Nuclear Delivery of Nanoparticle-Based Drug Delivery Systems by Nuclear Localization Signals
Abstract
:1. Introduction
2. Structure of NPCs and Nuclear Import
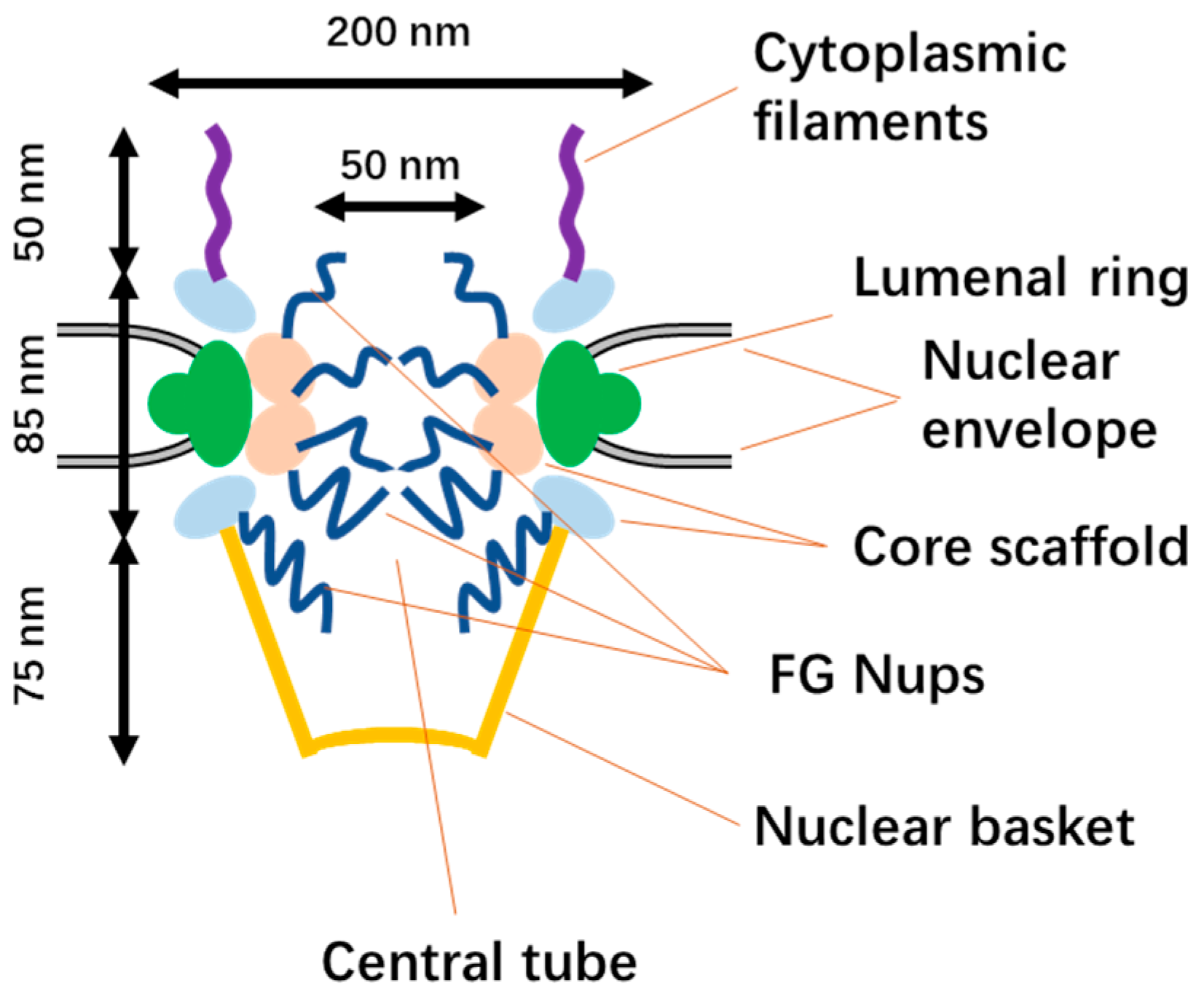
2.1. The Structure of NPCs
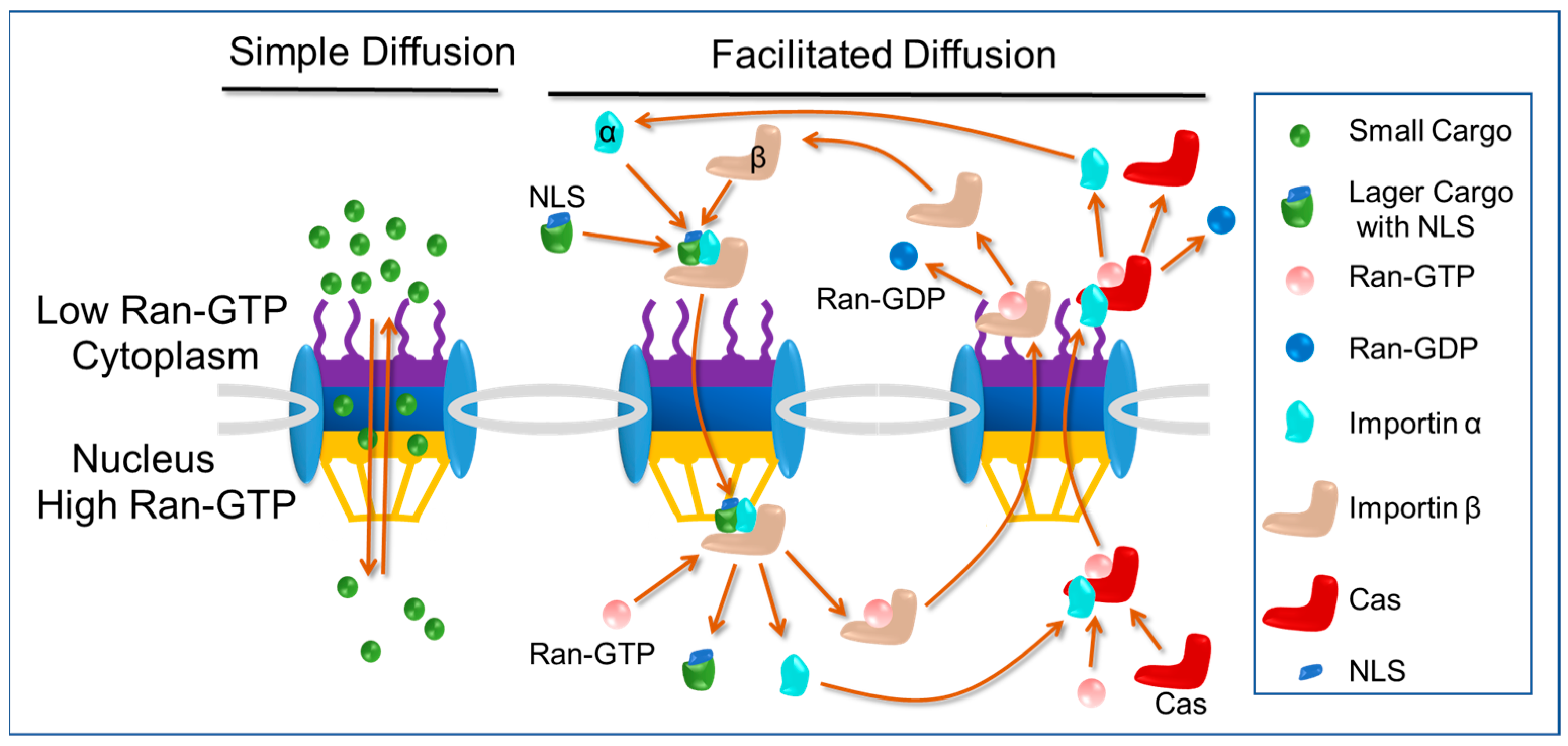
2.2. Nuclear Import Pathways: Free Diffusion and Classical Nuclear Import Pathway
2.3. Classical Nuclear Localization Signals (cNLSs) and Importin α
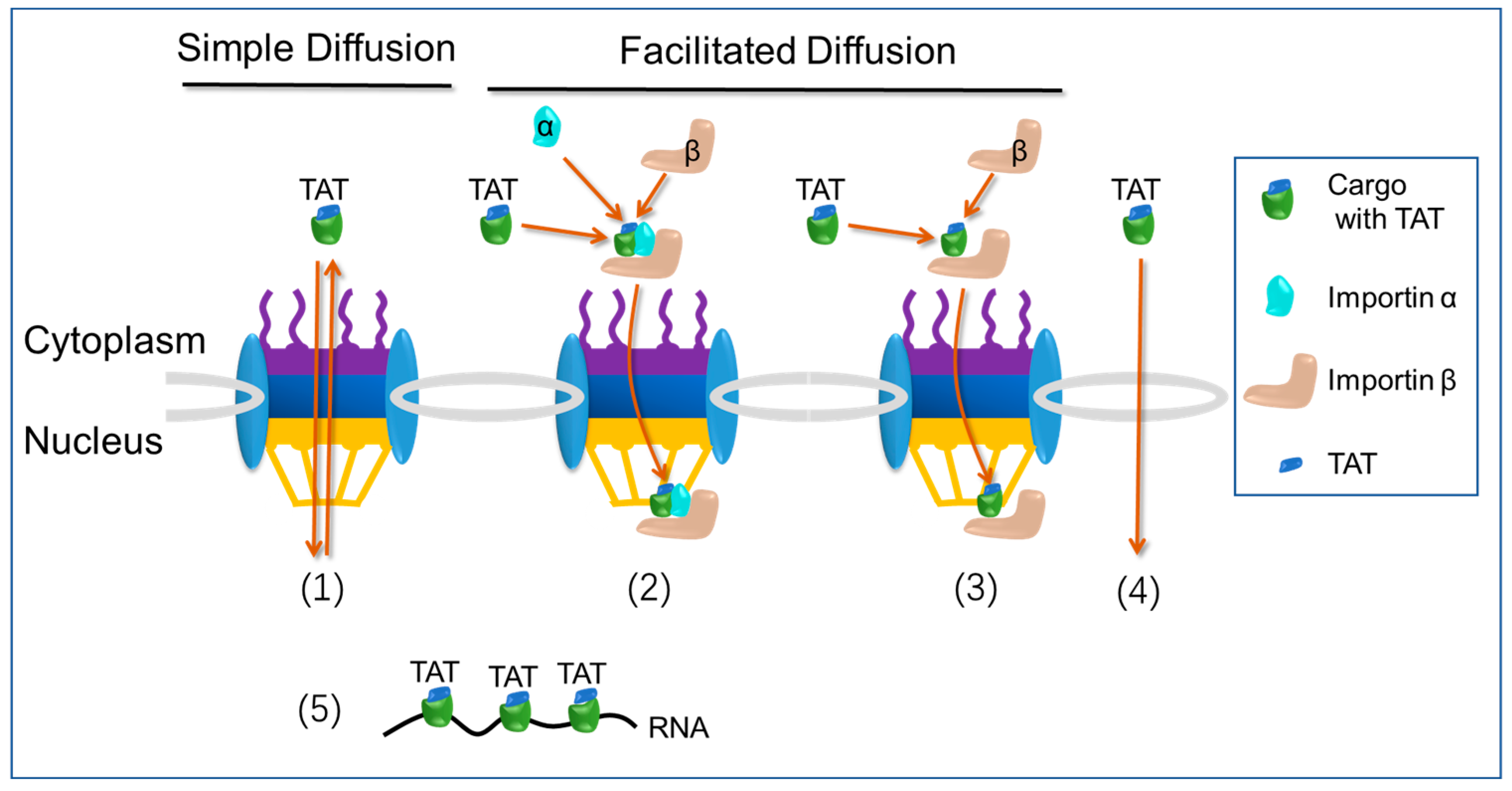
2.4. Nuclear Import Pathways: Transactivator of Transcription (TAT) Pathway
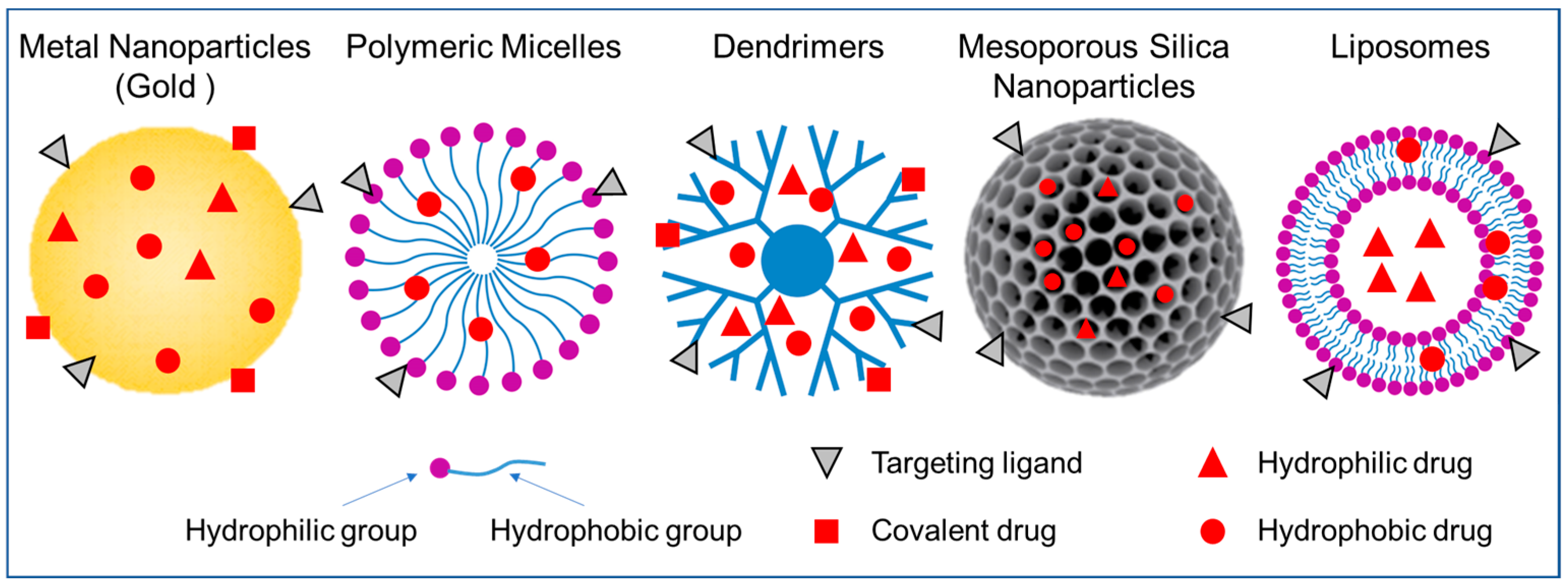
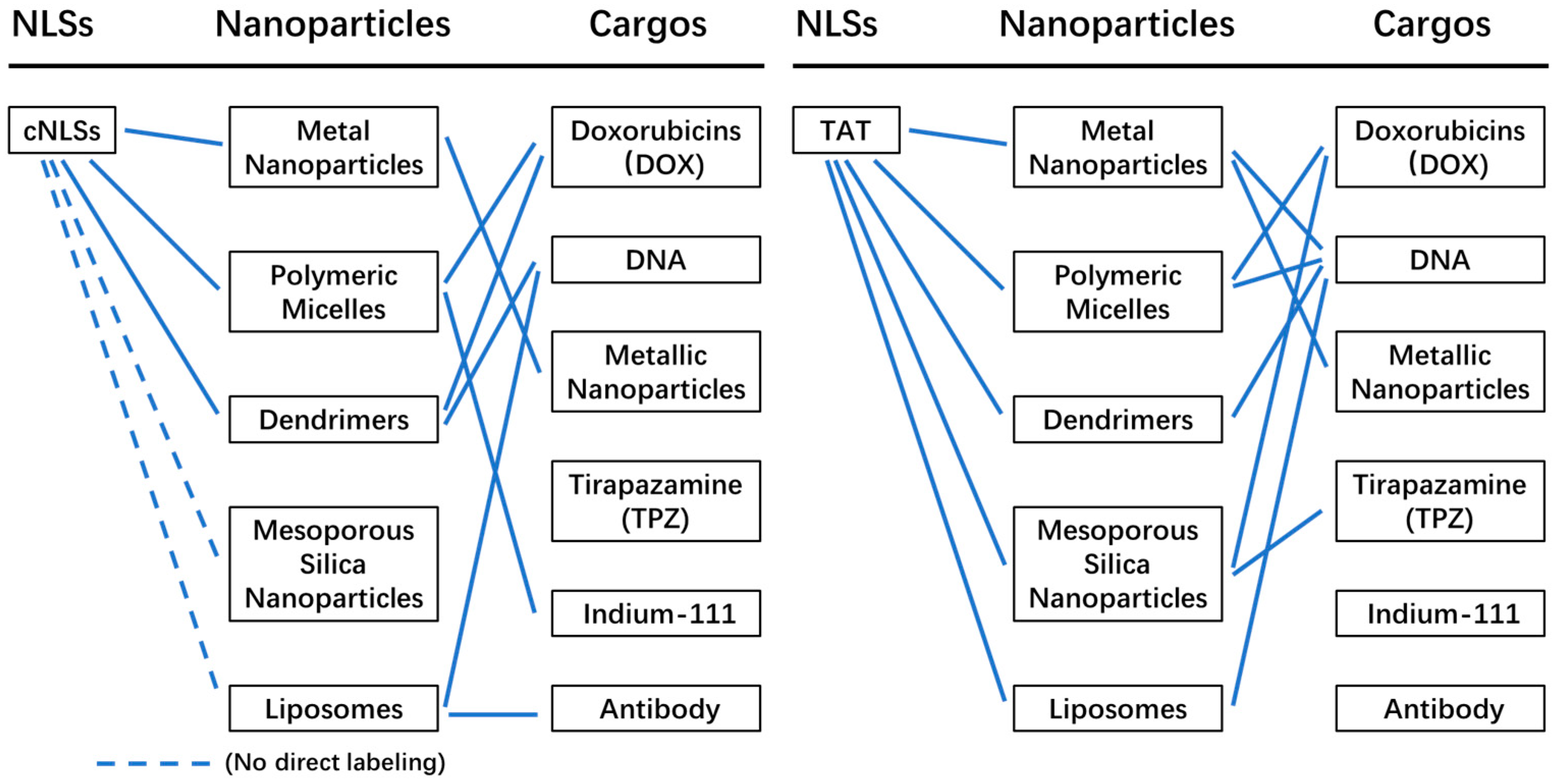
3. Nanoparticles for Nucleus Targeting
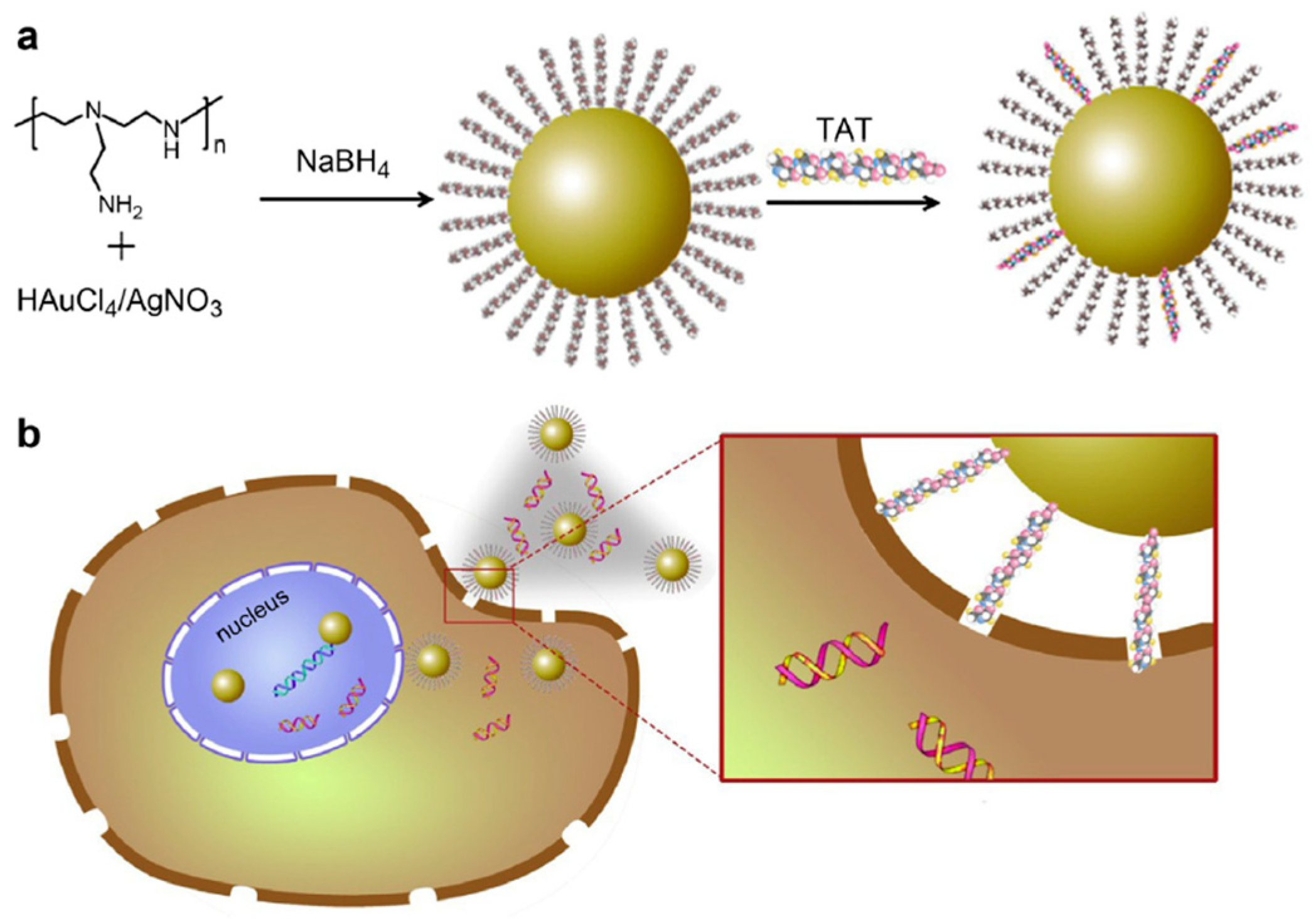
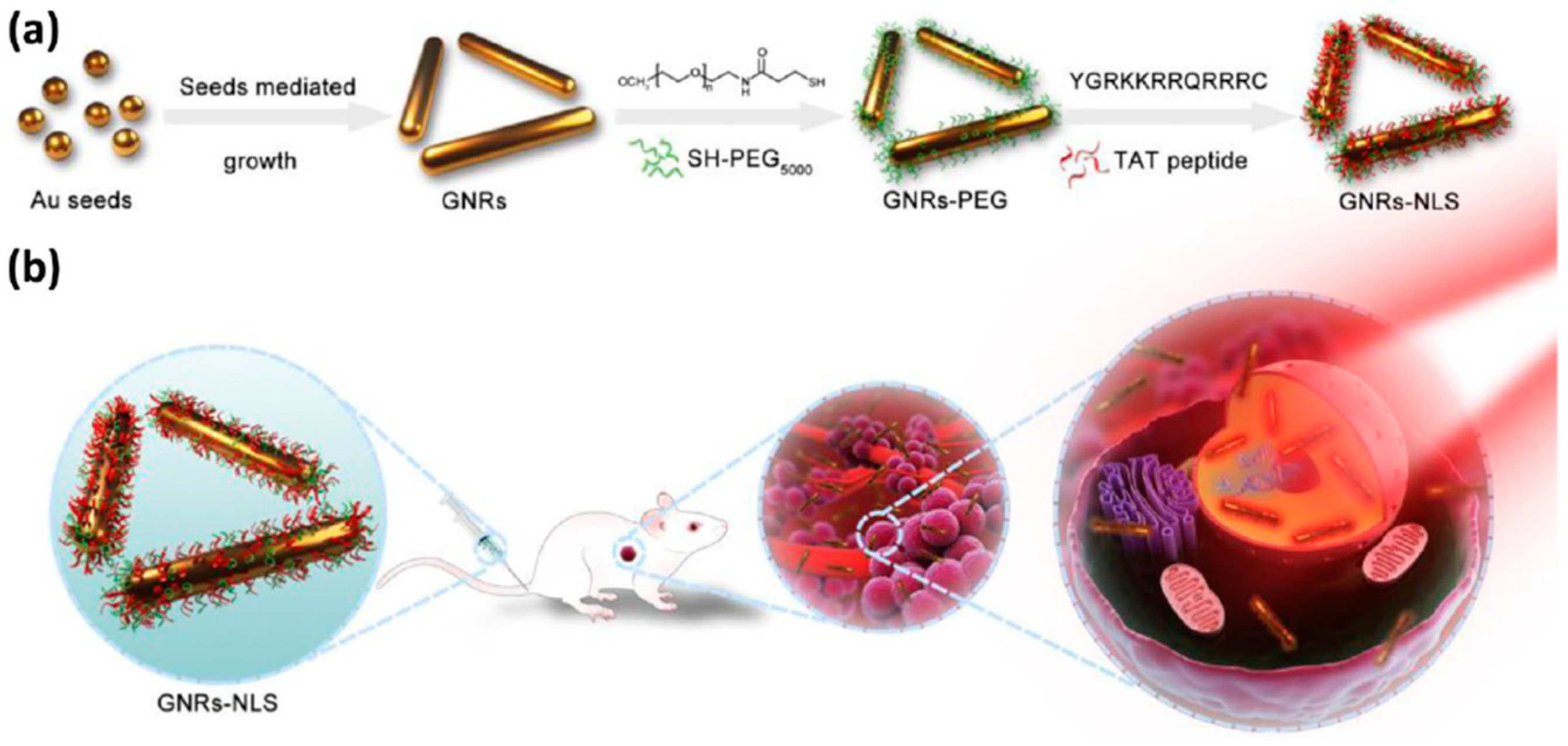
3.1. Metal Nanoparticles
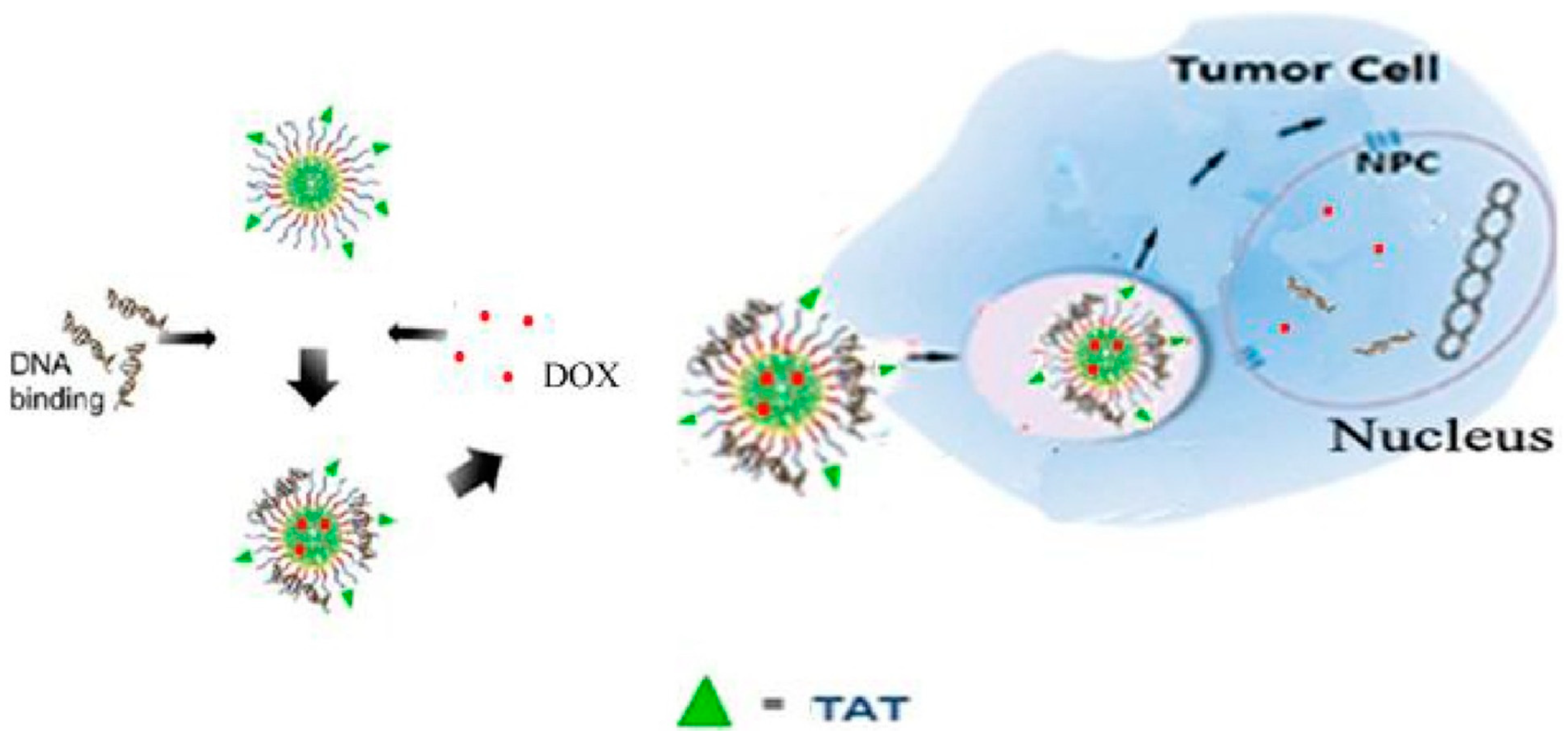
3.2. Polymeric Micelles
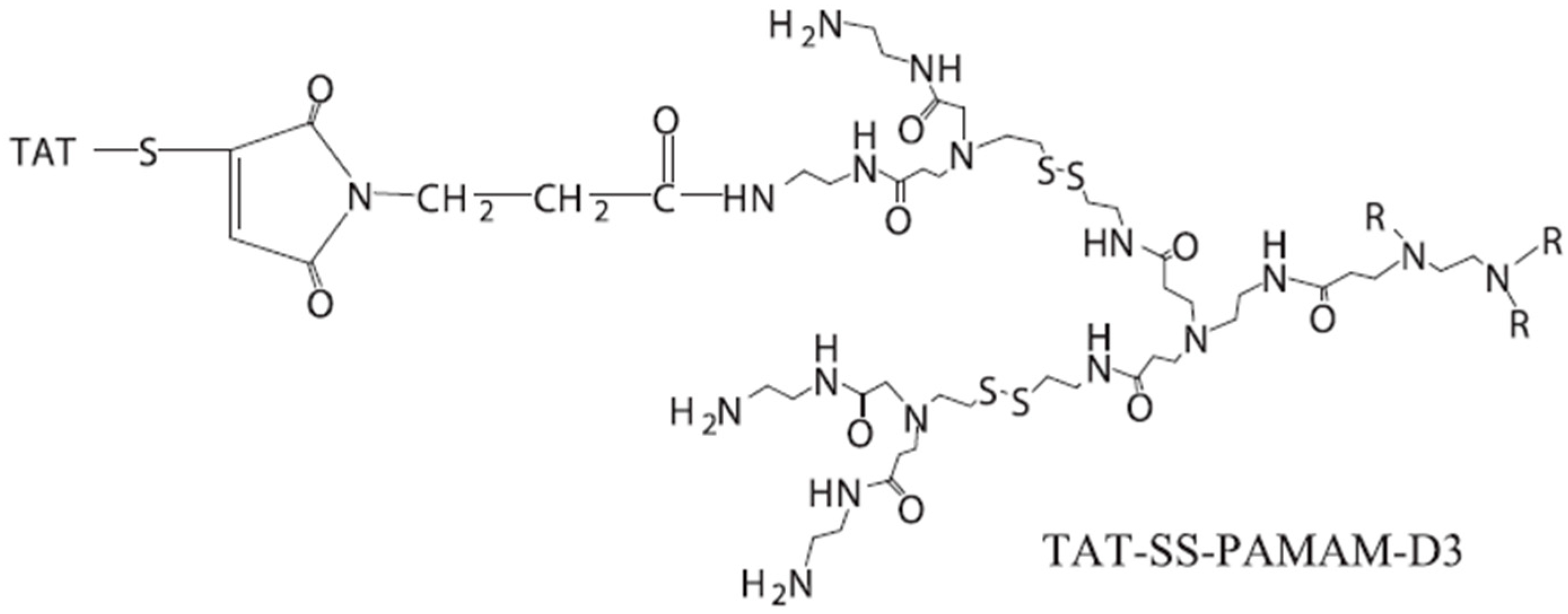
3.3. Dendrimers
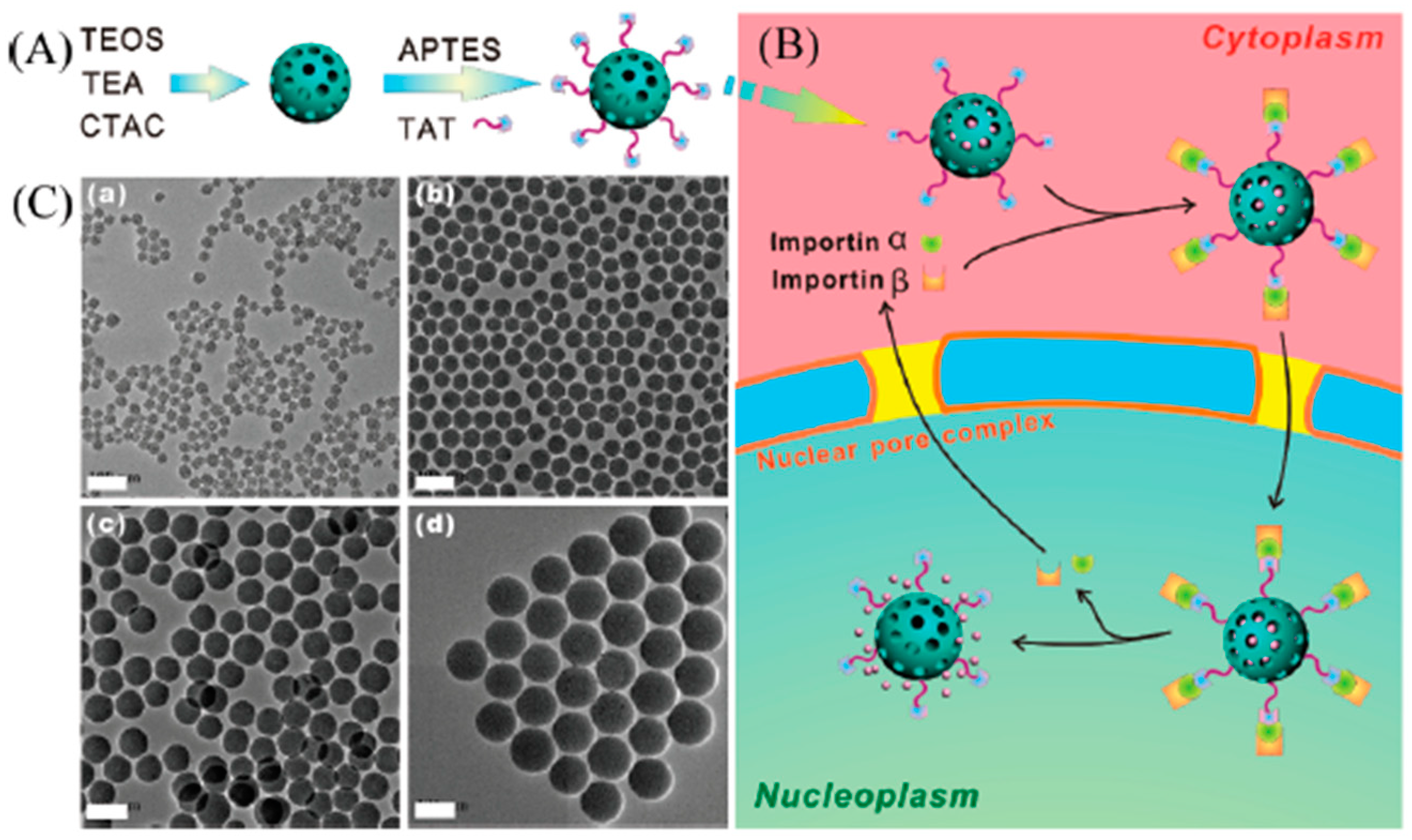
3.4. Mesoporous Silica Nanoparticles
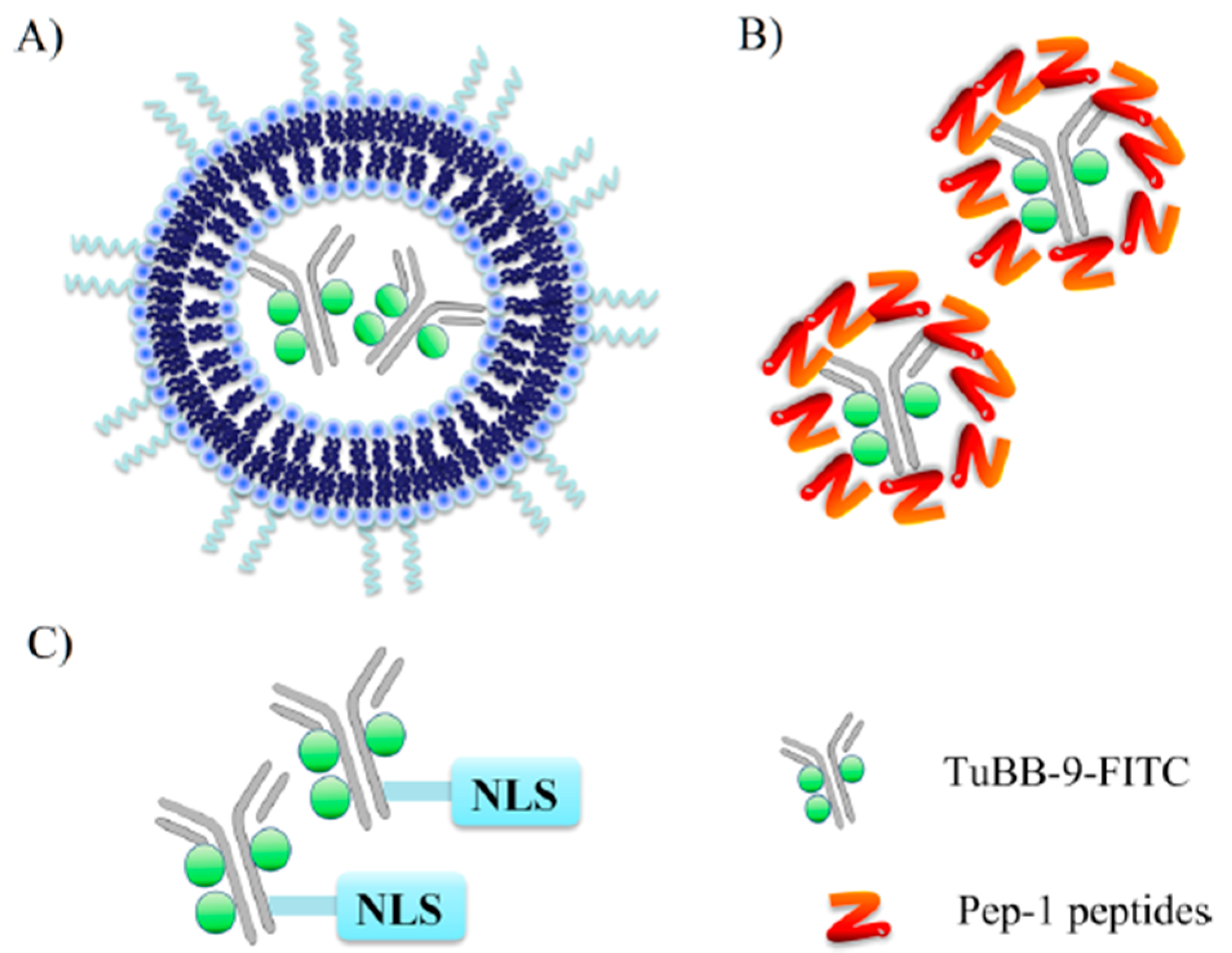
3.5. Liposomes
4. Summary and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fu, X.; Shi, Y.; Qi, T.; Qiu, S.; Huang, Y.; Zhao, X.; Sun, Q.; Lin, G. Precise design strategies of nanomedicine for improving cancer therapeutic efficacy using subcellular targeting. Signal Transduct. Target Ther. 2020, 5, 262. [Google Scholar] [CrossRef]
- Shi, J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17, 20–37. [Google Scholar] [CrossRef] [Green Version]
- Sakhrani, N.M.; Padh, H. Organelle targeting: Third level of drug targeting. Drug Des. Devel. Ther. 2013, 7, 585–599. [Google Scholar] [CrossRef] [Green Version]
- Dam, D.H.; Lee, J.H.; Sisco, P.N.; Co, D.T.; Zhang, M.; Wasielewski, M.R.; Odom, T.W. Direct observation of nanoparticle-cancer cell nucleus interactions. ACS Nano 2012, 6, 3318–3326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.H.; Meta, M.; Qiao, X.; Frost, D.; Bauch, J.; Coffinier, C.; Majumdar, S.; Bergo, M.O.; Young, S.G.; Fong, L.G. A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson-Gilford progeria syndrome mutation. J. Clin. Investig. 2006, 116, 2115–2121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tammam, S.N.; Azzazy, H.M.E.; Lamprecht, A. How successful is nuclear targeting by nanocarriers? J. Control Release 2016, 229, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Kvackajova-Kisucka, J.; Barancik, M.; Breier, A. Drug transporters and their role in multidrug resistance of neoplastic cells. Gen. Physiol. Biophys. 2001, 20, 215–237. [Google Scholar] [PubMed]
- Dai, Y.; Xu, C.; Sun, X.; Chen, X. Nanoparticle design strategies for enhanced anticancer therapy by exploiting the tumour microenvironment. Chem. Soc. Rev. 2017, 46, 3830–3852. [Google Scholar] [CrossRef]
- Du, W.; Zhang, L.; Li, X.; Ling, G.; Zhang, P. Nuclear targeting Subcellular-delivery nanosystems for precise cancer treatment. Int. J. Pharm. 2022, 619, 121735. [Google Scholar] [CrossRef]
- Stoffler, D.; Fahrenkrog, B.; Aebi, U. The nuclear pore complex: From molecular architecture to functional dynamics. Curr. Opin. Cell Biol. 1999, 11, 391–401. [Google Scholar] [CrossRef]
