MIA/CD-RAP Regulates MMP13 and Is a Potential New Disease-Modifying Target for Osteoarthritis Therapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Studies
2.2. Cell Culture
2.3. High Density Micromass Cultures
2.4. High Density Spheroid Assay
2.5. siRNA Transfection
2.6. Transfection of Plasmid DNA
2.7. Luciferase Reporter Gene Assay
2.8. Actinomycin Treatment and mRNA Decay
2.9. In Vitro Cultivation and Stimulation of Murine Articular Cartilage Explants
2.10. Histology and TUNEL Staining
2.11. DMMB Assay and MMP13 Activity Assay
2.12. ELISA
2.13. Analysis of mRNA Expression Using Real-Time PCR
2.14. Western Blot Protein Analysis
2.15. Statistical Analysis
3. Results
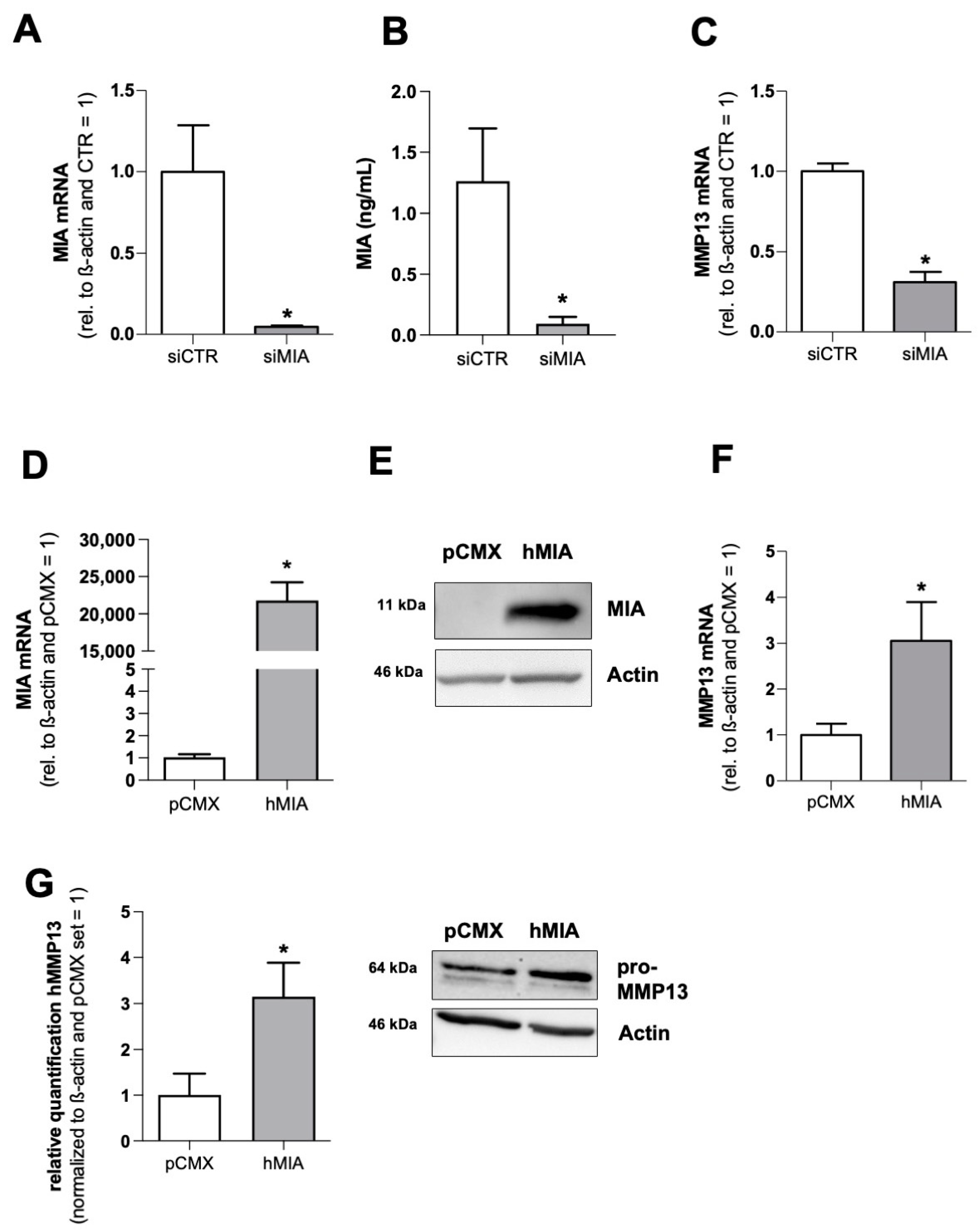
3.1. MIA/CD-RAP Regulates MMP13
3.2. MMP13 Expression and Activity Are Significantly Reduced due to the Loss of MIA/CD-RAP
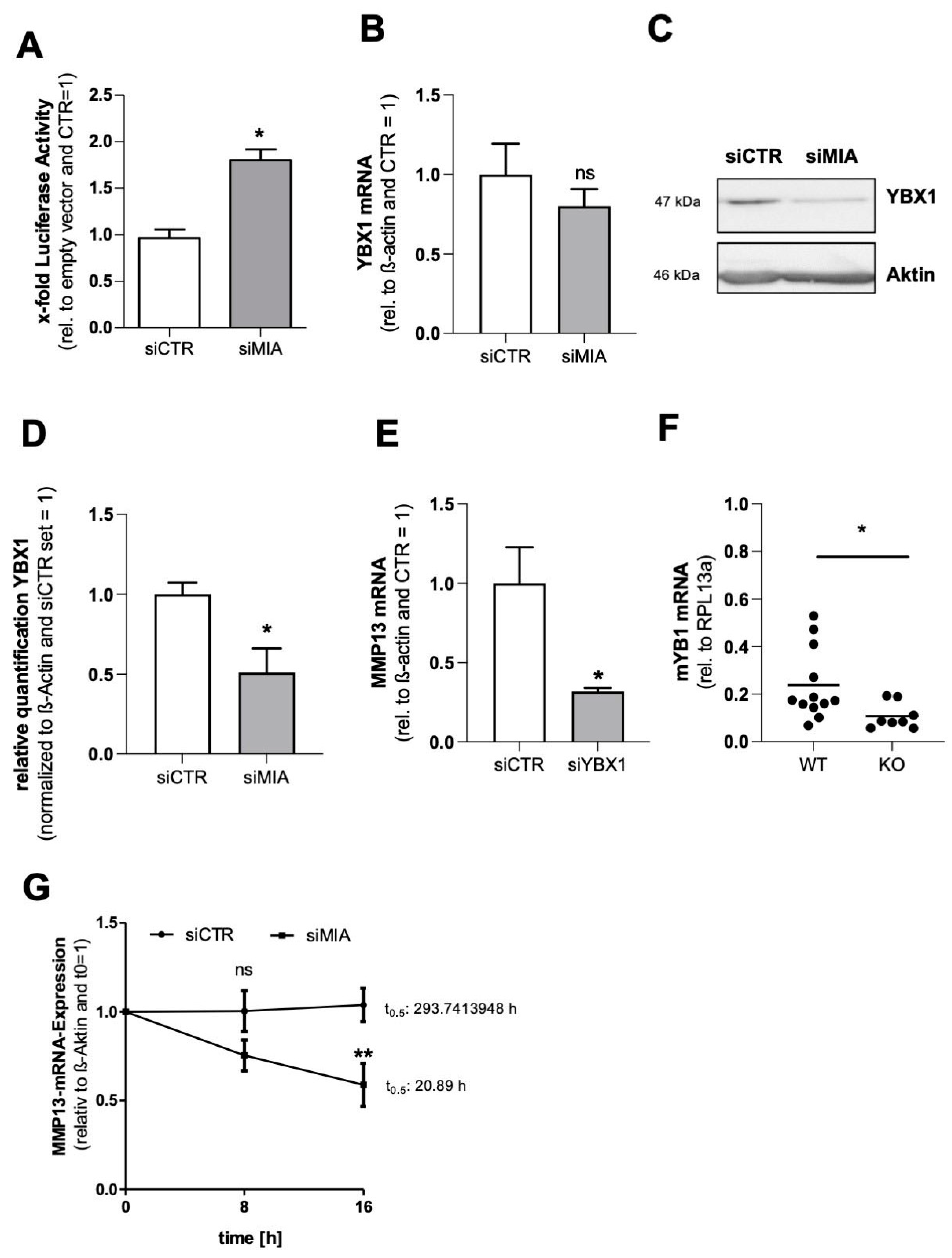
3.3. MIA/CD-RAP Regulates MMP13 via YBX1
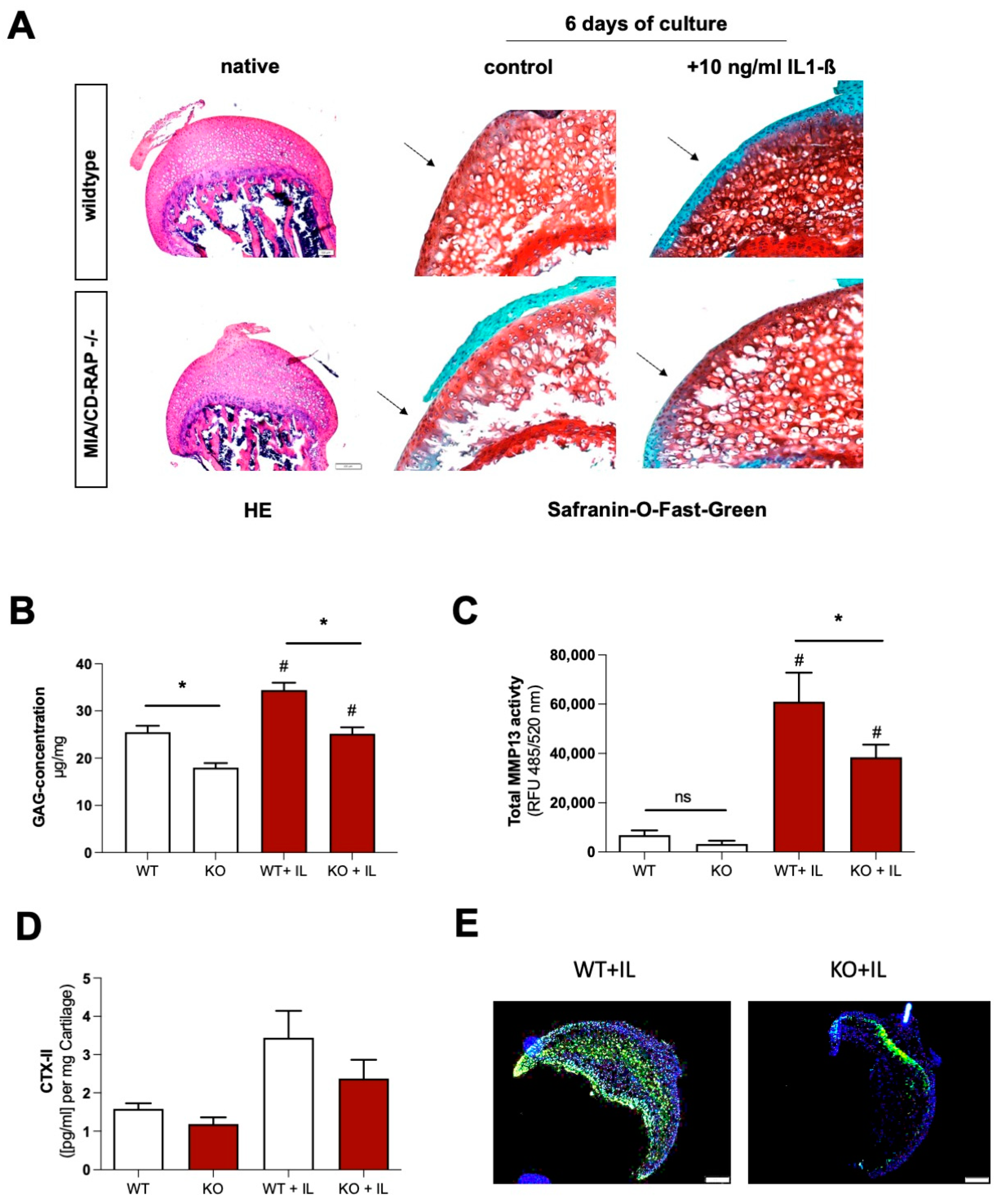
3.4. Decreased Cartilage Degeneration of MIA/CD-RAP −/− Articular Cartilage
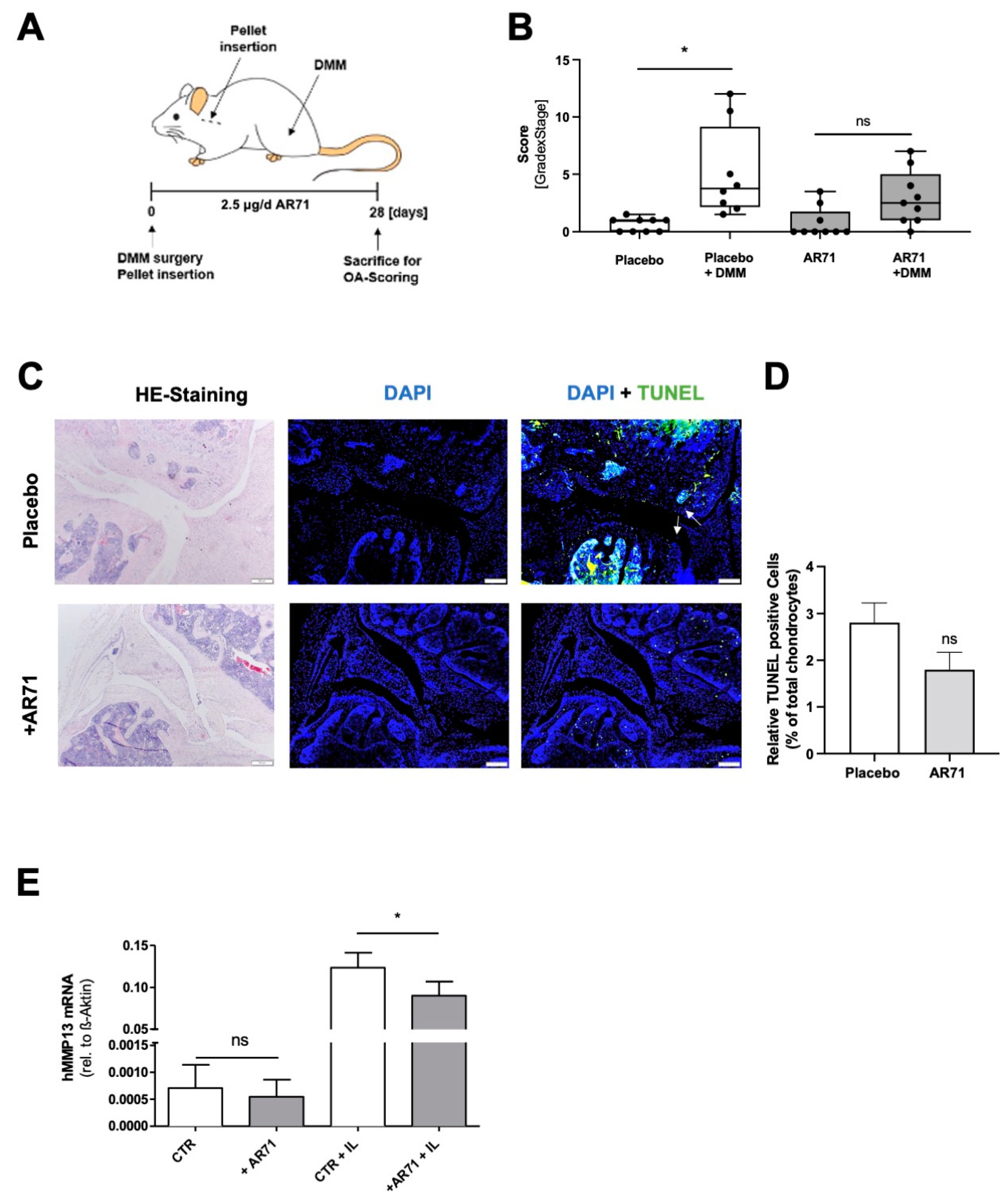
3.5. Inhibition of MIA/CD-RAP Reduces OA Development
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ghouri, A.; Conaghan, P.G. Update on novel pharmacological therapies for osteoarthritis. Ther. Adv. Musculoskelet Dis. 2019, 11, 1759720X19864492. [Google Scholar] [CrossRef] [PubMed]
- Oo, W.M.; Little, C.; Duong, V.; Hunter, D.J. The Development of Disease-Modifying Therapies for Osteoarthritis (DMOADs): The Evidence to Date. Drug Des. Devel. Ther. 2021, 15, 2921–2945. [Google Scholar] [CrossRef] [PubMed]
