Calcitonin Related Polypeptide Alpha Mediates Oral Cancer Pain
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Multiplex Immunofluorescence Imaging of Cancer Innervation
2.2.1. Generation of Mouse Orthotopic Xenografts
2.2.2. Multiplex Immunofluorescence Staining and Imaging of Mouse Orthotopic Xenografts
2.3. Quantitative Real-Time RT-PCR (qRT-PCR) Analysis
2.4. Sciatic Nerve Ligation and Behavior Assays in Cancer Models
2.4.1. Sciatic Nerve Ligation Model
2.4.2. Facial Mechanical Nociception Assay in the Tongue Orthotopic Xenograft Cancer Model
2.4.3. Mechanical Nociception Assay in the Hind Paw Xenograft Cancer Model
2.5. Single-Cell RNA-Sequencing (scRNA-Seq) of Oral Cancer Cells and Oral Cancer Xenografts
2.5.1. Preparation of Single-Cell Samples from Cultured Cells
2.5.2. Preparation of Single-Cell Samples from Orthotopic Xenografts
2.5.3. 10× Genomics Library Preparation and Sequencing
2.6. Statistical Analysis
3. Results
3.1. Oral Cancer Xenografts Are Innervated by CGRP-Expressing Neurons
3.2. CGRP Is Transported Anterograde along the Nerve Innervating the Cancer
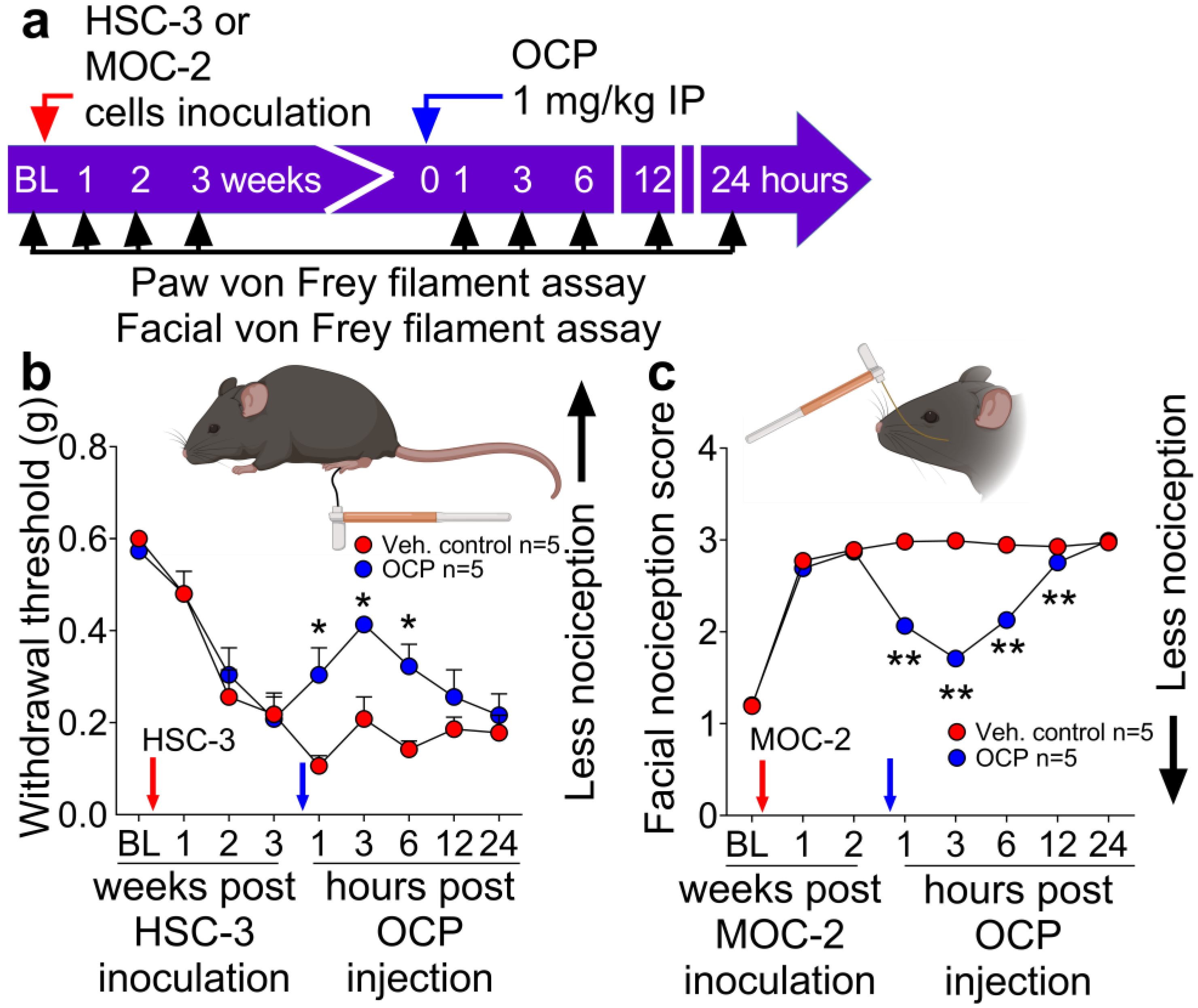
3.3. The CGRP Receptor Antagonist, Olcegepant, Reduces Nociception in Mice with Human or Mouse Oral Cancer Xenografts or Orthotopic Allografts
3.4. Expression of CGRP Receptor Components in Oral Cancer Cells and Orthotopic Xenografts
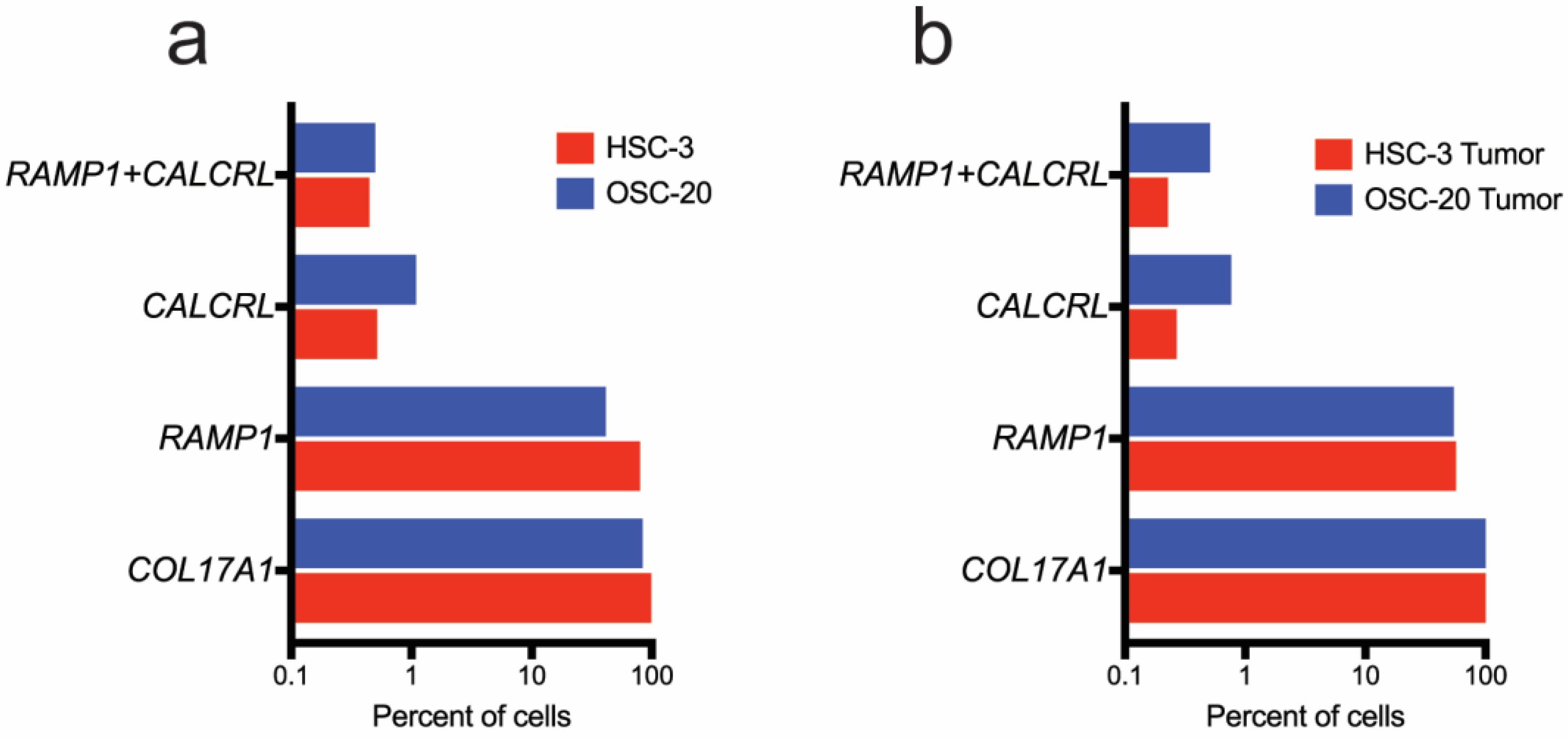
3.4.1. Oral Cancer Cells in Culture Express RAMP1 and CALCRL
3.4.2. Ramp1- and Calcrl-Expressing Cells Are Enriched in Fibroblasts and Immune Cells in the Xenograft Microenvironment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Everdingen, M.V.D.B.-V.; de Rijke, J.; Kessels, A.; Schouten, H.; van Kleef, M.; Patijn, J. Prevalence of pain in patients with cancer: A systematic review of the past 40 years. Ann. Oncol. 2007, 18, 1437–1449. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, B.L.; Pickering, V.; Liu, S.; Quang, P.; Dolan, J.; Connelly, S.T.; Jordan, R.C. Peripheral endothelin A receptor antagonism attenuates carcinoma-induced pain. Eur. J. Pain 2007, 11, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Kolokythas, A.; Connelly, S.T.; Schmidt, B.L. Validation of the University of California San Francisco