- Stoffler, D.; Goldie, K.N.; Feja, B.; Aebi, U. Calcium-mediated structural changes of native nuclear pore complexes monitored by time-lapse atomic force microscopy. J. Mol. Biol. 1999, 287, 741–752. [Google Scholar] [CrossRef]
- Fahrenkrog, B.; Aebi, U. The nuclear pore complex: Nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell Biol. 2003, 4, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Maimon, T.; Elad, N.; Dahan, I.; Medalia, O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure 2012, 20, 998–1006. [Google Scholar] [CrossRef] [Green Version]
- Stoffler, D.; Feja, B.; Fahrenkrog, B.; Walz, J.; Typke, D.; Aebi, U. Cryo-electron tomography provides novel insights into nuclear pore architecture: Implications for nucleocytoplasmic transport. J. Mol. Biol. 2003, 328, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Tu, L.C.; Zilman, A.; Musser, S.M. Investigating molecular crowding within nuclear pores using polarization-PALM. eLife 2017, 6, 28716. [Google Scholar] [CrossRef] [PubMed]
- Lim, R.Y.; Ullman, K.S.; Fahrenkrog, B. Biology and biophysics of the nuclear pore complex and its components. Int. Rev. Cell Mol. Biol. 2008, 267, 299–342. [Google Scholar] [CrossRef] [Green Version]
- Ori, A.; Banterle, N.; Iskar, M.; Andrés-Pons, A.; Escher, C.; Khanh Bui, H.; Sparks, L.; Solis-Mezarino, V.; Rinner, O.; Bork, P.; et al. Cell type-specific nuclear pores: A case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol. 2013, 9, 648. [Google Scholar] [CrossRef]
- Peleg, O.; Lim, R.Y. Converging on the function of intrinsically disordered nucleoporins in the nuclear pore complex. Biol. Chem. 2010, 391, 719–730. [Google Scholar] [CrossRef] [Green Version]
- Suntharalingam, M.; Wente, S.R. Peering through the pore: Nuclear pore complex structure, assembly, and function. Dev. Cell 2003, 4, 775–789. [Google Scholar] [CrossRef]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [Green Version]
- Moroianu, J. Nuclear import and export: Transport factors, mechanisms and regulation. Crit. Rev. Eukaryot. Gene Expr. 1999, 9, 89–106. [Google Scholar] [CrossRef]
- Kutay, U.; Bischoff, F.R.; Kostka, S.; Kraft, R.; Görlich, D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 1997, 90, 1061–1071. [Google Scholar] [CrossRef] [Green Version]
- Frey, S.; Görlich, D. FG/FxFG as well as GLFG repeats form a selective permeability barrier with self-healing properties. EMBO J. 2009, 28, 2554–2567. [Google Scholar] [CrossRef]
- Hough, L.E.; Dutta, K.; Sparks, S.; Temel, D.B.; Kamal, A.; Tetenbaum-Novatt, J.; Rout, M.P.; Cowburn, D. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. eLife 2015, 4, 10027. [Google Scholar] [CrossRef]
- Lim, R.Y.; Fahrenkrog, B.; Köser, J.; Schwarz-Herion, K.; Deng, J.; Aebi, U. Nanomechanical basis of selective gating by the nuclear pore complex. Science 2007, 318, 640–643. [Google Scholar] [CrossRef] [PubMed]
