A Systematic Review on Quiescent State Research Approaches in S. cerevisiae
Abstract
:1. Introduction
2. Methods
2.1. Literature Search
2.2. Data Analysis and Visualisation
3. Results
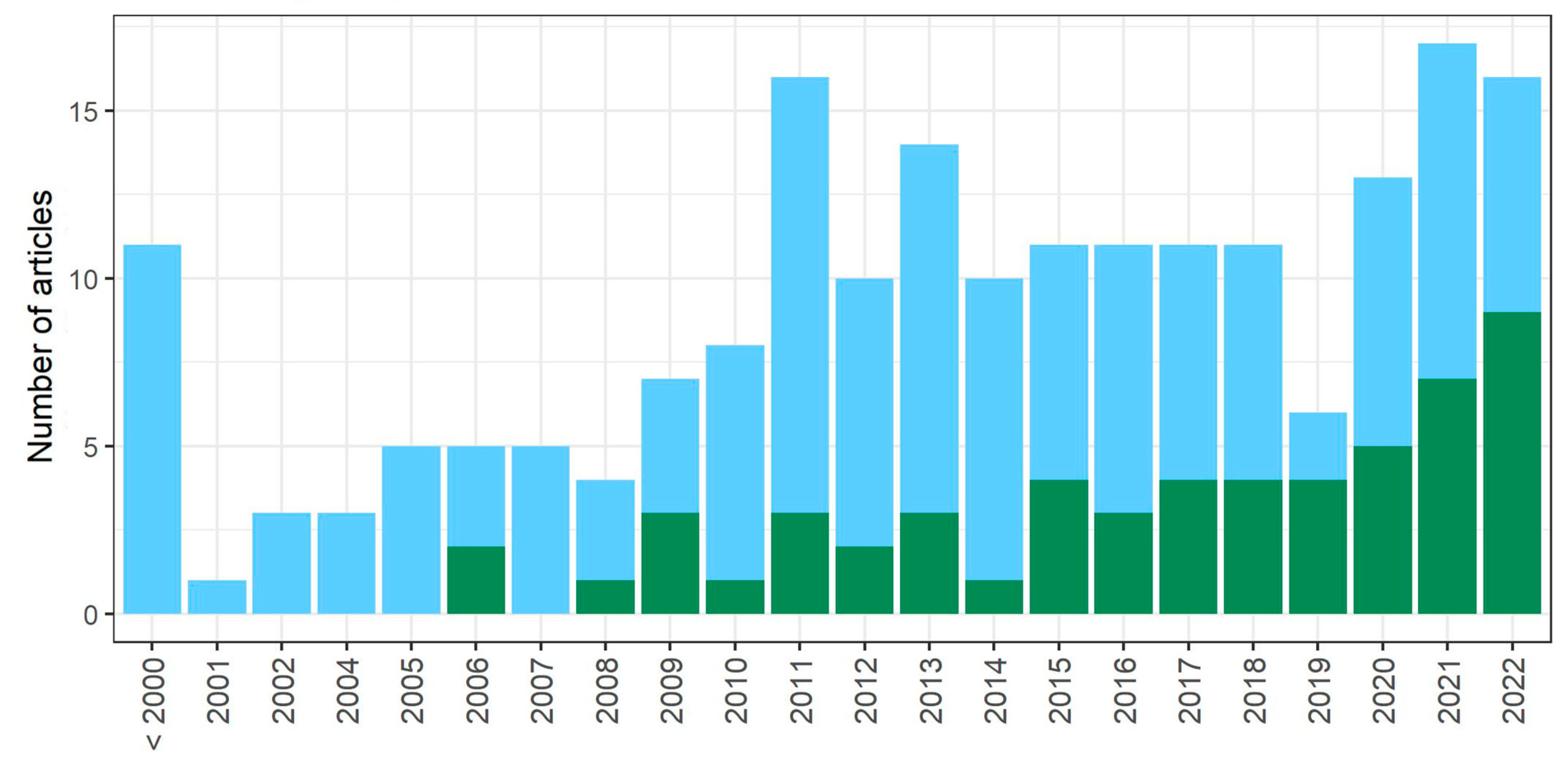
3.1. Literature Search
3.2. The Meaning of “Quiescence”
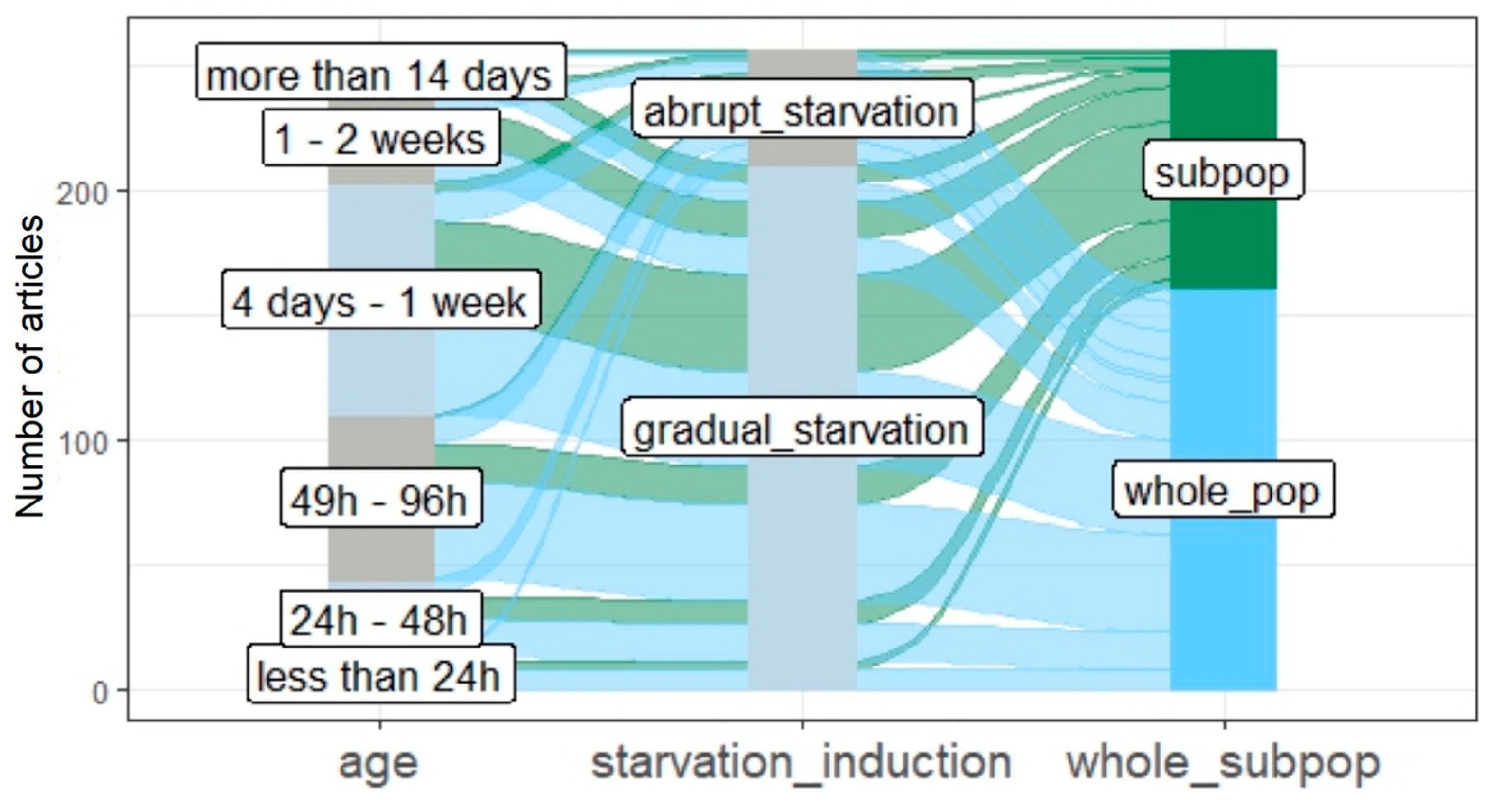
3.3. The Age of Studied Quiescent Populations or Cells
3.4. Starvation Induction
3.5. The Metabolic Profile and Origin of Studied Strains
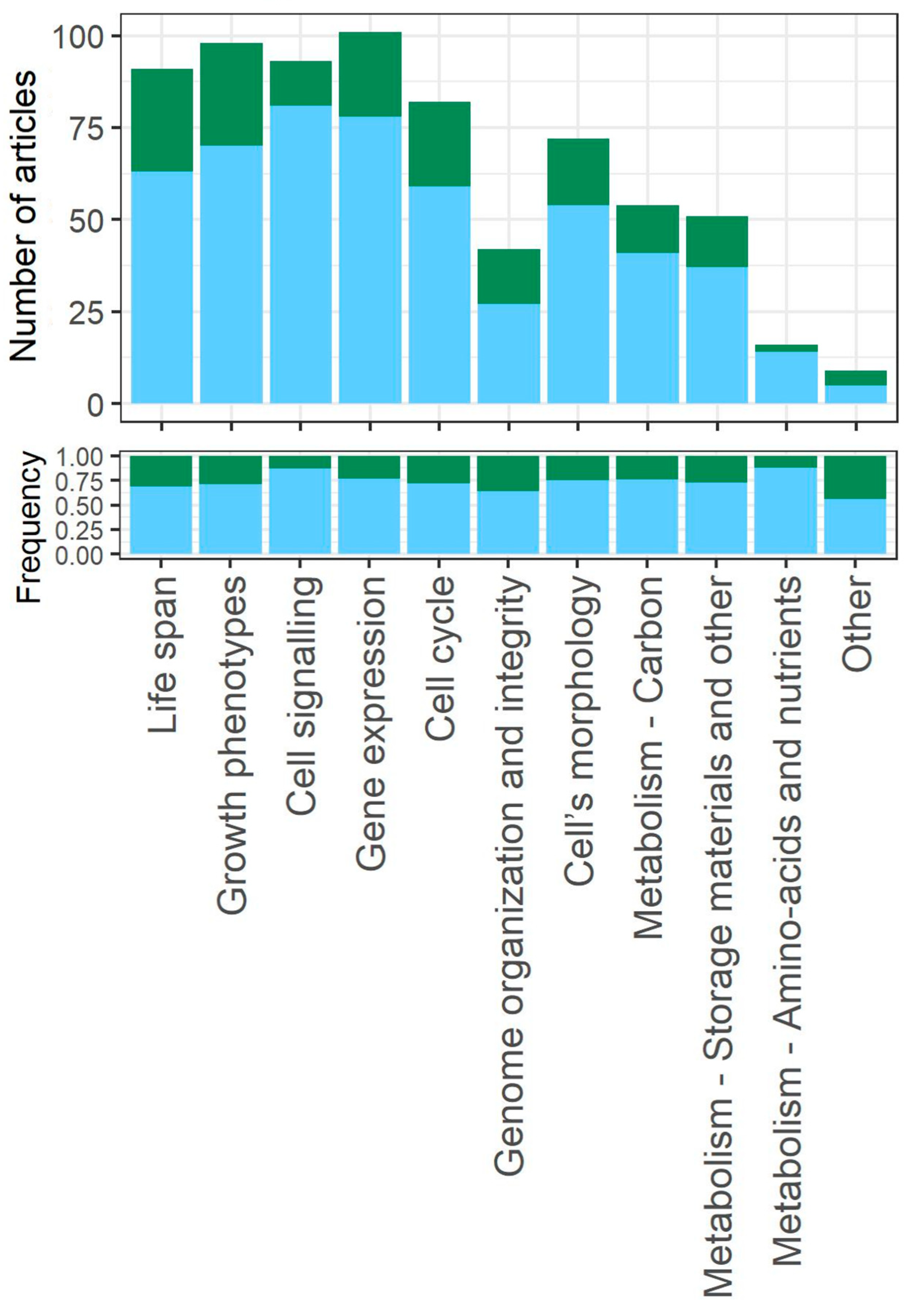
3.6. Biological Aspects
3.7. Experimental Set-Up
4. Discussion
| Guidelines for the research on yeast quiescence |
| 1. Define studied quiescent cells |
|
| 2. Specify how quiescence is induced and the time when you assume the population entered quiescence |
|
| 3. DEFINE THE METABOLIC PROFILE AND GENETIC BACKGROUND OF USED STRAINS |
|
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Farrell, P.H. Quiescence: Early evolutionary origins and universality do not imply uniformity. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 3498–3507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marescal, O.; Cheeseman, I.M. Cellular Mechanisms and Regulation of Quiescence. Dev. Cell 2020, 55, 259–271. [Google Scholar] [CrossRef] [PubMed]
- De Virgilio, C. The essence of yeast quiescence. FEMS Microbiol. Rev. 2012, 36, 306–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karathia, H.; Vilaprinyo, E.; Sorribas, A.; Alves, R. Saccharomyces cerevisiae as a model organism: A comparative study. PLoS ONE 2011, 6, e16015. [Google Scholar] [CrossRef] [Green Version]
- Barnett, J.A. A history of research on yeasts 10: Foundations of yeast genetics1. Yeast 2007, 24, 799–845. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. Nobel yeast research. FEMS Yeast Res. 2016, 16, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Breeden, L.L.; Tsukiyama, T. Quiescence in Saccharomyces cerevisiae. Annu. Rev. Genet. 2022, 56, 253–278. [Google Scholar] [CrossRef]
- Gray, J.V.; Petsko, G.A.; Johnston, G.C.; Ringe, D.; Singer, R.A.; Werner-Washburne, M. “Sleeping Beauty”: Quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2004, 68, 187–206. [Google Scholar] [CrossRef] [Green Version]
- Honigberg, S.M. Similar environments but diverse fates: Responses of budding yeast to nutrient deprivation. Microb. Cell 2016, 3, 302–328. [Google Scholar] [CrossRef]
- Allen, C.; Büttner, S.; Aragon, A.D.; Thomas, J.A.; Meirelles, O.; Jaetao, J.E.; Benn, D.; Ruby, S.W.; Veenhuis, M.; Madeo, F.; et al. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 2006, 174, 89–100. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Ren, Q.; Zhang, Z. Chromosome or chromatin condensation leads to meiosis or apoptosis in stationary yeast (Saccharomyces cerevisiae) cells. FEMS Yeast Res. 2006, 6, 1254–1263. [Google Scholar] [CrossRef] [Green Version]
- Klosinska, M.M.; Crutchfield, C.A.; Bradley, P.H.; Rabinowitz, J.D.; Broach, J.R. Yeast cells can access distinct quiescent states. Genes Dev. 2011, 25, 336–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacquel, B.; Aspert, T.; Laporte, D.; Sagot, I.; Charvin, G. Monitoring single-cell dynamics of entry into quiescence during an unperturbed life cycle. eLife 2021, 10, e73186. [Google Scholar] [CrossRef]
- Boer, V.M.; Amini, S.; Botstein, D. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc. Natl. Acad. Sci. USA 2008, 105, 6930–6935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, S.M.; Laflin, S.; Broadway, A.; Burnet, C.; Hartheimer, J.; Rodgers, J.; Smith, D.L.; Hartman, J.L. High-resolution yeast quiescence profiling in human-like media reveals complex influences of auxotrophy and nutrient availability. GeroScience 2021, 43, 941–964. [Google Scholar] [CrossRef] [PubMed]
