’s disease. Alzheimer’s Dement. 2015, 12, 11–20. [Google Scholar] [CrossRef]
- Nogueira, M.L.; Hamraz, M.; Abolhassani, M.; Bigan, E.; Lafitte, O.; Steyaert, J.; Dubois, B.; Schwartz, L. Mechanical stress increases brain amyloid β, tau, and α-synuclein concentrations in wild-type mice. Alzheimer’s Dement. 2017, 14, 444–453. [Google Scholar] [CrossRef]
- Moeendarbary, E.; Weber, I.P.; Sheridan, G.K.; Koser, D.E.; Soleman, S.; Haenzi, B.; Bradbury, E.J.; Fawcett, J.; Franze, K. The soft mechanical signature of glial scars in the central nervous system. Nat. Commun. 2017, 8, 14787. [Google Scholar] [CrossRef]
- Weickenmeier, J.; de Rooij, R.; Budday, S.; Steinmann, P.; Ovaert, T.; Kuhl, E. Brain stiffness increases with myelin content. Acta Biomater. 2016, 42, 265–272. [Google Scholar] [CrossRef]
- Chen, X.; Wanggou, S.; Bodalia, A.; Zhu, M.; Dong, W.; Fan, J.J.; Yin, W.C.; Min, H.-K.; Hu, M.; Draghici, D.; et al. A Feedforward Mechanism Mediated by Mechanosensitive Ion Channel PIEZO1 and Tissue Mechanics Promotes Glioma Aggression. Neuron 2018, 100, 799–815.e7. [Google Scholar] [CrossRef]
- Parpura, V.; Basarsky, T.A.; Liu, F.; Jeftinija, K.; Jeftinija, S.; Haydon, P.G. Glutamate-mediated astrocyte–neuron signalling. Nature 1994, 369, 744–747. [Google Scholar] [CrossRef] [PubMed]
- Araque, A.; Parpura, V.; Sanzgiri, R.P.; Haydon, P.G. Tripartite synapses: Glia, the unacknowledged partner. Trends Neurosci. 1999, 22, 208–215. [Google Scholar] [CrossRef]
- Nedergaard, M. Direct signaling from astrocytes to neurons in cultures of mammalian brain cells. Science 1994, 263, 1768–1771. [Google Scholar] [CrossRef] [PubMed]
- Fields, R.D.; Woo, D.H.; Basser, P.J. Glial Regulation of the Neuronal Connectome through Local and Long-Distant Communication. Neuron 2015, 86, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Volterra, A.; Meldolesi, J. Astrocytes, from brain glue to communication elements: The revolution continues. Nat. Rev. Neurosci. 2005, 6, 626–640. [Google Scholar] [CrossRef]
- Rungta, R.L.; Bernier, L.-P.; Dissing-Olesen, L.; Groten, C.J.; LeDue, J.M.; Ko, R.; Drissler, S.; MacVicar, B.A. Ca2+ transients in astrocyte fine processes occur via Ca2+ influx in the adult mouse hippocampus. Glia 2016, 64, 2093–2103. [Google Scholar] [CrossRef]
