Horizontal Transfer of Malignant Traits and the Involvement of Extracellular Vesicles in Metastasis
Abstract
1. Introduction
2. Model of Primary Tumor Cell Migration and Growth: The “Seed and Soil” Model
3. Role of EV-Mediated Intercellular Communication in the “Seed and Soil” Model
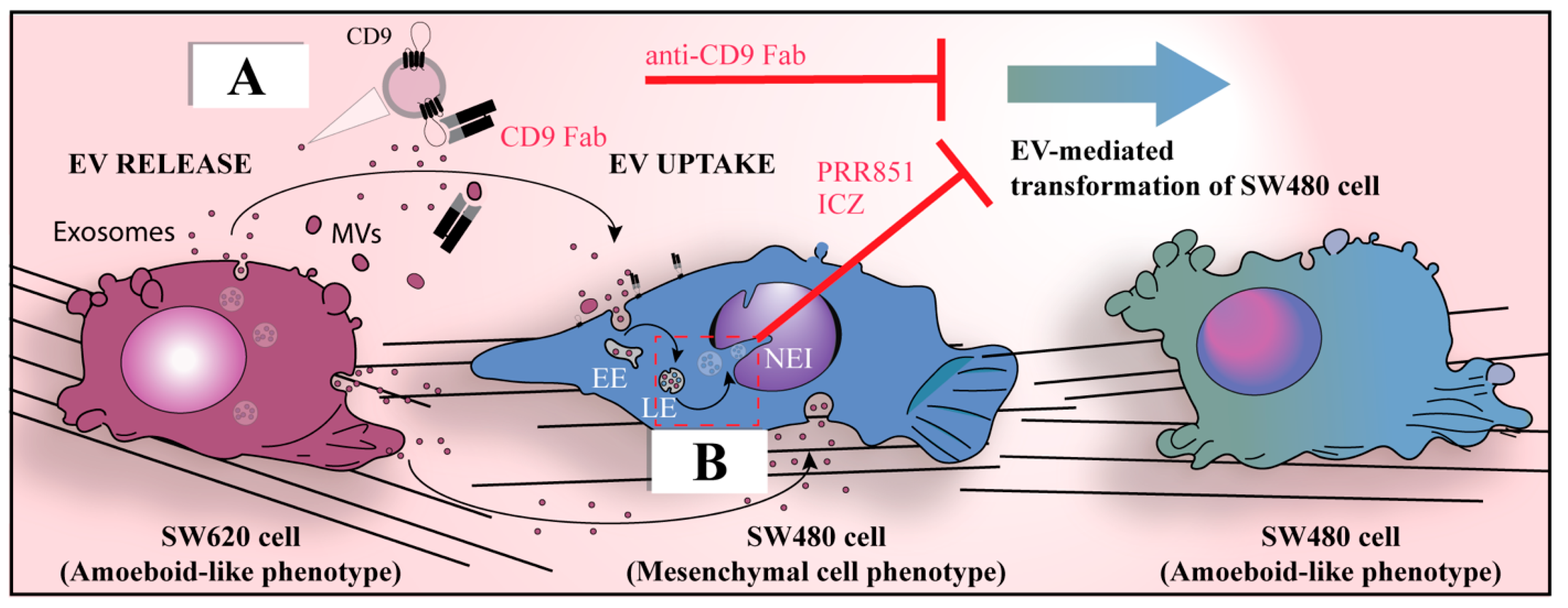
4. Limitations and Challenges of the Seed and Soil Model
5. Metastasis by Cellular Fusion
6. The Genometastatic Hypothesis
7. Horizontal Transfer of Malignant Traits Model
7.1. HTMT and Preneoplastic Lesions
7.2. Oncosuppressor Genes As Gatekeepers
7.3. The Malignant Metastatic Phenotype Is Dependent on the Cell Type That Uptakes the Cancer EV-Derived Oncogenic Message
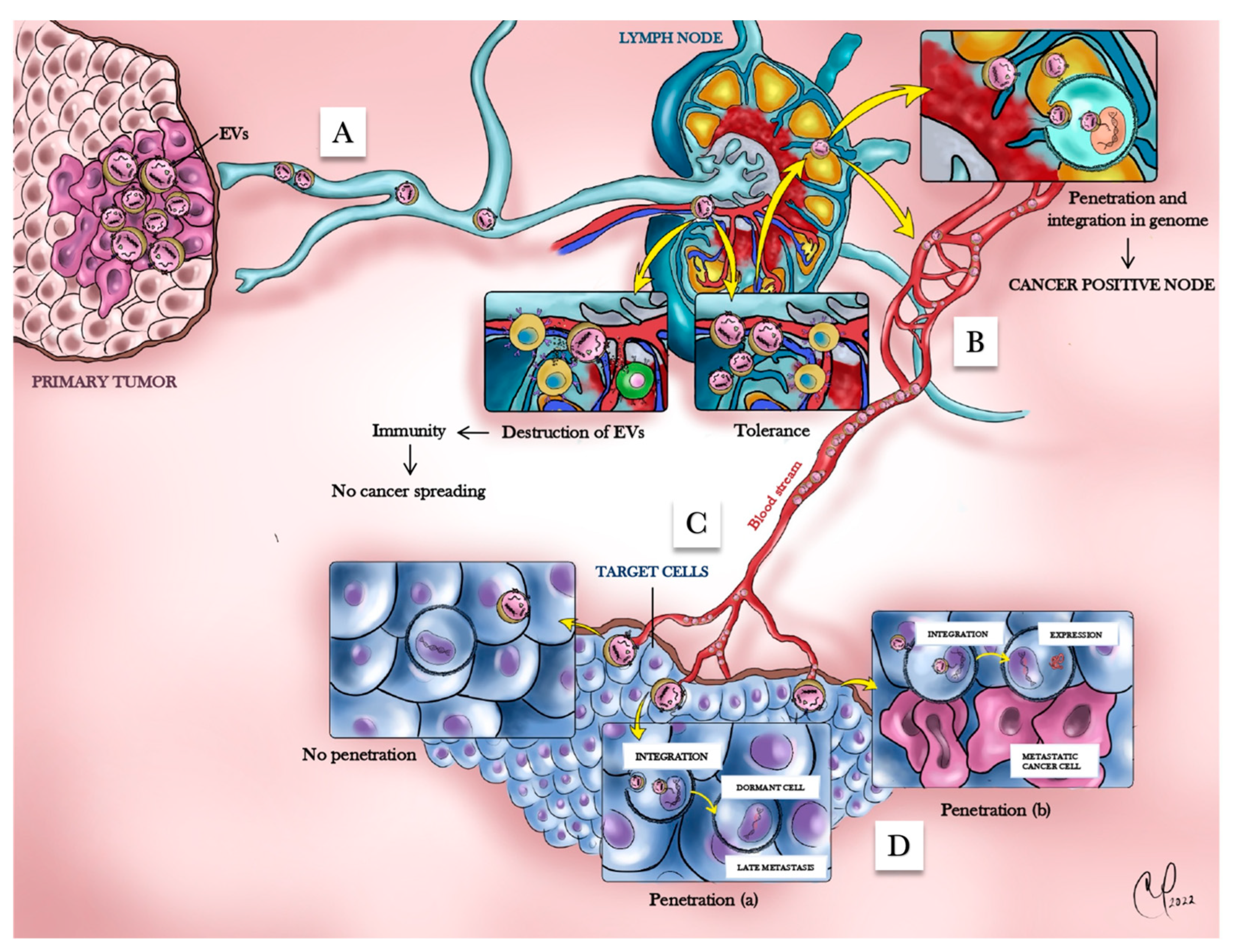
7.4. EV-Mediated Interactions of Cancer Cells with the Immune System
7.5. Cancer Dormancy According to the HTMT Model
7.6. Molecular Mechanisms Involved in the HTMT Model
8. Unanswered Questions Raised by the HTMT Model
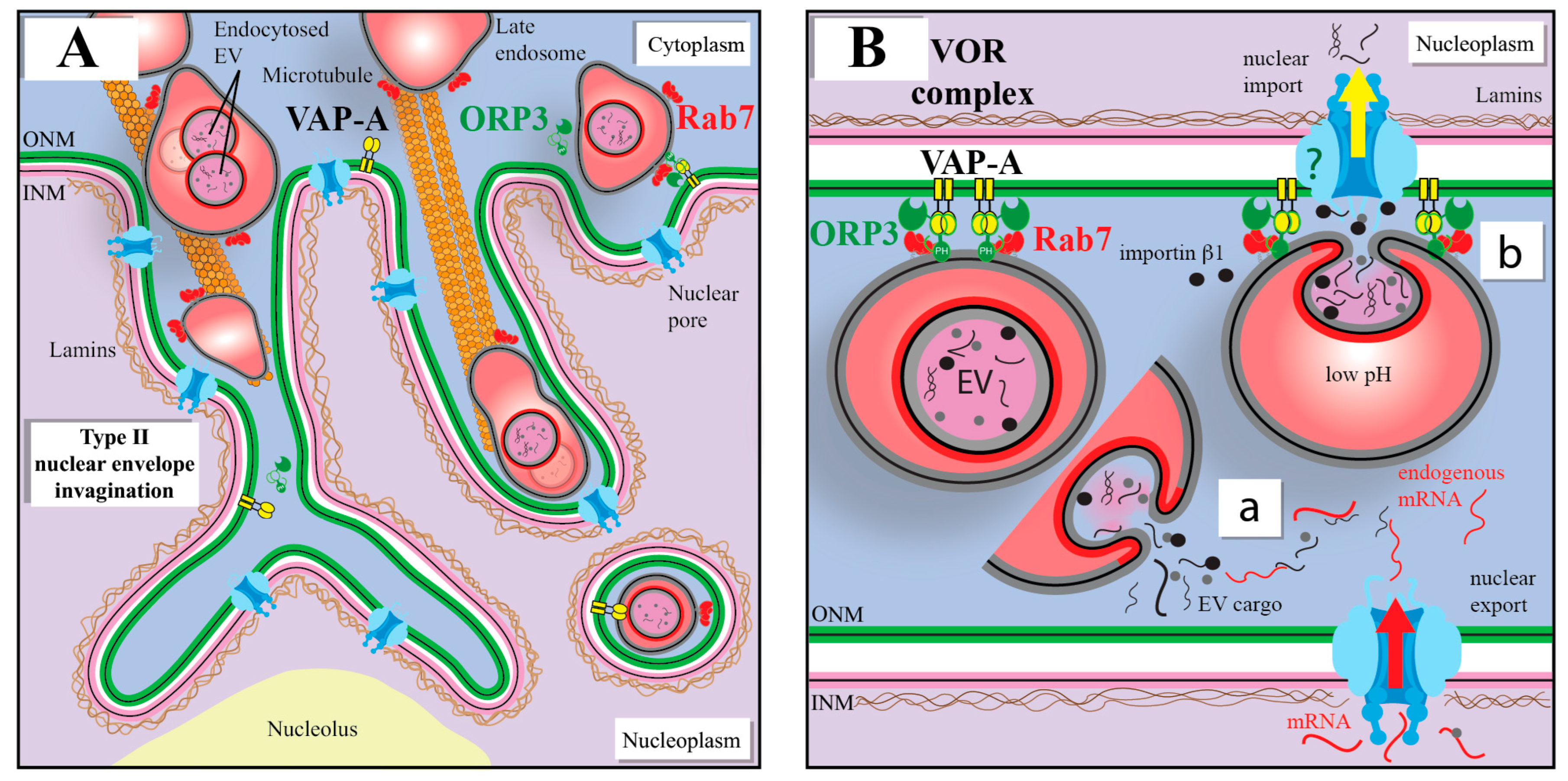
9. A Novel Intracellular Pathway Accounting for the Nuclear Transport of EV Cargo
10. VOR Complex Inhibition As a Means to Intercept the EV-Mediated Nuclear Transfer of Oncogenic Factors
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seyfried, T.N.; Huysentruyt, L.C. On the origin of cancer metastasis. Crit. Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef]
- Paget, S. The distribution of secondary growths in cancer of the breast. Cancer Metastasis Rev. 1989, 8, 98–101. [Google Scholar] [CrossRef]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Valcz, G.; Galamb, O.; Krenacs, T.; Spisak, S.; Kalmar, A.; Patai, A.V.; Wichmann, B.; Dede, K.; Tulassay, Z.; Molnar, B. Exosomes in colorectal carcinoma formation: ALIX under the magnifying glass. Mod. Pathol. 2016, 29, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Valcz, G.; Buzas, E.I.; Szallasi, Z.; Kalmar, A.; Krenacs, T.; Tulassay, Z.; Igaz, P.; Molnar, B. Perspective: Bidirectional exosomal transport between cancer stem cells and their fibroblast-rich microenvironment during metastasis formation. NPJ Breast Cancer 2018, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, R.; Maresh, G.; Zhang, X.; Salomon, C.; Hooper, J.; Margolin, D.; Li, L. The Emerging Roles of Extracellular Vesicles As Communication Vehicles within the Tumor Microenvironment and Beyond. Front. Endocrinol. 2017, 8, 194. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Abudula, M.; Li, C.; Chen, Z.; Zhang, Y.; Chen, Y. Icotinib-resistant HCC827 cells produce exosomes with mRNA MET oncogenes and mediate the migration and invasion of NSCLC. Respir. Res. 2019, 20, 217. [Google Scholar] [CrossRef] [PubMed]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef]
- Ratajczak, J.; Wysoczynski, M.; Hayek, F.; Janowska-Wieczorek, A.; Ratajczak, M.Z. Membrane-derived microvesicles: Important and underappreciated mediators of cell-to-cell communication. Leukemia 2006, 20, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Herrera, M.B.; Fonsato, V.; Gatti, S.; Deregibus, M.C.; Sordi, A.; Cantarella, D.; Calogero, R.; Bussolati, B.; Tetta, C.; Camussi, G. Human liver stem cell-derived microvesicles accelerate hepatic regeneration in hepatectomized rats. J. Cell Mol. Med. 2010, 14, 1605–1618. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef]
- Baj-Krzyworzeka, M.; Szatanek, R.; Weglarczyk, K.; Baran, J.; Urbanowicz, B.; Branski, P.; Ratajczak, M.Z.; Zembala, M. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol. Immunother. 2006, 55, 808–818. [Google Scholar] [CrossRef]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29C, 116–125. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Corbeil, D.; Santos, M.F.; Karbanová, J.; Kurth, T.; Rappa, G.; Lorico, A. Uptake and Fate of Extracellular Membrane Vesicles: Nucleoplasmic Reticulum-Associated Late Endosomes as a New Gate to Intercellular Communication. Cells 2020, 9, 1931. [Google Scholar] [CrossRef]
- Bergsmedh, A.; Szeles, A.; Henriksson, M.; Bratt, A.; Folkman, M.J.; Spetz, A.L.; Holmgren, L. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc. Natl. Acad. Sci. USA 2001, 98, 6407–6411. [Google Scholar] [CrossRef] [PubMed]
- Meehan, B.; Rak, J.; Di Vizio, D. Oncosomes—Large and small: What are they, where they came from? J. Extracell. Vesicles 2016, 5, 33109. [Google Scholar] [CrossRef]
- Lynch, C.; Panagopoulou, M.; Gregory, C.D. Extracellular Vesicles Arising from Apoptotic Cells in Tumors: Roles in Cancer Pathogenesis and Potential Clinical Applications. Front. Immunol. 2017, 8, 1174. [Google Scholar] [CrossRef] [PubMed]
- Minciacchi, V.R.; You, S.; Spinelli, C.; Morley, S.; Zandian, M.; Aspuria, P.J.; Cavallini, L.; Ciardiello, C.; Reis Sobreiro, M.; Morello, M.; et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget 2015, 6, 11327–11341. [Google Scholar] [CrossRef] [PubMed]
- Gregory, C.D.; Dransfield, I. Apoptotic Tumor Cell-Derived Extracellular Vesicles as Important Regulators of the Onco-Regenerative Niche. Front. Immunol. 2018, 9, 1111. [Google Scholar] [CrossRef] [PubMed]
- Di Vizio, D.; Kim, J.; Hager, M.H.; Morello, M.; Yang, W.; Lafargue, C.J.; True, L.D.; Rubin, M.A.; Adam, R.M.; Beroukhim, R.; et al. Oncosome formation in prostate cancer: Association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 2009, 69, 5601–5609. [Google Scholar] [CrossRef]
- Ma, L.; Li, Y.; Peng, J.; Wu, D.; Zhao, X.; Cui, Y.; Chen, L.; Yan, X.; Du, Y.; Yu, L. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 2015, 25, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Deniz, I.A.; Karbanová, J.; Wobus, M.; Bornhäuser, M.; Wimberger, P.; Kuhlmann, J.D.; Corbeil, D. Mesenchymal stromal cell-associated migrasomes: A new source of chemoattractant for cells of hematopoietic origin. Cell Commun. Signal. 2023, 21, 36. [Google Scholar] [CrossRef]