- Milles, S.; Mercadante, D.; Aramburu, I.V.; Jensen, M.R.; Banterle, N.; Koehler, C.; Tyagi, S.; Clarke, J.; Shammas, S.L.; Blackledge, M.; et al. Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell 2015, 163, 734–745. [Google Scholar] [CrossRef] [Green Version]
- Panté, N.; Kann, M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell 2002, 13, 425–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, M.; Fang, T.; Li, S.; Meng, K.; Guo, D. Bipartite Nuclear Localization Signal Controls Nuclear Import and DNA-Binding Activity of IFN Regulatory Factor 3. J. Immunol. 2015, 195, 289–297. [Google Scholar] [CrossRef] [Green Version]
- Freitas, N.; Cunha, C. Mechanisms and signals for the nuclear import of proteins. Curr. Genom. 2009, 10, 550–557. [Google Scholar] [CrossRef]
- Lu, J.; Wu, T.; Zhang, B.; Liu, S.; Song, W.; Qiao, J.; Ruan, H. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun. Signal 2021, 19, 60. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, K.M.; Jans, D.A. Importins and beyond: Non-conventional nuclear transport mechanisms. Traffic 2009, 10, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.F.; Zhang, H.Y.; Xing, X.; Wang, P.; Yan, J.Q.; Liu, D.S.; Gong, Q.Y.; Zhang, R.S.; Zhang, H.B. Robust strategies in nuclear-targeted cancer therapy based on functional nanomaterials. Mater. Des. 2022, 221, 110999. [Google Scholar] [CrossRef]
- Smith, K.M.; Himiari, Z.; Tsimbalyuk, S.; Forwood, J.K. Structural Basis for Importin-α Binding of the Human Immunodeficiency Virus Tat. Sci. Rep. 2017, 7, 1650. [Google Scholar] [CrossRef] [Green Version]
- Truant, R.; Cullen, B.R. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol. Cell. Biol. 1999, 19, 1210–1217. [Google Scholar] [CrossRef] [Green Version]
- Efthymiadis, A.; Briggs, L.J.; Jans, D.A. The Hiv-1 Tat Nuclear Localization Sequence Confers Novel Nuclear Import Properties. J. Biol. Chem. 1998, 273, 1623–1628. [Google Scholar] [CrossRef] [Green Version]
- Kurnaeva, M.A.; Zalevsky, A.O.; Arifulin, E.A.; Lisitsyna, O.M.; Tvorogova, A.V.; Shubina, M.Y.; Bourenkov, G.P.; Tikhomirova, M.A.; Potashnikova, D.M.; Kachalova, A.I.; et al. Molecular Coevolution of Nuclear and Nucleolar Localization Signals inside the Basic Domain of HIV-1 Tat. J. Virol. 2022, 96, e0150521. [Google Scholar] [CrossRef]
- Cardarelli, F.; Serresi, M.; Bizzarri, R.; Giacca, M.; Beltram, F. In vivo study of HIV-1 Tat arginine-rich motif unveils its transport properties. Mol. Ther. 2007, 15, 1313–1322. [Google Scholar] [CrossRef]
- Cardarelli, F.; Serresi, M.; Albanese, A.; Bizzarri, R.; Beltram, F. Quantitative analysis of Tat peptide binding to import carriers reveals unconventional nuclear transport properties. J. Biol. Chem. 2011, 286, 12292–12299. [Google Scholar] [CrossRef] [Green Version]
- Nitin, N.; LaConte, L.; Rhee, W.J.; Bao, G. Tat peptide is capable of importing large nanoparticles across nuclear membrane in digitonin permeabilized cells. Ann. Biomed. Eng. 2009, 37, 2018–2027. [Google Scholar] [CrossRef] [Green Version]
- Quan, X.; Sun, D.; Zhou, J. Molecular mechanism of HIV-1 TAT peptide and its conjugated gold nanoparticles translocating across lipid membranes. Phys. Chem. Chem. Phys. 2019, 21, 10300–10310. [Google Scholar] [CrossRef] [PubMed]