- Verisokin, A.Y.; Verveyko, D.V.; Postnov, D.E.; Brazhe, A.R. Modeling of Astrocyte Networks: Toward Realistic Topology and Dynamics. Front. Cell. Neurosci. 2021, 15, 645068. [Google Scholar] [CrossRef]
- Houades, V.; Rouach, N.; Ezan, P.; Kirchhoff, F.; Koulakoff, A.; Giaume, C. Shapes of astrocyte networks in the juvenile brain. Neuron Glia Biol. 2006, 2, 3–14. [Google Scholar] [CrossRef]
- Oberheim, N.A.; Takano, T.; Han, X.; He, W.; Lin, J.H.C.; Wang, F.; Xu, Q.; Wyatt, J.D.; Pilcher, W.; Ojemann, J.; et al. Uniquely Hominid Features of Adult Human Astrocytes. J. Neurosci. 2009, 29, 3276–3287. [Google Scholar] [CrossRef]
- Volterra, A.; Liaudet, N.; Savtchouk, I. Astrocyte Ca2+ signalling: An unexpectedunexpected complexity. Nat. Rev. Neurosci. 2014, 15, 327–335. [Google Scholar] [CrossRef]
- Gasiorowski, J.Z.; Murphy, C.J.; Nealey, P.F. Biophysical Cues and Cell Behavior: The Big Impact of Little Things. Annu. Rev. Biomed. Eng. 2013, 15, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.; Lamanna, J.; Moro, A.S.; Piazzoni, C.; Borghi, F.; Chighizola, M.; Ortoleva, S.; Racchetti, G.; Lenardi, C.; Podestà, A.; et al. Neuronal Cells Confinement by Micropatterned Cluster-Assembled Dots with Mechanotransductive Nanotopography. ACS Biomater. Sci. Eng. 2018, 4, 4062–4075. [Google Scholar] [CrossRef] [PubMed]
- Previdi, A.; Piazzoni, C.; Borghi, F.; Schulte, C.; Lorenzelli, L.; Giacomozzi, F.; Bucciarelli, A.; Malgaroli, A.; Lamanna, J.; Moro, A.; et al. Micropatterning of Substrates for the Culture of Cell Networks by Stencil-Assisted Additive Nanofabrication. Micromachines 2021, 12, 94. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.; Podestà, A.; Lenardi, C.; Tedeschi, G.; Milani, P. Quantitative Control of Protein and Cell Interaction with Nanostructured Surfaces by Cluster Assembling. Accounts Chem. Res. 2017, 50, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.; Ripamonti, M.; Maffioli, E.; Cappelluti, M.A.; Nonnis, S.; Puricelli, L.; Lamanna, J.; Piazzoni, C.; Podestà, A.; Lenardi, C.; et al. Scale Invariant Disordered Nanotopography Promotes Hippocampal Neuron Development and Maturation with Involvement of Mechanotransductive Pathways. Front. Cell. Neurosci. 2016, 10, 267. [Google Scholar] [CrossRef] [PubMed]