- Oo, W.M.; Hunter, D. Repurposed and investigational disease-modifying drugs in osteoarthritis (DMOADs). Ther. Adv. Musculoskelet Dis. 2022, 14, 1759720X221090297. [Google Scholar] [CrossRef] [PubMed]
- Goldring, M.B.; Goldring, S. Osteoarthritis. J. Cell Physiol. 2007, 213, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Malemud, C.J. Biologic basis of osteoarthritis: State of the evidence. Curr. Opin. Rheumatol. 2015, 27, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.G.; Magna, H.A.; Reeves, L.M.; Lopresti-Morrow, L.L.; Yocum, S.A.; Rosner, P.J.; Geoghegan, K.F.; Hambor, J.E. Cloning, expression, and type II collagenolytic activity of matrix metalloproteinase-13 from human osteoarthritic cartilage. J. Clin. Investig. 1996, 97, 761–768. [Google Scholar] [CrossRef]
- Monfort, J.; Tardif, G.; Reboul, P.; Mineau, F.; Roughley, P.; Pelletier, J.P.; Martel-Pelletier, J. Degradation of small leucine-rich repeat proteoglycans by matrix metalloprotease-13: Identification of a new biglycan cleavage site. Arthritis Res. Ther. 2006, 8, R26. [Google Scholar] [CrossRef]
- Little, C.B.; Barai, A.; Burkhardt, D.; Smith, S.M.; Fosang, A.J.; Werb, Z.; Shah, M.; Thompson, E.W. Matrix metalloproteinase 13-deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 2009, 60, 3723–3733. [Google Scholar] [CrossRef]
- Blesch, A.; Boßerhoff, A.K.; Apfel, R.; Behl, C.; Hessdoerfer, B.; Schmitt, A.; Jachimczak, P.; Lottspeich, F.; Buettner, R.; Bogdahn, U. Cloning of a novel malignant melanoma-derived growth-regulatory protein, MIA. Cancer Res. 1994, 54, 5695–5701. [Google Scholar]
