Circulating Tumor DNA in the Management of Early-Stage Breast Cancer
Abstract
1. Introduction
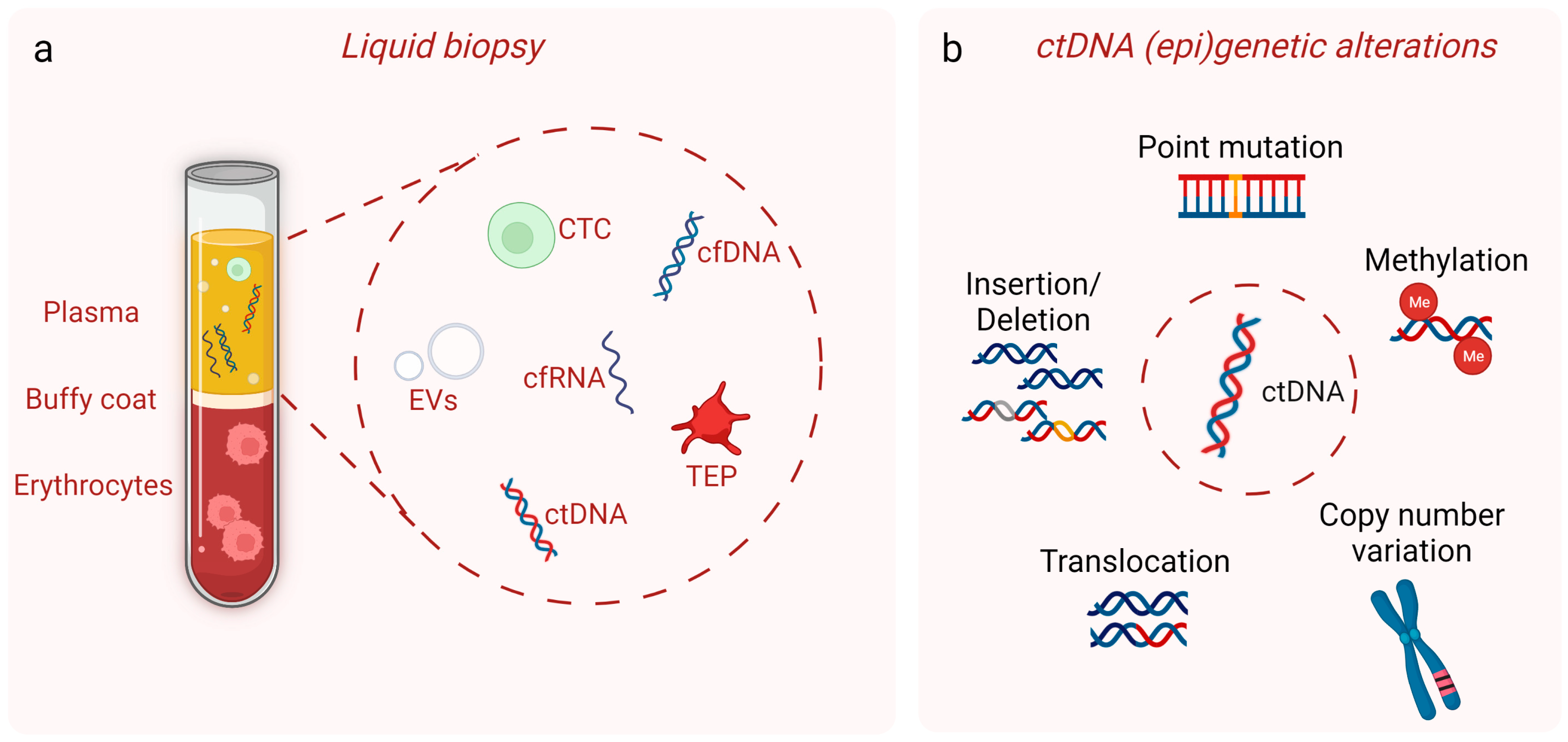
2. Circulating Tumor DNA
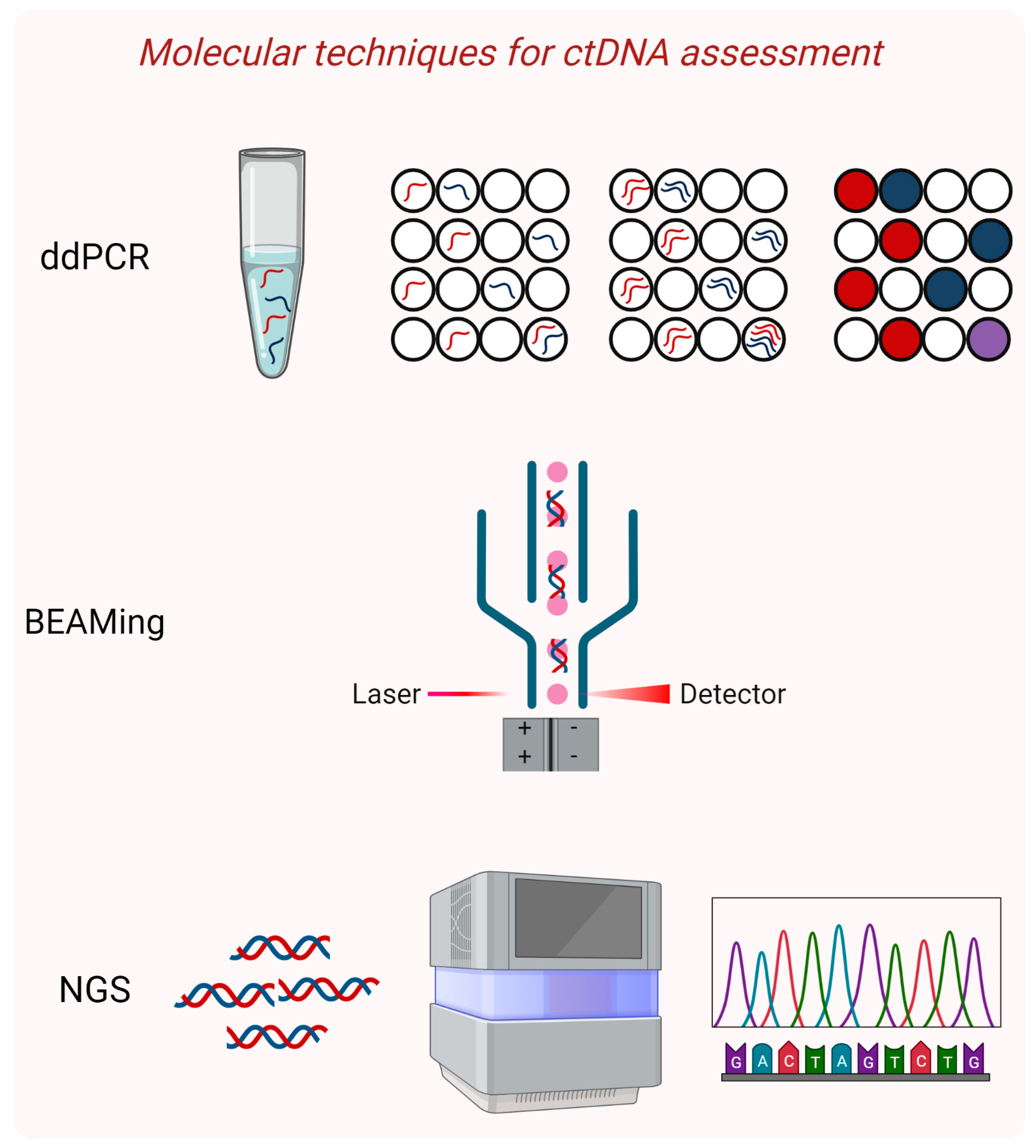
3. Techniques Used for ctDNA Detection and Analysis in Early-Stage Cancers
4. Circulating Tumor DNA in the Management of Early-Stage Breast Cancer
4.1. ctDNA in Early BrCa Diagnosis
4.2. ctDNA in Prognosis and Prediction of Therapy Response in the Neoadjuvant Setting
4.3. ctDNA in MRD Detection and Patient Surveillance
4.4. ctDNA in the Prediction of Relapse in TNBC Patients with Residual Disease
5. Conclusions-Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Giaquinto, A.Ν.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2022. CA. Cancer J. Clin. 2022, 72, 524–561. [Google Scholar] [CrossRef]
- Globocan. 2020. Available online: https://www.uicc.org/news/globocan-2020-new-global-cancer-data (accessed on 28 April 2023).
- International Agency for Research on Cancer. Available online: https://gco.iarc.fr/tomorrow/en (accessed on 28 April 2023).
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA. Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/breast-cancer-facts-and-figures/breast-cancer-facts-and-figures-2019-2020.pdf (accessed on 28 April 2023).
- Available online: https://seer.cancer.gov/ (accessed on 23 March 2023).
