The Fibrotic Effects of LINC00663 in Human Hepatic Stellate LX-2 Cells and in Bile Duct-Ligated Cholestasis Mice Are Mediated through the Splicing Factor 2-Fibronectin
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Preprocessing
2.2. WGCNA
2.3. Identification and Functional Annotation of Key Genes in Co-Expression Networks
2.4. Identification of Hub Genes
2.5. Construction of ceRNA Network
2.6. Cell culture and LX-2 Cells Activated by TGF-β
2.7. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
2.8. Western Blot Analysis
2.9. Cell Transfection
2.10. Dual-Luciferase Reporter Assay
2.11. RNA Fluorescence In Situ Hybridization Assay
2.12. RNA Immunoprecipitation
2.13. Chromatin Immunoprecipitation
2.14. Animal Studies
2.15. Histology and Immunohistochemistry
2.16. Hydroxyproline Assay and Serum ALT and AST
2.17. Wound Healing Assay
2.18. Statistical Analysis
3. Results
3.1. Data Preprocessing and Co-Expression Network Construction
3.2. Identification of Significant Modules
3.3. Functional Annotation
3.4. PPI Network Construction and Hub Gene Identification
3.5. Construction and Validation of ceRNA Network
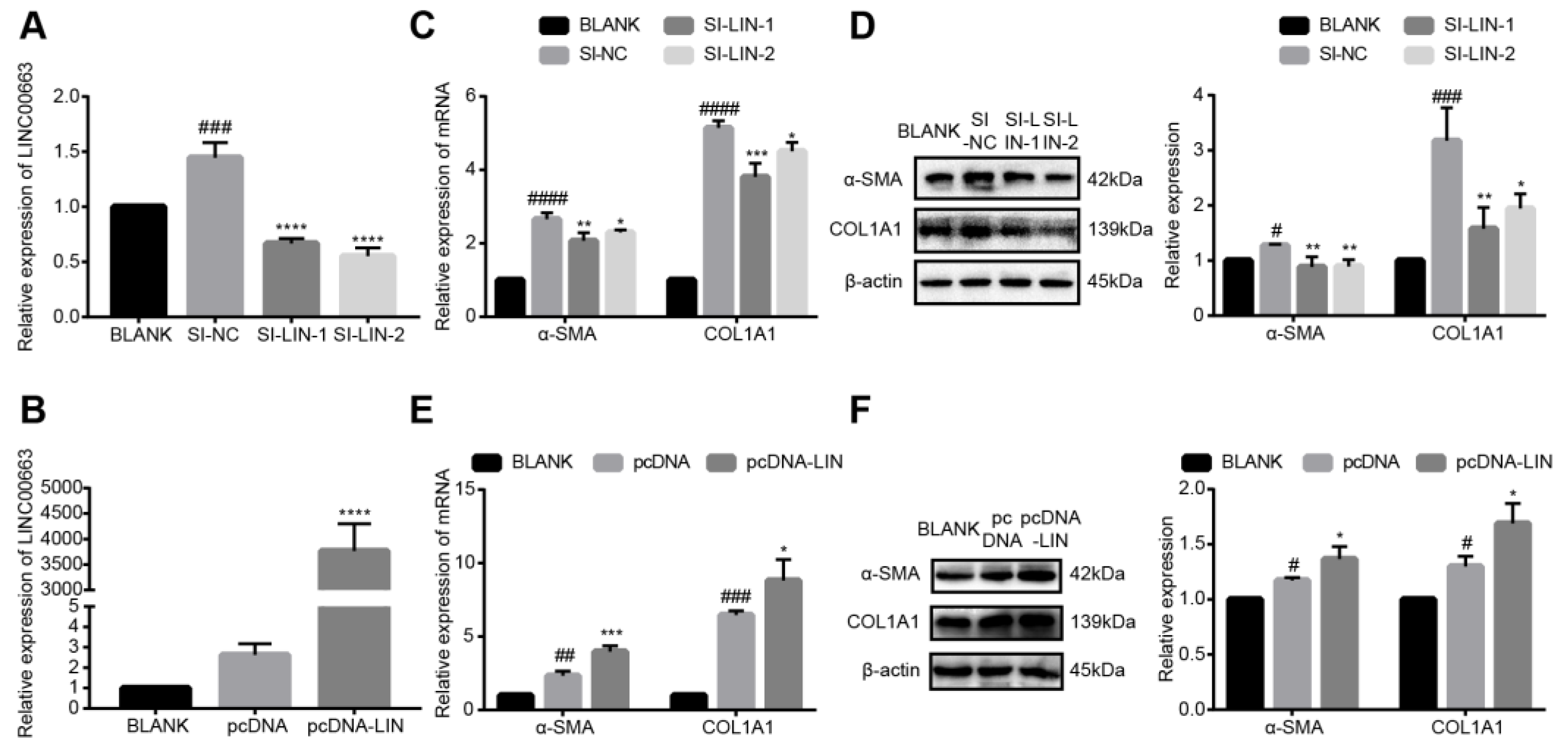
3.6. LINC00663 Regulated the Activation of LX-2 Cells
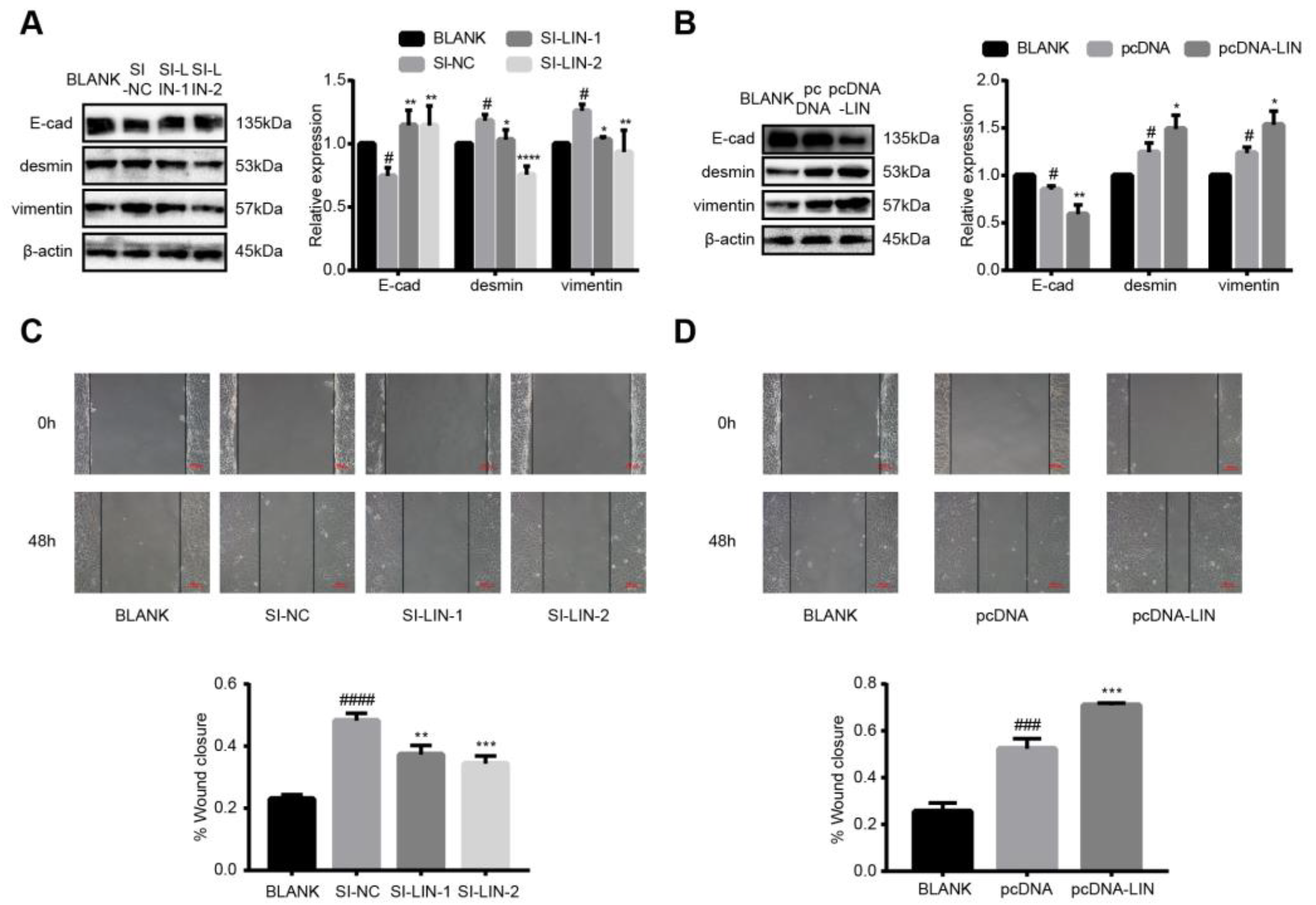
3.7. LINC00663 Regulated Epithelial–Mesenchymal Transition and Migration of LX-2 Cells
3.8. Overexpression of LINC00663 Aggravated BDL-Induced Hepatic Fibrosis In Vivo
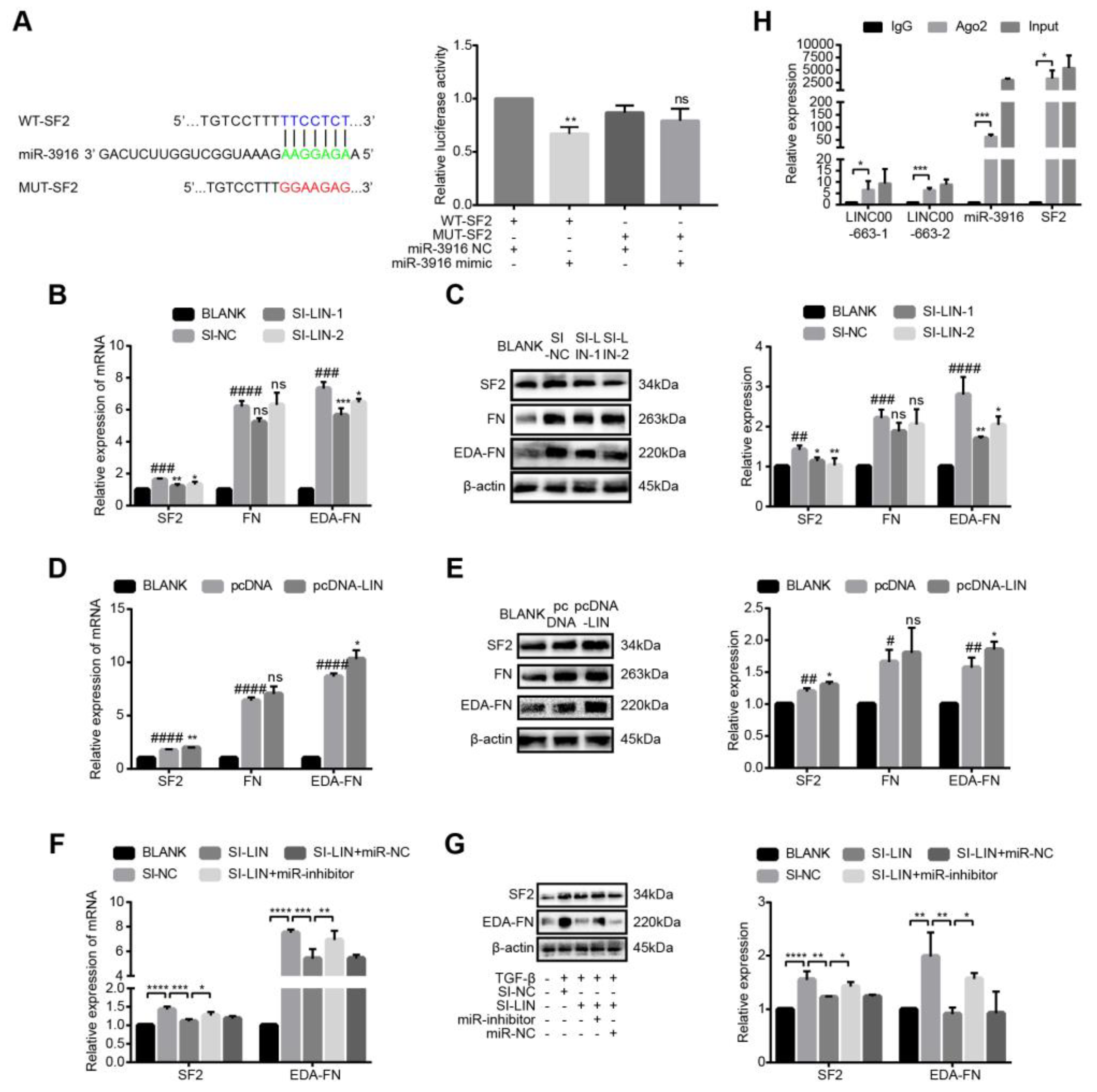
3.9. LINC00663 Regulated Activation, EMT, and Migration of LX-2 Cells through Hsa-miR-3916
3.10. LINC00663 Regulated SF2-FN Alternative Splicing by Sponging Hsa-miR-3916
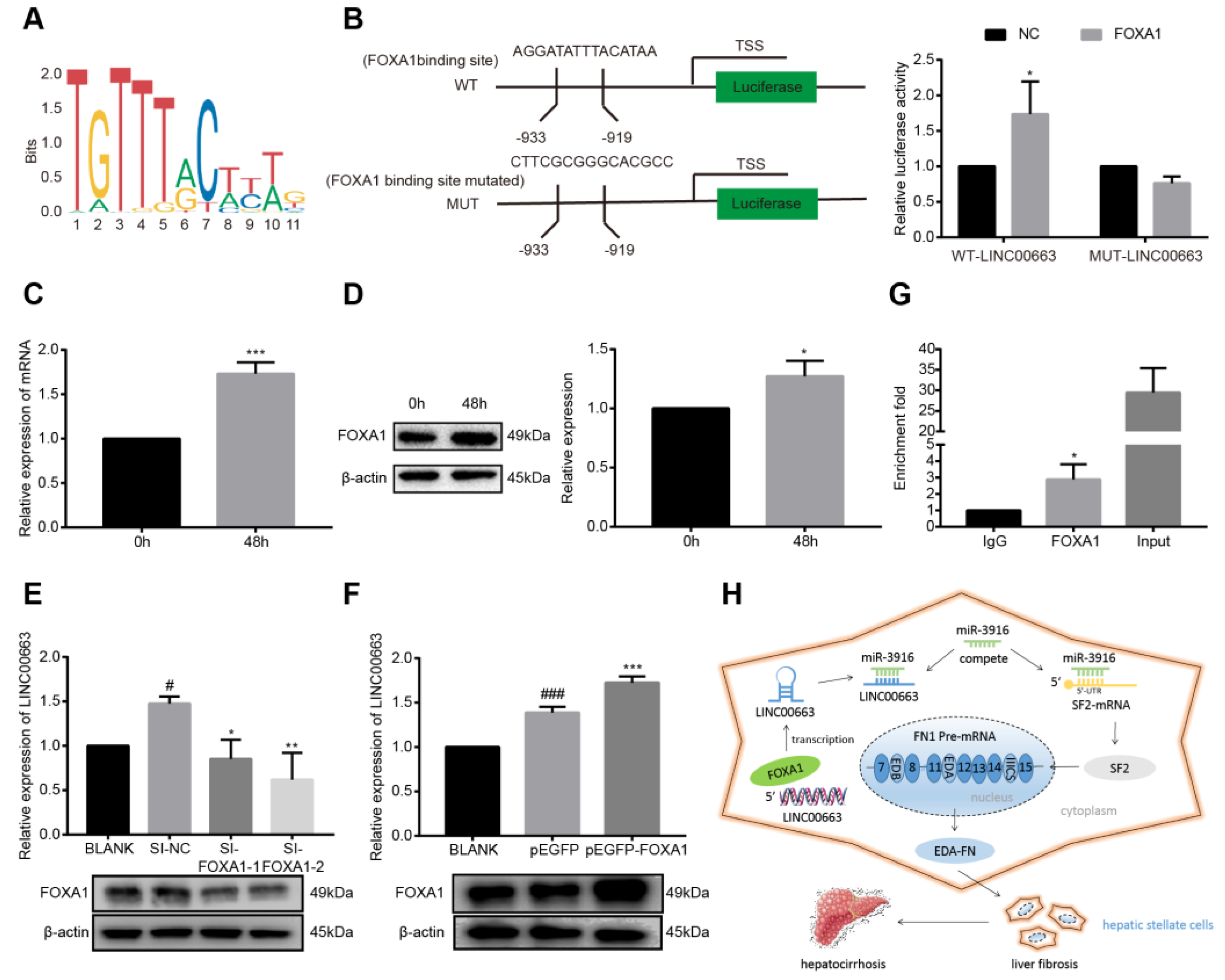