- Dietz, U.H.; Sandell, L. Cloning of a retinoic acid-sensitive mRNA expressed in cartilage and during chondrogenesis. J. Biol. Chem. 1996, 271, 3311–3316. [Google Scholar] [CrossRef]
- Schmid, R.; Meyer, K.; Spang, R.; Schittek, B.; Bosserhoff, A.K. YBX1 is a modulator of MIA/CD-RAP-dependent chondrogenesis. PLoS ONE 2013, 8, e82166. [Google Scholar] [CrossRef]
- Schubert, T.; Schlegel, J.; Schmid, R.; Opolka, A.; Grässel, S.; Humphries, M.; Bosserhoff, A.K. Modulation of cartilage differentiation by melanoma inhibiting activity/cartilage-derived retinoic acid-sensitive protein (MIA/CD-RAP). Exp. Mol. Med. 2010, 42, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Moser, M.; Bosserhoff, A.K.; Hunziker, E.B.; Sandell, L.; Fässler, R.; Buettner, R. Ultrastructural cartilage abnormalities in MIA/CD-RAP-deficient mice. Mol. Cell Biol. 2002, 22, 1438–1445. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schmid, R.; Schiffner, S.; Opolka, A.; Grässel, S.; Schubert, T.; Moser, M.; Bosserhoff, A.K. Enhanced cartilage regeneration in MIA/CD-RAP deficient mice. Cell Death Dis. 2010, 1, e97. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.; Riechers, A.; Stoll, R.; Amann, T.; Fink, F.; Spruss, T.; Gronwald, W.; König, B.; Hellerbrand, C.; Bosserhoff, A.K. Targeting melanoma metastasis and immunosuppression with a new mode of melanoma inhibitory activity (MIA) protein inhibition. PLoS ONE 2012, 7, e37941. [Google Scholar] [CrossRef]