Oral Cancer Pain Questionnaire. J. Pain 2007, 8, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Raju, B.; Haug, S.R.; Ibrahim, S.O.; Heyeraas, K.J. Sympathectomy decreases size and invasiveness of tongue cancer in rats. Neuroscience 2007, 149, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Magnon, C.; Hall, S.J.; Lin, J.; Xue, X.; Gerber, L.; Freedland, S.J.; Frenette, P.S. Autonomic nerve development contributes to prostate cancer progression. Science 2013, 341, 1236361. [Google Scholar] [CrossRef]
- Kamiya, A.; Hayama, Y.; Kato, S.; Shimomura, A.; Shimomura, T.; Irie, K.; Kaneko, R.; Yanagawa, Y.; Kobayashi, K.; Ochiya, T. Genetic manipulation of autonomic nerve fiber innervation and activity and its effect on breast cancer progression. Nat. Neurosci. 2019, 22, 1289–1305. [Google Scholar] [CrossRef]
- Madeo, M.; Colbert, P.L.; Vermeer, D.W.; Lucido, C.T.; Cain, J.T.; Vichaya, E.G.; Grossberg, A.J.; Muirhead, D.; Rickel, A.P.; Hong, Z.; et al. Cancer exosomes induce tumor innervation. Nat. Commun. 2018, 9, 4284. [Google Scholar] [CrossRef]
- Amit, M.; Takahashi, H.; Dragomir, M.P.; Lindemann, A.; Gleber-Netto, F.O.; Pickering, C.R.; Anfossi, S.; Osman, A.A.; Cai, Y.; Wang, R.; et al. Loss of p53 drives neuron reprogramming in head and neck cancer. Nature 2020, 578, 449–454. [Google Scholar] [CrossRef]
- McIlvried, L.A.; Atherton, M.A.; Horan, N.L.; Goch, T.N.; Scheff, N.N. Sensory Neurotransmitter Calcitonin Gene-Related Peptide Modulates Tumor Growth and Lymphocyte Infiltration in Oral Squamous Cell Carcinoma. Adv. Biol. 2022, 6, e2200019. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, C.; Liu, Z.; Sun, Y.; Chen, M.; Guo, Y.; Liu, W.; Zhang, C.; Chen, W.; Sun, J.; et al. Cancer cells co-opt nociceptive nerves to thrive in nutrient-poor environments and upon nutrient-starvation therapies. Cell Metab. 2022, 34, 1999–2017.e1910. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, M.; Liu, Z.; Wang, X.; Ji, T. The neuropeptide calcitonin gene-related peptide links perineural invasion with lymph node metastasis in oral squamous cell carcinoma. BMC Cancer 2021, 21, 1254. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Janal, M.N.; Veeramachaneni, R.; Dolgalev, I.; Dubeykovskaya, Z.; Tu, N.H.; Kim, H.; Zhang, S.; Wu, A.K.; Hagiwara, M.; et al. Oncogenes overexpressed in metastatic oral cancers from patients with pain: Potential pain mediators released in exosomes. Sci. Rep. 2020, 10, 14724. [Google Scholar] [CrossRef]