3.11. LINC00663 Regulated by the Transcription Factor FOXA1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Teran-Hinojosa, E.; Sobral, H.; Sánchez-Pérez, C.; Pérez-García, A.; Alemán-García, N.; Hernández-Ruiz, J. Differentiation of fibrotic liver tissue using laser-induced breakdown spectroscopy. Biomed. Opt. Express 2017, 8, 3816–3827. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, I.; Caballero-Díaz, D. Transforming Growth Factor-β-Induced Cell Plasticity in Liver Fibrosis and Hepatocarcinogenesis. Front. Oncol. 2018, 8, 357. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Atzori, L.; Poli, G.; Perra, A. Hepatic stellate cell: A star cell in the liver. Int. J. Biochem. Cell Biol. 2009, 41, 1639–1642. [Google Scholar] [CrossRef] [PubMed]
- Minato, Y.; Hasumura, Y.; Takeuchi, J. The Role of Fat-Storing Cells in Disse Space Fibrogenesis in Alcoholic Liver Disease. Hepatology 1983, 3, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, C.R. Hepatic stellate cell activation and pro-fibrogenic signals. J. Hepatol. 2017, 67, 1104–1105. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Shang, M.; Shi, M.; Zhao, L.; Lin, Z.; Chen, T.; Wu, Y.; Tang, Z.; Sun, H.; Yu, J.; et al. Clonorchis sinensis lysophospholipase inhibits TGF-β1-induced expression of pro-fibrogenic genes through attenuating the activations of Smad3, JNK2, and ERK1/2 in hepatic stellate cell line LX-2. Parasitol. Res. 2016, 115, 643–650. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Tamaki, Z.; Wang, W.; Hinchcliff, M.; Hoover, P.; Getsios, S.; White, E.S.; Varga, J. Fibronectin EDA Promotes Chronic Cutaneous Fibrosis Through Toll-Like Receptor Signaling. Sci. Transl. Med. 2014, 6, 232ra50. [Google Scholar] [CrossRef]