- Pritzker, K.P.; Gay, S.; Jimenez, S.A.; Ostergaard, K.; Pelletier, J.P.; Revell, P.A.; Salter, M.D.; Berg, W.B. Osteoarthritis cartilage histopathology: Grading and staging. Osteoarthr. Cartil. 2006, 14, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Greco, K.V.; Iqbal, A.J.; Rattazzi, L.; Nalesso, G.; Moradi-Bidhendi, N.; Moore, A.R.; Goldring, M.B.; Dell’Accio, F.; Perretti, M. High density micromass cultures of a human chondrocyte cell line: A reliable assay system to reveal the modulatory functions of pharmacological agents. Biochem. Pharmacol. 2011, 82, 1919–1929. [Google Scholar] [CrossRef]
- Ruedel, A.; Hofmeister, S.; Bosserhoff, A.-K. Development of a model system to analyze chondrogenic differentiation of mesenchymal stem cells. Int. J. Clin. Exp. Pathol. 2013, 6, 3042–3048. [Google Scholar] [PubMed]
- Schiffner, S.; Zimara, N.; Schmid, R.; Bosserhoff, A.K. p54nrb is a new regulator of progression of malignant melanoma. Carcinogenesis 2011, 32, 1176–1182. [Google Scholar] [CrossRef]
- Zimmermann, T.; Pommer, M.; Kluge, V.; Chiheb, C.; Muehlich, S.; Bosserhoff, A.K. Detection of Cellular Senescence in Human Primary Melanocytes and Malignant Melanoma Cells In Vitro. Cells 2022, 11, 1489. [Google Scholar] [CrossRef]
- Riechers, A.; Bosserhoff, A. Pitfalls in immunohistochemistry—A recent example. Int. J. Clin. Exp. Pathol. 2012, 5, 137–139. [Google Scholar]
- Samuel, S.; Beifuss, K.K.; Bernstein, L.R. YB-1 binds to the MMP-13 promoter sequence and represses MMP-13 transactivation via the AP-1 site. Biochim. Biophys. Acta 2007, 1769, 525–531. [Google Scholar] [CrossRef]