- Saphner, T.; Tormey, D.C.; Gray, R. Annual Hazard Rates of Recurrence for Breast Cancer after Primary Therapy. J. Clin. Oncol. 2016, 14, 2738–2746. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rubio, I.T.; Zackrisson, S.; Senkus, E. Early Breast Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2019, 30, 1194–1220. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating Liquid Biopsies into the Management of Cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef]
- Corvigno, S.; Johnson, A.M.; Wong, K.K.; Cho, M.S.; Afshar-Kharghan, V.; Menter, D.G.; Sood, A.K. Novel Markers for Liquid Biopsies in Cancer Management: Circulating Platelets and Extracellular Vesicles. Mol. Cancer Ther. 2022, 21, 1067–1075. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid Biopsy Enters the Clinic—Implementation Issues and Future Challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Heidrich, I.; Ačkar, L.; Mossahebi Mohammadi, P.; Pantel, K. Liquid Biopsies: Potential and Challenges. Int. J. Cancer 2021, 148, 528–545. [Google Scholar] [CrossRef]
- Dang, D.K.; Park, B.H. Vanderbilt-Ingram Circulating Tumor DNA: Current Challenges for Clinical Utility. J. Clin. Investig. 2022, 132, 1–9. [Google Scholar] [CrossRef]
- Vidal, J.; Fernández-Rodríguez, M.C.; Casadevall, D.; García-Alfonso, P.; Páez, D.; Guix, M.; Alonso, V.; Cano, M.T.; Santos, C.; Durán, G.; et al. Liquid Biopsy Detects Early Molecular Response and Predicts Benefit to First-Line Chemotherapy plus Cetuximab in Metastatic Colorectal Cancer: PLATFORM-B Study. Clin. Cancer Res. 2023, 29, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Chen, Y.; Yang, J.; Zhuo, C.; Huang, S.; Zhang, H.; Shi, Y. Clinical Perspectives on Liquid Biopsy in Metastatic Colorectal Cancer. Front. Genet. 2021, 12, 634642. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement from the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662. [Google Scholar] [CrossRef] [PubMed]
- Reichert, Z.R.; Morgan, T.M.; Li, G.; Castellanos, E.; Snow, T.; Dall’Olio, F.G.; Madison, R.W.; Fine, A.D.; Oxnard, G.R.; Graf, R.P.; et al. Prognostic Value of Plasma Circulating Tumor DNA Fraction across Four Common Cancer Types: A Real-World Outcomes Study. Ann. Oncol. 2023, 34, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Attard, G.; Bidard, F.C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.D.; Montagut, C.; et al. ESMO Recommendations on the Use of Circulating Tumour DNA Assays for Patients with Cancer: A Report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef] [PubMed]
- Mandel, P.; Metais, P. Les Acides Nucleiques Du Plasma Sanguin Chez l’homme. CR Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic Characteristics of the DNA Found in the Plasma of Cancer Patients. Oncology 1989, 46, 318–322. [Google Scholar] [CrossRef]
- Sánchez-Herrero, E.; Serna-Blasco, R.; Robado de Lope, L.; González-Rumayor, V.; Romero, A.; Provencio, M. Circulating Tumor DNA as a Cancer Biomarker: An Overview of Biological Features and Factors That May Impact on CtDNA Analysis. Front. Oncol. 2022, 12, 3410. [Google Scholar] [CrossRef]
- Anker, P.; Stroun, M.M.P. Spontaneous Release of DNA by Human Blood Lymphocytes as Shown in an in Vitro System. Cancer Res. 1975, 35, 2375–2382. [Google Scholar]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Diaz, L.A.; Bardelli, A. Liquid Biopsies: Genotyping Circulating Tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical Relevance of Blood-Based CtDNA Analysis: Mutation Detection and Beyond. Br. J. Cancer 2021, 124, 345–358. [Google Scholar] [CrossRef]
