Independent Membrane Binding Properties of the Caspase Generated Fragments of the Beaded Filament Structural Protein 1 (BFSP1) Involves an Amphipathic Helix
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analyses
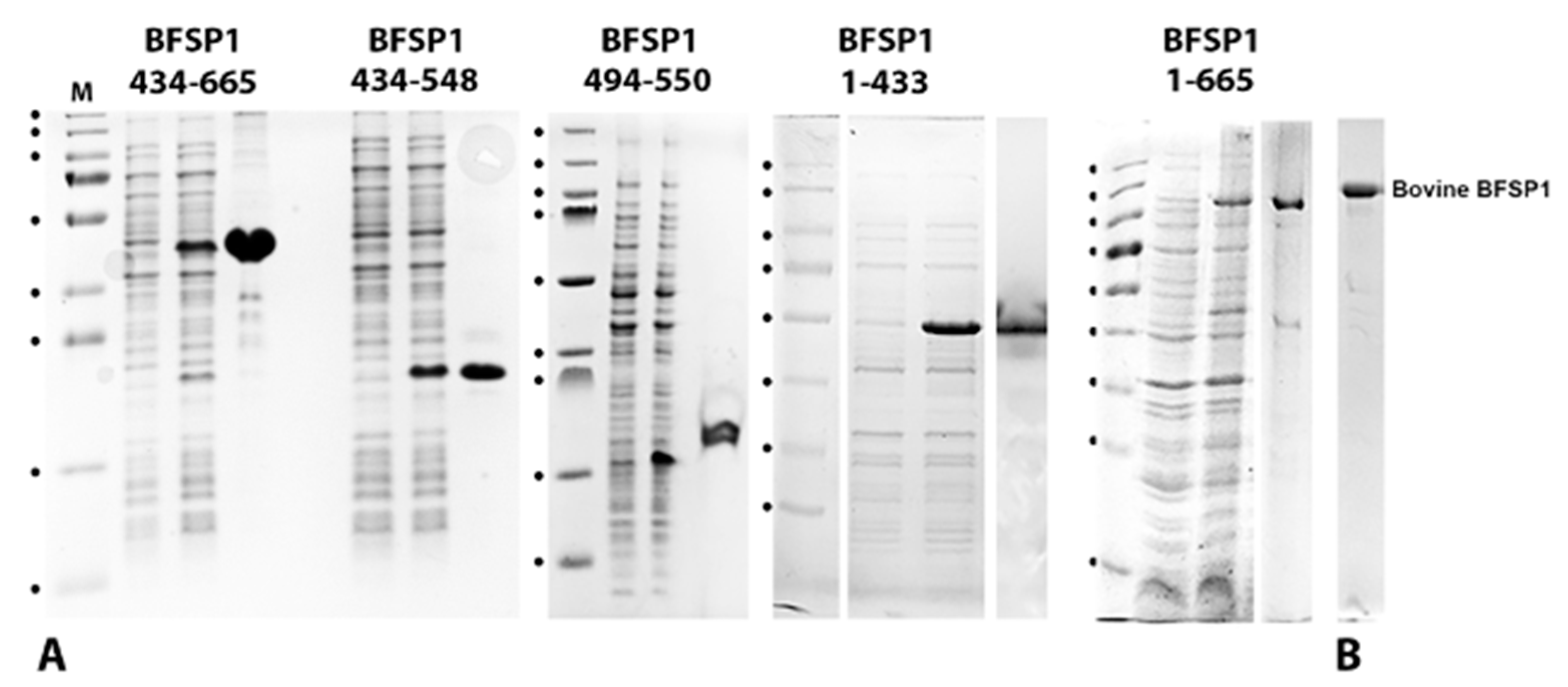
2.2. Cloning, Expression and Purification of Human BFSP1, Its Fragments and CRYAB
2.3. Extraction of Lipids from Bovine Lens Cell Membranes
2.4. Purification of Lens Membrane Lipids for SPR
2.5. Surface Plasmon Resonance to Measure Lipid-Binding of BFSP1 Constructs
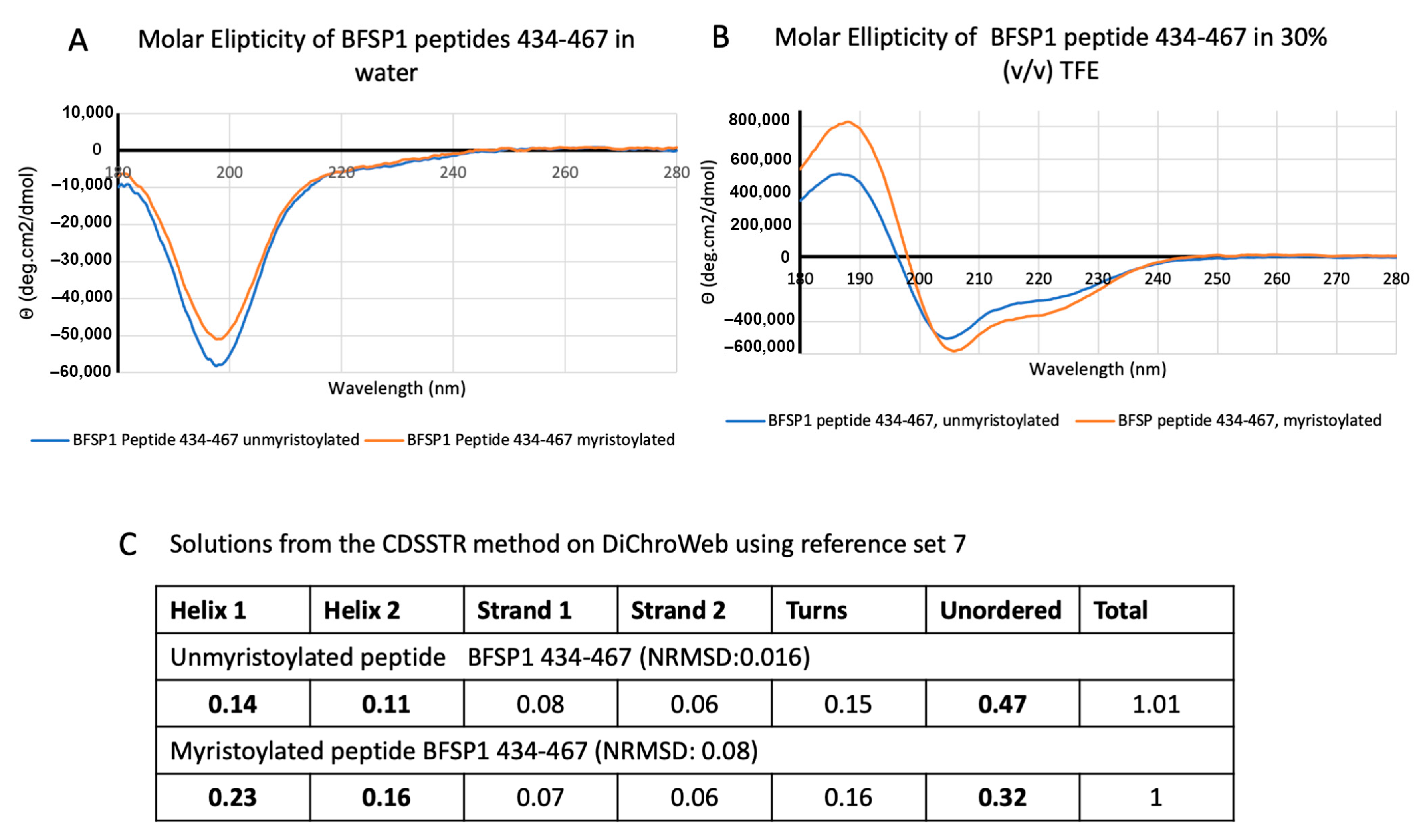
2.6. Circular Dichroism Measurements
2.7. Proteomic Analyses of Bovine Lens Membrane Preparations
2.8. SDS PAGE and Immunoblotting
2.9. Eukaryotic Cell Culture, Incubation with Recombinant BFSP1 Fragments, Transient Transfections of BFSP1 Constructs and Live Cell Imaging
3. Results
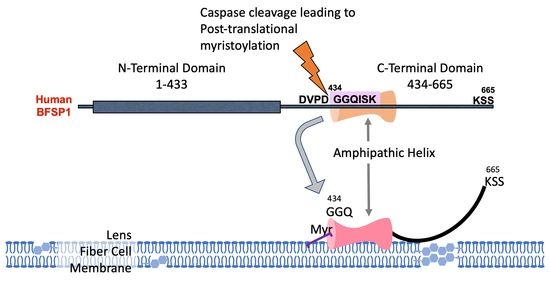
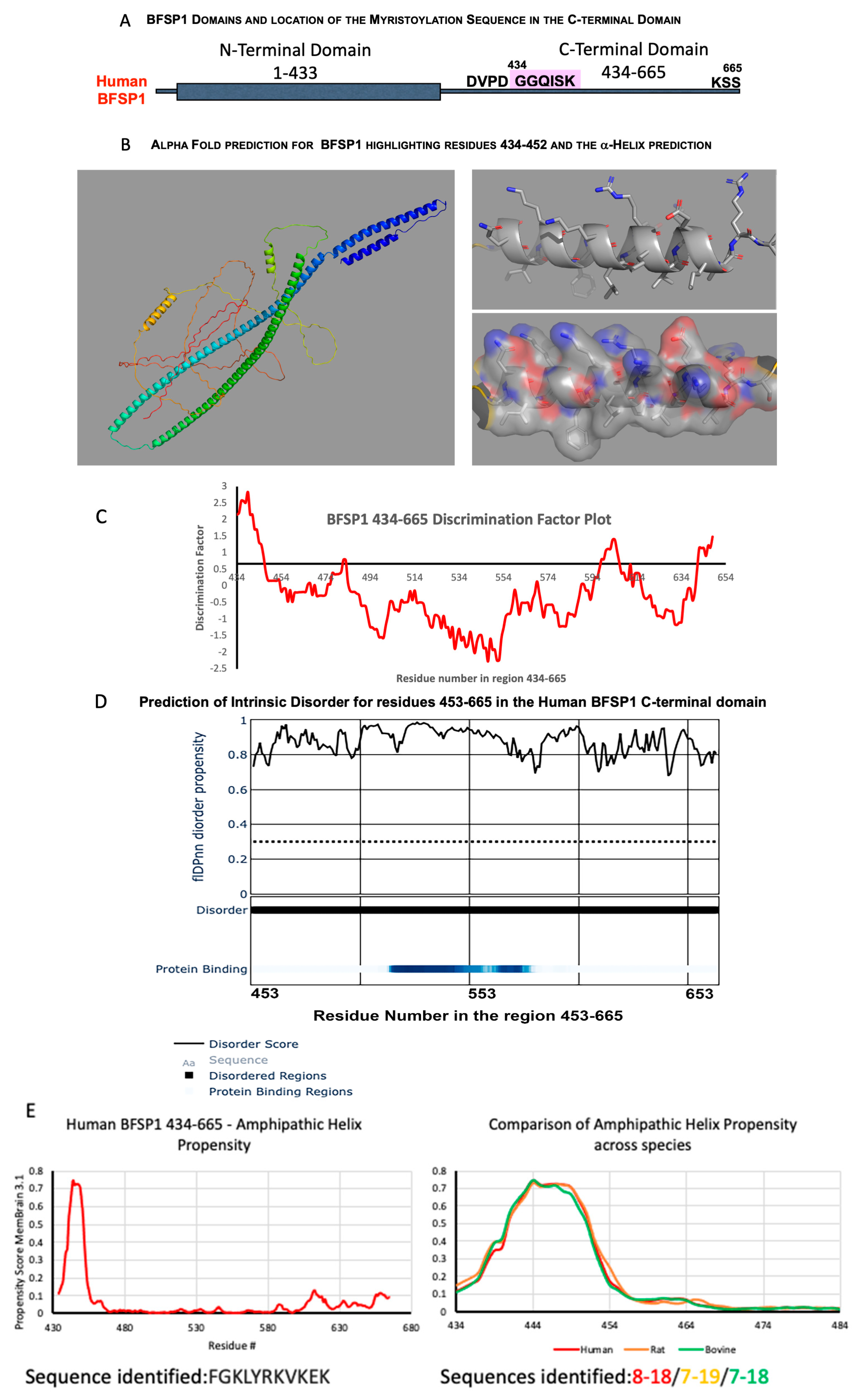
3.1. The C-Terminal Sequences of BFSP1 Contain an Amphipathic Helix That Spans the Myristoylation Motif and Predicted α-Helix
3.2. The BFSP1 C-Terminal Sequences Distal to 434 Bind Lipid Membranes Independently of AQP0
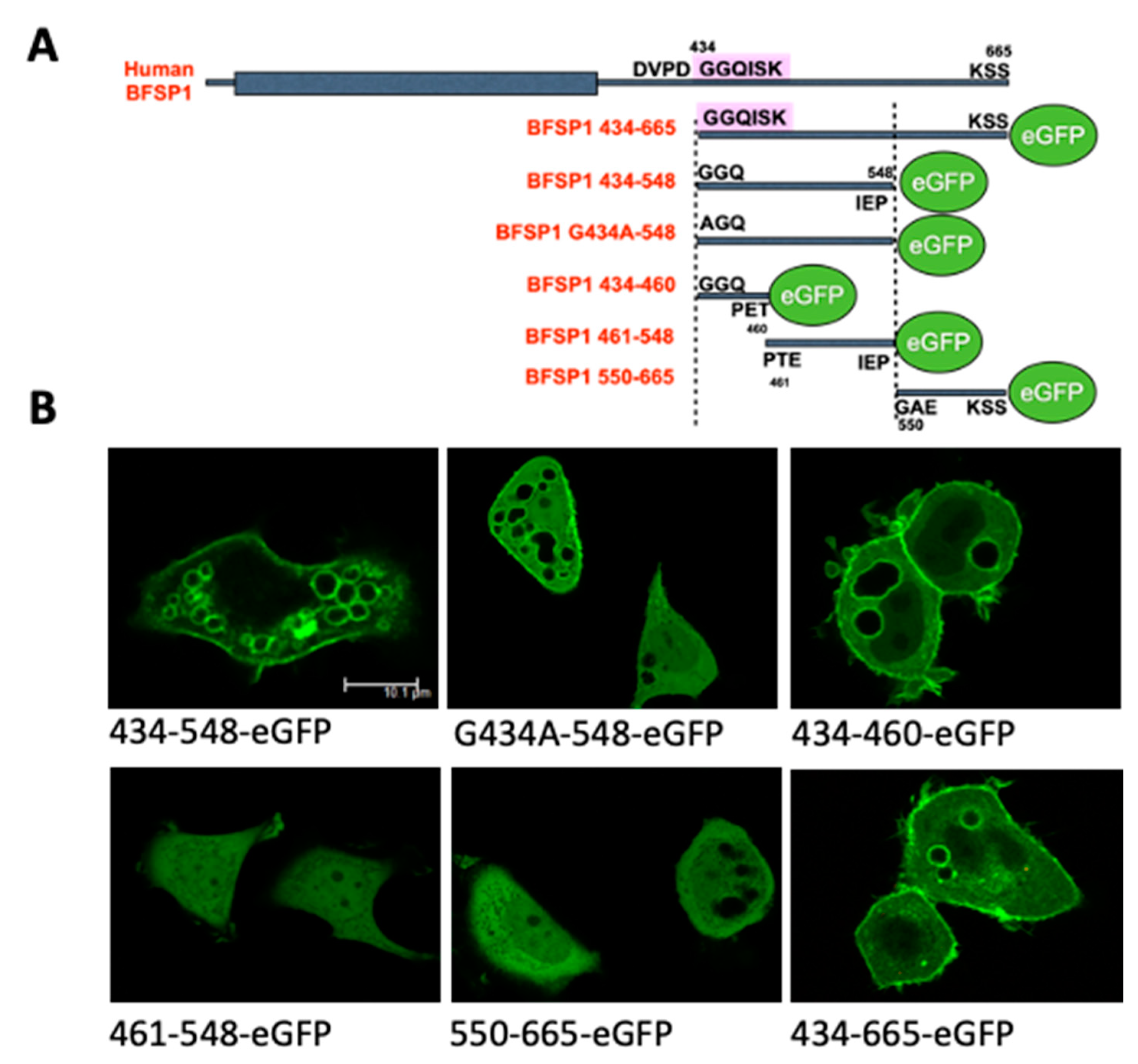
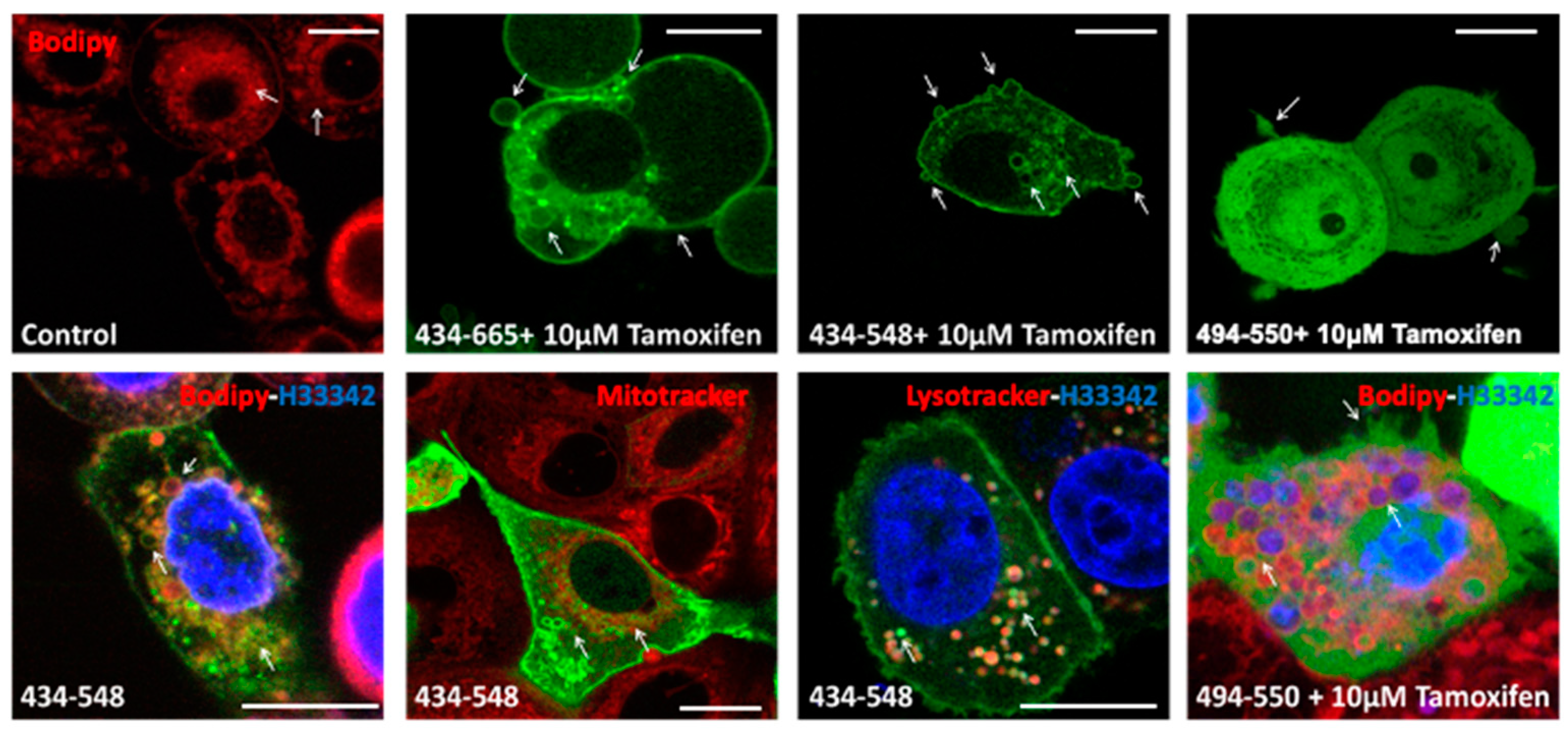
3.3. Transient Transfections of eGFP Labelled BFSP1 Constructs Confirms Membrane Association for Fragments Containing the Amphipathic Helix
4. Discussion
4.1. BFSP1 Contains an Amphipathic Helix at Residues 434–452
4.2. Potential Contribution of the Amphipathic Helix and Myristoylation to BFSP1 Structure and Function in the Lens
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quinlan, R.A.; Clark, J.I. Insights into the Biochemical and Biophysical Mechanisms Mediating The Longevity of the Transparent Optics of the Eye Lens. J. Biol. Chem. 2022, 298, 102537. [Google Scholar] [CrossRef] [PubMed]
- Perng, M.D.; Quinlan, R.A. Seeing is believing! The optical properties of the eye lens are dependent upon a functional intermediate filament cytoskeleton. Exp. Cell. Res. 2005, 305, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Carter, J.M.; Hutcheson, A.M.; Quinlan, R.A. In vitro studies on the assembly properties of the lens beaded filament proteins: Co-assembly with a-crystallin but not with vimentin. Exp. Eye Res. 1995, 60, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Maisel, H.; Perry, M.M. Electron microscope observations on some structural proteins of the chick lens. Exp. Eye Res. 1972, 14, 7–12. [Google Scholar] [CrossRef]
- Song, S.; Landsbury, A.; Dahm, R.; Liu, Y.; Zhang, Q.; Quinlan, R.A. Functions of the intermediate filament cytoskeleton in the eye lens. J. Clin. Investig. 2009, 119, 1837–1848. [Google Scholar] [CrossRef]
- Shiels, A.; Hejtmancik, J.F. Inherited cataracts: Genetic mechanisms and pathways new and old. Exp. Eye Res. 2021, 209, 108662. [Google Scholar] [CrossRef]