- Mòdol, T.; Brice, N.; de Galarreta, M.R.; Garzón, A.G.; Iraburu, M.J.; Martínez-Irujo, J.J.; López-Zabalza, M.J. Fibronectin Peptides as Potential Regulators of Hepatic Fibrosis Through Apoptosis of Hepatic Stellate Cells. J. Cell. Physiol. 2015, 230, 546–553. [Google Scholar] [CrossRef]
- Cramer, P.; Cáceres, J.F.; Cazalla, D.; Kadener, S.; Muro, A.F.; E Baralle, F.; Kornblihtt, A.R. Coupling of Transcription with Alternative Splicing: RNA Pol II Promoters Modulate SF2/ASF and 9G8 Effects on an Exonic Splicing Enhancer. Mol. Cell 1999, 4, 251–258. [Google Scholar] [CrossRef]
- White, E.S.; Sagana, R.L.; Booth, A.J.; Yan, M.; Cornett, A.M.; Bloomheart, C.A.; Tsui, J.L.; Wilke, C.A.; Moore, B.B.; Ritzenthaler, J.D.; et al. Control of fibroblast fibronectin expression and alternative splicing via the PI3K/Akt/mTOR pathway. Exp. Cell Res. 2010, 316, 2644–2653. [Google Scholar] [CrossRef] [PubMed]
- Lavigueur, A.; La Branche, H.; Kornblihtt, A.; Chabot, B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993, 7, 2405–2417. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yan, I.; Haga, H.; Patel, T. Long noncoding RNA in liver diseases. Hepatology 2014, 60, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Tang, L.; Zhang, Z.; Li, S.; Liang, S.; Ji, L.; Yang, B.; Liu, Y.; Wei, W. Long Noncoding RNA TUG1/miR-29c Axis Affects Cell Proliferation, Invasion, and Migration in Human Pancreatic Cancer. Dis. Markers 2018, 2018, 6857042. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Wang, P.; Yang, T.; Li, G.; Teng, X.; Huang, W.; Yu, H. Identification of key modules and genes associated with breast cancer prognosis using WGCNA and ceRNA network analysis. Aging 2020, 13, 2519–2538. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2–27. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Chang, T.-H.; Huang, H.-Y.; Hsu, J.B.-K.; Weng, S.-L.; Horng, J.-T.; Huang, H.-D. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinform. 2013, 14, S4. [Google Scholar] [CrossRef]
- Bossi, L.; Figueroa-Bossi, N. Competing endogenous RNAs: A target-centric view of small RNA regulation in bacteria. Nat. Rev. Genet. 2016, 14, 775–784. [Google Scholar] [CrossRef]
- Messeguer, X.; Escudero, R.; Farre, D.; Núñez, O.; Martínez, J.; Albà, M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef]
- Farré, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Roselló, L.; Albà, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R.B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Pérez, N.M.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef]
- Song, J.; Shu, H.; Zhang, L.; Xiong, J. Long noncoding RNA GAS5 inhibits angiogenesis and metastasis of colorectal cancer through the Wnt/β-catenin signaling pathway. J. Cell. Biochem. 2019. [Google Scholar] [CrossRef]
- Zhang, K.; Han, X.; Zhang, Z.; Zheng, L.; Hu, Z.; Yao, Q.; Cui, H.; Shu, G.; Si, M.; Li, C.; et al. The liver-enriched lnc-LFAR1 promotes liver fibrosis by activating TGFβ and Notch pathways. Nat. Commun. 2017, 8, 144. [Google Scholar] [CrossRef]
- Trivedi, P.J.; Hirschfield, G.M. Recent advances in clinical practice: Epidemiology of autoimmune liver diseases. Gut 2021, 70, 1989–2003. [Google Scholar] [CrossRef]