- Lyabin, D.N.; Eliseeva, I.; Ovchinnikov, L. YB-1 protein: Functions and regulation. Wiley Interdiscip. Rev. RNA 2014, 5, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Niebler, S.; Schubert, T.; Hunziker, E.B.; Bosserhoff, A.K. Activating enhancer binding protein 2 epsilon (AP-2epsilon)-deficient mice exhibit increased matrix metalloproteinase 13 expression and progressive osteoarthritis development. Arthritis Res. Ther. 2015, 17, 119. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.I. Matrix metalloproteinases: A multifunctional group of molecules. J. Pathol. 2001, 195, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Roach, H.I.; Yamada, N.; Cheung, K.S.; Tilley, S.; Clarke, N.M.; Oreffo, R.O.; Kokubun, S.; Bronner, F. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 2005, 52, 3110–3124. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.H.; Wu, C.H.; Jou, I.M.; Tu, Y.K.; Hung, C.H.; Hsieh, P.L.; Tsai, K.L. PKR activation causes inflammation and MMP-13 secretion in human degenerated articular chondrocytes. Redox Biol. 2018, 14, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Ruan, G.; Xu, J.; Wang, K.; Wu, J.; Zhu, Q.; Ren, J.; Bian, F.; Chang, B.; Bai, X.; Han, W. Associations between knee structural measures, circulating inflammatory factors and MMP13 in patients with knee osteoarthritis. Osteoarthr. Cartil. 2018, 26, 1063–1069. [Google Scholar] [CrossRef]
- McDermott, B.T.; Ellis, S.; Bou-Gharios, G.; Clegg, P.D.; Tew SMcDermott, B.T.; Ellis, S.; Bou-Gharios, G.; Clegg, P.D.; Tew, S. RNA binding proteins regulate anabolic and catabolic gene expression in chondrocytes. Osteoarthr. Cartil. 2016, 24, 1263–1273. [Google Scholar] [CrossRef]