- Fudge, D.S.; McCuaig, J.V.; Van Stralen, S.; Hess, J.F.; Wang, H.; Mathias, R.T.; FitzGerald, P.G. Intermediate filaments regulate tissue size and stiffness in the murine lens. Investig. Ophthalmol. Vis. Sci. 2011, 52, 3860–3867. [Google Scholar] [CrossRef]
- Sandilands, A.; Prescott, A.R.; Wegener, A.; Zoltoski, R.K.; Hutcheson, A.M.; Masaki, S.; Kuszak, J.R.; Quinlan, R.A. Knockout of the intermediate filament protein CP49 destabilises the lens fibre cell cytoskeleton and decreases lens optical quality, but does not induce cataract. Exp. Eye Res. 2003, 76, 385–391. [Google Scholar] [CrossRef]
- Sandilands, A.; Wang, X.; Hutcheson, A.M.; James, J.; Prescott, A.R.; Wegener, A.; Pekny, M.; Gong, X.; Quinlan, R.A. Bfsp2 mutation found in mouse 129 strains causes the loss of CP49 and induces vimentin-dependent changes in the lens fibre cell cytoskeleton. Exp. Eye Res. 2004, 78, 109–123. [Google Scholar] [CrossRef]
- Alizadeh, A.; Clark, J.; Seeberger, T.; Hess, J.; Blankenship, T.; FitzGerald, P.G. Targeted deletion of the lens fiber cell-specific intermediate filament protein filensin. Investig. Ophthalmol. Vis. Sci. 2003, 44, 5252–5258. [Google Scholar] [CrossRef]
- Alizadeh, A.; Clark, J.I.; Seeberger, T.; Hess, J.; Blankenship, T.; Spicer, A.; FitzGerald, P.G. Targeted genomic deletion of the lens-specific intermediate filament protein CP49. Investig. Ophthalmol. Vis. Sci. 2002, 43, 3722–3727. [Google Scholar]
- Kwon, H.J.; Shikinaka, K.; Kakugo, A.; Furukawa, H.; Osada, Y.; Gong, J.P. Motility and structural polymorphism of polymer-actin complex gel. J. Nanosci. Nanotechnol. 2007, 7, 844–847. [Google Scholar] [CrossRef] [PubMed]
- FitzGerald, P.G. Lens intermediate filaments. Exp. Eye Res. 2009, 88, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Landsbury, A.; Perng, M.D.; Pohl, E.; Quinlan, R.A. Functional symbiosis between the intermediate filament cytoskeleton and small heat shock proteins. In Small Stress Proteins and Human Diseases; Nova Science Publishers Inc.: Hauppauge, NY, USA, 2010; pp. 55–87. [Google Scholar]
- Kalligeraki, A.A.; Isted, A.; Jarrin, M.; Uwineza, A.; Pal, R.; Saunter, C.D.; Girkin, J.M.; Obara, B.; Quinlan, R.A. Three-dimensional data capture and analysis of intact eye lenses evidences emmetropia-associated changes in epithelial cell organization. Sci. Rep. 2020, 10, 16898. [Google Scholar] [CrossRef] [PubMed]
- FitzGerald, P.G.; Casselman, J. Immunologic conservation of the fiber cell beaded filament. Curr. Eye Res. 1991, 10, 471–478. [Google Scholar] [CrossRef]
- Der Perng, M.; Zhang, Q.; Quinlan, R.A. Insights into the beaded filament of the eye lens. Exp. Cell. Res. 2007, 313, 2180–2188. [Google Scholar] [CrossRef]
- Tapodi, A.; Clemens, D.M.; Uwineza, A.; Jarrin, M.; Goldberg, M.W.; Thinon, E.; Heal, W.P.; Tate, E.W.; Nemeth-Cahalan, K.; Vorontsova, I.; et al. BFSP1 C-terminal domains released by post-translational processing events can alter significantly the calcium regulation of AQP0 water permeability. Exp. Eye Res. 2019, 185, 107585. [Google Scholar] [CrossRef]