- Chinese Society of Hepatology. Guidelines on the management of cholestasis liver diseases (2021). Chin. J. Hepatol. 2021, 60, 1075–1087. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Guo, Y.; Chen, B.; Shi, L.; Dong, P.; Zhou, M.; Zheng, J. LincRNA-p21 Inhibits the Wnt/β-Catenin Pathway in Activated Hepatic Stellate Cells via Sponging MicroRNA-17-5p. Cell. Physiol. Biochem. 2017, 41, 1970–1980. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zheng, J.; Mao, Y.; Dong, P.; Lu, Z.; Li, G.; Guo, C.; Liu, Z.; Fan, X. Long Non-coding RNA Growth Arrest-specific Transcript 5 (GAS5) Inhibits Liver Fibrogenesis through a Mechanism of Competing Endogenous RNA. J. Biol. Chem. 2015, 290, 28286–28298. [Google Scholar] [CrossRef]
- Barsotti, A.M.; Beckerman, R.; Laptenko, O.; Huppi, K.; Caplen, N.J.; Prives, C. p53-Dependent induction of PVT1 and miR-1204*. J. Biol. Chem. 2012, 287, 2509–2519. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Shi, J.; Yin, S.; Meng, H.; He, C.; Wang, Y. The Effect and Mechanism of LINC00663 on the Biological Behavior of Glioma. Neurochem. Res. 2021, 46, 1737–1746. [Google Scholar] [CrossRef]
- Subramaniam, S.; Jeet, V.; Gunter, J.; Clements, J.; Batra, J. Allele-Specific MicroRNA-Mediated Regulation of a Glycolysis Gatekeeper PDK1 in Cancer Metabolism. Cancers 2021, 13, 3582. [Google Scholar] [CrossRef]
- Larsen, A.-C.; Mikkelsen, L.H.; Borup, R.; Kiss, K.; Toft, P.B.; von Buchwald, C.; Coupland, S.; Prause, J.U.; Heegaard, S. MicroRNA Expression Profile in Conjunctival Melanoma. Investig. Opthalmology Vis. Sci. 2016, 57, 4205–4212. [Google Scholar] [CrossRef]