- Underhill, H.R. Leveraging the Fragment Length of Circulating Tumour DNA to Improve Molecular Profiling of Solid Tumour Malignancies with Next-Generation Sequencing: A Pathway to Advanced Non-Invasive Diagnostics in Precision Oncology? Mol. Diagn. Ther. 2021, 25, 389–408. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating Mutant DNA to Assess Tumor Dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; Mouliere, F.; El Messaoudi, S.; Mollevi, C.; Lopez-Crapez, E.; Rolet, F.; Gillet, B.; Gongora, C.; Dechelotte, P.; Robert, B.; et al. Clinical Validation of the Detection of KRAS and BRAF Mutations from Circulating Tumor DNA. Nat. Med. 2014, 20, 430–435. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid Biopsy: Monitoring Cancer-Genetics in the Blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef]
- Diehl, F.; Li, M.; He, Y.; Kinzler, K.W.; Vogelstein, B.; Dressman, D. BEAMing: Single-Molecule PCR on Microparticles in Water-in-Oil Emulsions. Nat. Methods 2006, 3, 551–559. [Google Scholar] [CrossRef]
- Li, M.; Diehl, F.; Dressman, D.; Vogelstein, B.; Kinzler, K.W. BEAMing up for Detection and Quantification of Rare Sequence Variants. Nat. Methods 2006, 3, 95–97. [Google Scholar] [CrossRef]
- Vidal, J.; Muinelo, L.; Dalmases, A.; Jones, F.; Edelstein, D.; Iglesias, M.; Orrillo, M.; Abalo, A.; Rodríguez, C.; Brozos, E.; et al. Plasma CtDNA RAS Mutation Analysis for the Diagnosis and Treatment Monitoring of Metastatic Colorectal Cancer Patients. Ann. Oncol. 2017, 28, 1325–1332. [Google Scholar] [CrossRef] [PubMed]
- Higgins, M.J.; Jelovac, D.; Barnathan, E.; Blair, B.; Slater, S.; Powers, P.; Zorzi, J.; Jeter, S.C.; Oliver, G.R.; Fetting, J.; et al. Detection of Tumor PIK3CA Status in Metastatic Breast Cancer Using Peripheral Blood. Clin. Cancer Res. 2012, 18, 3462–3469. [Google Scholar] [CrossRef] [PubMed]
- Thress, K.S.; Brant, R.; Carr, T.H.; Dearden, S.; Jenkins, S.; Brown, H.; Hammett, T.; Cantarini, M.; Barrett, J.C. EGFR Mutation Detection in CtDNA from NSCLC Patient Plasma: A Cross-Platform Comparison of Leading Technologies to Support the Clinical Development of AZD9291. Lung Cancer 2015, 90, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Kastrisiou, M.; Zarkavelis, G.; Kougioumtzi, A.; Sakaloglou, P.; Kostoulas, C.; Georgiou, I.; Batistatou, A.; Pentheroudakis, G.; Magklara, A. Development and Validation of a Targeted ‘Liquid’ Ngs Panel for Treatment Customization in Patients with Metastatic Colorectal Cancer. Diagnostics 2021, 11, 2375. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. 2011, 83, 8604. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, T.; Yamamoto, Y.; Yamamoto-Ibusuki, M.; Inao, T.; Sueta, A.; Fujiwara, S.; Omoto, Y.; Iwase, H. Prognostic Role of PIK3CA Mutations of Cell-Free DNA in Early-Stage Triple Negative Breast Cancer. Cancer Sci. 2015, 106, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Oshiro, C.; Kagara, N.; Naoi, Y.; Shimoda, M.; Shimomura, A.; Maruyama, N.; Shimazu, K.; Kim, S.J.; Noguchi, S. PIK3CA Mutations in Serum DNA Are Predictive of Recurrence in Primary Breast Cancer Patients. Breast Cancer Res. Treat. 2015, 150, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Beaver, J.A.; Jelovac, D.; Balukrishna, S.; Cochran, R.L.; Croessmann, S.; Zabransky, D.J.; Wong, H.Y.; Toro, P.V.; Cidado, J.; Blair, B.G.; et al. Detection of Cancer DNA in Plasma of Patients with Early-Stage Breast Cancer. Clin. Cancer Res. 2014, 20, 2643–2650. [Google Scholar] [CrossRef]
- Denis, J.A.; Guillerm, E.; Coulet, F.; Larsen, A.K.; Lacorte, J.M. The Role of BEAMing and Digital PCR for Multiplexed Analysis in Molecular Oncology in the Era of Next-Generation Sequencing. Mol. Diagn. Ther. 2017, 21, 587–600. [Google Scholar] [CrossRef] [PubMed]