- Sagot, I.; Laporte, D. Quiescence, an individual journey. Curr. Genet. 2019, 65, 695–699. [Google Scholar] [CrossRef] [Green Version]
- Sagot, I.; Laporte, D. The cell biology of quiescent yeast—A diversity of individual scenarios. J. Cell Sci. 2019, 132, jcs213025. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-Y.Y.; Cheng, K.-Y.Y.; Chao, J.-C.C.; Leu, J.-Y.Y. Differentiated cytoplasmic granule formation in quiescent and non-quiescent cells upon chronological aging. Microb. Cell 2016, 3, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Laporte, D.; Gouleme, L.; Jimenez, L.; Khemiri, I.; Sagot, I. Mitochondria reorganization upon proliferation arrest predicts individual yeast cell fate. eLife 2018, 7, e35685. [Google Scholar] [CrossRef]
- Swygert, S.G.; Kim, S.; Wu, X.; Fu, T.; Hsieh, T.H.; Rando, O.J.; Eisenman, R.N.; Shendure, J.; McKnight, J.N.; Tsukiyama, T. Condensin-Dependent Chromatin Compaction Represses Transcription Globally during Quiescence. Mol. Cell 2019, 73, 533–546.e4. [Google Scholar] [CrossRef] [Green Version]
- Guidi, M.; Ruault, M.; Marbouty, M.; Loïodice, I.; Cournac, A.; Billaudeau, C.; Hocher, A.; Mozziconacci, J.; Koszul, R.; Taddei, A. Spatial reorganization of telomeres in long-lived quiescent cells. Genome Biol. 2015, 16, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laporte, D.; Courtout, F.; Tollis, S.; Sagot, I. Quiescent Saccharomyces cerevisiae forms telomere hyperclusters at the nuclear membrane vicinity through a multifaceted mechanism involving Esc1, the Sir complex, and chromatin condensation. Mol. Biol. Cell 2016, 27, 1875–1884. [Google Scholar] [CrossRef]
- Laporte, D.; Jimenez, L.; Gouleme, L.; Sagot, I. Yeast quiescence exit swiftness is influenced by cell volume and chronological age. Microb. Cell 2018, 5, 104–111. [Google Scholar] [CrossRef] [Green Version]
- Bojsen, R.; Regenberg, B.; Folkesson, A. Saccharomyces cerevisiae biofilm tolerance towards systemic antifungals depends on growth phase. BMC Microbiol. 2014, 14, 305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opalek, M.; Smug, B.; Doebeli, M.; Wloch-Salamon, D. On the Ecological Significance of Phenotypic Heterogeneity in Microbial Populations Undergoing Starvation. Microbiol. Spectr. 2022, 10, e00450-21. [Google Scholar] [CrossRef] [PubMed]
- Foo, Y.Z.; O’Dea, R.E.; Koricheva, J.; Nakagawa, S.; Lagisz, M. A practical guide to question formation, systematic searching and study screening for literature reviews in ecology and evolution. Methods Ecol. Evol. 2021, 12, 1705–1720. [Google Scholar] [CrossRef]
- O’Dea, R.E.; Lagisz, M.; Jennions, M.D.; Koricheva, J.; Noble, D.W.A.; Parker, T.H.; Gurevitch, J.; Page, M.J.; Stewart, G.; Moher, D.; et al. Preferred reporting items for systematic reviews and meta-analyses in ecology and evolutionary biology: A PRISMA extension. Biol. Rev. 2021, 96, 1695–1722. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing 2021. Available online: https://www.r-project.org/ (accessed on 1 February 2023).
- Wickham, H.; François, R.; Henry, L.; Müller, K. Dplyr: A Grammar of Data Manipulation 2022. Available online: https://cran.r-project.org/package=dplyr (accessed on 1 February 2023).
- Wickham, H.; Bryan, J. Readxl: Read Excel Files 2022. Available online: https://cran.r-project.org/package=readxl (accessed on 1 February 2023).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis 2016. Available online: https://ggplot2.tidyverse.org (accessed on 1 February 2023).