- Ventura, E.; Weller, M.; Macnair, W.; Eschbach, K.; Beisel, C.; Cordazzo, C.; Claassen, M.; Zardi, L.; Burghardt, I. TGF-β induces oncofetal fibronectin, which in turn modulates TGF-β superfamily signaling in endothelial cells. J. Cell Sci. 2018, 131, jcs209619. [Google Scholar] [CrossRef] [PubMed]
- Wight, T.N.; Potter-Perigo, S. The extracellular matrix: An active or passive player in fibrosis? Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G950–G955. [Google Scholar] [CrossRef] [PubMed]
- Klingberg, F.; Chau, G.; Walraven, M.; Boo, S.; Koehler, A.; Chow, M.L.; Olsen, A.L.; Im, M.; Lodyga, M.; Wells, R.G.; et al. The ED-A domain enhances the capacity of fibronectin to store latent TGF-β binding protein-1 in the fibroblast matrix. J. Cell Sci. 2018, 131, jcs201293. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. Fibroblast—Extracellular Matrix Interactions in Tissue Fibrosis. Curr. Pathobiol. Rep. 2016, 4, 11–18. [Google Scholar] [CrossRef]
- Zhou, Z.; Fu, X.-D. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 2013, 122, 191–207. [Google Scholar] [CrossRef]
- Karni, R.; De Stanchina, E.; Lowe, S.W.; Sinha, R.; Mu, D.; Krainer, A.R. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 2007, 14, 185–193. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef]
- Friedman, J.; Kaestner, K.H. The Foxa family of transcription factors in development and metabolism. Cell. Mol. Life Sci. 2006, 63, 2317–2328. [Google Scholar] [CrossRef]
- Li, Z.; White, P.; Tuteja, G.; Rubins, N.; Sackett, S.; Kaestner, K.H. Foxa1 and Foxa2 regulate bile duct development in mice. J. Clin. Investig. 2009, 119, 1537–1545. [Google Scholar] [CrossRef]