- Masaki, S.; Quinlan, R.A. Gene structure and sequence comparisons of the eye lens specific protein, filensin, from rat and mouse: Implications for protein classification and assembly. Gene 1997, 201, 11–20. [Google Scholar] [CrossRef]
- Wang, Z.; Obidike, J.E.; Schey, K.L. Posttranslational modifications of the bovine lens beaded filament proteins filensin and CP49. Investig. Ophthalmol. Vis. Sci. 2010, 51, 1565–1574. [Google Scholar] [CrossRef]
- Martin, D.D.; Beauchamp, E.; Berthiaume, L.G. Post-translational myristoylation: Fat matters in cellular life and death. Biochimie 2011, 93, 18–31. [Google Scholar] [CrossRef]
- Lemarié, F.L.; Sanders, S.S.; Nguyen, Y.; Martin, D.D.O.; Hayden, M.R. Full-length huntingtin is palmitoylated at multiple sites and post-translationally myristoylated following caspase-cleavage. Front. Physiol. 2023, 14, 1086112. [Google Scholar] [CrossRef] [PubMed]
- Madeo, G.; Savojardo, C.; Martelli, P.L.; Casadio, R. SVMyr: A Web Server Detecting Co- and Post-translational Myristoylation in Proteins. J. Mol. Biol. 2022, 434, 167605. [Google Scholar] [CrossRef] [PubMed]
- Sandilands, A.; Prescott, A.R.; Hutcheson, A.M.; Quinlan, R.A.; Casselman, J.T.; FitzGerald, P.G. Filensin is proteolytically processed during lens fiber cell differentiation by multiple independant pathways. Eur. J. Cell. Biol. 1995, 67, 238–253. [Google Scholar] [PubMed]
- Wang, Z.; Ryan, D.J.; Schey, K.L. Localization of the lens intermediate filament switch by imaging mass spectrometry. Exp. Eye Res. 2020, 198, 108134. [Google Scholar] [CrossRef] [PubMed]
- Sandilands, A.; Prescott, A.R.; Carter, J.M.; Hutcheson, A.M.; Quinlan, R.A.; Richards, J.; FitzGerald, P.G. Vimentin and CP49/Filensin form distinct networks in the lens which are independantly modulated during lens fibre cell differentiation. J. Cell. Sci. 1995, 108, 1397–1406. [Google Scholar] [CrossRef]
- Griesser, E.; Vemula, V.; Mónico, A.; Pérez-Sala, D.; Fedorova, M. Dynamic posttranslational modifications of cytoskeletal proteins unveil hot spots under nitroxidative stress. Redox Biol. 2021, 44, 102014. [Google Scholar] [CrossRef]
- Kaus-Drobek, M.; Mücke, N.; Szczepanowski, R.H.; Wedig, T.; Czarnocki-Cieciura, M.; Polakowska, M.; Herrmann, H.; Wysłouch-Cieszyńska, A.; Dadlez, M. Vimentin S-glutathionylation at Cys328 inhibits filament elongation and induces severing of mature filaments in vitro. Febs J. 2020, 287, 5304–5322. [Google Scholar] [CrossRef]
- MacTaggart, B.; Kashina, A. Posttranslational modifications of the cytoskeleton. Cytoskeleton 2021, 78, 142–173. [Google Scholar] [CrossRef]
- Snider, N.T.; Ku, N.O.; Omary, M.B. The sweet side of vimentin. Elife 2018, 7, e35336. [Google Scholar] [CrossRef]
- Wang, Z.; Schey, K.L. Identification of a direct Aquaporin-0 binding site in the lens-specific cytoskeletal protein filensin. Exp. Eye Res. 2017, 159, 23–29. [Google Scholar] [CrossRef]
- Schey, K.L.; Petrova, R.S.; Gletten, R.B.; Donaldson, P.J. The Role of Aquaporins in Ocular Lens Homeostasis. Int. J. Mol. Sci. 2017, 18, 2693. [Google Scholar] [CrossRef] [PubMed]