- Jiang, D.; Jiang, Z.; Lu, D.; Wang, X.; Liang, H.; Zhang, J.; Meng, Y.; Li, Y.; Wu, D.; Huang, Y.; et al. Migrasomes provide regional cues for organ morphogenesis during zebrafish gastrulation. Nat. Cell Biol. 2019, 21, 966–977. [Google Scholar] [CrossRef]
- Qin, Y.; Yang, J.; Liang, C.; Liu, J.; Deng, Z.; Yan, B.; Fu, Y.; Luo, Y.; Li, X.; Wei, X.; et al. Pan-cancer analysis identifies migrasome-related genes as a potential immunotherapeutic target: A bulk omics research and single cell sequencing validation. Front. Immunol. 2022, 13, 994828. [Google Scholar] [CrossRef]
- Rezaie, J.; Ahmadi, M.; Ravanbakhsh, R.; Mojarad, B.; Mahbubfam, S.; Shaban, S.A.; Shadi, K.; Berenjabad, N.J.; Etemadi, T. Tumor-derived extracellular vesicles: The metastatic organotropism drivers. Life Sci. 2022, 289, 120216. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Zhu, M.; Zou, Q.; Huang, R.; Li, Y.; Xing, X.; Fang, J.; Ma, L.; Li, L.; Yang, X.; Yu, L. Lateral transfer of mRNA and protein by migrasomes modifies the recipient cells. Cell Res. 2021, 31, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Meldolesi, J. Exosomes and Ectosomes in Intercellular Communication. Curr. Biol. 2018, 28, R435–R444. [Google Scholar] [CrossRef]
- Morad, G.; Carman, C.V.; Hagedorn, E.J.; Perlin, J.R.; Zon, L.I.; Mustafaoglu, N.; Park, T.E.; Ingber, D.E.; Daisy, C.C.; Moses, M.A. Tumor-Derived Extracellular Vesicles Breach the Intact Blood-Brain Barrier via Transcytosis. ACS Nano 2019, 13, 13853–13865. [Google Scholar] [CrossRef] [PubMed]
- Heidarzadeh, M.; Gursoy-Ozdemir, Y.; Kaya, M.; Eslami Abriz, A.; Zarebkohan, A.; Rahbarghazi, R.; Sokullu, E. Exosomal delivery of therapeutic modulators through the blood-brain barrier; promise and pitfalls. Cell Biosci. 2021, 11, 142. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, X. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell 2016, 30, 668–681. [Google Scholar] [CrossRef]
- Malkin, E.Z.; Bratman, S.V. Bioactive DNA from extracellular vesicles and particles. Cell Death Dis. 2020, 11, 584. [Google Scholar] [CrossRef]
- Grange, C.; Tapparo, M.; Collino, F.; Vitillo, L.; Damasco, C.; Deregibus, M.C.; Tetta, C.; Bussolati, B.; Camussi, G. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011, 71, 5346–5356. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Chow, A.; Zhou, W.; Liu, L.; Fong, M.Y.; Champer, J.; Van Haute, D.; Chin, A.R.; Ren, X.; Gugiu, B.G.; Meng, Z.; et al. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-kappaB. Sci. Rep. 2014, 4, 5750. [Google Scholar] [CrossRef] [PubMed]
- Ricklefs, F.L.; Alayo, Q.; Krenzlin, H.; Mahmoud, A.B.; Speranza, M.C.; Nakashima, H.; Hayes, J.L.; Lee, K.; Balaj, L.; Passaro, C.; et al. Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci. Adv. 2018, 4, eaar2766. [Google Scholar] [CrossRef] [PubMed]
- Marar, C.; Starich, B.; Wirtz, D. Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 2021, 22, 560–570. [Google Scholar] [CrossRef]
- Gao, Y.; Bado, I.; Wang, H.; Zhang, W.; Rosen, J.M.; Zhang, X.H. Metastasis Organotropism: Redefining the Congenial Soil. Dev. Cell 2019, 49, 375–391. [Google Scholar] [CrossRef]
- Chen, W.; Hoffmann, A.D.; Liu, H.; Liu, X. Organotropism: New insights into molecular mechanisms of breast cancer metastasis. NPJ Precis. Oncol. 2018, 2, 4. [Google Scholar] [CrossRef]
- Menck, K.; Scharf, C.; Bleckmann, A.; Dyck, L.; Rost, U.; Wenzel, D.; Dhople, V.M.; Siam, L.; Pukrop, T.; Binder, C.; et al. Tumor-derived microvesicles mediate human breast cancer invasion through differentially glycosylated EMMPRIN. J. Mol. Cell Biol. 2015, 7, 143–153. [Google Scholar] [CrossRef]
- Lorico, A.; Lorico-Rappa, M.; Karbanová, J.; Corbeil, D.; Pizzorno, G. CD9, a tetraspanin target for cancer therapy? Exp. Biol. Med. 2021, 246, 1121–1138. [Google Scholar] [CrossRef]
- Mercer, T.R.; Mattick, J.S. Understanding the regulatory and transcriptional complexity of the genome through structure. Genome Res. 2013, 23, 1081–1088. [Google Scholar] [CrossRef]
- Dragomir, M.; Chen, B.; Calin, G.A. Exosomal lncRNAs as new players in cell-to-cell communication. Transl. Cancer Res. 2018, 7, S243–S252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.L.; Liu, Y.; Jiang, J.; Tang, Y.J.; Tang, Y.L.; Liang, X.H. Extracellular vesicle long non-coding RNA-mediated crosstalk in the tumor microenvironment: Tiny molecules, huge roles. Cancer Sci. 2020, 111, 2726–2735. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Liao, Z.; Wang, Y.; Zhou, J.; He, X.; Ou, C. Exosomal long non-coding RNAs: Emerging players in cancer metastasis and potential diagnostic biomarkers for personalized oncology. Genes. Dis. 2021, 8, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Magistri, M.; Faghihi, M.A.; St Laurent, G., 3rd; Wahlestedt, C. Regulation of chromatin structure by long noncoding RNAs: Focus on natural antisense transcripts. Trends Genet. 2012, 28, 389–396. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Vagner, T.; Spinelli, C.; Minciacchi, V.R.; Balaj, L.; Zandian, M.; Conley, A.; Zijlstra, A.; Freeman, M.R.; Demichelis, F.; De, S.; et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell. Vesicles 2018, 7, 1505403. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef]
- Yokoi, A.; Villar-Prados, A.; Oliphint, P.A.; Zhang, J.; Song, X.; De Hoff, P.; Morey, R.; Liu, J.; Roszik, J.; Clise-Dwyer, K.; et al. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 2019, 5, eaax8849. [Google Scholar] [CrossRef]