- Zelmer, C.; Zweifel, L.P.; Kapinos, L.E.; Craciun, I.; Güven, Z.P.; Palivan, C.G.; Lim, R.Y.H. Organelle-specific targeting of polymersomes into the cell nucleus. Proc. Natl. Acad. Sci. USA 2020, 117, 2770–2778. [Google Scholar] [CrossRef] [Green Version]
- Wei, D.; Huang, Y.; Wang, B.; Ma, L.; Karges, J.; Xiao, H. Photo-Reduction with NIR Light of Nucleus-Targeting Pt(IV) Nanoparticles for Combined Tumor-Targeted Chemotherapy and Photodynamic Immunotherapy. Angew. Chem. Int. Ed. Engl. 2022, 61, e202201486. [Google Scholar] [CrossRef] [PubMed]
- Ekozin, A.; Otuechere, C.A.; Adewuyi, A. Apocynin loaded silver nanoparticles displays potent in vitro biological activities and mitigates pyrogallol-induced hepatotoxicity. Chem. Biol. Interact. 2022, 365, 110069. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.Y.; Zhang, J.W.; Li, R.F.; Wang, Z.X.; Wang, W.J.; Wang, W. Unique Roles of Gold Nanoparticles in Drug Delivery, Targeting and Imaging Applications. Molecules 2017, 22, 1445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.X.; Zuo, J.; Li, Q.Q.; Chang, Y.L.; Zhang, Y.L.; Tu, L.P.; Liu, X.M.; Xue, B.; Zhao, H.Y.; Zhang, H.; et al. One-step in situ solid-substrate-based whole blood immunoassay based on FRET between upconversion and gold nanoparticles. Biosens. Bioelectron. 2017, 92, 335–341. [Google Scholar] [CrossRef]
- Duncan, B.; Kim, C.; Rotello, V.M. Gold nanoparticle platforms as drug and biomacromolecule delivery systems. J. Control. Release 2010, 148, 122–127. [Google Scholar] [CrossRef] [Green Version]
- Peng, L.H.; Niu, J.; Zhang, C.Z.; Yu, W.; Wu, J.H.; Shan, Y.H.; Wang, X.R.; Shen, Y.Q.; Mao, Z.W.; Liang, W.Q.; et al. TAT conjugated cationic noble metal nanoparticles for gene delivery to epidermal stem cells. Biomaterials 2014, 35, 5605–5618. [Google Scholar] [CrossRef]
- Pan, L.; Liu, J.; Shi, J. Nuclear-Targeting Gold Nanorods for Extremely Low NIR Activated Photothermal Therapy. ACS Appl. Mater. Interfaces 2017, 9, 15952–15961. [Google Scholar] [CrossRef]
- Jhaveri, A.M.; Torchilin, V.P. Multifunctional polymeric micelles for delivery of drugs and siRNA. Front. Pharmacol. 2014, 5, 77. [Google Scholar] [CrossRef] [Green Version]
- Oerlemans, C.; Bult, W.; Bos, M.; Storm, G.; Nijsen, J.F.; Hennink, W.E. Polymeric micelles in anticancer therapy: Targeting, imaging and triggered release. Pharm. Res. 2010, 27, 2569–2589. [Google Scholar] [CrossRef] [Green Version]
- Topete, A.; Barbosa, S.; Taboada, P. Intelligent micellar polymeric nanocarriers for therapeutics and diagnosis. J. Appl. Polym. Sci. 2015, 132, 42650. [Google Scholar] [CrossRef] [Green Version]
- Ghezzi, M.; Pescina, S.; Padula, C.; Santi, P.; Del Favero, E.; Cantù, L.; Nicoli, S. Polymeric micelles in drug delivery: An insight of the techniques for their characterization and assessment in biorelevant conditions. J. Control. Release 2021, 332, 312–336. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Xie, X.; Xu, X.; Zhang, L.; Zhou, X.; Yu, H.; Wu, P.; Wang, T.; Che, X.; Hu, Z. Development of dual ligand-targeted polymeric micelles as drug carriers for cancer therapy in vitro and in vivo. J. Mater. Chem. B 2014, 2, 2114–2126. [Google Scholar] [CrossRef]