- Koneswarakantha, B. Easyalluvial: Generate Alluvial Plots with a Single Line of Code 2022. Available online: https://github.com/erblast/easyalluvial/ (accessed on 1 February 2023).
- Inkscape Project Inkscape 2020. Available online: https://inkscape.org/ (accessed on 1 February 2023).
- Lang, D.; Chien, G. Package ‘wordcloud2′: Create Word Cloud by “htmlwidget” 2022. Available online: https://github.com/lchiffon/wordcloud2 (accessed on 1 February 2023).
- An, Z.; Tassa, A.; Thomas, C.; Zhong, R.; Xiao, G.; Fotedar, R.; Tu, B.P.; Klionsky, D.J.; Levine, B. Autophagy is required for G1/G0quiescence in response to nitrogen starvation in Saccharomyces cerevisiae. Autophagy 2014, 10, 1702–1711. [Google Scholar] [CrossRef] [Green Version]
- Nussbaum, I.; Weindling, E.; Jubran, R.; Cohen, A.; Bar-Nun, S. Deteriorated Stress Response in Stationary-Phase Yeast: Sir2 and Yap1 Are Essential for Hsf1 Activation by Heat Shock and Oxidative Stress, Respectively. PLoS ONE 2014, 9, e111505. [Google Scholar] [CrossRef]
- Garay, E.; Campos, S.E.; González de la Cruz, J.; Gaspar, A.P.; Jinich, A.; DeLuna, A. High-Resolution Profiling of Stationary-Phase Survival Reveals Yeast Longevity Factors and Their Genetic Interactions. PLoS Genet. 2014, 10, e1004168. [Google Scholar] [CrossRef] [Green Version]
- Mews, P.; Zee, B.M.; Liu, S.; Donahue, G.; Garcia, B.A.; Berger, S.L. Histone Methylation Has Dynamics Distinct from Those of Histone Acetylation in Cell Cycle Reentry from Quiescence. Mol. Cell. Biol. 2014, 34, 3968–3980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porzoor, A.; Caine, J.M.; Macreadie, I.G. Pretreatment of chemically-synthesized Aβ42 affects its biological activity in yeast. Prion 2014, 8, 404–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, K.H.; Nostramo, R.; Zhang, B.; Varia, S.N.; Klett, B.M.; Herman, P.K. Protein kinases are associated with multiple, distinct cytoplasmic granules in quiescent yeast cells. Genetics 2014, 198, 1495–1512. [Google Scholar] [CrossRef] [PubMed]
- Martins, D.; English, A.M. SOD1 oxidation and formation of soluble aggregates in yeast: Relevance to sporadic ALS development. Redox Biol. 2014, 2, 632–639. [Google Scholar] [CrossRef] [Green Version]
- Acker, J.; Nguyen, N.T.T.; Vamme, M.; Tavenet, A.; Bri-Suleau, A.; Conesa, C. Sub1 and Maf1, two effectors of RNA polymerase III, are involved in the yeast quiescence cycle. PLoS ONE 2014, 9, e114587. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, S.; Dalgaard, J.Z.; Millar, J.B.A.; Arumugam, P. The Rim15-Endosulfine-PP2ACdc55 Signalling Module Regulates Entry into Gametogenesis and Quiescence via Distinct Mechanisms in Budding Yeast. PLoS Genet. 2014, 10, e1004456. [Google Scholar] [CrossRef]
- Quan, Z.; Cao, L.; Tang, Y.; Yan, Y.; Oliver, S.G.; Zhang, N. The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological Lifespan. PLoS Genet. 2015, 11, e1005282. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Miles, S.; Breeden, L.L. A Genetic Screen for Saccharomyces cerevisiae Mutants That Fail to Enter Quiescence. G3 Genes Genomes Genet. 2015, 5, 1783–1795. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.; Han, G.S.; Mileykovskaya, E.; Garrett, T.A.; Carman, G.M. Altered lipid synthesis by lack of yeast Pah1 phosphatidate phosphatase reduces chronological life span. J. Biol. Chem. 2015, 290, 25382–25394. [Google Scholar] [CrossRef] [Green Version]
- Carbó, R.; Ginovart, M.; Carta, A.; Portell, X.; del Valle, L.J. Effect of aerobic and microaerophilic culture in the growth dynamics of Saccharomyces cerevisiae and in training of quiescent and non-quiescent subpopulations. Arch. Microbiol. 2015, 197, 991–999. [Google Scholar] [CrossRef]
- Wanichthanarak, K.; Wongtosrad, N.; Petranovic, D. Genome-wide expression analyses of the stationary phase model of ageing in yeast. Mech. Ageing Dev. 2015, 149, 65–74. [Google Scholar] [CrossRef] [PubMed]
- McKnight, J.N.; Boerma, J.W.; Breeden, L.L.; Tsukiyama, T. Global Promoter Targeting of a Conserved Lysine Deacetylase for Transcriptional Shutoff during Quiescence Entry. Mol. Cell 2015, 59, 732–743. [Google Scholar] [CrossRef] [Green Version]
- Hughes Hallett, J.E.; Luo, X.; Capaldi, A.P. Snf1/AMPK promotes the formation of Kog1/raptor-bodies to increase the activation threshold of TORC1 in budding yeast. eLife 2015, 4, e09181. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Schmidt, O.; Angelova, M.; Faserl, K.; Weys, S.; Kremser, L.; Pfaffenwimmer, T.; Dalik, T.; Kraft, C.; Trajanoski, Z.; et al. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. eLife 2015, 4, e07736. [Google Scholar] [CrossRef] [PubMed]
- Vasicova, P.; Lejskova, R.; Malcova, I.; Hasek, J. The Stationary-Phase Cells of Saccharomyces cerevisiae Display Dynamic Actin Filaments Required for Processes Extending Chronological Life Span. Mol. Cell. Biol. 2015, 35, 3892–3908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutledge, M.T.; Russo, M.; Belton, J.M.; Dekker, J.; Broach, J.R. The yeast genome undergoes significant topological reorganization in quiescence. Nucleic Acids Res. 2015, 43, 8299–8313. [Google Scholar] [CrossRef] [Green Version]
- Sundaram, V.; Petkova, M.I.; Pujol-Carrion, N.; Boada, J.; de la Torre-Ruiz, M.A. Tor1, Sch9 and PKA downregulation in quiescence rely on Mtl1 to preserve mitochondrial integrity and cell survival. Mol. Microbiol. 2015, 97, 93–109. [Google Scholar] [CrossRef] [Green Version]
- Boucherie, H. Protein synthesis during transition and stationary phases under glucose limitation in Saccharomyces cerevisiae. J. Bacteriol. 1985, 161, 385–392. [Google Scholar] [CrossRef] [Green Version]
- Munder, M.C.; Midtvedt, D.; Franzmann, T.; Nüske, E.; Otto, O.; Herbig, M.; Ulbricht, E.; Müller, P.; Taubenberger, A.; Maharana, S.; et al. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife 2016, 5, e09347. [Google Scholar] [CrossRef]
- Cao, L.; Tang, Y.; Quan, Z.; Zhang, Z.; Oliver, S.G.; Zhang, N. Chronological Lifespan in Yeast Is Dependent on the Accumulation of Storage Carbohydrates Mediated by Yak1, Mck1 and Rim15 Kinases. PLoS Genet. 2016, 12, e1006458. [Google Scholar] [CrossRef] [Green Version]
- Oomuro, M.; Kato, T.; Zhou, Y.; Watanabe, D.; Motoyama, Y.; Yamagishi, H.; Akao, T.; Aizawa, M. Defective quiescence entry promotes the fermentation performance of bottom-fermenting brewer’s yeast. J. Biosci. Bioeng. 2016, 122, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Bisschops, M.M.M.; Luttik, M.A.H.; Doerr, A.; Verheijen, P.J.T.; Bruggeman, F.; Pronk, J.T.; Daran-Lapujade, P. Extreme calorie restriction in yeast retentostats induces uniform non-quiescent growth arrest. Biochim. Biophys. Acta-Mol. Cell Res. 2017, 1864, 231–242. [Google Scholar] [CrossRef]
- Klukovich, R.; Courchesne, W.E. Functions of Saccharomyces cerevisiae Ecm27p, a putative Na+/Ca2+ exchanger, in calcium homeostasis, carbohydrate storage and cell cycle reentry from the quiescent phase. Microbiol. Res. 2016, 186–187, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Miles, S.; Croxford, M.W.; Abeysinghe, A.P.; Breeden, L.L. Msa1 and Msa2 Modulate G1-Specific Transcription to Promote G1 Arrest and the Transition to Quiescence in Budding Yeast. PLoS Genet. 2016, 12, e1006088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, R.; Srivastava, S. Quantitative proteomic comparison of stationary/G 0 phase cells and tetrads in budding yeast. Sci. Rep. 2016, 6, 32031. [Google Scholar] [CrossRef] [Green Version]
- Svenkrtova, A.; Belicova, L.; Volejnikova, A.; Sigler, K.; Jazwinski, S.M.; Pichova, A. Stratification of yeast cells during chronological aging by size points to the role of trehalose in cell vitality. Biogerontology 2016, 17, 395–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nostramo, R.; Varia, S.N.; Zhang, B.; Emerson, M.M.; Herman, P.K. The Catalytic Activity of the Ubp3 Deubiquitinating Protease Is Required for Efficient Stress Granule Assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 2016, 36, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Melki, R.; Kabani, M. A prolonged chronological lifespan is an unexpected benefit of the [PSI+] prion in yeast. PLoS ONE 2017, 12, e184905. [Google Scholar] [CrossRef] [Green Version]