- Varadaraj, K.; FitzGerald, P.G.; Kumari, S.S. Deletion of beaded filament proteins or the C-terminal end of Aquaporin 0 causes analogous abnormal distortion aberrations in mouse lens. Exp. Eye Res. 2021, 209, 108645. [Google Scholar] [CrossRef] [PubMed]
- Brunkener, M.; Georgatos, S.D. Membrane-binding properties of filensin, a cytoskeletal protein of the lens fibre cells. J. Cell. Sci. 1992, 103, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Schey, K.L. Proteomic Analysis of Lipid Raft-Like Detergent-Resistant Membranes of Lens Fiber Cells. Investig. Ophthalmol. Vis. Sci. 2015, 56, 8349–8360. [Google Scholar] [CrossRef]
- Milks, L.C.; Kumar, N.M.; Houghten, R.; Unwin, N.; Gilula, N.B. Topology of the 32-kd liver gap junction protein determined by site-directed antibody localizations. EMBO J. 1988, 7, 2967–2975. [Google Scholar] [CrossRef] [PubMed]
- Giglione, C.; Meinnel, T. Mapping the myristoylome through a complete understanding of protein myristoylation biochemistry. Progress. Lipid Res. 2022, 85, 101139. [Google Scholar] [CrossRef]
- Wright, M.H.; Heal, W.P.; Mann, D.J.; Tate, E.W. Protein myristoylation in health and disease. J. Chem. Biol. 2010, 3, 19–35. [Google Scholar] [CrossRef]
- Harroun, T.A.; Bradshaw, J.P.; Balali-Mood, K.; Katsaras, J. A structural study of the myristoylated N-terminus of ARF1. Biochim. Biophys. Acta 2005, 1668, 138–144. [Google Scholar] [CrossRef]
- Liu, Y.; Kahn, R.A.; Prestegard, J.H. Structure and membrane interaction of myristoylated ARF1. Structure 2009, 17, 79–87. [Google Scholar] [CrossRef]
- Yang, F.; Li, T.; Peng, Z.; Liu, Y.; Guo, Y. The amphipathic helices of Arfrp1 and Arl14 are sufficient to determine subcellular localizations. J. Biol. Chem. 2020, 295, 16643–16654. [Google Scholar] [CrossRef]
- Brass, V.; Bieck, E.; Montserret, R.; Wölk, B.; Hellings, J.A.; Blum, H.E.; Penin, F.; Moradpour, D. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 2002, 277, 8130–8139. [Google Scholar] [CrossRef] [PubMed]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific alpha-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef] [PubMed]
- Keller, R.C. New user-friendly approach to obtain an eisenberg plot and its use as a practical tool in protein sequence analysis. Int. J. Mol. Sci. 2011, 12, 5577–5591. [Google Scholar] [CrossRef] [PubMed]
- Keller, R.C. Prediction of lipid-binding regions in cytoplasmic and extracellular loops of membrane proteins as exemplified by protein translocation membrane proteins. J. Membr. Biol. 2013, 246, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Katuwawala, A.; Wang, K.; Wu, Z.; Ghadermarzi, S.; Gao, J.; Kurgan, L. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat. Commun. 2021, 12, 4438. [Google Scholar] [CrossRef]
- Feng, S.H.; Xia, C.Q.; Zhang, P.D.; Shen, H.B. Ab-Initio Membrane Protein Amphipathic Helix Structure Prediction Using Deep Neural Networks. IEEE/ACM Trans. Comput. Biol. Bioinform./IEEE ACM 2022, 19, 795–805. [Google Scholar] [CrossRef]
- UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [CrossRef]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Wang, J.; Youkharibache, P.; Marchler-Bauer, A.; Lanczycki, C.; Zhang, D.; Lu, S.; Madej, T.; Marchler, G.H.; Cheng, T.; Chong, L.C.; et al. iCn3D: From Web-Based 3D Viewer to Structural Analysis Tool in Batch Mode. Front. Mol. Biosci. 2022, 9, 831740. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Conover, G.; Elliott, J.L.; Perng, M.D.; Herrmann, H.; Quinlan, R.A. αB-crystallin is a sensor for assembly intermediates and for the subunit topology of desmin intermediate filaments. Cell. Stress. Chaperones 2017, 22, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Perng, M.D.; Sandilands, A.; Kuszak, J.; Dahm, R.; Wegener, A.; Prescott, A.R.; Quinlan, R.A. The intermediate filament systems in the eye lens. Intermed. Filam. Cytoskelet. 2004, 78, 597–624. [Google Scholar]
- Uwineza, A.; Cummins, I.; Jarrin, M.; Kalligeraki, A.A.; Barnard, S.; Mol, M.; Degani, G.; Altomare, A.A.; Aldini, G.; Schreurs, A.; et al. Identification and quantification of ionising radiation-induced oxysterol formation in membranes of lens fibre cells. Adv. Redox Res. 2023, 7, 100057. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Cobb, B.A.; Petrash, J.M. alpha-Crystallin Chaperone-like Activity and Membrane Binding in Age-Related Cataracts. Biochemistry 2002, 41, 483–490. [Google Scholar] [CrossRef]
- Borchman, D.; Tang, D. Binding capacity of alpha-crystallin to bovine lens lipids. Exp. Eye Res. 1996, 63, 407–410. [Google Scholar] [CrossRef]
- Roccatano, D.; Colombo, G.; Fioroni, M.; Mark, A.E. Mechanism by which 2,2,2-trifluoroethanol/water mixtures stabilize secondary-structure formation in peptides: A molecular dynamics study. Proc. Natl. Acad. Sci. USA 2002, 99, 12179–12184. [Google Scholar] [CrossRef]
- Konno, K.; Hisada, M.; Fontana, R.; Lorenzi, C.C.; Naoki, H.; Itagaki, Y.; Miwa, A.; Kawai, N.; Nakata, Y.; Yasuhara, T.; et al. Anoplin, a novel antimicrobial peptide from the venom of the solitary wasp Anoplius samariensis. Biochim. Biophys. Acta 2001, 1550, 70–80. [Google Scholar] [CrossRef]
- Miles, A.J.; Ramalli, S.G.; Wallace, B.A. DichroWeb, a website for calculating protein secondary structure from circular dichroism spectroscopic data. Protein Sci. 2022, 31, 37–46. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Venyaminov, S.Y.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Inclusion of denatured proteins with native proteins in the analysis. Anal. Biochem. 2000, 287, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 277, 680–685. [Google Scholar] [CrossRef]