- Hoang, B.; Ekdawi, S.N.; Reilly, R.M.; Allen, C. Active targeting of block copolymer micelles with trastuzumab Fab fragments and nuclear localization signal leads to increased tumor uptake and nuclear localization in HER2-overexpressing xenografts. Mol. Pharm. 2013, 10, 4229–4241. [Google Scholar] [CrossRef]
- Wang, G.H.; Cai, Y.Y.; Du, J.K.; Li, L.; Li, Q.; Yang, H.K.; Lin, J.T. TAT-conjugated chitosan cationic micelle for nuclear-targeted drug and gene co-delivery. Colloids Surf. B Biointerfaces 2018, 162, 326–334. [Google Scholar] [CrossRef]
- Alibakhshi, A.; Abarghooi Kahaki, F.; Ahangarzadeh, S.; Yaghoobi, H.; Yarian, F.; Arezumand, R.; Ranjbari, J.; Mokhtarzadeh, A.; de la Guardia, M. Targeted cancer therapy through antibody fragments-decorated nanomedicines. J. Control Release 2017, 268, 323–334. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Hedstrand, D.M.; Ferritto, M.S. Comb-burst dendrimer topology—New macromolecular architecture derived from dendritic grafting. Macromolecules 1991, 24, 1435–1438. [Google Scholar] [CrossRef]
- Wang, J.; Li, B.X.; Qiu, L.; Qiao, X.; Yang, H. Dendrimer-based drug delivery systems: History, challenges, and latest developments. J. Biol. Eng. 2022, 16, 18. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, S.; Gupta, L.; Rani, S.; Dave, K.; Gupta, U. Impact of Dendrimers on Solubility of Hydrophobic Drug Molecules. Front. Pharmacol. 2017, 8, 261. [Google Scholar] [CrossRef] [Green Version]
- Santos, S.D.; Xavier, M.; Leite, D.M.; Moreira, D.A.; Custódio, B.; Torrado, M.; Castro, R.; Leiro, V.; Rodrigues, J.; Tomás, H.; et al. PAMAM dendrimers: Blood-brain barrier transport and neuronal uptake after focal brain ischemia. J. Control Release 2018, 291, 65–79. [Google Scholar] [CrossRef] [Green Version]
- Launay, N.; Caminade, A.M.; Majoral, J.P. Synthesis and reactivity of unusual phosphorus dendrimers—A useful divergent growth approach up to the 7th generation. J. Am. Chem. Soc. 1995, 117, 3282–3283. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Baker, H.; Dewald, J.; Hall, M.; Kallos, G.; Martin, S.; Roeck, J.; Ryder, J.; Smith, P. A New Class of Polymers—Starburst-Dendritic Macromolecules. Polym. J. 1985, 17, 117–132. [Google Scholar] [CrossRef] [Green Version]
- Hawker, C.J.; Frechet, J.M.J. A new convergent approach to monodisperse dendritic macromolecules. J. Chem. Soc. Chem. Commun. 1990, 1010–1013. [Google Scholar] [CrossRef]
- Lee, J.; Kwon, Y.E.; Kim, J.; Kim, D.W.; Guim, H.; Yeon, J.; Kim, J.C.; Choi, J.S. Nonviral gene delivery using PAMAM dendrimer conjugated with the nuclear localization signal peptide derived from human papillomavirus type 11 E2 protein. J. Biomater. Sci. Polym. Ed. 2021, 32, 1140–1160. [Google Scholar] [CrossRef]