- Saul, D.J.; Walton, E.F.; Sudbery, P.E.; Carter, B.L.A. Saccharomyces cerevisiae whi2 mutants in stationary phase retain the properties of exponentially growing cells. J. Gen. Microbiol. 1985, 131, 2245–2251. [Google Scholar] [CrossRef] [Green Version]
- Wloch-Salamon, D.M.; Tomala, K.; Aggeli, D.; Dunn, B. Adaptive roles of SSY1 and SIR3 during cycles of growth and starvation in saccharomyces cerevisiae populations enriched for quiescent or nonquiescent cells. G3 Genes Genomes Genet. 2017, 7, 1899–1911. [Google Scholar] [CrossRef] [Green Version]
- Leonov, A.; Feldman, R.; Piano, A.; Arlia-Ciommo, A.; Lutchman, V.; Ahmadi, M.; Elsaser, S.; Fakim, H.; Heshmati-Moghaddam, M.; Hussain, A.; et al. Caloric restriction extends yeast chronological lifespan via a mechanism linking cellular aging to cell cycle regulation, maintenance of a quiescent state, entry into a non-quiescent state and survival in the non-quiescent state. Oncotarget 2017, 8, 69328–69350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, C.A.; Denison, C.; Burkenstock, A.; Nutter, C.; Gordon, D.M. Cellular conditions that modulate the fungicidal activity of occidiofungin. J. Appl. Microbiol. 2017, 123, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Quasem, I.; Luby, C.; Mace, C.; Fuchs, S. Density separation of quiescent yeast using iodixanol. Biotechniques 2017, 63, 169–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, C.P.; Hillyer, C.; Hokamp, K.; Fitzpatrick, D.J.; Konstantinov, N.K.; Welty, J.S.; Ness, S.A.; Werner-Washburne, M.; Fleming, A.B.; Osley, M.A. Distinct histone methylation and transcription profiles are established during the development of cellular quiescence in yeast. BMC Genom. 2017, 18, 107. [Google Scholar] [CrossRef] [Green Version]
- Narayanan, V.; Schelin, J.; Gorwa-Grauslund, M.; Van Niel, E.W.J.; Carlquist, M. Increased lignocellulosic inhibitor tolerance of Saccharomyces cerevisiae cell populations in early stationary phase. Biotechnol. Biofuels 2017, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Kuang, Z.; Pinglay, S.; Ji, H.; Boeke, J.D. Msn2/4 regulate expression of glycolytic enzymes and control transition from quiescence to growth. eLife 2017, 6, e29938. [Google Scholar] [CrossRef]
- McCleary, D.F.; Rine, J. Nutritional control of chronological aging and heterochromatin in Saccharomyces cerevisiae. Genetics 2017, 205, 1179–1193. [Google Scholar] [CrossRef] [Green Version]
- Fazal, Z.; Pelowitz, J.; Johnson, P.E.; Harper, J.C.; Brinker, C.J.; Jakobsson, E. Three-Dimensional Encapsulation of Saccharomyces cerevisiae in Silicate Matrices Creates Distinct Metabolic States as Revealed by Gene Chip Analysis. ACS Nano 2017, 11, 3560–3575. [Google Scholar] [CrossRef]
- Gu, Z.C.; Wu, E.; Sailer, C.; Jando, J.; Styles, E.; Eisenkolb, I.; Kuschel, M.; Bitschar, K.; Wang, X.; Huang, L.; et al. Ubiquitin orchestrates proteasome dynamics between proliferation and quiescence in yeast. Mol. Biol. Cell 2017, 28, 2479–2491. [Google Scholar] [CrossRef]
- Plesset, J.; Ludwig, J.R.; Cox, B.S.; McLaughlin, C.S. Effect of cell cycle position on thermotolerance in Saccharomyces cerevisiae. J. Bacteriol. 1987, 169, 779–784. [Google Scholar] [CrossRef] [Green Version]
- Krishna, S.; Laxman, S.; Lew, D.J. A minimal “push–pull” bistability model explains oscillations between quiescent and proliferative cell states. Mol. Biol. Cell 2018, 29, 2243–2258. [Google Scholar] [CrossRef] [PubMed]
- Baroni, M.D.; Colombo, S.; Martegani, E. Antagonism between salicylate and the cAMP signal controls yeast cell survival and growth recovery from quiescence. Microb. Cell 2018, 5, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Nevers, A.; Doyen, A.; Malabat, C.; Néron, B.; Kergrohen, T.; Jacquier, A.; Badis, G. Antisense transcriptional interference mediates condition-specific gene repression in budding yeast. Nucleic Acids Res. 2018, 46, 6009–6025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argüello-Miranda, O.; Liu, Y.; Wood, N.E.; Kositangool, P.; Doncic, A. Integration of Multiple Metabolic Signals Determines Cell Fate Prior to Commitment. Mol. Cell 2018, 71, 733–744.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, E.M.; Maxwell, P.H. Low doses of DNA damaging agents extend Saccharomyces cerevisiae chronological lifespan by promoting entry into quiescence. Exp. Gerontol. 2018, 108, 189–200. [Google Scholar] [CrossRef]
- Lee, H.Y.; Chao, J.C.; Cheng, K.Y.; Leu, J.Y. Misfolding-prone proteins are reversibly sequestered to an Hsp42-associated granule upon chronological aging. J. Cell Sci. 2018, 131, jcs220202. [Google Scholar] [CrossRef] [Green Version]
- Becker-Kettern, J.; Paczia, N.; Conrotte, J.F.; Zhu, C.; Fiehn, O.; Jung, P.P.; Steinmetz, L.M.; Linster, C.L. NAD(P)HX repair deficiency causes central metabolic perturbations in yeast and human cells. FEBS J. 2018, 285, 3376–3401. [Google Scholar] [CrossRef] [Green Version]
- Peifer, A.C.; Maxwell, P.H. Preferential Ty1 retromobility in mother cells and nonquiescent stationary phase cells is associated with increased concentrations of total Gag or processed Gag and is inhibited by exposure to a high concentration of calcium. Aging (Albany NY) 2018, 10, 402–424. [Google Scholar] [CrossRef]
- Maqani, N.; Fine, R.D.; Shahid, M.; Li, M.; Enriquez-Hesles, E.; Smith, J.S. Spontaneous mutations in CYC8 and MIG1 suppress the short chronological lifespan of budding yeast lacking SNF1/AMPK. Microb. Cell 2018, 5, 233–248. [Google Scholar] [CrossRef] [Green Version]
- Tomova, A.A.; Kujumdzieva, A.V.; Petrova, V.Y. Carbon source influences Saccharomyces cerevisiae yeast cell survival strategies: Quiescence or sporulation. Biotechnol. Biotechnol. Equip. 2019, 33, 1464–1470. [Google Scholar] [CrossRef] [Green Version]
- Seufert, W.; McGrath, J.P.; Jentsch, S. UBC1 encodes a novel member of an essential subfamily of yeast ubiquitin-conjugating enzymes involved in protein degradation. EMBO J. 1990, 9, 4535–4541. [Google Scholar] [CrossRef]
- Cho, J.E.; Jinks-Robertson, S. Deletions associated with stabilization of the Top1 cleavage complex in yeast are products of the nonhomologous end-joining pathway. Proc. Natl. Acad. Sci. USA 2019, 116, 22683–22691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Martinez-Gomez, K.; Heinzle, E.; Wahl, S.A. Metabolic switches from quiescence to growth in synchronized Saccharomyces cerevisiae. Metabolomics 2019, 15, 121. [Google Scholar] [CrossRef] [Green Version]
- Miles, S.; Li, L.H.; Melville, Z.; Breeden, L.L. Ssd1 and the cell wall integrity pathway promote entry, maintenance, and recovery from quiescence in budding yeast. Mol. Biol. Cell 2019, 30, 2205–2217. [Google Scholar] [CrossRef] [PubMed]
- Bramasole, L.; Sinha, A.; Harshuk, D.; Cirigliano, A.; Gurevich, S.; Yu, Z.; Carmeli, R.L.; Glickman, M.H.; Rinaldi, T.; Pick, E. The proteasome lid triggers COP9 signalosome activity during the transition of Sachharomyces cerevisiae cells into quiescence. Biomolecules 2019, 9, 449. [Google Scholar] [CrossRef] [Green Version]
- Barraza, C.E.; Solari, C.A.; Rinaldi, J.; Ojeda, L.; Rossi, S.; Ashe, M.P.; Portela, P. A prion-like domain of Tpk2 catalytic subunit of protein kinase A modulates P-body formation in response to stress in budding yeast. Biochim. Biophys. Acta-Mol. Cell Res. 2021, 1868, 118884. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Cao, G.; Gao, H.; Li, M.; Peng, G.; Ji, Y.; Zhang, Y.; Zhang, W.; Li, W.; Dou, F. A Single Site Phosphorylation on Hsp82 Ensures Cell Survival during Starvation in Saccharomyces cerevisiae. J. Mol. Biol. 2020, 432, 5809–5824. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Morshed, S.; Mase, S.; Hosoyamada, S.; Kobayashi, T.; Ushimaru, T. Cdc14 protein phosphatase and topoisomerase II mediate rDNA dynamics and nucleophagic degradation of nucleolar proteins after TORC1 inactivation. Cell. Signal. 2021, 79, 109884. [Google Scholar] [CrossRef]
- Yang, R.; Bogdan, P. Controlling the Multifractal Generating Measures of Complex Networks. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Baryshnikova, A.; Brandt, N.; Gresham, D. Genetic interaction profiles of regulatory kinases differ between environmental conditions and cellular states. Mol. Syst. Biol. 2020, 16, 1–18. [Google Scholar] [CrossRef]