- Zhang, Q.; Jeppesen, D.K.; Higginbotham, J.N.; Demory Beckler, M.; Poulin, E.J.; Walsh, A.J.; Skala, M.C.; McKinley, E.T.; Manning, H.C.; Hight, M.R.; et al. Mutant KRAS Exosomes Alter the Metabolic State of Recipient Colonic Epithelial Cells. Cell Mol. Gastroenterol. Hepatol. 2018, 5, 627–629.e626. [Google Scholar] [CrossRef]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Chennakrishnaiah, S.; Audemard, E.; Montermini, L.; Meehan, B.; Rak, J. Oncogenic ras-driven cancer cell vesiculation leads to emission of double-stranded DNA capable of interacting with target cells. Biochem. Biophys. Res. Commun. 2014, 451, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Chennakrishnaiah, S.; Meehan, B.; Montermini, L.; Garnier, D.; D’Asti, E.; Hou, W.; Magnus, N.; Gayden, T.; Jabado, N.; et al. Barriers to horizontal cell transformation by extracellular vesicles containing oncogenic H-ras. Oncotarget 2016, 7, 51991–52002. [Google Scholar] [CrossRef] [PubMed]
- Paget, S. The distribution of secondary growths in cancer of the breast. Lancet 1889, 133, 571–573. [Google Scholar] [CrossRef]
- Fidler, I.J.; Poste, G. The “seed and soil” hypothesis revisited. Lancet Oncol. 2008, 9, 808. [Google Scholar] [CrossRef]
- Hart, I.R.; Fidler, I.J. Role of organ selectivity in the determination of metastatic patterns of B16 melanoma. Cancer Res. 1980, 40, 2281–2287. [Google Scholar]
- Langley, R.R.; Fidler, I.J. The seed and soil hypothesis revisited—The role of tumor-stroma interactions in metastasis to different organs. Int. J. Cancer 2011, 128, 2527–2535. [Google Scholar] [CrossRef]
- Ewing, J. Neoplastic Diseases, 3rd ed.; W.B. Saunders Company: Philadelphia, PA, USA, 1928; p. 1127. [Google Scholar]
- Goldenberg, D.M.; Pavia, R.A.; Tsao, M.C. In vivo hybridisation of human tumour and normal hamster cells. Nature 1974, 250, 649–651. [Google Scholar] [CrossRef]
- Hamam, D.; Abdouh, M.; Gao, Z.H.; Arena, V.; Arena, M.; Arena, G.O. Transfer of malignant trait to BRCA1 deficient human fibroblasts following exposure to serum of cancer patients. J. Exp. Clin. Cancer Res. 2016, 35, 80. [Google Scholar] [CrossRef]
- Garcia-Olmo, D.; Garcia-Olmo, D.C.; Ontanon, J.; Martinez, E.; Vallejo, M. Tumor DNA circulating in the plasma might play a role in metastasis. The hypothesis of the genometastasis. Histol. Histopathol. 1999, 14, 1159–1164. [Google Scholar] [CrossRef]
- Santos, M.F.; Rappa, G.; Karbanová, J.; Kurth, T.; Corbeil, D.; Lorico, A. VAMP-associated protein-A and oxysterol-binding protein-related protein 3 promote the entry of late endosomes into the nucleoplasmic reticulum. J. Biol. Chem. 2018, 293, 13834–13848. [Google Scholar] [CrossRef] [PubMed]
- Rappa, G.; Santos, M.F.; Green, T.M.; Karbanová, J.; Hassler, J.; Bai, Y.; Barsky, S.H.; Corbeil, D.; Lorico, A. Nuclear transport of cancer extracellular vesicle-derived biomaterials through nuclear envelope invagination-associated late endosomes. Oncotarget 2017, 8, 14443–14461. [Google Scholar] [CrossRef] [PubMed]
- Massague, J.; Obenauf, A.C. Metastatic colonization by circulating tumour cells. Nature 2016, 529, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Obenauf, A.C.; Massague, J. Surviving at a Distance: Organ-Specific Metastasis. Trends Cancer 2015, 1, 76–91. [Google Scholar] [CrossRef] [PubMed]
- Kinsey, D.L. An experimental study of preferential metastasis. Cancer 1960, 13, 674–676. [Google Scholar] [CrossRef]
- Auerbach, R.; Lu, W.C.; Pardon, E.; Gumkowski, F.; Kaminska, G.; Kaminski, M. Specificity of adhesion between murine tumor cells and capillary endothelium: An in vitro correlate of preferential metastasis in vivo. Cancer Res. 1987, 47, 1492–1496. [Google Scholar]
- Hedley, B.D.; Chambers, A.F. Tumor dormancy and metastasis. Adv. Cancer Res. 2009, 102, 67–101. [Google Scholar] [CrossRef]
- Budczies, J.; von Winterfeld, M.; Klauschen, F.; Bockmayr, M.; Lennerz, J.K.; Denkert, C.; Wolf, T.; Warth, A.; Dietel, M.; Anagnostopoulos, I.; et al. The landscape of metastatic progression patterns across major human cancers. Oncotarget 2015, 6, 570–583. [Google Scholar] [CrossRef]
- Roussos, E.T.; Condeelis, J.S.; Patsialou, A. Chemotaxis in cancer. Nat. Rev. Cancer 2011, 11, 573–587. [Google Scholar] [CrossRef]
- Muller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef]
- Balkwill, F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin. Cancer Biol. 2004, 14, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Kryczek, I.; Wei, S.; Keller, E.; Liu, R.; Zou, W. Stroma-derived factor (SDF-1/CXCL12) and human tumor pathogenesis. Am. J. Physiol. Cell Physiol. 2007, 292, C987–C995. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Behnam Azad, B.; Nimmagadda, S. The intricate role of CXCR4 in cancer. Adv. Cancer Res. 2014, 124, 31–82. [Google Scholar] [CrossRef]
- Mannavola, F.; Tucci, M.; Felici, C.; Passarelli, A.; D’Oronzo, S.; Silvestris, F. Tumor-derived exosomes promote the in vitro osteotropism of melanoma cells by activating the SDF-1/CXCR4/CXCR7 axis. J. Transl. Med. 2019, 17, 230. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, P.A.; Devreotes, P.N. Navigating through models of chemotaxis. Curr. Opin. Cell Biol. 2008, 20, 35–40. [Google Scholar] [CrossRef]
- Shi, Y.; Riese, D.J., 2nd; Shen, J. The Role of the CXCL12/CXCR4/CXCR7 Chemokine Axis in Cancer. Front. Pharmacol. 2020, 11, 574667. [Google Scholar] [CrossRef]