- Maffioli, E.; Schulte, C.; Nonnis, S.; Scalvini, F.G.; Piazzoni, C.; Lenardi, C.; Negri, A.; Milani, P.; Tedeschi, G. Proteomic Dissection of Nanotopography-Sensitive Mechanotransductive Signaling Hubs that Foster Neuronal Differentiation in PC12 Cells. Front. Cell. Neurosci. 2018, 11, 417. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.; Rodighiero, S.; Cappelluti, M.A.; Puricelli, L.; Maffioli, E.; Borghi, F.; Negri, A.; Sogne, E.; Galluzzi, M.; Piazzoni, C.; et al. Conversion of nanoscale topographical information of cluster-assembled zirconia surfaces into mechanotransductive events promotes neuronal differentiation. J. Nanobiotech. 2016, 14, 18. [Google Scholar] [CrossRef]
- Schulte, C. Cluster-assembled nanostructured materials for cell biology. Front. Nanosci. 2020, 15, 271–289. [Google Scholar] [CrossRef]
- Chighizola, M.; Previdi, A.; Dini, T.; Piazzoni, C.; Lenardi, C.; Milani, P.; Schulte, C.; Podestà, A. Adhesion force spectroscopy with nanostructured colloidal probes reveals nanotopography-dependent early mechanotransductive interactions at the cell membrane level. Nanoscale 2020, 12, 14708–14723. [Google Scholar] [CrossRef]
- Piseri, P.; Vahedi Tafreshi, H.; Milani, P. Manipulation of nanoparticles in supersonic beams for the production of nanostructured materials. Curr. Opin. Solid State Mater. Sci. 2004, 8, 195–202. [Google Scholar] [CrossRef]
- Wegner, K.; Piseri, P.; Tafreshi, H.V.; Milani, P. Cluster beam deposition: A tool for nanoscale science and technology. J. Phys. D Appl. Phys. 2006, 39, R439–R459. [Google Scholar] [CrossRef]
- Borghi, F.; Sogne, E.; Lenardi, C.; Podestà, A.; Merlini, M.; Ducati, C.; Milani, P. Cluster-assembled cubic zirconia films with tunable and stable nanoscale morphology against thermal annealing. J. Appl. Phys. 2016, 120, 055302. [Google Scholar] [CrossRef]
- Borghi, F.; Podestà, A.; Piazzoni, C.; Milani, P. Growth Mechanism of Cluster-Assembled Surfaces: From Submonolayer to Thin-Film Regime. Phys. Rev. Appl. 2018, 9, 044016. [Google Scholar] [CrossRef]
- Podestà, A.; Borghi, F.; Indrieri, M.; Bovio, S.; Piazzoni, C.; Milani, P. Nanomanufacturing of titania interfaces with controlled structural and functional properties by supersonic cluster beam deposition. J. Appl. Phys. 2015, 118, 234309. [Google Scholar] [CrossRef]