- Marek, A.; Opalek, M.; Kałdon, A.; Mickowska, B.; Wloch-Salamon, D. Hypersensitive SSY1 mutations negatively influence transition to quiescence in yeast Saccharomyces cerevisiae. Yeast 2021, 38, 102–116. [Google Scholar] [CrossRef] [PubMed]
- Poon, P.P.; Storms, R.K. The periodically expressed TMP1 gene of saccharomyces cerevisiae is subject to START-dependent and START-independent regulation. J. Biol. Chem. 1991, 266, 16808–16812. [Google Scholar] [CrossRef] [PubMed]
- Baroni, M.D.; Colombo, S.; Libens, O.; Pallavi, R.; Giorgio, M.; Martegani, E.; In, S. cerevisiae hydroxycitric acid antagonizes chronological aging and apoptosis regardless of citrate lyase. Apoptosis 2020, 25, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Poramba-Liyanage, D.W.; Korthout, T.; Cucinotta, C.E.; van Kruijsbergen, I.; van Welsem, T.; El Atmioui, D.; Ovaa, H.; Tsukiyama, T.; van Leeuwen, F. Inhibition of transcription leads to rewiring of locus-specific chromatin proteomes. Genome Res. 2020, 30, 635–646. [Google Scholar] [CrossRef]
- Barré, B.P.; Hallin, J.; Yue, J.X.; Persson, K.; Mikhalev, E.; Irizar, A.; Holt, S.; Thompson, D.; Molin, M.; Warringer, J.; et al. Intragenic repeat expansion in the cell wall protein gene HPF1 controls yeast chronological aging. Genome Res. 2020, 30, 697–710. [Google Scholar] [CrossRef] [Green Version]
- Kwon, Y.Y.; Kim, S.S.; Lee, H.J.; Sheen, S.H.; Kim, K.H.; Lee, C.K. Long-living budding yeast cell subpopulation induced by ethanol/acetate and respiration. Journals Gerontol.-Ser. A Biol. Sci. Med. Sci. 2020, 75, 1448–1456. [Google Scholar] [CrossRef]
- Wood, N.E.; Kositangool, P.; Hariri, H.; Marchand, A.J.; Henne, W.M. Nutrient Signaling, Stress Response, and Inter-organelle Communication Are Non-canonical Determinants of Cell Fate. Cell Rep. 2020, 33, 108446. [Google Scholar] [CrossRef]
- Long, L.J.; Lee, P.-H.; Small, E.M.; Hillyer, C.; Guo, Y.; Osley, M.A. Regulation of UV damage repair in quiescent yeast cells. DNA Repair 2020, 90, 102861. [Google Scholar] [CrossRef]
- Montella-Manuel, S.; Pujol-Carrion, N.; Mechoud, M.A.; de la Torre-Ruiz, M.A. Bulk autophagy induction and life extension is achieved when iron is the only limited nutrient in Saccharomyces cerevisiae. Biochem. J. 2021, 478, 811–837. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, K.; Orfanos, E.; Titorenko, V.I. Caloric restriction causes a distinct reorganization of the lipidome in quiescent and non-quiescent cells of budding yeast. Oncotarget 2021, 12, 2351–2374. [Google Scholar] [CrossRef]
- Argüello-Miranda, O.; Marchand, A.J.; Kennedy, T.; Russo, M.A.X.; Noh, J. Cell cycle–independent integration of stress signals by Xbp1 promotes Non-G1/G0 quiescence entry. J. Cell Biol. 2021, 221, e202103171. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.H.; Osley, M.A. Chromatin structure restricts origin utilization when quiescent cells re-enter the cell cycle. Nucleic Acids Res. 2021, 49, 864–878. [Google Scholar] [CrossRef] [PubMed]
- Nurse, P.; Broek, D. Yeast Cells Can Enter a Quiescent State through G, S, G2, or M Phase of the Cell Cycle. Cancer Res. 1993, 53, 1867–1870. [Google Scholar]
- Wang, R.; Huang, A.; Wang, Y.; Mei, P.; Zhu, H.; Chen, Q.; Xu, S. High-Resolution Microscopy to Learn the Nuclear Organization of the Living Yeast Cells. Stem Cells Int. 2021, 2021, 9951114. [Google Scholar] [CrossRef]
- Nicastro, R.; Raucci, S.; Michel, A.H.; Stumpe, M.; Osuna, G.M.G.; Jaquenoud, M.; Kornmann, B.; de Virgilio, C. Indole-3-acetic acid is a physiological inhibitor of TORC1 in yeast. PLoS Genet. 2021, 17, e1009414. [Google Scholar] [CrossRef]
- Daskalova, A.; Petrova, V.; Velkova, L.; Kujumdzieva, A.; Tomova, A.; Voelter, W.; Dolashka, P. Investigation of protein expression of Saccharomyces cerevisiae cells in quiescent and proliferating state before and after toxic stress. Biotechnol. Biotechnol. Equip. 2021, 35, 366–376. [Google Scholar] [CrossRef]
- Swygert, S.G.; Lin, D.; Portillo-Ledesma, S.; Lin, P.Y.; Hunt, D.R.; Kao, C.F.; Schlick, T.; Noble, W.S.; Tsukiyama, T. Local chromatin fiber folding represses transcription and loop extrusion in quiescent cells. eLife 2021, 10, e72062. [Google Scholar] [CrossRef]
- Daskalova, A.V.; Tomova, A.A.; Kujumdzieva, A.V.; Velkova, L.G.; Dolashka, P.A.; Petrova, V.Y. Menadione and hydrogen peroxide trigger specific alterations in RNA polymerases profiles in quiescent Saccharomyces cerevisiae cells. Biotechnol. Biotechnol. Equip. 2021, 35, 1190–1199. [Google Scholar] [CrossRef]
- Lesage, E.; Perez-Fernandez, J.; Queille, S.; Dez, C.; Gadal, O.; Kwapisz, M. Non-Coding, RNAPII-Dependent Transcription at the Promoters of rRNA Genes Regulates Their Chromatin State in S. cerevisiae. Non-Coding RNA 2021, 7, 41. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Habernig, L.; Broeskamp, F.; Aufschnaiter, A.; Diessl, J.; Atienza, I.; Matz, S.; Ruiz, F.A.; Büttner, S. Phosphate restriction promotes longevity via activation of autophagy and the multivesicular body pathway. Cells 2021, 10, 3161. [Google Scholar] [CrossRef]
- Dokládal, L.; Stumpe, M.; Hu, Z.; Jaquenoud, M.; Dengjel, J.; De Virgilio, C. Phosphoproteomic responses of TORC1 target kinases reveal discrete and convergent mechanisms that orchestrate the quiescence program in yeast. Cell Rep. 2021, 37, 110149. [Google Scholar] [CrossRef]
- Neil, A.J.; Hisey, J.A.; Quasem, I.; McGinty, R.J.; Hitczenko, M.; Khristich, A.N.; Mirkin, S.M. Replication-independent instability of Friedreich’s ataxia GAA repeats during chronological aging. Proc. Natl. Acad. Sci. USA 2021, 118, e2013080118. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, C.E.; Dell, R.H.; Braceros, K.C.A.; Tsukiyama, T. RSC primes the quiescent genome for hypertranscription upon cell-cycle re-entry. eLife 2021, 10, e67033. [Google Scholar] [CrossRef]
- Lennon, K.; Pretel, R.; Kesselheim, J.; Hessen, S.T.; Kukuruzinska, M.A. Proliferation-dependent differential regulation of the dolichol pathway genes in saccharomyces cerevisiae. Glycobiology 1995, 5, 633–642. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.; Ahn, H.; Duan, R.; Liu, Y.; Ryu, H.Y.; Ahn, S.H. The Spt7 subunit of the SAGA complex is required for the regulation of lifespan in both dividing and nondividing yeast cells. Mech. Ageing Dev. 2021, 196, 111480. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.; Egervari, G.; Sidoli, S.; Donahue, G.; Alexander, D.C.; Sen, P.; Garcia, B.A.; Berger, S.L. Enzymatic transfer of acetate on histones from lysine reservoir sites to lysine activating sites. Sci. Adv. 2022, 8, eabj5688. [Google Scholar] [CrossRef] [PubMed]
- Pisareva, E.I.; Tomova, A.A.; Petrova, V.Y. Saccharomyces cerevisiae quiescent cells: Cadmium resistance and adaptive response. Biotechnol. Biotechnol. Equip. 2021, 35, 1827–1837. [Google Scholar] [CrossRef]
- Bailey, T.B.; Whitty, P.A.; Selker, E.U.; McKnight, J.N.; McKnight, L.E. Tup1 is critical for transcriptional repression in Quiescence in S. cerevisiae. PLoS Genet. 2022, 18, e1010559. [Google Scholar] [CrossRef]
- Breeden, L.; Miles, S. A common SSD1 truncation is toxic to cells entering quiescence and promotes sporulation. Micropublication Biol. 2022, 3–7. [Google Scholar] [CrossRef]
- Cesur, M.F.; Çakır, T.; Pir, P. Genome-Wide Analysis of Yeast Metabolic Cycle through Metabolic Network Models Reveals Superiority of Integrated ATAC-seq Data over RNA-seq Data. mSystems 2022, 7, e01347-21. [Google Scholar] [CrossRef]
- Galkina, K.V.; Zubareva, V.M.; Kashko, N.D.; Lapashina, A.S.; Markova, O.V.; Feniouk, B.A.; Knorre, D.A. Heterogeneity of Starved Yeast Cells in IF1 Levels Suggests the Role of This Protein in vivo. Front. Microbiol. 2022, 13, 816622. [Google Scholar] [CrossRef]
- Irvali, D.; Schlottmann, F.P.; Muralidhara, P.; Nadelson, I.; Kleemann, K.; Wood, N.E.; Doncic, A.; Ewald, J.C. When yeast cells change their mind: Cell cycle “Start” is reversible under starvation. EMBO J. 2023, 42, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Leonov, A.; Feldman, R.; Piano, A.; Arlia-Ciommo, A.; Junio, J.A.B.; Orfanos, E.; Tafakori, T.; Lutchman, V.; Mohammad, K.; Elsaser, S.; et al. Diverse geroprotectors differently affect a mechanism linking cellular aging to cellular quiescence in budding yeast. Oncotarget 2022, 13, 918–943. [Google Scholar] [CrossRef] [PubMed]