- Harriott, A.M.; Gold, M.S.; Guo, Z.; Liu, P.; Ren, F.; Cao, Y.-Q.; Scheff, N.N. Electrophysiological properties of dural afferents in the absence and presence of inflammatory mediators. J. Neurophysiol. 2009, 101, 3126–3134. [Google Scholar] [CrossRef]
- Mohanty, D.; Lippmann, S. CGRP Inhibitors for Migraine. Innov. Clin. Neurosci. 2020, 17, 39–40. [Google Scholar]
- Judd, N.P.; Winkler, A.E.; Murillo-Sauca, O.; Brotman, J.J.; Law, J.H.; Lewis, J.S., Jr.; Dunn, G.P.; Bui, J.D.; Sunwoo, J.B.; Uppaluri, R. ERK1/2 regulation of CD44 modulates oral cancer aggressiveness. Cancer Res. 2012, 72, 365–374. [Google Scholar] [CrossRef]
- Decosterd, I.; Woolf, C.J. Spared nerve injury: An animal model of persistent peripheral neuropathic pain. Pain 2000, 87, 149–158. [Google Scholar] [CrossRef]
- Tu, N.H.; Katano, T.; Matsumura, S.; Pham, V.M.; Muratani, T.; Minami, T.; Ito, S. Role of c-Jun N-terminal kinase in late nerve regeneration monitored by in vivo imaging of thy1-yellow fluorescent protein transgenic mice. Eur. J. Neurosci. 2016, 43, 548–560. [Google Scholar] [CrossRef]
- Tu, N.H.; Inoue, K.; Chen, E.; Anderson, B.M.; Sawicki, C.M.; Scheff, N.N.; Tran, H.D.; Kim, D.H.; Alemu, R.G.; Yang, L.; et al. Cathepsin S Evokes PAR(2)-Dependent Pain in Oral Squamous Cell Carcinoma Patients and Preclinical Mouse Models. Cancers 2021, 13, 4697. [Google Scholar] [CrossRef]
- Ye, Y.; Bae, S.S.; Viet, C.T.; Troob, S.; Bernabé, D.; Schmidt, B.L. IB4(+) and TRPV1(+) sensory neurons mediate pain but not proliferation in a mouse model of squamous cell carcinoma. Behav. Brain Funct. 2014, 10, 5. [Google Scholar] [CrossRef]
- Ye, Y.; Dang, D.; Zhang, J.; Viet, C.T.; Lam, D.K.; Dolan, J.C.; Gibbs, J.L.; Schmidt, B.L. Nerve growth factor links oral cancer progression, pain, and cachexia. Mol. Cancer Ther. 2011, 10, 1667–1676. [Google Scholar] [CrossRef]