- Lee, J.; Kwon, Y.E.; Kim, Y.; Choi, J.S. Enhanced transfection efficiency of low generation PAMAM dendrimer conjugated with the nuclear localization signal peptide derived from herpesviridae. J. Biomater. Sci. Polym. Ed. 2021, 32, 22–41. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.; Kwon, Y.E.; Kim, Y.J.; Choi, J.S. Gene Delivery by PAMAM Dendrimer Conjugated with the Nuclear Localization Signal Peptide Derived from Influenza B Virus Nucleoprotein. Macromol. Res. 2019, 27, 360–368. [Google Scholar] [CrossRef]
- Lee, J.; Jung, J.; Kim, Y.J.; Lee, E.; Choi, J.S. Gene delivery of PAMAM dendrimer conjugated with the nuclear localization signal peptide originated from fibroblast growth factor 3. Int. J. Pharm. 2014, 459, 10–18. [Google Scholar] [CrossRef]
- Cooper, R.C.; Yang, H. Duplex of Polyamidoamine Dendrimer/Custom-Designed Nuclear-Localization Sequence Peptide for Enhanced Gene Delivery. Bioelectricity 2020, 2, 150–157. [Google Scholar] [CrossRef]
- Wu, Z.Y.; Shen, J.M.; Lang, H.; Yue, T.; Sun, C. pH/enzyme dual sensitive and nucleus-targeting dendrimer nanoparticles to enhance the antitumour activity of doxorubicin. Pharm. Dev. Technol. 2022, 27, 357–371. [Google Scholar] [CrossRef]
- Wang, G.H.; Chen, H.; Cai, Y.Y.; Li, L.; Yang, H.K.; Li, Q.; He, Z.J.; Lin, J.T. Efficient gene vector with size changeable and nucleus targeting in cancer therapy. Mater. Sci. Eng. C Mater. Biol. Appl. 2018, 90, 568–575. [Google Scholar] [CrossRef]
- Surekha, B.; Kommana, N.S.; Dubey, S.K.; Kumar, A.V.P.; Shukla, R.; Kesharwani, P. PAMAM dendrimer as a talented multifunctional biomimetic nanocarrier for cancer diagnosis and therapy. Colloids Surf. B Biointerfaces 2021, 204, 111837. [Google Scholar] [CrossRef] [PubMed]
- Thiagarajan, G.; Greish, K.; Ghandehari, H. Charge affects the oral toxicity of poly(amidoamine) dendrimers. Eur. J. Pharm. Biopharm. 2013, 84, 330–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tariq, I.; Ali, M.Y.; Sohail, M.F.; Amin, M.U.; Ali, S.; Bukhari, N.I.; Raza, A.; Pinnapireddy, S.R.; Schäfer, J.; Bakowsky, U. Lipodendriplexes mediated enhanced gene delivery: A cellular to pre-clinical investigation. Sci. Rep. 2020, 10, 21446. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhao, L.; Li, Y.; Xu, T. Design of biocompatible dendrimers for cancer diagnosis and therapy: Current status and future perspectives. Chem. Soc. Rev. 2011, 40, 2673–2703. [Google Scholar] [CrossRef] [PubMed]
- Larson, N.; Ghandehari, H. Polymeric conjugates for drug delivery. Chem. Mater. 2012, 24, 840–853. [Google Scholar] [CrossRef] [Green Version]
- Vivero-Escoto, J.L.; Slowing, I.; Trewyn, B.G.; Lin, V.S. Mesoporous silica nanoparticles for intracellular controlled drug delivery. Small 2010, 6, 1952–1967. [Google Scholar] [CrossRef]
- Liong, M.; Lu, J.; Kovochich, M.; Xia, T.; Ruehm, S.G.; Nel, A.E.; Tamanoi, F.; Zink, J.I. Multifunctional inorganic nanoparticles for imaging, targeting, and drug delivery. ACS Nano 2008, 2, 889–896. [Google Scholar] [CrossRef] [Green Version]