- Schmid, R.; Meyer, K.; Spang, R.; Schittek, B.; Bosserhoff, A.K. Melanoma inhibitory activity promotes melanoma development through activation of YBX1. Pigment Cell Melanoma Res. 2013, 26, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Wolffe, A.P. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. BioEssays 1994, 16, 245–251. [Google Scholar] [CrossRef]
- Evdokimova, V.; Ruzanov, P.; Imataka, H.; Raught, B.; Svitkin, Y.; Ovchinnikov, L.P.; Sonenberg, N. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 2001, 20, 5491–5502. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, W.N.; Doolaanea, A.; Rasad, M.B.A. Effect of shRNA Mediated Silencing of YB-1 Protein on the Expression of Matrix Collagenases in Malignant Melanoma Cell In Vitro. Cells 2018, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ma, Z.; Liu, H.; Zhang, Q.; Cai, X.; Qu, Y.; Xu, M.; Lu, L. Y-box Protein-1 Regulates the Expression of Collagen I in Hepatic Progenitor Cells via PDGFR-beta/ERK/p90RSK Signalling. Stem Cells Int. 2017, 2017, 6193106. [Google Scholar] [CrossRef] [PubMed]
- Koshy PJ, T.; Lundy, C.J.; Rowan, A.D.; Porter, S.; Edwards, D.R.; Hogan, A.; Clark, I.M.; Cawston, T.E. The modulation of matrix metalloproteinase and ADAM gene expression in human chondrocytes by interleukin-1 and oncostatin M: A time-course study using real-time quantitative reverse transcription-polymerase chain reaction. Arthritis Rheum 2002, 46, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Young, D.A.; Barter, M.; Wilkinson, D. Recent advances in understanding the regulation of metalloproteinases. F1000Research 2019, 8, 195. [Google Scholar] [CrossRef]