- Song, B.; Park, S.-H.; Zhao, J.C.; Fong, K.-W.; Li, S.; Lee, Y.; Yang, Y.A.; Sridhar, S.; Lu, X.; Abdulkadir, S.A.; et al. Targeting FOXA1-mediated repression of TGF-β signaling suppresses castration-resistant prostate cancer progression. J. Clin. Investig. 2019, 129, 569–582. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, Y.-S.; Wang, D.-L.; Yang, B.; Yan, H.-Y.; Lin, L.-H.; Li, Y.; Chen, J.; Xie, L.-M.; Liao, J.-Y.; et al. LncRNA DSCAM-AS1 interacts with YBX1 to promote cancer progression by forming a positive feedback loop that activates FOXA1 transcription network. Theranostics 2020, 10, 10823–10837. [Google Scholar] [CrossRef] [PubMed]




| Genes | Sequence (5′-3′) |
|---|---|
| LINC00663 | F: GCAGCGATGATGACCGTAA |
| R: GGTGTTCTCAACTCATCCAC | |
| α-SMA | F: GGCAAGTGATCACCATCGGA |
| R: GTGGTTTCATGGATGCCAGC | |
| COL1A1 | F: GAGGGCCAAGACGAAGACATC |
| R: CAGATCACGTCATCGCACAAC | |
| FN | F: GGTGACACTTATGAGCGTCCTAAA |
| R: AACATGTAACCACCAGTCTCATGTG | |
| EDA-FN | F: GGAGAGAGTCAGCCTCTGGTTCAG |
| R: TGTCCACTGGGCGCTCAGGCTTGTG | |
| SF2 | F: TGTCGGGAGGTGGTGTGATTCGT |
| R: CTTGGTTCGGATGTCTGGAGGTAAGTT | |
| β-actin | F: AACTGGGACGACATGGAGAAAA |
| R: GGATAGCACAGCCTGGATAGCAA | |
| hsa-miR-3916 | F: CGAAGAGGAAGAAATGGCTGGT |
| R: AGTGCAGGGTCCGAGGTATT | |
| RT primer: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTGAGA | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT |
| Targets | Sequence (5′-3′) |
|---|---|
| LINC00663-1 | F: TTGGACTTGTGTAGGCTTGTAG |
| R: GGTATAGGCATTGGATCAACATTC | |
| LINC00663-2 | F: GTGAGAGAGTTTGCATGACATCT |
| R: TGAGCAGGAGGAAGTGTG | |
| SF2 | F: CAATTGGCTATTGCCGTTT |
| R: CTGATAGACCCAGTTAGAATCT |
| Hub Gene | Full Name | Degree |
|---|---|---|
| FN | fibronectin | 23 |
| Alb | albumin | 21 |
| Mki67 | marker of proliferation Ki-67 | 16 |
| CDK1 | cyclin dependent kinase 1 | 15 |