- Kaplan, R.N.; Riba, R.D.; Zacharoulis, S.; Bramley, A.H.; Vincent, L.; Costa, C.; MacDonald, D.D.; Jin, D.K.; Shido, K.; Kerns, S.A.; et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 2005, 438, 820–827. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Johansson-Percival, A.; Backhouse, J.; Li, J.; Lee, G.Y.F.; Hamzah, J.; Ganss, R. Remodeling of Metastatic Vasculature Reduces Lung Colonization and Sensitizes Overt Metastases to Immunotherapy. Cell Rep. 2020, 30, 714–724.e715. [Google Scholar] [CrossRef]
- Padua, D.; Zhang, X.H.; Wang, Q.; Nadal, C.; Gerald, W.L.; Gomis, R.R.; Massague, J. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell 2008, 133, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Song, N.; Ding, Y.; Yuan, S.; Li, X.; Cai, H.; Shi, H.; Luo, Y. Pulmonary vascular destabilization in the premetastatic phase facilitates lung metastasis. Cancer Res. 2009, 69, 7529–7537. [Google Scholar] [CrossRef]
- Gupta, G.P.; Nguyen, D.X.; Chiang, A.C.; Bos, P.D.; Kim, J.Y.; Nadal, C.; Gomis, R.R.; Manova-Todorova, K.; Massague, J. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature 2007, 446, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T.; Doughty-Shenton, D.; Cassetta, L.; Fragkogianni, S.; Brownlie, D.; Kato, Y.; Carragher, N.; Pollard, J.W. Monocytes Differentiate to Immune Suppressive Precursors of Metastasis-Associated Macrophages in Mouse Models of Metastatic Breast Cancer. Front. Immunol. 2017, 8, 2004. [Google Scholar] [CrossRef] [PubMed]
- Lowy, C.M.; Oskarsson, T. Tenascin C in metastasis: A view from the invasive front. Cell Adh Migr. 2015, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Gonzalez, L.; Alonso, J. Periostin: A Matricellular Protein with Multiple Functions in Cancer Development and Progression. Front. Oncol. 2018, 8, 225. [Google Scholar] [CrossRef]
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 2017, 17, 302–317. [Google Scholar] [CrossRef]
- Kalluri, R.; McAndrews, K.M. The role of extracellular vesicles in cancer. Cell 2023, 186, 1610–1626. [Google Scholar] [CrossRef]
- Osti, D.; Del Bene, M.; Rappa, G.; Santos, M.; Matafora, V.; Richichi, C.; Faletti, S.; Beznoussenko, G.V.; Mironov, A.; Bachi, A.; et al. Clinical Significance of Extracellular Vesicles in Plasma from Glioblastoma Patients. Clin. Cancer Res. 2019, 25, 266–276. [Google Scholar] [CrossRef]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol 2013, 113, 1–11. [Google Scholar] [CrossRef]
- Duijvesz, D.; Versluis, C.Y.; van der Fels, C.A.; Vredenbregt-van den Berg, M.S.; Leivo, J.; Peltola, M.T.; Bangma, C.H.; Pettersson, K.S.; Jenster, G. Immuno-based detection of extracellular vesicles in urine as diagnostic marker for prostate cancer. Int. J. Cancer 2015, 137, 2869–2878. [Google Scholar] [CrossRef] [PubMed]
- Fais, S.; Venturi, G.; Gatenby, B. Microenvironmental acidosis in carcinogenesis and metastases: New strategies in prevention and therapy. Cancer Metastasis Rev. 2014, 33, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [PubMed]
- Logozzi, M.; Mizzoni, D.; Angelini, D.F.; Di Raimo, R.; Falchi, M.; Battistini, L.; Fais, S. Microenvironmental pH and Exosome Levels Interplay in Human Cancer Cell Lines of Different Histotypes. Cancers 2018, 10, 370. [Google Scholar] [CrossRef]
- Ban, J.J.; Lee, M.; Im, W.; Kim, M. Low pH increases the yield of exosome isolation. Biochem. Biophys. Res. Commun. 2015, 461, 76–79. [Google Scholar] [CrossRef]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef]
- Hurwitz, S.N.; Meckes, D.G., Jr. Extracellular Vesicle Integrins Distinguish Unique Cancers. Proteomes 2019, 7, 14. [Google Scholar] [CrossRef]
- Lu, H.; Bowler, N.; Harshyne, L.A.; Craig Hooper, D.; Krishn, S.R.; Kurtoglu, S.; Fedele, C.; Liu, Q.; Tang, H.Y.; Kossenkov, A.V.; et al. Exosomal alphavbeta6 integrin is required for monocyte M2 polarization in prostate cancer. Matrix Biol. 2018, 70, 20–35. [Google Scholar] [CrossRef]
- DeRita, R.M.; Sayeed, A.; Garcia, V.; Krishn, S.R.; Shields, C.D.; Sarker, S.; Friedman, A.; McCue, P.; Molugu, S.K.; Rodeck, U.; et al. Tumor-Derived Extracellular Vesicles Require beta1 Integrins to Promote Anchorage-Independent Growth. iScience 2019, 14, 199–209. [Google Scholar] [CrossRef]
- Fedele, C.; Singh, A.; Zerlanko, B.J.; Iozzo, R.V.; Languino, L.R. The alphavbeta6 integrin is transferred intercellularly via exosomes. J. Biol. Chem. 2015, 290, 4545–4551. [Google Scholar] [CrossRef] [PubMed]
- Krishn, S.R.; Salem, I.; Quaglia, F.; Naranjo, N.M.; Agarwal, E.; Liu, Q.; Sarker, S.; Kopenhaver, J.; McCue, P.A.; Weinreb, P.H.; et al. The alphavbeta6 integrin in cancer cell-derived small extracellular vesicles enhances angiogenesis. J. Extracell. Vesicles 2020, 9, 1763594. [Google Scholar] [CrossRef]
- Quaglia, F.; Krishn, S.R.; Daaboul, G.G.; Sarker, S.; Pippa, R.; Domingo-Domenech, J.; Kumar, G.; Fortina, P.; McCue, P.; Kelly, W.K.; et al. Small extracellular vesicles modulated by alphaVbeta3 integrin induce neuroendocrine differentiation in recipient cancer cells. J. Extracell. Vesicles 2020, 9, 1761072. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Yoshioka, Y.; Ochiya, T. Intercellular crosstalk between cancer cells and cancer-associated fibroblasts via extracellular vesicles. Cancer Cell Int. 2022, 22, 367. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yoshimura, T.; Tian, M.; Wang, Y.; Kondo, T.; Yamamoto, K.-I.; Fujisawa, M.; Ohara, T.; Sakaguchi, M.; Matsukawa, A. Exosomal Wnt7a from a low metastatic subclone promotes lung metastasis of a highly metastatic subclone in the murine 4t1 breast cancer. Breast Cancer Res. 2022, 24, 60. [Google Scholar] [CrossRef]
- Schillaci, O.; Fontana, S.; Monteleone, F.; Taverna, S.; Di Bella, M.A.; Di Vizio, D.; Alessandro, R. Exosomes from metastatic cancer cells transfer amoeboid phenotype to non-metastatic cells and increase endothelial permeability: Their emerging role in tumor heterogeneity. Sci. Rep. 2017, 7, 4711. [Google Scholar] [CrossRef]