- Conaghan, P.G. IS IL-1 still the “OA cytokine”. Osteoarthr. Cartil. 2021, 29, S6–S7. [Google Scholar] [CrossRef]
- Catterall, J.; Parr, S.D.; Fagerlund, K.; Caterson, B. CTX-II is a marker of cartilage degradation but not of bone turnover. Osteoarthr. Cartil. 2013, 21, S77. [Google Scholar] [CrossRef]
- Hwang, H.S.; Kim, H. Chondrocyte Apoptosis in the Pathogenesis of Osteoarthritis. Int. J. Mol. Sci. 2015, 16, 26035–26054. [Google Scholar] [CrossRef]
- Bosserhoff, A.K.; Kondo, S.; Moser, M.; Dietz, U.H.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Buettner, R.; Sandell, L.J. Mouse CD-RAP/MIA Gene: Structure, Chromosomal Localization, and Expression in Cartilage and Chondrosarcoma. Dev. Dyn. 1997, 208, 516–525. [Google Scholar] [CrossRef]
- Stoll, R.; Renner, C.; Zweckstetter, M.; Brüggert, M.; Ambrosius, D.; Palme, S.; Emgh, R.A.; Golob, M.; Breibach, I.; Buettner, R. The extracellular human melanoma inhibitory activity (MIA) protein adopts an SH3 domain-like fold. EMBO J. 2001, 20, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Riechers, A.; Schmidt, J.; König, B.; Bosserhoff, A.K. Heterogeneous transition metal-based fluorescence polarization (HTFP) assay for probing protein interactions. Biotechniques 2009, 47, 837–844. [Google Scholar] [CrossRef] [PubMed]

| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| ACTB (human) | CTACGTCGCCCTGGACTTCGAGC | GATGGAGCCGCCGATCCACACGG |
| MMP13 (human) | TACCAGACTTCACGATGGCATTGCTG | AAAGTGGCTTTTGCCGGTGTAGGTG |