- Chaplan, S.R.; Bach, F.W.; Pogrel, J.W.; Chung, J.M.; Yaksh, T.L. Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods 1994, 53, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Pickering, V.; Jay Gupta, R.; Quang, P.; Jordan, R.C.; Schmidt, B.L. Effect of peripheral endothelin-1 concentration on carcinoma-induced pain in mice. Eur. J. Pain 2008, 12, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.L.; Petersen, S.; Kristensen, D.M.; Olesen, J.; Munro, G. Targeting CGRP via receptor antagonism and antibody neutralisation in two distinct rodent models of migraine-like pain. Cephalalgia 2019, 39, 1827–1837. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.F.; Hay, D.L. CGRP physiology, pharmacology, and therapeutic targets: Migraine and beyond. Physiol. Rev. 2023, 103, 1565–1644. [Google Scholar] [CrossRef] [PubMed]
- Nagamine, K.; Ozaki, N.; Shinoda, M.; Asai, H.; Nishiguchi, H.; Mitsudo, K.; Tohnai, I.; Ueda, M.; Sugiura, Y. Mechanical allodynia and thermal hyperalgesia induced by experimental squamous cell carcinoma of the lower gingiva in rats. J. Pain 2006, 7, 659–670. [Google Scholar] [CrossRef]
- Balood, M.; Ahmadi, M.; Eichwald, T.; Ahmadi, A.; Majdoubi, A.; Roversi, K.; Roversi, K.; Lucido, C.T.; Restaino, A.C.; Huang, S.; et al. Nociceptor neurons affect cancer immunosurveillance. Nature 2022, 611, 405–412. [Google Scholar] [CrossRef]
- Lorenzen, E.; Dodig-Crnković, T.; Kotliar, I.B.; Pin, E.; Ceraudo, E.; Vaughan, R.D.; Uhlèn, M.; Huber, T.; Schwenk, J.M.; Sakmar, T.P. Multiplexed analysis of the secretin-like GPCR-RAMP interactome. Sci. Adv. 2019, 5, eaaw2778. [Google Scholar] [CrossRef]
- Kotliar, I.B.; Lorenzen, E.; Schwenk, J.M.; Hay, D.L.; Sakmar, T.P. Elucidating the Interactome of G Protein-Coupled Receptors and Receptor Activity-Modifying Proteins. Pharmacol. Rev. 2023, 75, 1–34. [Google Scholar] [CrossRef]
- Yin, S.; Song, R.; Ma, J.; Liu, C.; Wu, Z.; Cao, G.; Liu, J.; Zhang, G.; Zhang, H.; Sun, R.; et al. Receptor activity-modifying protein 1 regulates mouse skin fibroblast proliferation via the Gαi3-PKA-CREB-YAP axis. Cell Commun. Signal. 2022, 20, 52. [Google Scholar] [CrossRef]
- Snijders, A.M.; Schmidt, B.L.; Fridlyand, J.; Dekker, N.; Pinkel, D.; Jordan, R.C.; Albertson, D.G. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene 2005, 24, 4232–4242. [Google Scholar] [CrossRef]