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and Quantification of Rare Mutations with Massively Parallel Sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9530–9535. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.L.; Christie, M.; et al. Circulating Tumor DNA Analysis Detects Minimal Residual Disease and Predicts Recurrence in Patients with Stage II Colon Cancer. Sci. Transl. Med. 2016, 8, 346ra92. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Cohen, J.D.; Lahouel, K.; Lo, S.N.; Wang, Y.; Kosmider, S.; Wong, R.; Shapiro, J.; Lee, M.; Harris, S.; et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N. Engl. J. Med. 2022, 386, 2261–2272. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An Ultrasensitive Method for Quantitating Circulating Tumor DNA with Broad Patient Coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.Y.; Kaper, F.; Dawson, S.J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive Identification and Monitoring of Cancer Mutations by Targeted Deep Sequencing of Plasma DNA. Sci. Transl. Med. 2012, 4, 136ra68. [Google Scholar] [CrossRef]
- Gale, D.; Lawson, A.R.J.; Howarth, K.; Madi, M.; Durham, B.; Smalley, S.; Calaway, J.; Blais, S.; Jones, G.; Clark, J.; et al. Development of a Highly Sensitive Liquid Biopsy Platform to Detect Clinically-Relevant Cancer Mutations at Low Allele Fractions in Cell-Free DNA. PLoS ONE 2018, 13, e0194630. [Google Scholar] [CrossRef]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct Detection of Early-Stage Cancers Using Circulating Tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415. [Google Scholar] [CrossRef]
- Rodriguez, B.J.; Córdoba, G.D.; Aranda, A.G.; Álvarez, M.; Vicioso, L.; Pérez, C.L.; Hernando, C.; Bermejo, B.; Parreño, A.J.; Lluch, A.; et al. Detection of TP53 and PIK3CA Mutations in Circulating Tumor DNA Using Next-Generation Sequencing in the Screening Process for Early Breast Cancer Diagnosis. J. Clin. Med. 2019, 8, 1183. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, W.; Wei, W.; You, Z.; Ou, X.; Sun, M.; Yin, Y.; Tang, X.; Zhao, Z.; Hu, C.; et al. Parallel Analyses of Somatic Mutations in Plasma Circulating Tumor DNA (CtDNA) and Matched Tumor Tissues in Early-Stage Breast Cancer. Clin. Cancer Res. 2019, 25, 6546–6553. [Google Scholar] [CrossRef]
- Chan, H.T.; Chin, Y.M.; Nakamura, Y.; Low, S.K. Clonal Hematopoiesis in Liquid Biopsy: From Biological Noise to Valuable Clinical Implications. Cancers 2020, 12, 2277. [Google Scholar] [CrossRef]
- Korde, L.A.; Somerfield, M.R.; Carey, L.A.; Crews, J.R.; Denduluri, N.; Hwang, E.S.; Khan, S.A.; Loibl, S.; Morris, E.A.; Perez, A.; et al. Neoadjuvant Chemotherapy, Endocrine Therapy, and Targeted Therapy for Breast Cancer: ASCO Guideline. J. Clin. Oncol. 2021, 39, 1485–1505. [Google Scholar] [CrossRef]
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological Complete Response and Long-Term Clinical Benefit in Breast Cancer: The CTNeoBC Pooled Analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Riva, F.; Bidard, F.C.; Houy, A.; Saliou, A.; Madic, J.; Rampanou, A.; Hego, C.; Milder, M.; Cottu, P.; Sablin, M.P.; et al. Patient-Specific Circulating Tumor DNA Detection during Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer. Clin. Chem. 2017, 63, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Cavallone, L.; Aguilar-Mahecha, A.; Lafleur, J.; Brousse, S.; Aldamry, M.; Roseshter, T.; Lan, C.; Alirezaie, N.; Bareke, E.; Majewski, J.; et al. Prognostic and Predictive Value of Circulating Tumor DNA during Neoadjuvant Chemotherapy for Triple Negative Breast Cancer. Sci. Rep. 2020, 10, 14704. [Google Scholar] [CrossRef] [PubMed]
- Rothé, F.; Silva, M.J.; Venet, D.; Campbell, C.; Bradburry, I.; Rouas, G.; De Azambuja, E.; Maetens, M.; Fumagalli, D.; Rodrik-Outmezguine, V.; et al. Precision Medicine and Imaging Circulating Tumor DNA in HER2-Amplified Breast Cancer: A Translational Research Substudy of the NeoALTTO Phase III Trial. Clin. Cancer Res. 2019, 25, 3581–3588. [Google Scholar] [CrossRef]
- Magbanua, M.J.M.; Swigart, L.B.; Wu, H.T.; Hirst, G.L.; Yau, C.; Wolf, D.M.; Tin, A.; Salari, R.; Shchegrova, S.; Pawar, H.; et al. Circulating Tumor DNA in Neoadjuvant-Treated Breast Cancer Reflects Response and Survival. Ann. Oncol. 2021, 32, 229–239. [Google Scholar] [CrossRef]