| YBX1 (human) | GGGACAAGAAGGTCATCGCA | GTAACATTTGCTGCCTCCGC |
| MIA (human) | CATGCATGCGGTCCTATGCCCAAGCTG | GATAAGCTTTCACTGGCAGTAGAAATC |
| RPL13a (murine) | CCGGAAGCGGATGAATACCA | ACAACCTTGAGAGCAGCAGG |
| YBX1 (murine) | ATCGCAACGAAGGTTTTGGG | GTAACATTTGCTGCCTCCGC |
| MMP13 (murine) | CAGGAGCCCTGATGTTTCCC | TCCTCGGAGACTGGTAATGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Staebler, S.; Lichtblau, A.; Gurbiel, S.; Schubert, T.; Riechers, A.; Rottensteiner-Brandl, U.; Bosserhoff, A. MIA/CD-RAP Regulates MMP13 and Is a Potential New Disease-Modifying Target for Osteoarthritis Therapy. Cells 2023, 12, 229. https://doi.org/10.3390/cells12020229
Staebler S, Lichtblau A, Gurbiel S, Schubert T, Riechers A, Rottensteiner-Brandl U, Bosserhoff A. MIA/CD-RAP Regulates MMP13 and Is a Potential New Disease-Modifying Target for Osteoarthritis Therapy. Cells. 2023; 12(2):229. https://doi.org/10.3390/cells12020229
Chicago/Turabian StyleStaebler, Sebastian, Adrian Lichtblau, Slavyana Gurbiel, Thomas Schubert, Alexander Riechers, Ulrike Rottensteiner-Brandl, and Anja Bosserhoff. 2023. "MIA/CD-RAP Regulates MMP13 and Is a Potential New Disease-Modifying Target for Osteoarthritis Therapy" Cells 12, no. 2: 229. https://doi.org/10.3390/cells12020229
APA StyleStaebler, S., Lichtblau, A., Gurbiel, S., Schubert, T., Riechers, A., Rottensteiner-Brandl, U., & Bosserhoff, A. (2023). MIA/CD-RAP Regulates MMP13 and Is a Potential New Disease-Modifying Target for Osteoarthritis Therapy. Cells, 12(2), 229. https://doi.org/10.3390/cells12020229