- Pang, H.; Liu, L.; Sun, X.; Xi, W.; Bao, Y.; Wu, L.; Shan, J.; Wang, Z.; Guo, Y.; Zhao, C. Exosomes derived from colon cancer cells and plasma of colon cancer patients promote migration of SW480 cells through Akt/mTOR pathway. Pathol. Res. Pract. 2021, 222, 153454. [Google Scholar] [CrossRef]
- Fais, S. Cannibalism: A way to feed on metastatic tumors. Cancer Lett. 2007, 258, 155–164. [Google Scholar] [CrossRef]
- Ono, M.; Kosaka, N.; Tominaga, N.; Yoshioka, Y.; Takeshita, F.; Takahashi, R.U.; Yoshida, M.; Tsuda, H.; Tamura, K.; Ochiya, T. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci. Signal. 2014, 7, ra63. [Google Scholar] [CrossRef]
- Bartosh, T.J.; Ullah, M.; Zeitouni, S.; Beaver, J.; Prockop, D.J. Cancer cells enter dormancy after cannibalizing mesenchymal stem/stromal cells (MSCs). Proc. Natl. Acad. Sci. USA 2016, 113, E6447–E6456. [Google Scholar] [CrossRef]
- Rappa, G.; Green, T.M.; Karbanová, J.; Corbeil, D.; Lorico, A. Tetraspanin CD9 determines invasiveness and tumorigenicity of human breast cancer cells. Oncotarget 2015, 6, 7970–7991. [Google Scholar] [CrossRef]
- Santos, M.F.; Rappa, G.; Karbanová, J.; Vanier, C.; Morimoto, C.; Corbeil, D.; Lorico, A. Anti-human CD9 antibody Fab fragment impairs the internalization of extracellular vesicles and the nuclear transfer of their cargo proteins. J. Cell Mol. Med. 2019, 23, 4408–4421. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.F.; Rappa, G.; Fontana, S.; Karbanová, J.; Aalam, F.; Tai, D.; Li, Z.; Pucci, M.; Alessandro, R.; Morimoto, C.; et al. Anti-Human CD9 Fab Fragment Antibody Blocks the Extracellular Vesicle-Mediated Increase in Malignancy of Colon Cancer Cells. Cells 2022, 11, 2474. [Google Scholar] [CrossRef] [PubMed]
- Nigri, J.; Leca, J.; Tubiana, S.S.; Finetti, P.; Guillaumond, F.; Martinez, S.; Lac, S.; Iovanna, J.L.; Audebert, S.; Camoin, L.; et al. CD9 mediates the uptake of extracellular vesicles from cancer-associated fibroblasts that promote pancreatic cancer cell aggressiveness. Sci. Signal. 2022, 15, eabg8191. [Google Scholar] [CrossRef]
- Miki, Y.; Yashiro, M.; Okuno, T.; Kitayama, K.; Masuda, G.; Hirakawa, K.; Ohira, M. CD9-positive exosomes from cancer-associated fibroblasts stimulate the migration ability of scirrhous-type gastric cancer cells. Br. J. Cancer 2018, 118, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Leibovitz, A.; Stinson, J.C.; McCombs, W.B., 3rd; McCoy, C.E.; Mazur, K.C.; Mabry, N.D. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976, 36, 4562–4569. [Google Scholar]
- Hewitt, R.E.; McMarlin, A.; Kleiner, D.; Wersto, R.; Martin, P.; Tsokos, M.; Stamp, G.W.; Stetler-Stevenson, W.G. Validation of a model of colon cancer progression. J. Pathol. 2000, 192, 446–454. [Google Scholar] [CrossRef]
- Lugini, L.; Valtieri, M.; Federici, C.; Cecchetti, S.; Meschini, S.; Condello, M.; Signore, M.; Fais, S. Exosomes from human colorectal cancer induce a tumor-like behavior in colonic mesenchymal stromal cells. Oncotarget 2016, 7, 50086–50098. [Google Scholar] [CrossRef]
- Kim, J.; Afshari, A.; Sengupta, R.; Sebastiano, V.; Gupta, A.; Kim, Y.H.; Reproducibility Project: Cancer, B.; Iorns, E.; Tsui, R.; Denis, A.; et al. Replication study: Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Elife 2018, 7, 39944. [Google Scholar] [CrossRef]
- Liu, Y. Tumor Exosomal RNAs Promote Lung Pre-metastatic Niche Formation by Activating Alveolar Epithelial TLR3 to Recruit Neutrophils. Cancer Cell 2016, 30, 243–256. [Google Scholar] [CrossRef]
- Plebanek, M.P.; Angeloni, N.L.; Vinokour, E.; Li, J.; Henkin, A.; Martinez-Marin, D.; Filleur, S.; Bhowmick, R.; Henkin, J.; Miller, S.D.; et al. Pre-metastatic cancer exosomes induce immune surveillance by patrolling monocytes at the metastatic niche. Nat. Commun. 2017, 8, 1319. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef]
- Javle, M.; Li, Y.; Tan, D.; Dong, X.; Chang, P.; Kar, S.; Li, D. Biomarkers of TGF-beta signaling pathway and prognosis of pancreatic cancer. PLoS ONE 2014, 9, e85942. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, N.J.; Li, L.; Kapoun, A.M.; Medicherla, S.; Reddy, M.; Li, G.; O’Young, G.; Quon, D.; Henson, M.; Damm, D.L.; et al. Inhibition of transforming growth factor beta signaling reduces pancreatic adenocarcinoma growth and invasiveness. Mol. Pharmacol. 2007, 72, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Ellermeier, J.; Wei, J.; Duewell, P.; Hoves, S.; Stieg, M.R.; Adunka, T.; Noerenberg, D.; Anders, H.J.; Mayr, D.; Poeck, H.; et al. Therapeutic efficacy of bifunctional siRNA combining TGF-beta1 silencing with RIG-I activation in pancreatic cancer. Cancer Res. 2013, 73, 1709–1720. [Google Scholar] [CrossRef] [PubMed]
- Luzzi, K.J.; MacDonald, I.C.; Schmidt, E.E.; Kerkvliet, N.; Morris, V.L.; Chambers, A.F.; Groom, A.C. Multistep nature of metastatic inefficiency: Dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am. J. Pathol. 1998, 153, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Fidler, I.J. Metastasis: Quantitative analysis of distribution and fate of tumor emboli labeled with 125 I-5-iodo-2’-deoxyuridine. J. Natl. Cancer Inst. 1970, 45, 773–782. [Google Scholar]
- Nguyen, D.X.; Bos, P.D.; Massagué, J. Metastasis: From dissemination to organ-specific colonization. Nat. Rev. Cancer 2009, 9, 274–284. [Google Scholar] [CrossRef]
- Phan, T.G.; Croucher, P.I. The dormant cancer cell life cycle. Nat. Rev. Cancer 2020, 20, 398–411. [Google Scholar] [CrossRef]
- Lazebnik, Y. What are the hallmarks of cancer? Nat. Rev. Cancer 2010, 10, 232–233. [Google Scholar] [CrossRef]