- Lamanna, J.; Signorini, M.G.; Cerutti, S.; Malgaroli, A. A pre-docking source for the power-law behavior of spontaneous quantal release: Application to the analysis of LTP. Front. Cell. Neurosci. 2015, 9, 44. [Google Scholar] [CrossRef]
- Kang, M.; Othmer, H.G. Spatiotemporal characteristics of calcium dynamics in astrocytes. Chaos 2009, 19, 037116. [Google Scholar] [CrossRef]
- Wang, Z.; Haydon, P.G.; Yeung, E.S. Direct Observation of Calcium-Independent Intercellular ATP Signaling in Astrocytes. Anal. Chem. 2000, 72, 2001–2007. [Google Scholar] [CrossRef]
- Newman, E.A. Propagation of Intercellular Calcium Waves in Retinal Astrocytes and Müller Cells. J. Neurosci. 2001, 21, 2215–2223. [Google Scholar] [CrossRef]
- Venance, L.; Stella, N.; Glowinski, J.; Giaume, C. Mechanism Involved in Initiation and Propagation of Receptor-Induced Intercellular Calcium Signaling in Cultured Rat Astrocytes. J. Neurosci. 1997, 17, 1981–1992. [Google Scholar] [CrossRef]
- Scemes, E.; Giaume, C. Astrocyte calcium waves: What they are and what they do. Glia 2006, 54, 716–725. [Google Scholar] [CrossRef]
- Arcuino, G.; Lin, J.H.-C.; Takano, T.; Liu, C.; Jiang, L.; Gao, Q.; Kang, J.; Nedergaard, M. Intercellular calcium signaling mediated by point-source burst release of ATP. Proc. Natl. Acad. Sci. USA 2002, 99, 9840–9845. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Maekawa, S.; Morita, M. Astrocyte calcium waves propagate proximally by gap junction and distally by extracellular diffusion of ATP released from volume-regulated anion channels. Sci. Rep. 2017, 7, 13115. [Google Scholar] [CrossRef] [PubMed]
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple Comparisons among Means. J. Am. Stat. Assoc. 1961, 56, 52–64. [Google Scholar] [CrossRef]
- Kimelberg, H.K. The problem of astrocyte identity. Neurochem. Int. 2004, 45, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Haim, L.B.; Rowitch, D.H. Functional diversity of astrocytes in neural circuit regulation. Nat. Rev. Neurosci. 2016, 18, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Spallazzi, M.; Dobisch, L.; Becke, A.; Berron, D.; Stucht, D.; Oeltze-Jafra, S.; Caffarra, P.; Speck, O.; Düzel, E. Hippocampal vascularization patterns: A high-resolution 7 Tesla time-of-flight magnetic resonance angiography study. NeuroImage Clin. 2018, 21, 101609. [Google Scholar] [CrossRef]