- Snijders, A.M.; Huey, B.; Connelly, S.T.; Roy, R.; Jordan, R.C.; Schmidt, B.L.; Albertson, D.G. Stromal control of oncogenic traits expressed in response to the overexpression of GLI2, a pleiotropic oncogene. Oncogene 2009, 28, 625–637. [Google Scholar] [CrossRef][Green Version]
- Toda, M.; Suzuki, T.; Hosono, K.; Hayashi, I.; Hashiba, S.; Onuma, Y.; Amano, H.; Kurihara, Y.; Kurihara, H.; Okamoto, H.; et al. Neuronal system-dependent facilitation of tumor angiogenesis and tumor growth by calcitonin gene-related peptide. Proc. Natl. Acad. Sci. USA 2008, 105, 13550–13555. [Google Scholar] [CrossRef]
- De Logu, F.; Nassini, R.; Hegron, A.; Landini, L.; Jensen, D.D.; Latorre, R.; Ding, J.; Marini, M.; de Araujo, D.S.M.; Ramírez-Garcia, P.; et al. Schwann cell endosome CGRP signals elicit periorbital mechanical allodynia in mice. Nat. Commun. 2022, 13, 646. [Google Scholar] [CrossRef]
- Kajiwara, I.; Sano, M.; Ichimaru, Y.; Oshima, Y.; Kitajima, O.; Hao, H.; Masamune, A.; Kim, J.; Ishii, Y.; Ijichi, H.; et al. Duloxetine improves cancer-associated pain in a mouse model of pancreatic cancer through stimulation of noradrenaline pathway and its antitumor effects. Pain 2020, 161, 2909–2919. [Google Scholar] [CrossRef]
- Pinho-Ribeiro, F.A.; Deng, L.; Neel, D.V.; Erdogan, O.; Basu, H.; Yang, D.; Choi, S.; Walker, A.J.; Carneiro-Nascimento, S.; He, K.; et al. Bacteria hijack a meningeal neuroimmune axis to facilitate brain invasion. Nature 2023, 615, 472–481. [Google Scholar] [CrossRef]
- Scheff, N.N.; Alemu, R.G.; Klares, R., 3rd; Wall, I.M.; Yang, S.C.; Dolan, J.C.; Schmidt, B.L. Granulocyte-Colony Stimulating Factor-Induced Neutrophil Recruitment Provides Opioid-Mediated Endogenous Anti-nociception in Female Mice with Oral Squamous Cell Carcinoma. Front. Mol. Neurosci. 2019, 12, 217. [Google Scholar] [CrossRef]
- Scheff, N.N.; Bhattacharya, A.; Dowse, E.; Dang, R.X.; Dolan, J.C.; Wang, S.; Kim, H.; Albertson, D.G.; Schmidt, B.L. Neutrophil-Mediated Endogenous Analgesia Contributes to Sex Differences in Oral Cancer Pain. Front. Integr. Neurosci. 2018, 12, 52. [Google Scholar] [CrossRef]
- Olesen, J.; Diener, H.-C.; Husstedt, I.W.; Goadsby, P.J.; Hall, D.; Meier, U.; Pollentier, S.; Lesko, L.M. Calcitonin Gene–Related Peptide Receptor Antagonist BIBN 4096 BS for the Acute Treatment of Migraine. N. Engl. J. Med. 2004, 350, 1104–1110. [Google Scholar] [CrossRef]