- Chambers, A.F.; Groom, A.C.; MacDonald, I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2002, 2, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Tarin, D. Gene expression profiling of human lymph node metastases and matched primary breast carcinomas: Clinical implications. Mol. Oncol. 2007, 1, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Micalizzi, D.S.; Maheswaran, S.; Haber, D.A. A conduit to metastasis: Circulating tumor cell biology. Genes. Dev. 2017, 31, 1827–1840. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Yao, D.; Dai, C.; Peng, S. Mechanism of the mesenchymal-epithelial transition and its relationship with metastatic tumor formation. Mol. Cancer Res. 2011, 9, 1608–1620. [Google Scholar] [CrossRef]
- Jolly, M.K.; Ware, K.E.; Gilja, S.; Somarelli, J.A.; Levine, H. EMT and MET: Necessary or permissive for metastasis? Mol. Oncol. 2017, 11, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef]
- Christiansen, J.J.; Rajasekaran, A.K. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006, 66, 8319–8326. [Google Scholar] [CrossRef]
- Chui, M.H. Insights into cancer metastasis from a clinicopathologic perspective: Epithelial-Mesenchymal Transition is not a necessary step. Int. J. Cancer 2013, 132, 1487–1495. [Google Scholar] [CrossRef]
- Jie, X.X.; Zhang, X.Y.; Xu, C.J. Epithelial-to-mesenchymal transition, circulating tumor cells and cancer metastasis: Mechanisms and clinical applications. Oncotarget 2017, 8, 81558–81571. [Google Scholar] [CrossRef] [PubMed]
- Arena, G.O.; Arena, V.; Arena, M.; Abdouh, M. Transfer of malignant traits as opposed to migration of cells: A novel concept to explain metastatic disease. Med. Hypotheses 2017, 100, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Sun, B.; Hu, L.; Lahdesmaki, H.; Dunmire, V.; Feng, Y.; Zhang, S.W.; Wang, H.; Wu, C.; Wang, H.; et al. Differential gene and protein expression in primary breast malignancies and their lymph node metastases as revealed by combined cDNA microarray and tissue microarray analysis. Cancer 2004, 100, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, B.; Wessels, L.F.; Bosma, A.J.; Glas, A.M.; Nuyten, D.S.; He, Y.D.; Dai, H.; Peterse, J.L.; van’t Veer, L.J. No common denominator for breast cancer lymph node metastasis. Br. J. Cancer 2005, 93, 924–932. [Google Scholar] [CrossRef]
- Magbanua, M.J.M.; Rugo, H.S.; Hauranieh, L.; Roy, R.; Scott, J.H.; Lee, J.C.; Hsiao, F.; Sosa, E.V.; Van’t Veer, L.; Esserman, L.J.; et al. Genomic and expression profiling reveal molecular heterogeneity of disseminated tumor cells in bone marrow of early breast cancer. NPJ Breast Cancer 2018, 4, 31. [Google Scholar] [CrossRef]
- Brukman, N.G.; Uygur, B.; Podbilewicz, B.; Chernomordik, L.V. How cells fuse. J. Cell Biol. 2019, 218, 1436–1451. [Google Scholar] [CrossRef]
- Aichel, O. Über zellverschmelzung mit quantitativ abnormer chromosomenverteilung als ursache der geschwulstbildung. In Vorträge und Aufsätze über Entwicklungsmechanik der Organismen. In Vorträge und Aufsätze über Entvickelungsmechanik Der Organismen; Roux, W., Ed.; Wilhelm Engelmann: Leipzig, Germany, 1911; pp. 92–111. [Google Scholar]
- Pawelek, J.M.; Chakraborty, A.K. Fusion of tumour cells with bone marrow-derived cells: A unifying explanation for metastasis. Nat. Rev. Cancer 2008, 8, 377–386. [Google Scholar] [CrossRef]
- Laberge, G.S.; Duvall, E.; Haedicke, K.; Pawelek, J. Leukocyte(-)Cancer Cell Fusion-Genesis of a Deadly Journey. Cells 2019, 8, 170. [Google Scholar] [CrossRef]
- Weiler, J.; Dittmar, T. Cell Fusion in Human Cancer: The Dark Matter Hypothesis. Cells 2019, 8, 132. [Google Scholar] [CrossRef]
- Barski, G.; Cornefert, F. Characteristics of “hybrid”-type clonal cell lines obtained from mixed cultures in vitro. J. Natl. Cancer Inst. 1962, 28, 801–821. [Google Scholar]
- Goldenberg, D.M.; Pavia, R.A. In vivo horizontal oncogenesis by a human tumor in nude mice. Proc. Natl. Acad. Sci. USA 1982, 79, 2389–2392. [Google Scholar] [CrossRef] [PubMed]
- Pawelek, J.M.; Chakraborty, A.K. The cancer cell--leukocyte fusion theory of metastasis. Adv. Cancer Res. 2008, 101, 397–444. [Google Scholar] [CrossRef] [PubMed]
- Rappa, G.; Mercapide, J.; Lorico, A. Spontaneous formation of tumorigenic hybrids between breast cancer and multipotent stromal cells is a source of tumor heterogeneity. Am. J. Pathol. 2012, 180, 2504–2515. [Google Scholar] [CrossRef]
- Ehrlich, P.; Apolant, H. Beobachtungen über maligne Mäusetumoren [Observations on malignant murine tumors]. Berl. Klin. Wochenschr. 1905, 42, 871–874. [Google Scholar]
- Beattie, G.M.; Knowles, A.F.; Jensen, F.C.; Baird, S.M.; Kaplan, N.O. Induction of sarcomas in athymic mice. Proc. Natl. Acad. Sci. USA 1982, 79, 3033–3036. [Google Scholar] [CrossRef] [PubMed]
- Bowen, J.M.; Cailleau, R.; Giovanella, B.C.; Pathak, S.; Siciliano, M.J. A retrovirus-producing transformed mouse cell line derived from a human breast adenocarcinoma transplanted in a nude mouse. Vitr. Cell Dev. Biol.-Plant 1983, 19, 635–641. [Google Scholar] [CrossRef]
- Staab, H.J.; Heilbronner, H.; Schrader, M.; Anderer, F.A. In vivo induction of neoplastic growth in nude mouse connective tissue adjacent to xenografted human tumors. J. Cancer Res. Clin. Oncol. 1983, 106, 27–35. [Google Scholar] [CrossRef]
- Mishra, N.C. Gene transfer in fungi. Adv. Genet. 1985, 23, 73–178. [Google Scholar] [CrossRef]
- Low, K.B.; Porter, D.D. Modes of gene transfer and recombination in bacteria. Annu. Rev. Genet. 1978, 12, 249–287. [Google Scholar] [CrossRef]