- Xiong, B.; Li, A.; Lou, Y.; Chen, S.; Long, B.; Peng, J.; Yang, Z.; Xu, T.; Yang, X.; Li, X.; et al. Precise Cerebral Vascular Atlas in Stereotaxic Coordinates of Whole Mouse Brain. Front. Neuroanat. 2017, 11, 128. [Google Scholar] [CrossRef]
- Liewald, D.; Miller, R.; Logothetis, N.; Wagner, H.-J.; Schüz, A. Distribution of axon diameters in cortical white matter: An electron-microscopic study on three human brains and a macaque. Biol. Cybern. 2014, 108, 541–557. [Google Scholar] [CrossRef]
- Leclech, C.; Natale, C.F.; Barakat, A.I. The basement membrane as a structured surface-role in vascular health and disease. J. Cell Sci. 2020, 133, jcs239889. [Google Scholar] [CrossRef]
- Zamanian, J.L.; Xu, L.; Foo, L.C.; Nouri, N.; Zhou, L.; Giffard, R.G.; Barres, B.A. Genomic Analysis of Reactive Astrogliosis. J. Neurosci. 2012, 32, 6391–6410. [Google Scholar] [CrossRef] [PubMed]
- East, E.; Golding, J.P.; Phillips, J.B. A versatile 3D culture model facilitates monitoring of astrocytes undergoing reactive gliosis. J. Tissue Eng. Regen. Med. 2009, 3, 634–646. [Google Scholar] [CrossRef]
- Cao, H.; Marcy, G.; Goh, E.L.K.; Wang, F.; Wang, J.; Chew, S.Y. The effects of nanofiber topography on astrocyte behavior and gene silencing efficiency. Macromol. Biosci. 2012, 12, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Puschmann, T.B.; Zandén, C.; De Pablo, Y.; Kirchhoff, F.; Pekna, M.; Liu, J.; Pekny, M. Bioactive 3D cell culture system minimizes cellular stress and maintains the in vivo-like morphological complexity of astroglial cells. Glia 2013, 61, 432–440. [Google Scholar] [CrossRef]
- Hu, W.-W.; Wang, Z.; Zhang, S.-S.; Jiang, L.; Zhang, J.; Zhang, X.; Lei, Q.-F.; Park, H.-J.; Fang, W.-J.; Chen, Z. Morphology and functions of astrocytes cultured on water-repellent fractal tripalmitin surfaces. Biomaterials 2014, 35, 7386–7397. [Google Scholar] [CrossRef] [PubMed]
- Moshayedi, P.; Ng, G.; Kwok, J.C.; Yeo, G.S.; Bryant, C.E.; Fawcett, J.W.; Franze, K.; Guck, J. The relationship between glial cell mechanosensitivity and foreign body reactions in the central nervous system. Biomaterials 2014, 35, 3919–3925. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.L.; Hayward, S.L.; Kidambi, S. Astrogliosis in a dish: Substrate stiffness induces astrogliosis in primary rat astrocytes. RSC Adv. 2016, 6, 34447–34457. [Google Scholar] [CrossRef] [PubMed]