- Marinovska, P.G.; Todorova, T.I.; Boyadzhiev, K.P.; Pisareva, E.I.; Tomova, A.A.; Parvanova, P.N.; Dimitrova, M.; Chankova, S.G.; Petrova, V.Y. Cellular susceptibility and oxidative stress response to menadione of logarithmic, quiescent, and nonquiescent Saccharomyces cerevisiae cell populations. BioRisk 2022, 17, 127–138. [Google Scholar] [CrossRef]
- Ohtani, K.; DeGregori, J.; Leone, G.; Herendeen, D.R.; Kelly, T.J.; Nevins, J.R. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol. Cell. Biol. 1996, 16, 6977–6984. [Google Scholar] [CrossRef] [Green Version]
- Marinovska, P.; Todorova, T.; Tomova, A.; Pisareva, E.; Boyadzhiev, K.; Dimitrov, M.; Parvanova, P.; Todorova, M.; Chankova, S.; Petrova, V. Saccharomyces cerevisiae yeast cells as a test system for assessing Zeocin toxicity. BioRisk 2022, 17, 105–116. [Google Scholar] [CrossRef]
- Miles, S.; Breeden, L.L. Whi7/Srl3 polymorphisms reveal its role in cell size and quiescence. microPublication Biol. 2022, 2022, 5–8. [Google Scholar] [CrossRef]
- Miyata, N.; Ito, T.; Nakashima, M.; Fujii, S.; Kuge, O. Mitochondrial phosphatidylethanolamine synthesis affects mitochondrial energy metabolism and quiescence entry through attenuation of Snf1/AMPK signaling in yeast. FASEB J. 2022, 36, 1–19. [Google Scholar] [CrossRef]
- Peselj, C.; Ebrahimi, M.; Broeskamp, F.; Prokisch, S.; Habernig, L.; Alvarez-Guerra, I.; Kohler, V.; Vögtle, F.N.; Büttner, S. Sterol Metabolism Differentially Contributes to Maintenance and Exit of Quiescence. Front. Cell Dev. Biol. 2022, 10, 1–17. [Google Scholar] [CrossRef]
- Willis, S.D.; Hanley, S.E.; Doyle, S.J.; Beluch, K.; Strich, R.; Cooper, K.F. Cyclin C-Cdk8 Kinase Phosphorylation of Rim15 Prevents the Aberrant Activation of Stress Response Genes. Front. Cell Dev. Biol. 2022, 10, 1–17. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, G.R. Chromosome-condensed G1 phase yeast cells are tolerant to desiccation stress. Microb. Cell 2022, 9, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Davis, C.; Broach, J.R. Efficient transition to growth on fermentable carbon sources in Saccharomyces cerevisiae requires signaling through the Ras pathway. EMBO J. 1998, 17, 6942–6951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paz, I.; Meunier, J.R.; Choder, M. Monitoring dynamics of gene expression in yeast during stationary phase. Gene 1999, 236, 33–42. [Google Scholar] [CrossRef]
- McHugh, P.J.; Sones, W.R.; Hartley, J.A. Repair of Intermediate Structures Produced at DNA Interstrand Cross-Links in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000, 20, 3425–3433. [Google Scholar] [CrossRef] [PubMed]
- Paz, I.; Choder, M. Eukaryotic translation initiation factor 4E-dependent translation is not essential for survival of starved yeast cells. J. Bacteriol. 2001, 183, 4477–4483. [Google Scholar] [CrossRef] [Green Version]
- Krause, S.A.; Gray, J.V. The protein kinase C pathway is required for viability in quiescence in Saccharomyces cerevisiae. Curr. Biol. 2002, 12, 588–593. [Google Scholar] [CrossRef] [Green Version]
- Werner-Washburne, M.; Wylie, B.; Boyack, K.; Fuge, E.; Galbraith, J.; Weber, J.; Davidson, G. Comparative analysis of multiple genome-scale data sets. Genome Res. 2002, 12, 1564–1573. [Google Scholar] [CrossRef] [Green Version]
- Piper, P.W.; Bringloe, D. Loss of prohibitins, though it shortens the replicative life span of yeast cells undergoing division, does not shorten the chronological life span of G0-arrested cells. Mech. Ageing Dev. 2002, 123, 287–295. [Google Scholar] [CrossRef]
- Sousa-Lopes, A.; Antunes, F.; Cyrne, L.; Marinho, H.S. Decreased cellular permeability to H 2O 2 protects Saccharomyces cerevisiae cells in stationary phase against oxidative stress. FEBS Lett. 2004, 578, 152–156. [Google Scholar] [CrossRef] [Green Version]
- Hossain, M.S.; Kurokawa, K.; Akimitsu, N.; Sekimizu, K. DNA topoisomerase II is required for the G0-to-S phase transition in Drosophila Schneider cells, but not in yeast. Genes to Cells 2004, 9, 905–917. [Google Scholar] [CrossRef]
- Martinez, M.J.; Roy, S.; Archuletta, A.B.; Wentzell, P.D.; Anna-Arriola, S.S.; Rodriguez, A.L.; Aragon, A.D.; Quiñones, G.A.; Allen, C.; Werner-Washburne, M. Genomic Analysis of Stationary-Phase and Exit in Saccharomyces cerevisiae: Gene Expression and Identification of Novel Essential Genes. Mol. Biol. Cell 2004, 15, 5295–5305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frankenberg-Schwager, M.; Kirchermeier, D.; Greif, G.; Baer, K.; Becker, M.; Frankenberg, D. Cisplatin-mediated DNA double-strand breaks in replicating but not in quiescent cells of the yeast Saccharomyces cerevisiae. Toxicology 2005, 212, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Sagot, I.; Schaeffer, J.; Daignan-Fornier, B. Guanylic nucleotide starvation affects Saccharomyces cerevisiae mother-daughter separation and may be a signal for entry into quiescence. BMC Cell Biol. 2005, 6, 24. [Google Scholar] [CrossRef] [Green Version]
- Radonjic, M.; Andrau, J.C.; Lijnzaad, P.; Kemmeren, P.; Kockelkorn, T.T.J.P.; Van Leenen, D.; Van Berkum, N.L.; Holstege, F.C.P. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell 2005, 18, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Wanke, V.; Pedruzzi, I.; Cameroni, E.; Dubouloz, F.; De Virgilio, C. Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J. 2005, 24, 4271–4278. [Google Scholar] [CrossRef] [Green Version]
- Dubouloz, F.; Deloche, O.; Wanke, V.; Cameroni, E.; De Virgilio, C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell 2005, 19, 15–26. [Google Scholar] [CrossRef]
- Sagot, I.; Pinson, B.; Salin, B.; Daignan-Fornier, B. Actin Bodies in Yeast Quiescent Cells: An Immediately Available Actin Reserve? Mol. Biol. Cell 2006, 17, 4645–4655. [Google Scholar] [CrossRef] [Green Version]
- Escusa, S.; Camblong, J.; Galan, J.M.; Pinson, B.; Daignan-Fornier, B. Proteasome- and SCF-dependent degradation of yeast adenine deaminase upon transition from proliferation to quiescence requires a new F-box protein named Saf1p. Mol. Microbiol. 2006, 60, 1014–1025. [Google Scholar] [CrossRef]
- Aragon, A.D.; Quiñones, G.A.; Thomas, E.V.; Roy, S.; Werner-Washburne, M. Release of extraction-resistant mRNA in stationary phase Saccharomyces cerevisiae produces a massive increase in transcript abundance in response to stress. Genome Biol. 2006, 7, R9. [Google Scholar] [CrossRef] [Green Version]
- Slattery, M.G.; Heideman, W. Coordinated regulation of growth genes in Saccharomyces cerevisiae. Cell Cycle 2007, 6, 1210–1219. [Google Scholar] [CrossRef]
- Weinberger, M.; Feng, L.; Paul, A.; Smith, D.L.; Hontz, R.D.; Smith, J.S.; Vujcic, M.; Singh, K.K.; Huberman, J.A.; Burhans, W.C. DNA replication stress is a determinant of chronological lifespan in budding yeast. PLoS ONE 2007, 2, e748. [Google Scholar] [CrossRef] [PubMed]
- Gomes, P.; Sampaio-Marques, B.; Ludovico, P.; Rodrigues, F.; Leão, C. Low auxotrophy-complementing amino acid concentrations reduce yeast chronological life span. Mech. Ageing Dev. 2007, 128, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Escusa, S.; Laporte, D.; Massoni, A.; Boucherie, H.; Dautant, A.; Daignan-Fornier, B. Skp1-Cullin-F-box-dependent degradation of Aah1p requires its interaction with the F-box protein Saf1p. J. Biol. Chem. 2007, 282, 20097–20103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liko, D.; Slattery, M.G.; Heideman, W. Stb3 binds to ribosomal RNA processing element motifs that control transcriptional responses to growth in Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 26623–26628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, C.J.; Burtner, C.R.; Kennedy, B.K.; Kaeberlein, M. A method for high-throughput quantitative analysis of yeast chronological life span. J. Gerontol.-Ser. A Biol. Sci. Med. Sci. 2008, 63, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Aragon, A.D.; Rodriguez, A.L.; Meirelles, O.; Roy, S.; Davidson, G.S.; Tapia, P.H.; Allen, C.; Joe, R.; Benn, D.; Werner-Washburne, M. Characterization of Differentiated Quiescent and Nonquiescent Cells in Yeast Stationary-Phase Cultures. Mol. Biol. Cell 2008, 19, 1271–1280. [Google Scholar] [CrossRef] [Green Version]