- Edvinsson, L.; Haanes, K.A.; Warfvinge, K.; Krause, D.N. CGRP as the target of new migraine therapies—successful translation from bench to clinic. Nat. Rev. Neurol. 2018, 14, 338–350. [Google Scholar] [CrossRef]
- Goadsby, P.J.; Dodick, D.W.; Ailani, J.; Trugman, J.M.; Finnegan, M.; Lu, K.; Szegedi, A. Safety, tolerability, and efficacy of orally administered atogepant for the prevention of episodic migraine in adults: A double-blind, randomised phase 2b/3 trial. Lancet Neurol. 2020, 19, 727–737. [Google Scholar] [CrossRef]
- Yarwood, R.E.; Imlach, W.L.; Lieu, T.; Veldhuis, N.A.; Jensen, D.D.; Klein Herenbrink, C.; Aurelio, L.; Cai, Z.; Christie, M.J.; Poole, D.P.; et al. Endosomal signaling of the receptor for calcitonin gene-related peptide mediates pain transmission. Proc. Natl. Acad. Sci. USA 2017, 114, 12309–12314. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, N.H.; Inoue, K.; Lewis, P.K.; Khan, A.; Hwang, J.H.; Chokshi, V.; Dabovic, B.B.; Selvaraj, S.; Bhattacharya, A.; Dubeykovskaya, Z.; et al. Calcitonin Related Polypeptide Alpha Mediates Oral Cancer Pain. Cells 2023, 12, 1675. https://doi.org/10.3390/cells12131675
Tu NH, Inoue K, Lewis PK, Khan A, Hwang JH, Chokshi V, Dabovic BB, Selvaraj S, Bhattacharya A, Dubeykovskaya Z, et al. Calcitonin Related Polypeptide Alpha Mediates Oral Cancer Pain. Cells. 2023; 12(13):1675. https://doi.org/10.3390/cells12131675
Chicago/Turabian StyleTu, Nguyen Huu, Kenji Inoue, Parker K. Lewis, Ammar Khan, Jun Hyeong Hwang, Varun Chokshi, Branka Brukner Dabovic, Shanmugapriya Selvaraj, Aditi Bhattacharya, Zinaida Dubeykovskaya, and et al. 2023. "Calcitonin Related Polypeptide Alpha Mediates Oral Cancer Pain" Cells 12, no. 13: 1675. https://doi.org/10.3390/cells12131675
APA StyleTu, N. H., Inoue, K., Lewis, P. K., Khan, A., Hwang, J. H., Chokshi, V., Dabovic, B. B., Selvaraj, S., Bhattacharya, A., Dubeykovskaya, Z., Pinkerton, N. M., Bunnett, N. W., Loomis, C. A., Albertson, D. G., & Schmidt, B. L. (2023). Calcitonin Related Polypeptide Alpha Mediates Oral Cancer Pain. Cells, 12(13), 1675. https://doi.org/10.3390/cells12131675