- Holmgren, L.; Szeles, A.; Rajnavolgyi, E.; Folkman, J.; Klein, G.; Ernberg, I.; Falk, K.I. Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood 1999, 93, 3956–3963. [Google Scholar] [CrossRef]
- Garcia-Olmo, D.C.; Dominguez, C.; Garcia-Arranz, M.; Anker, P.; Stroun, M.; Garcia-Verdugo, J.M.; Garcia-Olmo, D. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 2010, 70, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Olmo, D.; Garcia-Olmo, D.C. Functionality of circulating DNA: The hypothesis of genometastasis. Ann. N. Y. Acad. Sci. 2001, 945, 265–275. [Google Scholar] [CrossRef]
- Trejo-Becerril, C.; Perez-Cardenas, E.; Taja-Chayeb, L.; Anker, P.; Herrera-Goepfert, R.; Medina-Velazquez, L.A.; Hidalgo-Miranda, A.; Perez-Montiel, D.; Chavez-Blanco, A.; Cruz-Velazquez, J.; et al. Cancer progression mediated by horizontal gene transfer in an in vivo model. PLoS ONE 2012, 7, e52754. [Google Scholar] [CrossRef]
- Todaro, G.J.; Green, H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963, 17, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Anker, P.; Lyautey, J.; Lefort, F.; Lederrey, C.; Stroun, M. Transformation of NIH/3T3 cells and SW 480 cells displaying K-ras mutation. C. R. Acad. Sci. III 1994, 317, 869–874. [Google Scholar] [PubMed]
- Abdouh, M.; Zhou, S.; Arena, V.; Arena, M.; Lazaris, A.; Onerheim, R.; Metrakos, P.; Arena, G.O. Transfer of malignant trait to immortalized human cells following exposure to human cancer serum. J. Exp. Clin. Cancer Res. 2014, 33, 86. [Google Scholar] [CrossRef]
- Abdouh, M.; Floris, M.; Gao, Z.H.; Arena, V.; Arena, M.; Arena, G.O. Colorectal cancer-derived extracellular vesicles induce transformation of fibroblasts into colon carcinoma cells. J. Exp. Clin. Cancer Res. 2019, 38, 257. [Google Scholar] [CrossRef]
- Abdouh, M.; Hamam, D.; Gao, Z.-H.; Arena, V.; Arena, M.; Arena, G.O. Exosomes isolated from cancer patients’ sera transfer malignant traits and confer the same phenotype of primary tumors to oncosuppressor-mutated cells. J. Exp. Clin. Cancer Res. 2017, 36, 113. [Google Scholar] [CrossRef]
- Abdouh, M.; Hamam, D.; Arena, V.; Arena, M.; Alamri, H.; Arena, G.O. Novel blood test to predict neoplastic activity in healthy patients and metastatic recurrence after primary tumor resection. J. Circ. Biomark. 2016, 5, 1849454416663661. [Google Scholar] [CrossRef]
- Abdouh, M.; Tabah, R.; Arena, V.; Arena, M.; Gao, Z.H.; Lorico, A.; Arena, G.O. Oncosuppressor-Mutated Cell-Based Diagnostic Platform for Liquid Biopsy Diagnoses Benign Head and Neck Masses and Predicts Malignancy in Thyroid Nodules: Results from a Consecutive Cohort of Patients. Eur. Thyroid. J. 2021, 10, 285–294. [Google Scholar] [CrossRef]
- Abdouh, M.; Gao, Z.-H.; Arena, V.; Arena, M.; Burnier, M.N.; Arena, G.O. Oncosuppressor-Mutated Cells as a Liquid Biopsy Test for Cancer-Screening. Sci. Rep. 2019, 9, 2384. [Google Scholar] [CrossRef]
- Fernandez, N.; Chavarriaga, J.; Ayala, P.; Pedraza, A.; Bolivar, J.; Prada, J.G.; Catano, J.G.; Garcia-Perdomo, H.A.; Villanueva, J.; Varela, D.; et al. MicroRNAs as Potential Liquid Biopsy Biomarker for Patients with Castration-Resistant Prostate Cancer. Res. Rep. Urol. 2022, 14, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Mauri, G.; Vitiello, P.P.; Sogari, A.; Crisafulli, G.; Sartore-Bianchi, A.; Marsoni, S.; Siena, S.; Bardelli, A. Liquid biopsies to monitor and direct cancer treatment in colorectal cancer. Br. J. Cancer 2022, 127, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Gattuso, G.; Crimi, S.; Lavoro, A.; Rizzo, R.; Musumarra, G.; Gallo, S.; Facciponte, F.; Paratore, S.; Russo, A.; Bordonaro, R.; et al. Liquid Biopsy and Circulating Biomarkers for the Diagnosis of Precancerous and Cancerous Oral Lesions. Noncoding RNA 2022, 8, 60. [Google Scholar] [CrossRef]
- Doglioni, G.; Parik, S.; Fendt, S.M. Interactions in the (Pre)metastatic Niche Support Metastasis Formation. Front. Oncol. 2019, 9, 219. [Google Scholar] [CrossRef]
- Souza, A.G.; Bastos, V.A.F.; Fujimura, P.T.; Ferreira, I.C.C.; Leal, L.F.; da Silva, L.S.; Laus, A.C.; Reis, R.M.; Martins, M.M.; Santos, P.S.; et al. Cell-free DNA promotes malignant transformation in non-tumor cells. Sci. Rep. 2020, 10, 21674. [Google Scholar] [CrossRef] [PubMed]
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.M.; Ding, W.; et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994, 266, 66–71. [Google Scholar] [CrossRef]
- Donin, N.; Filson, C.; Drakaki, A.; Tan, H.J.; Castillo, A.; Kwan, L.; Litwin, M.; Chamie, K. Risk of second primary malignancies among cancer survivors in the United States, 1992 through 2008. Cancer 2016, 122, 3075–3086. [Google Scholar] [CrossRef]
- Feller, A.; Matthes, K.L.; Bordoni, A.; Bouchardy, C.; Bulliard, J.L.; Herrmann, C.; Konzelmann, I.; Maspoli, M.; Mousavi, M.; Rohrmann, S.; et al. The relative risk of second primary cancers in Switzerland: A population-based retrospective cohort study. BMC Cancer 2020, 20, 51. [Google Scholar] [CrossRef]
- Allen, I.; Hassan, H.; Sofianopoulou, E.; Eccles, D.; Turnbull, C.; Tischkowitz, M.; Pharoah, P.; Antoniou, A.C. Risk of developing a second primary cancer in male breast cancer survivors: A systematic review and meta-analysis. Br. J. Cancer 2022, 127, 1660–1669. [Google Scholar] [CrossRef]
- Kerkar, S.P.; Restifo, N.P. Cellular constituents of immune escape within the tumor microenvironment. Cancer Res. 2012, 72, 3125–3130. [Google Scholar] [CrossRef] [PubMed]