- Galli, A.; Maffioli, E.; Sogne, E.; Moretti, S.; Di Cairano, E.S.; Negri, A.; Nonnis, S.; Norata, G.D.; Bonacina, F.; Borghi, F.; et al. Cluster-assembled zirconia substrates promote long-term differentiation and functioning of human islets of Langerhans. Sci. Rep. 2018, 8, 9979. [Google Scholar] [CrossRef]
- Chighizola, M.; Dini, T.; Marcotti, S.; D’Urso, M.; Piazzoni, C.; Borghi, F.; Previdi, A.; Ceriani, L.; Folliero, C.; Stramer, B.; et al. The glycocalyx affects the mechanotransductive perception of the topographical microenvironment. J. Nanobiotech. 2022, 20, 418. [Google Scholar] [CrossRef]
- Singh, A.V.; Raymond, M.; Pace, F.; Certo, A.; Zuidema, J.M.; McKay, C.A.; Gilbert, R.J.; Lu, X.L.; Wan, L.Q. Astrocytes Increase ATP Exocytosis Mediated Calcium Signaling in Response to Microgroove Structures. Sci. Rep. 2015, 5, srep07847. [Google Scholar] [CrossRef]
- Cornell-Bell, A.H.; Finkbeiner, S.M.; Cooper, M.S.; Smith, S.J. Glutamate Induces Calcium Waves in Cultured Astrocytes: Long-Range Glial Signaling. Science 1990, 247, 470–473. [Google Scholar] [CrossRef]
- Matias, I.; Morgado, J.; Gomes, F.C.A. Astrocyte Heterogeneity: Impact to Brain Aging and Disease. Front. Aging Neurosci. 2019, 11, 59. [Google Scholar] [CrossRef]
- Parri, R.; Gould, T.M.; Crunelli, V. Spontaneous astrocytic Ca2+ oscillations in situ drive NMDAR-mediated neuronal excitation. Nat. Neurosci. 2001, 4, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Elosegui-Artola, A.; Andreu, I.; Beedle, A.E.M.; Lezamiz, A.; Uroz, M.; Kosmalska, A.J.; Oria, R.; Kechagia, J.Z.; Rico-Lastres, P.; Le Roux, A.-L.; et al. Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 2017, 171, 1397–1410.e14. [Google Scholar] [CrossRef] [PubMed]
- Houades, V.; Koulakoff, A.; Ezan, P.; Seif, I.; Giaume, C. Gap junction-mediated astrocytic networks in the mouse barrel cortex. J. Neurosci. 2008, 28, 5207–5217. [Google Scholar] [CrossRef]
- Roux, L.; Benchenane, K.; Rothstein, J.D.; Bonvento, G.; Giaume, C. Plasticity of astroglial networks in olfactory glomeruli. Proc. Natl. Acad. Sci. USA 2011, 108, 18442–18446. [Google Scholar] [CrossRef] [PubMed]