- Zhou, Q.; Gampenrieder, S.P.; Frantal, S.; Rinnerthaler, G.; Singer, C.F.; Egle, D.; Pfeiler, G.; Bartsch, R.; Wette, V.; Pichler, A.; et al. Persistence of CtDNA in Patients with Breast Cancer During Neoadjuvant Treatment Is a Significant Predictor of Poor Tumor Response. Clin. Cancer Res. 2022, 28, 697–707. [Google Scholar] [CrossRef]
- Ståhlberg, A.; Krzyzanowski, P.M.; Egyud, M.; Filges, S.; Stein, L.; Godfrey, T.E. Simple Multiplexed PCR-Based Barcoding of DNA for Ultrasensitive Mutation Detection by next-Generation Sequencing. Nat. Protoc. 2017, 12, 664–682. [Google Scholar] [CrossRef]
- Papakonstantinou, A.; Gonzalez, N.S.; Pimentel, I.; Suñol, A.; Zamora, E.; Ortiz, C.; Espinosa-Bravo, M.; Peg, V.; Vivancos, A.; Saura, C.; et al. Prognostic Value of CtDNA Detection in Patients with Early Breast Cancer Undergoing Neoadjuvant Therapy: A Systematic Review and Meta-Analysis. Cancer Treat. Rev. 2022, 104, 102362. [Google Scholar] [CrossRef]
- Pondé, N.F.; Zardavas, D.; Piccart, M. Progress in Adjuvant Systemic Therapy for Breast Cancer. Nat. Rev. Clin. Oncol. 2019, 16, 27–44. [Google Scholar] [CrossRef]
- Kerr, A.J.; Dodwell, D.; McGale, P.; Holt, F.; Duane, F.; Mannu, G.; Darby, S.C.; Taylor, C.W. Adjuvant and Neoadjuvant Breast Cancer Treatments: A Systematic Review of Their Effects on Mortality. Cancer Treat. Rev. 2022, 105, 102375. [Google Scholar] [CrossRef]
- Olsson, E.; Winter, C.; George, A.; Chen, Y.; Howlin, J.; Tang, M.E.; Dahlgren, M.; Schulz, R.; Grabau, D.; Westen, D.; et al. Serial Monitoring of Circulating Tumor DNA in Patients with Primary Breast Cancer for Detection of Occult Metastatic Disease. EMBO Mol. Med. 2015, 7, 1034–1047. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Murillas, I.; Schiavon, G.; Weigelt, B.; Ng, C.; Hrebien, S.; Cutts, R.J.; Cheang, M.; Osin, P.; Nerurkar, A.; Kozarewa, I.; et al. Mutation Tracking in Circulating Tumor DNA Predicts Relapse in Early Breast Cancer. Sci. Transl. Med. 2015, 7, 302ra133. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Murillas, I.; Chopra, N.; Comino-Méndez, I.; Beaney, M.; Tovey, H.; Cutts, R.J.; Swift, C.; Kriplani, D.; Afentakis, M.; Hrebien, S.; et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol. 2019, 5, 1473–1478. [Google Scholar] [CrossRef] [PubMed]
- Coombes, R.C.; Page, K.; Salari, R.; Hastings, R.K.; Armstrong, A.; Ahmed, S.; Ali, S.; Cleator, S.; Kenny, L.; Stebbing, J.; et al. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clin. Cancer Res. 2019, 25, 4255–4263. [Google Scholar] [CrossRef]
- Lipsyc-Sharf, M.; De Bruin, E.C.; Santos, K.; Mcewen, R.; Stetson, D.; Patel, A.; Kirkner, G.J.; Hughes, M.E.; Tolaney, S.M.; Partridge, A.H.; et al. Circulating Tumor DNA and Late Recurrence in High-Risk Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Breast Cancer. J. Clin. Oncol. 2022, 40, 2408–2419. [Google Scholar] [CrossRef]
- Parsons, H.A.; Rhoades, J.; Reed, S.C.; Gydush, G.; Ram, P.; Exman, P.; Xiong, K.; Lo, C.C.; Li, T.; Fleharty, M.; et al. Sensitive Detection of Minimal Residual Disease in Patients Treated for Early-Stage Breast Cancer. Clin. Cancer Res. 2020, 26, 2556–2564. [Google Scholar] [CrossRef]
- McDonald, B.R.; Contente-Cuomo, T.; Sammut, S.J.; Odenheimer-Bergman, A.; Ernst, B.; Perdigones, N.; Chin, S.F.; Farooq, M.; Mejia, R.; Cronin, P.A.; et al. Personalized Circulating Tumor DNA Analysis to Detect Residual Disease after Neoadjuvant Therapy in Breast Cancer. Sci. Transl. Med. 2019, 11, eaax7392. [Google Scholar] [CrossRef]
- Chen, Y.H.; Hancock, B.A.; Solzak, J.P.; Brinza, D.; Scafe, C.; Miller, K.D.; Radovich, M. Next-Generation Sequencing of Circulating Tumor DNA to Predict Recurrence in Triple-Negative Breast Cancer Patients with Residual Disease after Neoadjuvant Chemotherapy. npj Breast Cancer 2017, 3, 24. [Google Scholar] [CrossRef]
- Radovich, M.; Jiang, G.; Hancock, B.A.; Chitambar, C.; Nanda, R.; Falkson, C.; Lynce, F.C.; Gallagher, C.; Isaacs, C.; Blaya, M.; et al. Association of Circulating Tumor DNA and Circulating Tumor Cells after Neoadjuvant Chemotherapy with Disease Recurrence in Patients with Triple-Negative Breast Cancer: Preplanned Secondary Analysis of the BRE12-158 Randomized Clinical Trial. JAMA Oncol. 2020, 6, 1410–1415. [Google Scholar] [CrossRef]