- Rosenholm, J.M.; Peuhu, E.; Eriksson, J.E.; Sahlgren, C.; Lindén, M. Targeted intracellular delivery of hydrophobic agents using mesoporous hybrid silica nanoparticles as carrier systems. Nano Lett. 2009, 9, 3308–3311. [Google Scholar] [CrossRef]
- Rosenholm, J.M.; Peuhu, E.; Bate-Eya, L.T.; Eriksson, J.E.; Sahlgren, C.; Lindén, M. Cancer-cell-specific induction of apoptosis using mesoporous silica nanoparticles as drug-delivery vectors. Small 2010, 6, 1234–1241. [Google Scholar] [CrossRef]
- Pan, L.; He, Q.; Liu, J.; Chen, Y.; Ma, M.; Zhang, L.; Shi, J. Nuclear-targeted drug delivery of TAT peptide-conjugated monodisperse mesoporous silica nanoparticles. J. Am. Chem. Soc. 2012, 134, 5722–5725. [Google Scholar] [CrossRef]
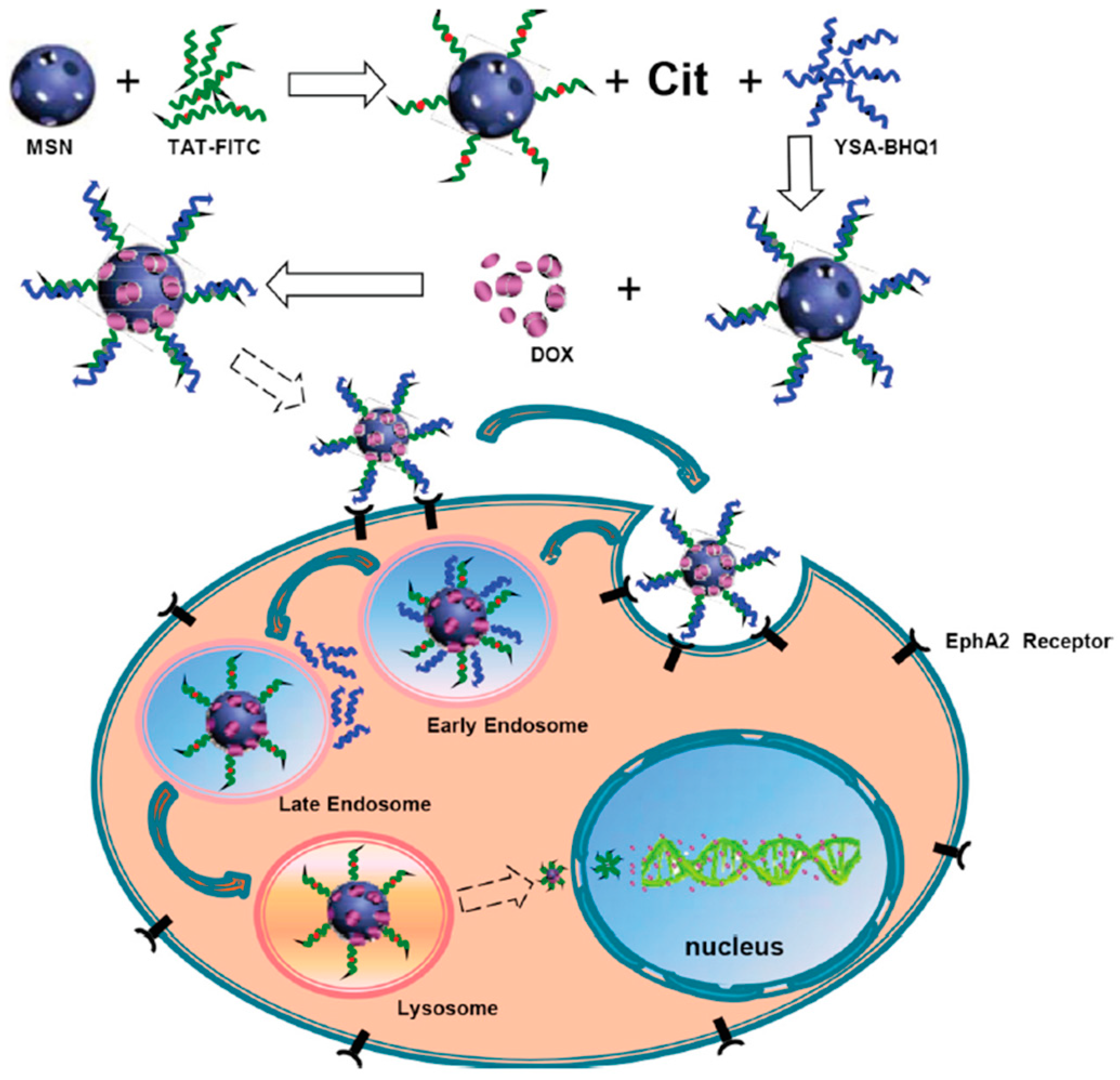
- Zhao, J.; Zhao, F.; Wang, X.; Fan, X.; Wu, G. Secondary nuclear targeting of mesoporous silica nano-particles for cancer-specific drug delivery based on charge inversion. Oncotarget 2016, 7, 70100–70112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Placzek, W.J.; Stebbins, J.L.; Mitra, S.; Noberini, R.; Koolpe, M.; Zhang, Z.; Dahl, R.; Pasquale, E.B.; Pellecchia, M. Novel targeted system to deliver chemotherapeutic drugs to EphA2-expressing cancer cells. J. Med. Chem. 2012, 55, 2427–2436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Yan, W.; Suo, X.; Peng, H.; Yang, X.; Li, Z.; Zhang, J.; Liu, D. Nucleus-targeted nano delivery system eradicates cancer stem cells by combined thermotherapy and hypoxia-activated chemotherapy. Biomaterials 2019, 200, 1–14. [Google Scholar] [CrossRef]
- Nsairat, H.; Khater, D.; Sayed, U.; Odeh, F.; Al Bawab, A.; Alshaer, W. Liposomes: Structure, composition, types, and clinical applications. Heliyon 2022, 8, e09394. [Google Scholar] [CrossRef]
- Bangham, A.D.; Standish, M.M.; Watkins, J.C. Diffusion of univalent ions across the lamellae of swollen phospholipids. J. Mol. Biol. 1965, 13, 238–252. [Google Scholar] [CrossRef]
- Li, B.; Shao, H.; Gao, L.; Li, H.; Sheng, H.; Zhu, L. Nano-drug co-delivery system of natural active ingredients and chemotherapy drugs for cancer treatment: A review. Drug Deliv. 2022, 29, 2130–2161. [Google Scholar] [CrossRef]
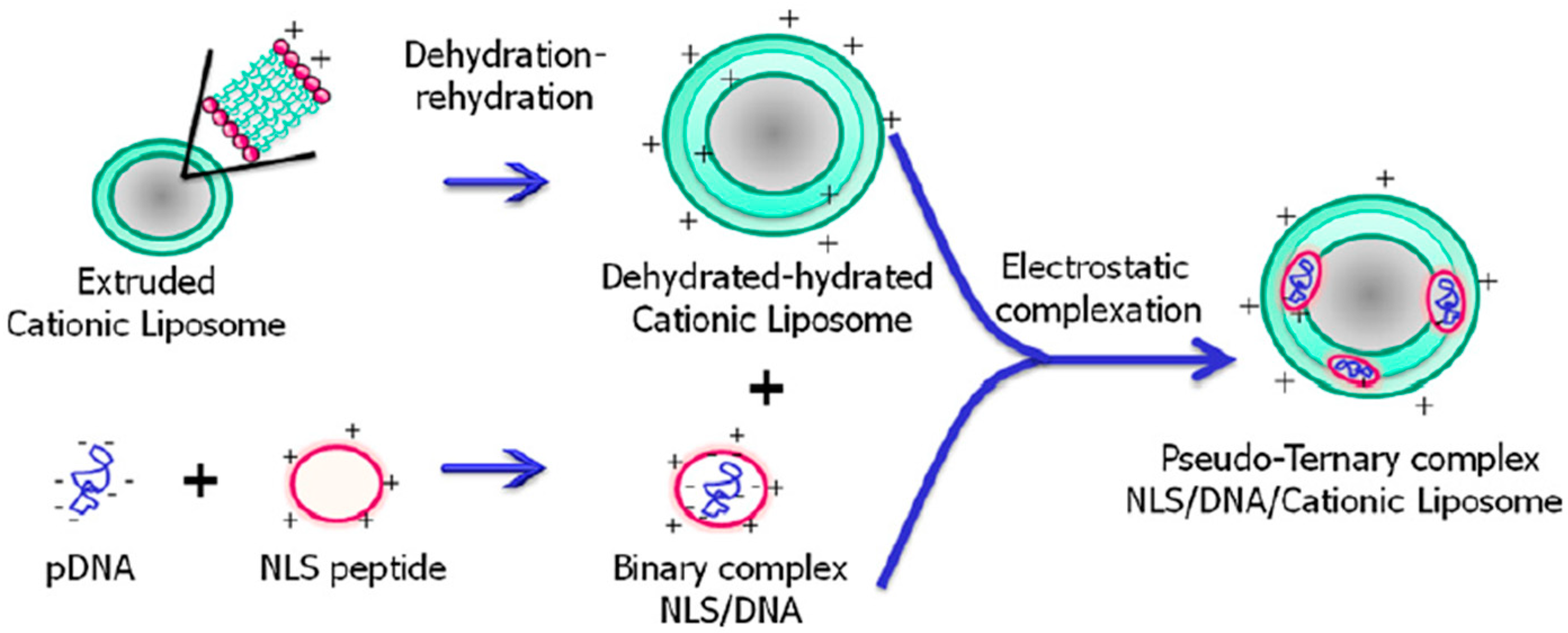
- Rosada, R.S.; Silva, C.L.; Santana, M.H.; Nakaie, C.R.; de la Torre, L.G. Effectiveness, against tuberculosis, of pseudo-ternary complexes: Peptide-DNA-cationic liposome. J. Colloid Interface Sci. 2012, 373, 102–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Hüttmann, G.; Zhang, Z.; Vogel, A.; Birngruber, R.; Tangutoori, S.; Hasan, T.; Rahmanzadeh, R. Light-Controlled Delivery of Monoclonal Antibodies for Targeted Photoinactivation of Ki-67. Mol. Pharm. 2015, 12, 3272–3281. [Google Scholar] [CrossRef]
- Li, X.; Wu, X.; Yang, H.; Li, L.; Ye, Z.; Rao, Y. A nuclear targeted Dox-aptamer loaded liposome delivery platform for the circumvention of drug resistance in breast cancer. Biomed. PharmacoTher. 2019, 117, 109072. [Google Scholar] [CrossRef] [PubMed]
- Aronsohn, A.I.; Hughes, J.A. Nuclear localization signal peptides enhance cationic liposome-mediated gene therapy. J. Drug Target. 1998, 5, 163–169. [Google Scholar] [CrossRef]