- Pannasch, U.; Rouach, N. Emerging role for astroglial networks in information processing: From synapse to behavior. Trends Neurosci. 2013, 36, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Dokukina, I.V.; Gracheva, M.E.; Grachev, E.A.; Gunton, J.D. Role of network connectivity in intercellular calcium signaling. Phys. D Nonlinear Phenom. 2008, 237, 745–754. [Google Scholar] [CrossRef]
- Lallouette, J.; De Pittà, M.; Ben-Jacob, E.; Berry, H. Sparse short-distance connections enhance calcium wave propagation in a 3D model of astrocyte networks. Front. Comput. Neurosci. 2014, 8, 45. [Google Scholar] [CrossRef]
- Semyanov, A.; Henneberger, C.; Agarwal, A. Making sense of astrocytic calcium signals—From acquisition to interpretation. Nat. Rev. Neurosci. 2020, 21, 551–564. [Google Scholar] [CrossRef]
- Wu, Y.-W.; Gordleeva, S.; Tang, X.; Shih, P.-Y.; Dembitskaya, Y.; Semyanov, A. Morphological profile determines the frequency of spontaneous calcium events in astrocytic processes. Glia 2018, 67, 246–262. [Google Scholar] [CrossRef] [PubMed]
- Semyanov, A. Spatiotemporal pattern of calcium activity in astrocytic network. Cell Calcium 2018, 78, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Denizot, A.; Arizono, M.; Nägerl, U.V.; Soula, H.; Berry, H. Simulation of calcium signaling in fine astrocytic processes: Effect of spatial properties on spontaneous activity. PLoS Comput. Biol. 2019, 15, e1006795. [Google Scholar] [CrossRef]
- Stobart, J.L.; Ferrari, K.D.; Barrett, M.J.P.; Stobart, M.J.; Looser, Z.J.; Saab, A.S.; Weber, B. Long-term In Vivo Calcium Imaging of Astrocytes Reveals Distinct Cellular Compartment Responses to Sensory Stimulation. Cereb. Cortex 2016, 28, 184–198. [Google Scholar] [CrossRef] [PubMed]
- Cotrina, M.L.; Lin, J.H.-C.; Nedergaard, M. Cytoskeletal Assembly and ATP Release Regulate Astrocytic Calcium Signaling. J. Neurosci. 1998, 18, 8794–8804. [Google Scholar] [CrossRef]
- Wheeler, B.C.; Corey, J.M.; Brewer, G.J.; Branch, D.W. Microcontact Printing for Precise Control of Nerve Cell Growth in Culture. J. Biomech. Eng. 1999, 121, 73–78. [Google Scholar] [CrossRef]