- Sahin, A.; Daignan-Fornier, B.; Sagot, I. Polarized growth in the absence of F-actin in Saccharomyces cerevisiae exiting quiescence. PLoS ONE 2008, 3, e2556. [Google Scholar] [CrossRef] [Green Version]
- Laporte, D.; Salin, B.; Daignan-Fornier, B.; Sagot, I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 2008, 181, 737–745. [Google Scholar] [CrossRef] [Green Version]
- Friis, R.M.N.; Wu, B.P.; Reinke, S.N.; Hockman, D.J.; Sykes, B.D.; Schultz, M.C. A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res. 2009, 37, 3969–3980. [Google Scholar] [CrossRef] [Green Version]
- Barea, F.; Bonatto, D. Aging defined by a chronologic-replicative protein network in Saccharomyces cerevisiae: An interactome analysis. Mech. Ageing Dev. 2009, 130, 444–460. [Google Scholar] [CrossRef]
- Li, L.; Lu, Y.; Qin, L.-X.; Bar-Joseph, Z.; Werner-Washburne, M.; Breeden, L.L. Budding Yeast SSD1-V Regulates Transcript Levels of Many Longevity Genes and Extends Chronological Life Span in Purified Quiescent Cells. Mol. Biol. Cell 2009, 20, 3851–3864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldberg, A.A.; Bourque, S.D.; Kyryakov, P.; Gregg, C.; Boukh-Viner, T.; Beach, A.; Burstein, M.T.; Machkalyan, G.; Richard, V.; Rampersad, S.; et al. Effect of calorie restriction on the metabolic history of chronologically aging yeast. Exp. Gerontol. 2009, 44, 555–571. [Google Scholar] [CrossRef] [PubMed]
- Benbadis, L.; Cot, M.; Rigoulet, M.; Francois, J. Isolation of two cell populations from yeast during high-level alcoholic fermentation that resemble quiescent and nonquiescent cells from the stationary phase on glucose. FEMS Yeast Res. 2009, 9, 1172–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minois, N.; Lagona, F.; Frajnt, M.; Vaupel, J.W. Plasticity of death rates in stationary phase in Saccharomyces cerevisiae. Aging Cell 2009, 8, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Tapia, H.; Morano, K.A. Hsp90 Nuclear Accumulation in Quiescence Is Linked to Chaperone Function and Spore Development in Yeast. Mol. Biol. Cell 2010, 21, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Fabrizio, P.; Hoon, S.; Shamalnasab, M.; Galbani, A.; Wei, M.; Giaever, G.; Nislow, C.; Longo, V.D. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet. 2010, 6, e1001024. [Google Scholar] [CrossRef] [Green Version]
- Weinberger, M.; Mesquita, A.; Carroll, T.; Marks, L.; Yang, H.; Zhang, Z.; Ludovico, P.; Burhans, W.C. Growth signaling promotes chronological aging in budding yeast by inducing superoxide anions that inhibit quiescence. Aging (Albany NY) 2010, 2, 709–726. [Google Scholar] [CrossRef] [Green Version]
- Talarek, N.; Cameroni, E.; Jaquenoud, M.; Luo, X.; Bontron, S.; Lippman, S.; Devgan, G.; Snyder, M.; Broach, J.R.; De Virgilio, C. Initiation of the TORC1-Regulated G0 Program Requires Igo1/2, which License Specific mRNAs to Evade Degradation via the 5′-3′ mRNA Decay Pathway. Mol. Cell 2010, 38, 345–355. [Google Scholar] [CrossRef] [Green Version]
- Petkova, M.I.; Pujol-Carrion, N.; Arroyo, J.; García-Cantalejo, J.; De La Torre-Ruiz, M.A. Mtl1 is required to activate general stress response through TOR1 and RAS2 inhibition under conditions of glucose starvation and oxidative stress. J. Biol. Chem. 2010, 285, 19521–19531. [Google Scholar] [CrossRef] [Green Version]
- Liko, D.; Conway, M.K.; Grunwald, D.S.; Heideman, W. Stb3 plays a role in the glucose-induced transition from quiescence to growth in Saccharomyces cerevisiae. Genetics 2010, 185, 797–810. [Google Scholar] [CrossRef] [Green Version]
- Gresham, D.; Boer, V.M.; Caudy, A.; Ziv, N.; Brandt, N.J.; Storey, J.D.; Botstein, D. System-level analysis of genes and functions affecting survival during nutrient starvation in Saccharomyces cerevisiae. Genetics 2011, 187, 299–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vodenicharov, M.D.; Laterreur, N.; Wellinger, R.J. Telomere capping in non-dividing yeast cells requires Yku and Rap1. EMBO J. 2010, 29, 3007–3019. [Google Scholar] [CrossRef] [Green Version]
- Emerman, A.B.; Zhang, Z.-R.; Chakrabarti, O.; Hegde, R.S. Trehalose Is a Key Determinant of the Quiescent Metabolic State That Fuels Cell Cycle Progression upon Return to Growth. Mol. Biol. Cell 2010, 21, 4325–4337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Werner-Washburne, M.; Lane, T. A multiple network learning approach to capture system-wide condition-specific responses. Bioinformatics 2011, 27, 1832–1838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boender, L.G.M.; van Maris, A.J.A.; de Hulster, E.A.F.; Almering, M.J.H.; van der Klei, I.J.; Veenhuis, M.; de Winde, J.H.; Pronk, J.T.; Daran-Lapujade, P. Cellular responses of Saccharomyces cerevisiae at near-zero growth rates: Transcriptome analysis of anaerobic retentostat cultures. FEMS Yeast Res. 2011, 11, 603–620. [Google Scholar] [CrossRef] [Green Version]
- Harper, J.C.; Lopez, D.M.; Larkin, E.C.; Economides, M.K.; McIntyre, S.K.; Alam, T.M.; Tartis, M.S.; Werner-Washburne, M.; Brinker, C.J.; Brozik, S.M.; et al. Encapsulation of S. cerevisiae in poly(glycerol) silicate derived matrices: Effect of matrix additives and cell metabolic phase on long-term viability and rate of gene expression. Chem. Mater. 2011, 23, 2555–2564. [Google Scholar] [CrossRef]
- Boender, L.G.M.; Almering, M.J.H.; Dijk, M.; van Maris, A.J.A.; de Winde, J.H.; Pronk, J.T.; Daran-Lapujade, P. Extreme calorie restriction and energy source starvation in Saccharomyces cerevisiae represent distinct physiological states. Biochim. Biophys. Acta-Mol. Cell Res. 2011, 1813, 2133–2144. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Talarek, N.; De Virgilio, C. Initiation of the yeast G0 program requires Igo1 and Igo2, which antagonize activation of decapping of specific nutrient-regulated mRNAs. RNA Biol. 2011, 8, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Slavov, N.; Macinskas, J.; Caudy, A.; Botstein, D. Metabolic cycling without cell division cycling in respiring yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 19090–19095. [Google Scholar] [CrossRef] [Green Version]
- Laporte, D.; Lebaudy, A.; Sahin, A.; Pinson, B.; Ceschin, J.; Daignan-Fornier, B.; Sagot, I. Metabolic status rather than cell cycle signals control quiescence entry and exit. J. Cell Biol. 2011, 192, 949–957. [Google Scholar] [CrossRef] [Green Version]
- Kelly, M.K.; Alver, B.; Kirkpatrick, D.T. Minisatellite alterations in ZRT1 mutants occur via RAD52-dependent and RAD52-independent mechanisms in quiescent stationary phase yeast cells. DNA Repair 2011, 10, 556–566. [Google Scholar] [CrossRef] [Green Version]
- Ngubo, M.; Kemp, G.; Patterton, H.G. Nano-electrospray tandem mass spectrometric analysis of the acetylation state of histones H3 and H4 in stationary phase in Saccharomyces cerevisiae. BMC Biochem. 2011, 12, 34. [Google Scholar] [CrossRef] [Green Version]
- Zakrajšek, T.; Raspor, P.; Jamnik, P. Saccharomyces cerevisiae in the stationary phase as a model organism—characterization at cellular and proteome level. J. Proteomics 2011, 74, 2837–2845. [Google Scholar] [CrossRef]
- Urbanczyk, H.; Noguchi, C.; Wu, H.; Watanabe, D.; Akao, T.; Takagi, H.; Shimoi, H. Sake yeast strains have difficulty in entering a quiescent state after cell growth cessation. J. Biosci. Bioeng. 2011, 112, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, V.; Shah, K.H.; Herman, P.K. The cAMP-Dependent Protein Kinase Signaling Pathway Is a Key Regulator of P Body Foci Formation. Mol. Cell 2011, 43, 973–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, G.S.; Joe, R.M.; Roy, S.; Meirelles, O.; Allen, C.P.; Wilson, M.R.; Tapia, P.H.; Manzanilla, E.E.; Dodson, A.E.; Chakraborty, S.; et al. The proteomics of quiescent and nonquiescent cell differentiation in yeast stationary-phase cultures. Mol. Biol. Cell 2011, 22, 988–998. [Google Scholar] [CrossRef]
- Bonzanni, N.; Zhang, N.; Oliver, S.G.; Fisher, J. The role of proteosome-mediated proteolysis in modulating potentially harmful transcription factor activity in Saccharomyces cerevisiae. Bioinformatics 2011, 27, 283–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escoté, X.; Miranda, M.; Rodríguez-Porrata, B.; Mas, A.; Cordero, R.; Posas, F.; Vendrell, J. The stress-activated protein kinase Hog1 develops a critical role after resting state. Mol. Microbiol. 2011, 80, 423–435. [Google Scholar] [CrossRef]