- Tachibana, R.; Harashima, H.; Shono, M.; Azumano, M.; Niwa, M.; Futaki, S.; Kiwada, H. Intracellular regulation of macromolecules using pH-sensitive liposomes and nuclear localization signal: Qualitative and quantitative evaluation of intracellular trafficking. BioChem. Biophys. Res. Commun. 1998, 251, 538–544. [Google Scholar] [CrossRef]
- Torchilin, V.P.; Levchenko, T.S.; Rammohan, R.; Volodina, N.; Papahadjopoulos-Sternberg, B.; D’Souza, G.G. Cell transfection in vitro and in vivo with nontoxic TAT peptide-liposome-DNA complexes. Proc. Natl. Acad. Sci. USA 2003, 100, 1972–1977. [Google Scholar] [CrossRef] [Green Version]
- Pan, Z.Z.; Wang, H.Y.; Zhang, M.; Lin, T.T.; Zhang, W.Y.; Zhao, P.F.; Tang, Y.S.; Xiong, Y.; Zeng, Y.E.; Huang, Y.Z. Nuclear-targeting TAT-PEG-Asp8-doxorubicin polymeric nanoassembly to overcome drug-resistant colon cancer. Acta Pharmacol. Sin. 2016, 37, 1110–1120. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.S.; Gao, Z.; Kim, D.; Park, K.; Kwon, I.C.; Bae, Y.H. Super pH-sensitive multifunctional polymeric micelle for tumor pH(e) specific TAT exposure and multidrug resistance. J. Control Release 2008, 129, 228–236. [Google Scholar] [CrossRef] [Green Version]
- Del Gaizo, V.; MacKenzie, J.A.; Payne, R.M. Targeting proteins to mitochondria using TAT. Mol. Genet. Metab. 2003, 80, 170–180. [Google Scholar] [CrossRef]
- Tu, L.C.; Fu, G.; Zilman, A.; Musser, S.M. Large cargo transport by nuclear pores: Implications for the spatial organization of FG-nucleoporins. EMBO J. 2013, 32, 3220–3230. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Fu, G.; Ciziene, D.; Stewart, M.; Musser, S.M. Choreography of importin-α/CAS complex assembly and disassembly at nuclear pores. Proc. Natl. Acad. Sci. USA 2013, 110, E1584–E1593. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, R.; Sau, A.; Musser, S.M. Super-resolved 3D tracking of cargo transport through nuclear pore complexes. Nat. Cell Biol. 2022, 24, 112–122. [Google Scholar] [CrossRef]
- Gwosch, K.C.; Pape, J.K.; Balzarotti, F.; Hoess, P.; Ellenberg, J.; Ries, J.; Hell, S.W. MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nat. Methods 2020, 17, 217–224. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, Y.; Fu, G.; Leng, Y. Nuclear Delivery of Nanoparticle-Based Drug Delivery Systems by Nuclear Localization Signals. Cells 2023, 12, 1637. https://doi.org/10.3390/cells12121637
Nie Y, Fu G, Leng Y. Nuclear Delivery of Nanoparticle-Based Drug Delivery Systems by Nuclear Localization Signals. Cells. 2023; 12(12):1637. https://doi.org/10.3390/cells12121637
Chicago/Turabian StyleNie, Yuhan, Guo Fu, and Yuxin Leng. 2023. "Nuclear Delivery of Nanoparticle-Based Drug Delivery Systems by Nuclear Localization Signals" Cells 12, no. 12: 1637. https://doi.org/10.3390/cells12121637
APA StyleNie, Y., Fu, G., & Leng, Y. (2023). Nuclear Delivery of Nanoparticle-Based Drug Delivery Systems by Nuclear Localization Signals. Cells, 12(12), 1637. https://doi.org/10.3390/cells12121637