- Hansson, E. Actin Filament Reorganization in Astrocyte Networks is a Key Functional Step in Neuroinflammation Resulting in Persistent Pain: Novel Findings on Network Restoration. Neurochem. Res. 2014, 40, 372–379. [Google Scholar] [CrossRef]
- Hansson, E.; Werner, T.; Björklund, U.; Skiöldebrand, E. Therapeutic innovation: Inflammatory-reactive astrocytes as targets of inflammation. IBRO Rep. 2016, 1, 1–9. [Google Scholar] [CrossRef]
- Sergeeva, M.; Ubl, J.; Reiser, G. Disruption of actin cytoskeleton in cultured rat astrocytes suppresses ATP- and bradykinin-induced [Ca2+]i oscillations by reducing the coupling efficiency between Ca2+ release, capacitative Ca2+ entry, and store refilling. Neuroscience 2000, 97, 765–769. [Google Scholar] [CrossRef]



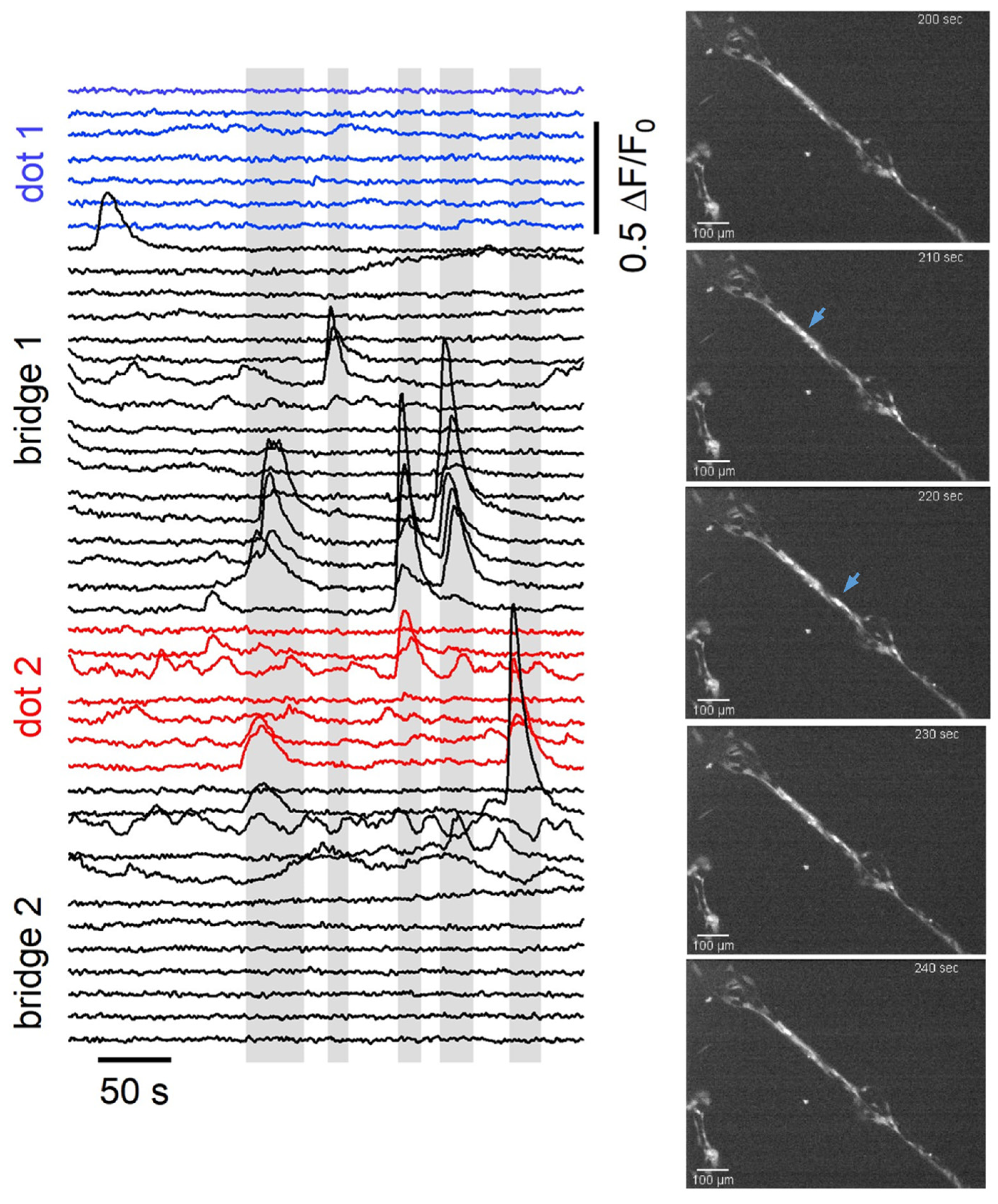

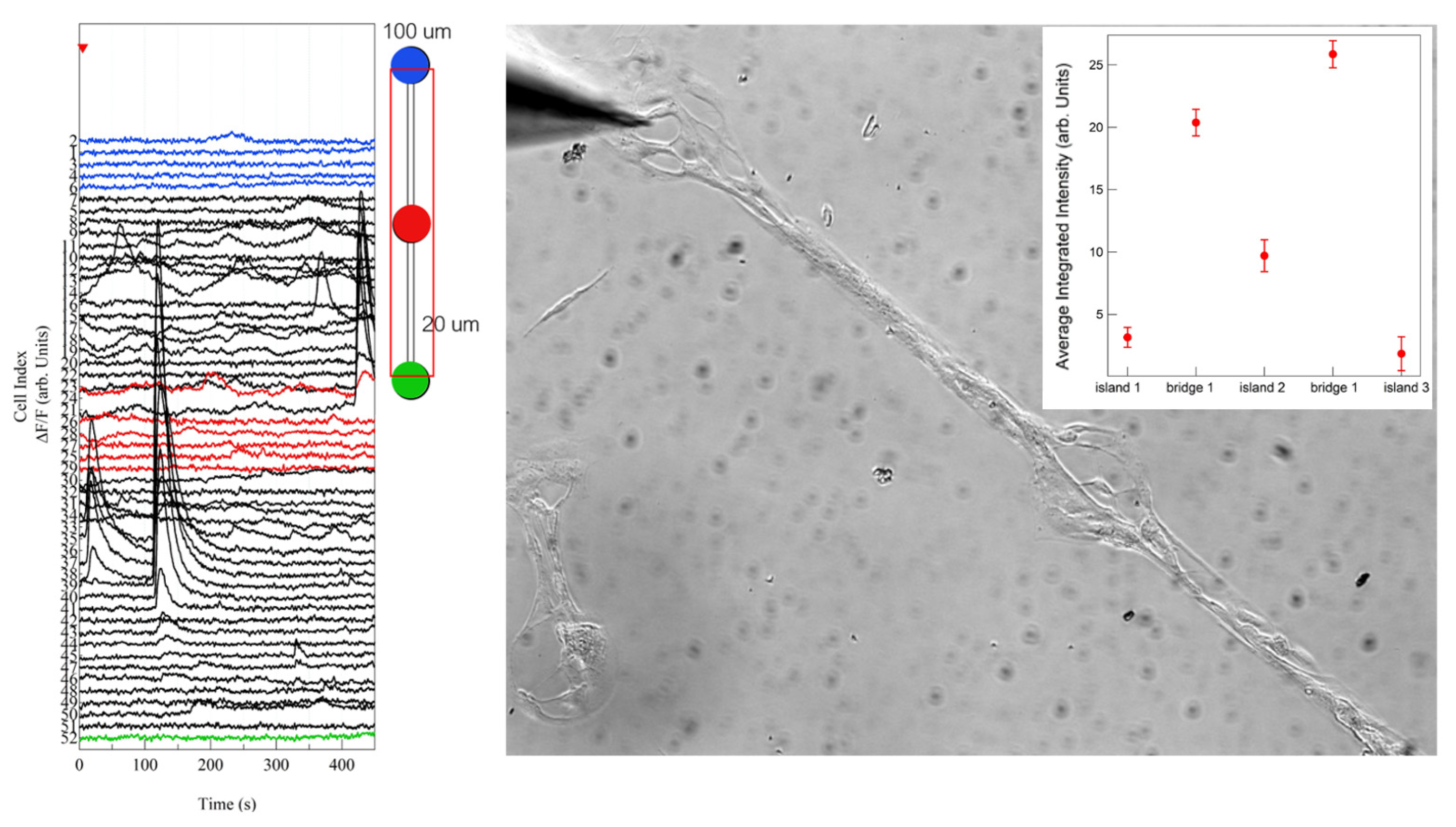
| Dot (100 µm Diameter) | 50 μm Bridge | 20 μm Bridge | |
|---|---|---|---|
| Eccentricity | 1.08 ± 0.23 | 0.80 ± 0.24 | 0.36 ± 0.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Previdi, A.; Borghi, F.; Profumo, F.; Schulte, C.; Piazzoni, C.; Lamanna, J.; Racchetti, G.; Malgaroli, A.; Milani, P. Nanotopography and Microconfinement Impact on Primary Hippocampal Astrocyte Morphology, Cytoskeleton and Spontaneous Calcium Wave Signalling. Cells 2023, 12, 293. https://doi.org/10.3390/cells12020293
Previdi A, Borghi F, Profumo F, Schulte C, Piazzoni C, Lamanna J, Racchetti G, Malgaroli A, Milani P. Nanotopography and Microconfinement Impact on Primary Hippocampal Astrocyte Morphology, Cytoskeleton and Spontaneous Calcium Wave Signalling. Cells. 2023; 12(2):293. https://doi.org/10.3390/cells12020293
Chicago/Turabian StylePrevidi, Anita, Francesca Borghi, Filippo Profumo, Carsten Schulte, Claudio Piazzoni, Jacopo Lamanna, Gabriella Racchetti, Antonio Malgaroli, and Paolo Milani. 2023. "Nanotopography and Microconfinement Impact on Primary Hippocampal Astrocyte Morphology, Cytoskeleton and Spontaneous Calcium Wave Signalling" Cells 12, no. 2: 293. https://doi.org/10.3390/cells12020293
APA StylePrevidi, A., Borghi, F., Profumo, F., Schulte, C., Piazzoni, C., Lamanna, J., Racchetti, G., Malgaroli, A., & Milani, P. (2023). Nanotopography and Microconfinement Impact on Primary Hippocampal Astrocyte Morphology, Cytoskeleton and Spontaneous Calcium Wave Signalling. Cells, 12(2), 293. https://doi.org/10.3390/cells12020293