- Watanabe, D.; Araki, Y.; Zhou, Y.; Maeya, N.; Akao, T.; Shimoi, H. A Loss-of-Function Mutation in the PAS Kinase Rim15p Is Related to Defective Quiescence Entry and High Fermentation Rates of Saccharomyces cerevisiae Sake Yeast Strains. Appl. Environ. Microbiol. 2012, 78, 4008–4016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyryakov, P.; Beach, A.; Richard, V.R.; Burstein, M.T.; Leonov, A.; Levy, S.; Titorenko, V.I. Caloric restriction extends yeast chronological lifespan by altering a pattern of age-related changes in trehalose concentration. Front. Physiol. 2012, 3, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conway, M.K.; Grunwald, D.; Heideman, W. Glucose, nitrogen, and phosphate repletion in saccharomyces cerevisiae: Common transcriptional responses to different nutrient signals. G3 Genes Genomes Genet. 2012, 2, 1003–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burstein, M.T.; Kyryakov, P.; Beach, A.; Richard, V.R.; Koupaki, O.; Gomez-Perez, A.; Leonov, A.; Levy, S.; Noohi, F.; Titorenko, V.I. Lithocholic acid extends longevity of chronologically aging yeast only if added at certain critical periods of their lifespan. Cell Cycle 2012, 11, 3443–3462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reimand, J.; Aun, A.; Vilo, J.; Vaquerizas, J.M.; Sedman, J.; Luscombe, N.M. M:Explorer: Multinomial regression models reveal positive and negative regulators of longevity in yeast quiescence. Genome Biol. 2012, 13, R55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petkova, M.I.; Pujol-Carrion, N.; de la Torre-Ruiz, M.A. Mtl1 O-mannosylation mediated by both Pmt1 and Pmt2 is important for cell survival under oxidative conditions and TOR blockade. Fungal Genet. Biol. 2012, 49, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.K.; Brosnan, L.; Jauert, P.A.; Dunham, M.J.; Kirkpatrick, D.T. Multiple pathways regulate minisatellite stability during stationary phase in yeast. G3 Genes Genomes Genet. 2012, 2, 1185–1195. [Google Scholar] [CrossRef] [Green Version]
- Murakami, C.; Delaney, J.R.; Chou, A.; Carr, D.; Schleit, J.; Sutphin, G.L.; An, E.H.; Castanza, A.S.; Fletcher, M.; Goswami, S.; et al. pH neutralization protects against reduction in replicative lifespan following chronological aging in yeast. Cell Cycle 2012, 11, 3087–3096. [Google Scholar] [CrossRef] [Green Version]
- Hanna, J.; Waterman, D.; Boselli, M.; Finley, D. Spg5 protein regulates the proteasome in quiescence. J. Biol. Chem. 2012, 287, 34400–34409. [Google Scholar] [CrossRef] [Green Version]
- Liu, I.C.; Chiu, S.W.; Lee, H.Y.; Leu, J.Y. The histone deacetylase Hos2 forms an Hsp42-dependent cytoplasmic granule in quiescent yeast cells. Mol. Biol. Cell 2012, 23, 1231–1242. [Google Scholar] [CrossRef]
- Alver, B.; Jauert, P.A.; Brosnan, L.; O’Hehir, M.; VanderSluis, B.; Myers, C.L.; Kirkpatrick, D.T. A whole genome screen for minisatellite stability genes in stationary-phase yeast cells. G3 Genes Genomes Genet. 2013, 3, 741–756. [Google Scholar] [CrossRef] [Green Version]
- Lei, S.; Tu, B.P. Acetyl-CoA induces transcription of the key G1 cyclin CLN3 to promote entry into the cell division cycle in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2013, 110, 7318–7323. [Google Scholar] [CrossRef] [Green Version]
- Laporte, D.; Courtout, F.; Salin, B.; Ceschin, J.; Sagot, I. An array of nuclear microtubules reorganizes the budding yeast nucleus during quiescence. J. Cell Biol. 2013, 203, 585–594. [Google Scholar] [CrossRef] [Green Version]
- Weinberger, M.; Sampaio-Marques, B.; Ludovico, P.; Burhans, W.C. DNA replication stress-induced loss of reproductive capacity in s. cerevisiae and its inhibition by caloric restriction. Cell Cycle 2013, 12, 1189–1200. [Google Scholar] [CrossRef] [Green Version]
- Saunier, R.; Esposito, M.; Dassa, E.P.; Delahodde, A. Integrity of the Saccharomyces cerevisiae Rpn11 Protein Is Critical for Formation of Proteasome Storage Granules (PSG) and Survival in Stationary Phase. PLoS ONE 2013, 8, e70357. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Miles, S.; Melville, Z.; Prasad, A.; Bradley, G.; Breeden, L.L. Key events during the transition from rapid growth to quiescence in budding yeast require posttranscriptional regulators. Mol. Biol. Cell 2013, 24, 3697–3709. [Google Scholar] [CrossRef] [PubMed]
- Casatta, N.; Porro, A.; Orlandi, I.; Brambilla, L.; Vai, M. Lack of Sir2 increases acetate consumption and decreases extracellular pro-aging factors. Biochim. Biophys. Acta-Mol. Cell Res. 2013, 1833, 593–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webb, K.J.; Xu, T.; Park, S.K.; Yates, J.R. Modified MuDPIT Separation Identified 4488 Proteins in a System-wide Analysis of Quiescence in Yeast. J. Proteome Res. 2013, 12, 2177–2184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alver, B.; Kelly, M.K.; Kirkpatrick, D.T. Novel Checkpoint Pathway Organization Promotes Genome Stability in Stationary-Phase Yeast Cells. Mol. Cell. Biol. 2013, 33, 457–472. [Google Scholar] [CrossRef] [Green Version]
- Shah, K.H.; Zhang, B.; Ramachandran, V.; Herman, P.K. Processing body and stress granule assembly occur by independent and Differentially regulated pathways in Saccharomyces cerevisiae. Genetics 2013, 193, 109–123. [Google Scholar] [CrossRef] [Green Version]
- Lee, P.; Kim, M.S.; Paik, S.M.; Choi, S.H.; Cho, B.R.; Hahn, J.S. Rim15-dependent activation of Hsf1 and Msn2/4 transcription factors by direct phosphorylation in Saccharomyces cerevisiae. FEBS Lett. 2013, 587, 3648–3655. [Google Scholar] [CrossRef] [Green Version]
- Miles, S.; Li, L.; Davison, J.; Breeden, L.L. Xbp1 Directs Global Repression of Budding Yeast Transcription during the Transition to Quiescence and Is Important for the Longevity and Reversibility of the Quiescent State. PLoS Genet. 2013, 9, e1003854. [Google Scholar] [CrossRef] [Green Version]
- Bontron, S.; Jaquenoud, M.; Vaga, S.; Talarek, N.; Bodenmiller, B.; Aebersold, R.; De Virgilio, C. Yeast Endosulfines Control Entry into Quiescence and Chronological Life Span by Inhibiting Protein Phosphatase 2A. Cell Rep. 2013, 3, 16–22. [Google Scholar] [CrossRef] [Green Version]
- Palkova, Z. Multicellular microorganisms: Laboratory versus nature. EMBO Rep. 2004, 5, 470–476. [Google Scholar] [CrossRef] [Green Version]
- Mozzachiodi, S.; Bai, F.Y.; Baldrian, P.; Bell, G.; Boundy-Mills, K.; Buzzini, P.; Čadež, N.; Cubillos, F.A.; Dashko, S.; Dimitrov, R.; et al. Yeasts from temperate forests. Yeast 2022, 39, 4–24. [Google Scholar] [CrossRef] [PubMed]
- Finkel, S.E.; Kolter, R. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. USA 1999, 96, 4023–4027. [Google Scholar] [CrossRef] [Green Version]
- Zambrano, M.M.; Kolter, R. GASPing for Life in Stationary Phase. Cell 1996, 86, 181–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aouizerat, T.; Gelman, D.; Szitenberg, A.; Gutman, I.; Glazer, S.; Reich, E.; Schoemann, M.; Kaplan, R.; Saragovi, A.; Hazan, R.; et al. Eukaryotic Adaptation to Years-Long Starvation Resembles that of Bacteria. iScience 2019, 19, 545–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Opalek, M.; Tutaj, H.; Pirog, A.; Smug, B.J.; Rutkowska, J.; Wloch-Salamon, D. A Systematic Review on Quiescent State Research Approaches in S. cerevisiae. Cells 2023, 12, 1608. https://doi.org/10.3390/cells12121608
Opalek M, Tutaj H, Pirog A, Smug BJ, Rutkowska J, Wloch-Salamon D. A Systematic Review on Quiescent State Research Approaches in S. cerevisiae. Cells. 2023; 12(12):1608. https://doi.org/10.3390/cells12121608
Chicago/Turabian StyleOpalek, Monika, Hanna Tutaj, Adrian Pirog, Bogna J. Smug, Joanna Rutkowska, and Dominika Wloch-Salamon. 2023. "A Systematic Review on Quiescent State Research Approaches in S. cerevisiae" Cells 12, no. 12: 1608. https://doi.org/10.3390/cells12121608
APA StyleOpalek, M., Tutaj, H., Pirog, A., Smug, B. J., Rutkowska, J., & Wloch-Salamon, D. (2023). A Systematic Review on Quiescent State Research Approaches in S. cerevisiae. Cells, 12(12), 1608. https://doi.org/10.3390/cells12121608