- Cui, Q.; Tashiro, S.; Onodera, S.; Minami, M.; Ikejima, T. Autophagy preceded apoptosis in oridonin-treated human breast cancer MCF-7 cells. Biol. Pharm. Bull. 2007, 30, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Backes, C.; Kuentzer, J.; Lenhof, H.P.; Comtesse, N.; Meese, E. GraBCas: A bioinformatics tool for score-based prediction of Caspase- and Granzyme B-cleavage sites in protein sequences. Nucleic Acids Res. 2005, 33, W208–W213. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, X.M.; Tan, H.; Akutsu, T.; Whisstock, J.C.; Song, J. Cascleave 2.0, a new approach for predicting caspase and granzyme cleavage targets. Bioinformatics 2014, 30, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Oka, M.; Furuki, K.; Mitsuishi, A.; Nakashima, E.; Takehana, M. The effect of the interaction between aquaporin 0 (AQP0) and the filensin tail region on AQP0 water permeability. Mol. Vis. 2011, 17, 3191–3199. [Google Scholar] [PubMed]
- Robustelli, J.; Baumgart, T. Membrane partitioning and lipid selectivity of the N-terminal amphipathic H0 helices of endophilin isoforms. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183660. [Google Scholar] [CrossRef]
- Zhukovsky, M.A.; Filograna, A.; Luini, A.; Corda, D.; Valente, C. Protein Amphipathic Helix Insertion: A Mechanism to Induce Membrane Fission. Front. Cell. Dev. Biol. 2019, 7, 291. [Google Scholar] [CrossRef]
- Higgins, M.K.; Oprian, D.D.; Schertler, G.F. Recoverin binds exclusively to an amphipathic peptide at the N terminus of rhodopsin kinase, inhibiting rhodopsin phosphorylation without affecting catalytic activity of the kinase. J. Biol. Chem. 2006, 281, 19426–19432. [Google Scholar] [CrossRef] [PubMed]
- Lottermoser, J.A.; Dittman, J.S. Complexin Membrane Interactions: Implications for Synapse Evolution and Function. J. Mol. Biol. 2023, 435, 167774. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, R.D.; Perumalsamy, V.; Hejtmancik, J.F. Autosomal recessive juvenile onset cataract associated with mutation in BFSP1. Hum. Genet. 2007, 121, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Borchman, D.; Yappert, M.C. Lipids and the ocular lens. J. Lipid Res. 2010, 51, 2473–2488. [Google Scholar] [CrossRef] [PubMed]
- Costello, M.J.; Brennan, L.A.; Mohamed, A.; Gilliland, K.O.; Johnsen, S.; Kantorow, M. Identification and Ultrastructural Characterization of a Novel Nuclear Degradation Complex in Differentiating Lens Fiber Cells. PLoS ONE 2016, 11, e0160785. [Google Scholar] [CrossRef]
- Bassnett, S. Lens organelle degradation. Exp. Eye Res. 2002, 74, 1–6. [Google Scholar] [CrossRef]
- Gletten, R.B.; Cantrell, L.S.; Bhattacharya, S.; Schey, K.L. Lens Aquaporin-5 Inserts into Bovine Fiber Cell Plasma Membranes via Unconventional Protein Secretion. Investig. Ophthalmol. Vis. Sci. 2022, 63, 5. [Google Scholar] [CrossRef]
- Bassnett, S.; Costello, M.J. The cause and consequence of fiber cell compaction in the vertebrate lens. Exp. Eye Res. 2017, 156, 50–57. [Google Scholar] [CrossRef]
- Shi, Y.; Barton, K.; De Maria, A.; Petrash, J.M.; Shiels, A.; Bassnett, S. The stratified syncytium of the vertebrate lens. J. Cell. Sci. 2009, 122, 1607–1615. [Google Scholar] [CrossRef]