| Birc5 | baculoviral IAP repeat containing 5 | 12 |
| Mad2l1 | mitotic arrest deficient 2 like 1 | 11 |
| Prc1 | protein regulator of cytokinesis 1 | 11 |
| Group | ALT (U/L) | AST (U/L) |
|---|---|---|
| Sham | 75.0 ± 34.43 | 182.4 ± 74.56 |
| BDL | 284.3 ± 80.21 #### | 427.9 ± 57.24 #### |
| BDL + pcDNA | 343.9 ± 78.34 | 488.6 ± 69.99 |
| BDL + pcDNA-LIN | 477.3 ± 139.2 * | 594.1 ± 101.1 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, Y.; Bao, L.; Teng, Y.; Yuan, B.; Ma, L.; Liu, Y.; Kang, H. The Fibrotic Effects of LINC00663 in Human Hepatic Stellate LX-2 Cells and in Bile Duct-Ligated Cholestasis Mice Are Mediated through the Splicing Factor 2-Fibronectin. Cells 2023, 12, 215. https://doi.org/10.3390/cells12020215
Chu Y, Bao L, Teng Y, Yuan B, Ma L, Liu Y, Kang H. The Fibrotic Effects of LINC00663 in Human Hepatic Stellate LX-2 Cells and in Bile Duct-Ligated Cholestasis Mice Are Mediated through the Splicing Factor 2-Fibronectin. Cells. 2023; 12(2):215. https://doi.org/10.3390/cells12020215
Chicago/Turabian StyleChu, Yang, Linan Bao, Yun Teng, Bo Yuan, Lijie Ma, Ying Liu, and Hui Kang. 2023. "The Fibrotic Effects of LINC00663 in Human Hepatic Stellate LX-2 Cells and in Bile Duct-Ligated Cholestasis Mice Are Mediated through the Splicing Factor 2-Fibronectin" Cells 12, no. 2: 215. https://doi.org/10.3390/cells12020215
APA StyleChu, Y., Bao, L., Teng, Y., Yuan, B., Ma, L., Liu, Y., & Kang, H. (2023). The Fibrotic Effects of LINC00663 in Human Hepatic Stellate LX-2 Cells and in Bile Duct-Ligated Cholestasis Mice Are Mediated through the Splicing Factor 2-Fibronectin. Cells, 12(2), 215. https://doi.org/10.3390/cells12020215