- Turner, N.C.; Swift, C.; Jenkins, B.; Kilburn, L.; Coakley, M.; Beaney, M.; Fox, L.; Goddard, K.; Garcia-Murillas, I.; Proszek, P.; et al. Results of the C-TRAK TN Trial: A Clinical Trial Utilising CtDNA Mutation Tracking to Detect Molecular Residual Disease and Trigger Intervention in Patients with Moderate- and High-Risk Early-Stage Triple-Negative Breast Cancer. Ann. Oncol. 2023, 34, 200–211. [Google Scholar] [CrossRef]
- Pan, H.; Gray, R.; Braybrooke, J.; Davies, C.; Taylor, C.; McGale, P.; Peto, R.; Pritchard, K.I.; Bergh, J.; Dowsett, M.; et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N. Engl. J. Med. 2017, 377, 1836–1846. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, R.N.; Esen, B.Ö.; Mellemkjær, L.; Christiansen, P.; Ejlertsen, B.; Lash, T.L.; Nørgaard, M.; Cronin-Fenton, D. The Incidence of Breast Cancer Recurrence 10–32 Years after Primary Diagnosis. JNCI J. Natl. Cancer Inst. 2022, 114, 391–399. [Google Scholar] [CrossRef]
- Li, X.; Yang, J.; Peng, L.; Sahin, A.A.; Huo, L.; Ward, K.C.; O’Regan, R.; Torres, M.A.; Meisel, J.L. Triple-Negative Breast Cancer Has Worse Overall Survival and Cause-Specific Survival than Non-Triple-Negative Breast Cancer. Breast Cancer Res. Treat. 2017, 161, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Liedtke, C.; Mazouni, C.; Hess, K.R.; André, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M.; et al. Response to Neoadjuvant Therapy and Long-Term Survival in Patients with Triple-Negative Breast Cancer. J. Clin. Oncol. 2008, 26, 1275–1281. [Google Scholar] [CrossRef]
- Clark, T.A.; Chung, J.H.; Kennedy, M.; Hughes, J.D.; Chennagiri, N.; Lieber, D.S.; Fendler, B.; Young, L.; Zhao, M.; Coyne, M.; et al. Analytical Validation of a Hybrid Capture–Based Next-Generation Sequencing Clinical Assay for Genomic Profiling of Cell-Free Circulating Tumor DNA. J. Mol. Diagn. 2018, 20, 686–702. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and Localization of Surgically Resectable Cancers with a Multi-Analyte Blood Test. Science 2018, 3247, 926–930. [Google Scholar] [CrossRef]
- Lennon, A.M.; Buchanan, A.H.; Kinde, I.; Warren, A.; Honushefsky, A.; Cohain, A.T.; Ledbetter, D.H.; Sanfilippo, F.; Sheridan, K.; Rosica, D.; et al. Feasibility of Blood Testing Combined with PET-CT to Screen for Cancer and Guide Intervention. Science 2020, 369, eabb9601. [Google Scholar] [CrossRef]
- Jamshidi, A.; Liu, M.C.; Klein, E.A.; Venn, O.; Hubbell, E.; Beausang, J.F.; Gross, S.; Melton, C.; Fields, A.P.; Liu, Q.; et al. Evaluation of Cell-Free DNA Approaches for Multi-Cancer Early Detection. Cancer Cell 2022, 40, 1537–1549. [Google Scholar] [CrossRef]

| ctDNA Assay | No. of Patients # | No. of Samples Analyzed | BrCa Subtype | Therapy Given | Lead Time before Clinical Relapse | Ref. |
|---|---|---|---|---|---|---|
| ddPCR (4–6 chromosomal rearrangements/patient) | 20 | 93 | mixed | AT | 11 months | [65] |
| ddPCR (1–2 mutations/patient) | 55 | n/s | mixed | NAT | 7.9 months | [66] |
| ddPCR (≥1 mutations/patient) | 101 | 695 | mixed | AT and/or NAT | 10.7 months | [67] |
| Personalized NGS (16 mutations/patient) | 49 | 208 | mixed | AT and/or NAT or none | 8.9 months | [68] |
| Personalized NGS (12–16 mutations/patient) | 84 | 291 | mixed | NAT | n/s | [59] |
| Personalized NGS (36 mutations/patient) * | 83 | 219 | HR+ | AT | 12.4 months | [69] |
| NGS (53 mutations/patient) * | 142 | 271 | mixed | AT and/or NAT | 18.9 months | [70] |
| NGS (TARDIS) (18 mutations/patient) * | 33 | 80 | mixed | NAT and/or AT | n/s | [71] |
| NGS (Ion Ampliseq Oncomine Research Panel) | 33 | n/s | TNBC | NAT and AT | 0.07–8.87 months | [72] |
| NGS (Foundation Medicine panels) | 142 | 142 | TNBC | NAT | n/s | [73] |