- FitzGerald, P.; Sun, N.; Shibata, B.; Hess, J.F. Expression of the type VI intermediate filament proteins CP49 and filensin in the mouse lens epithelium. Mol. Vis. 2016, 22, 970–989. [Google Scholar]
- Yoon, K.H.; Fitzgerald, P.G. Periplakin Serves as a Linker for Both Vimentin and the Highly Divergent Beaded Filament Proteins in the Ocular Lens. Investig. Ophthalmol. Vis. Sci. 2008, 50, 1283–1289. [Google Scholar] [CrossRef] [PubMed]
- Straub, B.K.; Boda, J.; Kuhn, C.; Schnoelzer, M.; Korf, U.; Kempf, T.; Spring, H.; Hatzfeld, M.; Franke, W.W. A novel cell-cell junction system: The cortex adhaerens mosaic of lens fiber cells. J. Cell. Sci. 2003, 116, 4985–4995. [Google Scholar] [CrossRef]
- Franke, W.W.; Kapprell, H.P.; Cowin, P. Plakoglobin is a component of the filamentous subplasmalemmal coat of lens cells. Eur. J. Cell. Biol. 1987, 43, 301–315. [Google Scholar] [PubMed]
- Quinlan, R.A.; Schwarz, N.; Windoffer, R.; Richardson, C.; Hawkins, T.; Broussard, J.A.; Green, K.J.; Leube, R.E. A rim-and-spoke hypothesis to explain the biomechanical roles for cytoplasmic intermediate filament networks. J. Cell. Sci. 2017, 130, 3437–3445. [Google Scholar] [CrossRef] [PubMed]
- Patteson, A.E.; Vahabikashi, A.; Goldman, R.D.; Janmey, P.A. Mechanical and Non-Mechanical Functions of Filamentous and Non-Filamentous Vimentin. Bioessays 2020, 42, e2000078. [Google Scholar] [CrossRef]
- Suprewicz, Ł.; Swoger, M.; Gupta, S.; Piktel, E.; Byfield, F.J.; Iwamoto, D.V.; Germann, D.; Reszeć, J.; Marcińczyk, N.; Carroll, R.J.; et al. Extracellular Vimentin as a Target against SARS-CoV-2 Host Cell Invasion. Small 2022, 18, e2105640. [Google Scholar] [CrossRef]
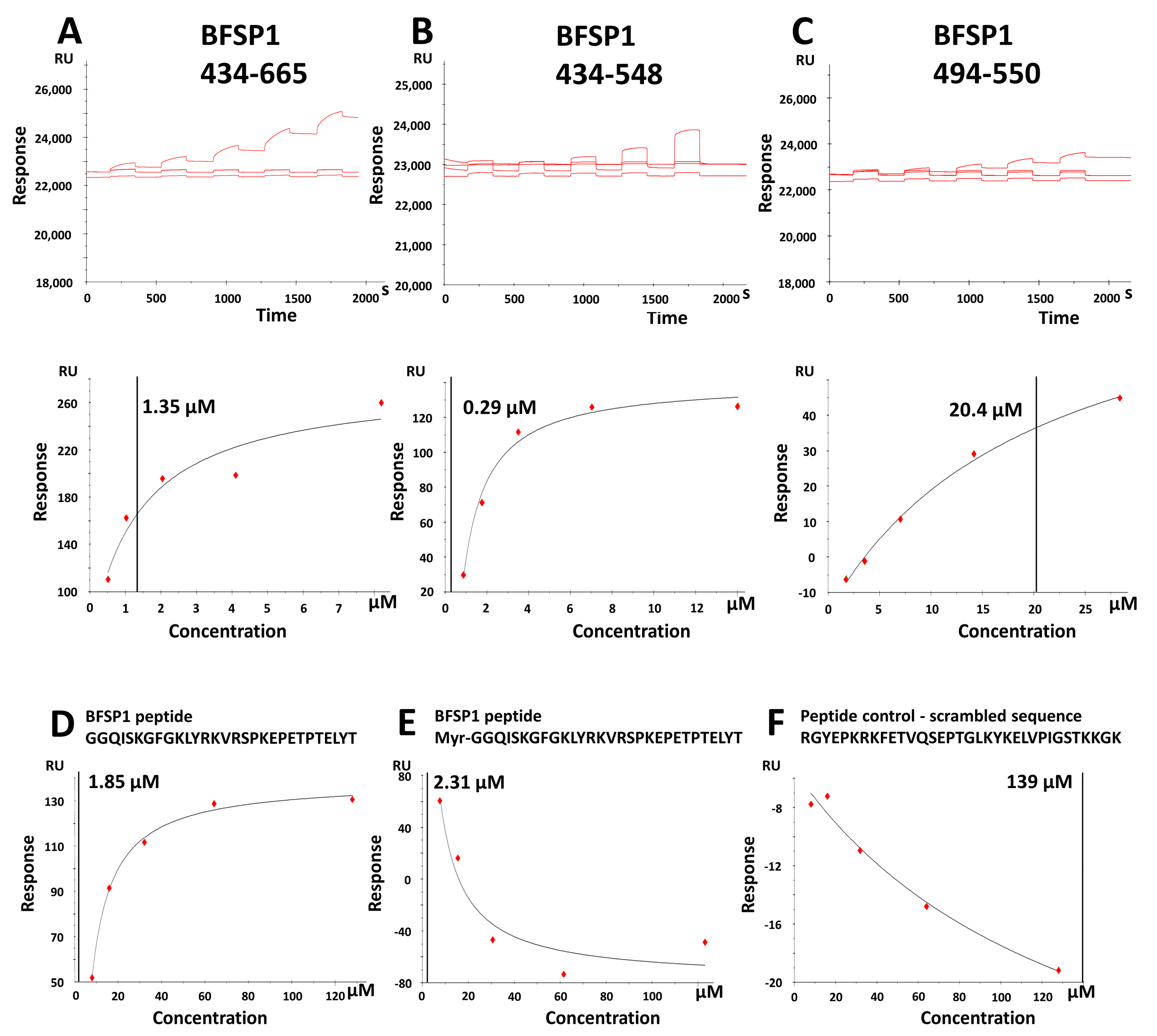
| Bovine BFSP1 (Native) | Human BFSP1 (Recomb.) | Human BFSP1 434–665 | Human BFSP1 434–548 | Human BFSP1 494–550 | |
|---|---|---|---|---|---|
| 6.02 × 10−6 | 1.32 × 10−6 | 3.06 × 10−6 | 1.60 × 10−7 | 1.05 × 10−5 | |
| 6.75 × 10−6 | 3.43 × 10−6 | 1.34 × 10−6 | 2.88 × 10−7 | 2.04 × 10−5 | |
| 7.52 × 10−6 | 3.94 × 10−6 | 2.19 × 10−6 | 2.24 × 10−7 | 1.53 × 10−5 | |
| 6.76 × 10−6 | 2.90 × 10−6 | 2.20 × 10−6 | 2.24 × 10−7 | 1.54 × 10−5 | Mean |
| 4.34 × 10−7 | 8.02 × 10−7 | 4.94 × 10−7 | 3.70 × 10−8 | 2.86 × 10−6 | SEM |
| Unmyristoylated BFSP1 434–467 | Myristoylated BFSP1 434–467 | Scrambled Polypeptide Control | |
|---|---|---|---|
| 1.85 × 10−6 | 2.19 × 10−6 | 1.39 × 10−4 | |
| 2.42 × 10−6 | 2.31 × 10−6 | 1.67 × 10−4 | |
| 2.13 × 10−6 | 2.25 × 10−6 | 1.53 × 10−4 | Mean |
| 2.83 × 10−7 | 5.85 × 10−8 | 1.37 × 10−5 | SEM |
| BFSP1 434–454 (Wild Type) | Wild Type | Q436R | Y445C | R446S | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide Sequence Scan | HM | z | DF | HM | z | DF | HM | z | DF | DM | z | DF |
| GGQISKGFGKLYRKVKEK | 0.53932 | 5 | 2.15911808 | 0.56706 | 6 | 2.51530464 | 0.55949 | 5 | 2.17815856 | 0.49211 | 4 | 1.78455184 |
| GQISKGFGKLYRKVKEKV | 0.59755 | 5 | 2.2140872 | 0.62717 | 6 | 2.57204848 | 0.61913 | 5 | 2.23445872 | 0.55201 | 4 | 1.84109744 |