- Friberg, S.; Nystrom, A. Cancer Metastases: Early Dissemination and Late Recurrences. Cancer Growth Metastasis 2015, 8, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Holmgren, L.; O’Reilly, M.S.; Folkman, J. Dormancy of micrometastases: Balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat. Med. 1995, 1, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.Y.; Hynes, R.O. Lymphatic or hematogenous dissemination: How does a metastatic tumor cell decide? Cell Cycle 2006, 5, 812–817. [Google Scholar] [CrossRef] [PubMed]
- Kinchington, P.R.; Leger, A.J.; Guedon, J.M.; Hendricks, R.L. Herpes simplex virus and varicella zoster virus, the house guests who never leave. Herpesviridae 2012, 3, 5. [Google Scholar] [CrossRef]
- Sugi, K.; Nawata, K.; Fujita, N.; Ueda, K.; Tanaka, T.; Matsuoka, T.; Kaneda, Y.; Esato, K. Systematic lymph node dissection for clinically diagnosed peripheral non-small-cell lung cancer less than 2 cm in diameter. World J. Surg. 1998, 22, 290–294, discussion 294–295. [Google Scholar] [CrossRef]
- Darling, G.E.; Allen, M.S.; Decker, P.A.; Ballman, K.; Malthaner, R.A.; Inculet, R.I.; Jones, D.R.; McKenna, R.J.; Landreneau, R.J.; Rusch, V.W.; et al. Randomized trial of mediastinal lymph node sampling versus complete lymphadenectomy during pulmonary resection in the patient with N0 or N1 (less than hilar) non-small cell carcinoma: Results of the American College of Surgery Oncology Group Z0030 Trial. J. Thorac. Cardiovasc. Surg. 2011, 141, 662–670. [Google Scholar] [CrossRef]
- Feuerstein, M.A.; Kent, M.; Bazzi, W.M.; Bernstein, M.; Russo, P. Analysis of lymph node dissection in patients with >/=7-cm renal tumors. World J. Urol. 2014, 32, 1531–1536. [Google Scholar] [CrossRef]
- Huang, X.; Wang, J.; Chen, Q.; Jiang, J. Mediastinal lymph node dissection versus mediastinal lymph node sampling for early stage non-small cell lung cancer: A systematic review and meta-analysis. PLoS ONE 2014, 9, e109979. [Google Scholar] [CrossRef]
- Valcz, G.; Ujvari, B.; Buzas, E.I.; Krenacs, T.; Spisak, S.; Kittel, A.; Tulassay, Z.; Igaz, P.; Takacs, I.; Molnar, B. Small extracellular vesicle DNA-mediated horizontal gene transfer as a driving force for tumor evolution: Facts and riddles. Front. Oncol. 2022, 12, 945376. [Google Scholar] [CrossRef]
- Cai, J.; Wu, G.; Tan, X.; Han, Y.; Chen, C.; Li, C.; Wang, N.; Zou, X.; Chen, X.; Zhou, F.; et al. Transferred BCR/ABL DNA from K562 extracellular vesicles causes chronic myeloid leukemia in immunodeficient mice. PLoS ONE 2014, 9, e105200. [Google Scholar] [CrossRef]
- Qian, Z.; Shen, Q.; Yang, X.; Qiu, Y.; Zhang, W. The Role of Extracellular Vesicles: An Epigenetic View of the Cancer Microenvironment. Biomed. Res. Int. 2015, 2015, 649161. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.S.; de Beer, M.A.; Giepmans, B.N.G.; Zuhorn, I.S. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020, 14, 4444–4455. [Google Scholar] [CrossRef] [PubMed]
- Malhas, A.; Goulbourne, C.; Vaux, D.J. The nucleoplasmic reticulum: Form and function. Trends Cell Biol. 2011, 21, 362–373. [Google Scholar] [CrossRef]
- Read, J.; Ingram, A.; Al Saleh, H.A.; Platko, K.; Gabriel, K.; Kapoor, A.; Pinthus, J.; Majeed, F.; Qureshi, T.; Al-Nedawi, K. Nuclear transportation of exogenous epidermal growth factor receptor and androgen receptor via extracellular vesicles. Eur. J. Cancer 2017, 70, 62–74. [Google Scholar] [CrossRef]
- Singer, D.; Thamm, K.; Zhuang, H.; Karbanová, J.; Gao, Y.; Walker, J.V.; Jin, H.; Wu, X.; Coveney, C.R.; Marangoni, P.; et al. Prominin-1 controls stem cell activation by orchestrating ciliary dynamics. EMBO J. 2019, 38, e99845. [Google Scholar] [CrossRef]
- Waldenstrom, A.; Genneback, N.; Hellman, U.; Ronquist, G. Cardiomyocyte microvesicles contain DNA/RNA and convey biological messages to target cells. PLoS ONE 2012, 7, e34653. [Google Scholar] [CrossRef]
- Santos, M.F.; Rappa, G.; Karbanová, J.; Fontana, S.; Di Bella, M.A.; Pope, M.R.; Parrino, B.; Cascioferro, S.M.; Vistoli, G.; Diana, P.; et al. Itraconazole inhibits nuclear delivery of extracellular vesicle cargo by disrupting the entry of late endosomes into the nucleoplasmic reticulum. J. Extracell. Vesicles 2021, 10, e12132. [Google Scholar] [CrossRef]
- Strating, J.R.; van der Linden, L.; Albulescu, L.; Bigay, J.; Arita, M.; Delang, L.; Leyssen, P.; van der Schaar, H.M.; Lanke, K.H.; Thibaut, H.J.; et al. Itraconazole inhibits enterovirus replication by targeting the oxysterol-binding protein. Cell Rep. 2015, 10, 600–615. [Google Scholar] [CrossRef] [PubMed]
- Retsas, S. Cancer and the arts: Metastasis–as perceived through the ages. ESMO Open. 2017, 2, e000226. [Google Scholar] [CrossRef] [PubMed]
- Liddell, H.G.; Scott, R.; Jones, H.S.; McKenzie, R.; Barber, E.A. A Greek-English Lexicon, 9th ed.; Clarendon Press: Oxford, UK, 1968. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arena, G.O.; Forte, S.; Abdouh, M.; Vanier, C.; Corbeil, D.; Lorico, A. Horizontal Transfer of Malignant Traits and the Involvement of Extracellular Vesicles in Metastasis. Cells 2023, 12, 1566. https://doi.org/10.3390/cells12121566
Arena GO, Forte S, Abdouh M, Vanier C, Corbeil D, Lorico A. Horizontal Transfer of Malignant Traits and the Involvement of Extracellular Vesicles in Metastasis. Cells. 2023; 12(12):1566. https://doi.org/10.3390/cells12121566
Chicago/Turabian StyleArena, Goffredo O., Stefano Forte, Mohamed Abdouh, Cheryl Vanier, Denis Corbeil, and Aurelio Lorico. 2023. "Horizontal Transfer of Malignant Traits and the Involvement of Extracellular Vesicles in Metastasis" Cells 12, no. 12: 1566. https://doi.org/10.3390/cells12121566
APA StyleArena, G. O., Forte, S., Abdouh, M., Vanier, C., Corbeil, D., & Lorico, A. (2023). Horizontal Transfer of Malignant Traits and the Involvement of Extracellular Vesicles in Metastasis. Cells, 12(12), 1566. https://doi.org/10.3390/cells12121566