| dPCR (1–2 mutations/patient) | 161 | n/s | TNBC | NAT and AT | 1.6 months (intervention group) 4.1 (observation group) | [74] |
| Title of Study | Condition | Study Objective | Study Type/Design | Estimated Enrollment |
|---|---|---|---|---|
| HARMONY (NCT05433753) | HER2+ receiving neoadjuvant therapy | MRD detection using ctDNA to predict recurrence | observational, prospective | 60 |
| NCT04353557 | stage I–III | assess the prognostic and predictive value of ctDNA | observational, prospective | 200 |
| NCT05649475 | stage I–III undergoing NAT | whether ctDNA is a marker of response to NAT | observational, prospective | 100 |
| SURVIVE (NCT05658172) | medium- and high-risk patients after completion of primary anti-tumor therapy | evaluate the potential benefits of intensified versus standard surveillance | partially double-blinded, interventional, randomized, controlled, superiority study | 3500 |
| CIPHER (NCT05333874) | stage II–III TNBC and HER2+ | examine the impact of ctDNA on treatment decision-making in patients after NAT and surgery | interventional, non-randomized | 34 ** |
| TRAK-ER (NCT04985266) | ER+ | demonstrate that palbociclib and fulvestrant can defer or prevent relapse in patients with ctDNA-detected molecular relapse | interventional, randomized, phase II study | 1100 |
| DARE (NCT04567420) | stage II–III HR+/HER2− | assess the incidence of ctDNA detection in patients who receive standard-of-care endocrine AT and have high risk of recurrence and assess whether palbociclib plus fulvestrant improves relapse-free survival in ctDNA+ patients | interventional, randomized | 100 |
| LEADER (NCT03285412) | ER+ | ctDNA for MRD detection and therapy guidance | interventional, randomized | 120 |
| NCT04768426 | TNBC | evaluate the use ctDNA to identify patients who will or will not benefit from treatment with capecitabine | interventional, phase II study | 25 |
| NCT03881384 | not specified | whether ctDNA detection can reflect the tumor response to NCT and detect MRD after surgery | observational, prospective | 200 |
| STRIVE (NCT03085888) | women undergoing mammography screening | validate the ability of the GRAIL Test to detect early-stage breast and other invasive cancers | observational, prospective | 99,481 ** |
| RENOVATE (NCT04781062) | women with radiologically identified lesions, BIRADS-3/4/5, smaller than 2 cm | develop a Horizontal Data Integration classifier enabling early noninvasive diagnosis | interventional, non-randomized | 367 ** |
| APOLLO (NCT04501523) | stage II-III TNBC | use ctDNA to identify patients with high relapse risk and randomize them to receive boost or standard therapy | interventional randomized, phase II | 460 |
| ARTEMIS (NCT04803539) | stage II-III TNBC | identify high-risk patients and initiate boost therapy | interventional randomized, phase II/III | 120 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vlataki, K.; Antonouli, S.; Kalyvioti, C.; Lampri, E.; Kamina, S.; Mauri, D.; Harissis, H.V.; Magklara, A. Circulating Tumor DNA in the Management of Early-Stage Breast Cancer. Cells 2023, 12, 1573. https://doi.org/10.3390/cells12121573
Vlataki K, Antonouli S, Kalyvioti C, Lampri E, Kamina S, Mauri D, Harissis HV, Magklara A. Circulating Tumor DNA in the Management of Early-Stage Breast Cancer. Cells. 2023; 12(12):1573. https://doi.org/10.3390/cells12121573
Chicago/Turabian StyleVlataki, Katerina, Sevastiani Antonouli, Christina Kalyvioti, Evangeli Lampri, Sevasti Kamina, Davide Mauri, Haralampos V. Harissis, and Angeliki Magklara. 2023. "Circulating Tumor DNA in the Management of Early-Stage Breast Cancer" Cells 12, no. 12: 1573. https://doi.org/10.3390/cells12121573
APA StyleVlataki, K., Antonouli, S., Kalyvioti, C., Lampri, E., Kamina, S., Mauri, D., Harissis, H. V., & Magklara, A. (2023). Circulating Tumor DNA in the Management of Early-Stage Breast Cancer. Cells, 12(12), 1573. https://doi.org/10.3390/cells12121573