| RISKGFGKLYRKVKEKVR | 0.63442 | 6 | 2.57889248 | 0.66175 | 7 | 2.934692 | 0.65431 | 6 | 2.59766864 | 0.58691 | 5 | 2.20404304 |
| ISKGFGKLYRKVKEKVRS | 0.62845 | 6 | 2.5732568 | 0.62845 | 6 | 2.5732568 | 0.64803 | 6 | 2.59174032 | 0.58061 | 5 | 2.19809584 |
| SKGFGKLYRKVKEKVRSP | 0.57526 | 6 | 2.52304544 | 0.57526 | 6 | 2.52304544 | 0.59602 | 6 | 2.54264288 | 0.52872 | 5 | 2.14911168 |
| KGFGKLYRKVKEKVRSPK | 0.56039 | 7 | 2.83900816 | 0.56039 | 7 | 2.83900816 | 0.57893 | 7 | 2.85650992 | 0.51154 | 6 | 2.46289376 |
| GFGKLYRKVKEKVRSPKE | 0.54099 | 5 | 2.16069456 | 0.54099 | 5 | 2.16069456 | 0.55961 | 5 | 2.17827184 | 0.49222 | 4 | 1.78465568 |
| FGKLYRKVKEKVRSPKEP | 0.54667 | 5 | 2.16605648 | 0.54667 | 5 | 2.16605648 | 0.56336 | 5 | 2.18181184 | 0.49629 | 4 | 1.78849776 |
| GKLYRKVKEKVRSPKEPE | 0.4226 | 4 | 1.7189344 | 0.4226 | 4 | 1.7189344 | 0.43641 | 4 | 1.73197104 | 0.37039 | 3 | 1.33964816 |
| KLYRKVKEKVRSPKEPET | 0.41416 | 4 | 1.71096704 | 0.41416 | 4 | 1.71096704 | 0.42877 | 4 | 1.72475888 | 0.36327 | 3 | 1.33292688 |
| LYRKVKEKVRSPKEPETP | 0.35341 | 3 | 1.32361904 | 0.35341 | 3 | 1.32361904 | 0.36259 | 3 | 1.33228496 | 0.29968 | 2 | 0.94289792 |
| YRKVKEKVRSPKEPETPT | 0.28225 | 3 | 1.256444 | 0.28225 | 3 | 1.256444 | 0.29517 | 3 | 1.26864048 | 0.22951 | 2 | 0.87665744 |
| RKVKEKVRSPKEPETPTE | 0.26427 | 2 | 0.90947088 | 0.26427 | 2 | 0.90947088 | 0.26427 | 2 | 0.90947088 | 0.21098 | 1 | 0.52916512 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jarrin, M.; Kalligeraki, A.A.; Uwineza, A.; Cawood, C.S.; Brown, A.P.; Ward, E.N.; Le, K.; Freitag-Pohl, S.; Pohl, E.; Kiss, B.; et al. Independent Membrane Binding Properties of the Caspase Generated Fragments of the Beaded Filament Structural Protein 1 (BFSP1) Involves an Amphipathic Helix. Cells 2023, 12, 1580. https://doi.org/10.3390/cells12121580
Jarrin M, Kalligeraki AA, Uwineza A, Cawood CS, Brown AP, Ward EN, Le K, Freitag-Pohl S, Pohl E, Kiss B, et al. Independent Membrane Binding Properties of the Caspase Generated Fragments of the Beaded Filament Structural Protein 1 (BFSP1) Involves an Amphipathic Helix. Cells. 2023; 12(12):1580. https://doi.org/10.3390/cells12121580
Chicago/Turabian StyleJarrin, Miguel, Alexia A. Kalligeraki, Alice Uwineza, Chris S. Cawood, Adrian P. Brown, Edward N. Ward, Khoa Le, Stefanie Freitag-Pohl, Ehmke Pohl, Bence Kiss, and et al. 2023. "Independent Membrane Binding Properties of the Caspase Generated Fragments of the Beaded Filament Structural Protein 1 (BFSP1) Involves an Amphipathic Helix" Cells 12, no. 12: 1580. https://doi.org/10.3390/cells12121580
APA StyleJarrin, M., Kalligeraki, A. A., Uwineza, A., Cawood, C. S., Brown, A. P., Ward, E. N., Le, K., Freitag-Pohl, S., Pohl, E., Kiss, B., Tapodi, A., & Quinlan, R. A. (2023). Independent Membrane Binding Properties of the Caspase Generated Fragments of the Beaded Filament Structural Protein 1 (BFSP1) Involves an Amphipathic Helix. Cells, 12(12), 1580. https://doi.org/10.3390/cells12121580