Generation of Red Blood Cells from Human Pluripotent Stem Cells—An Update
Abstract
1. Introduction
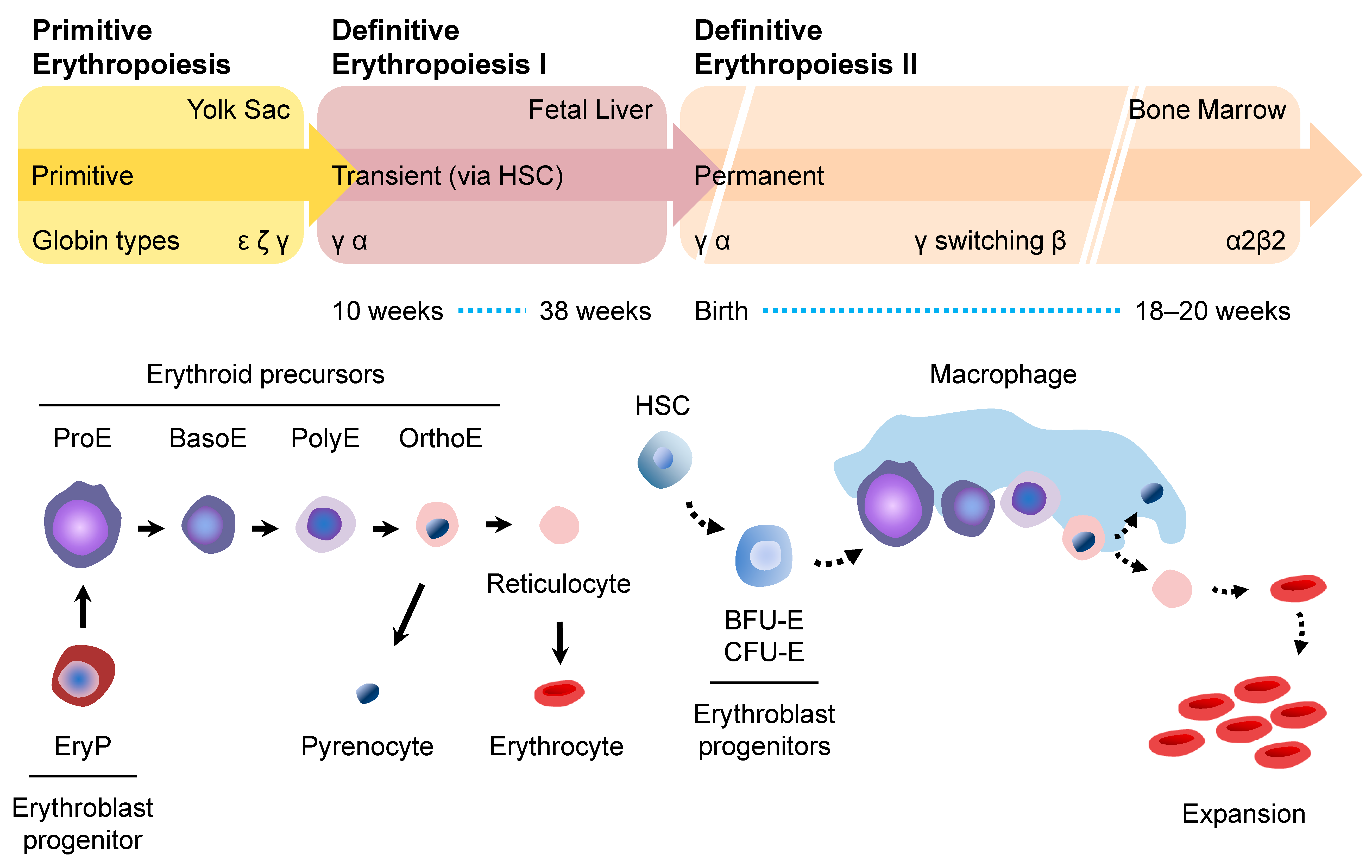
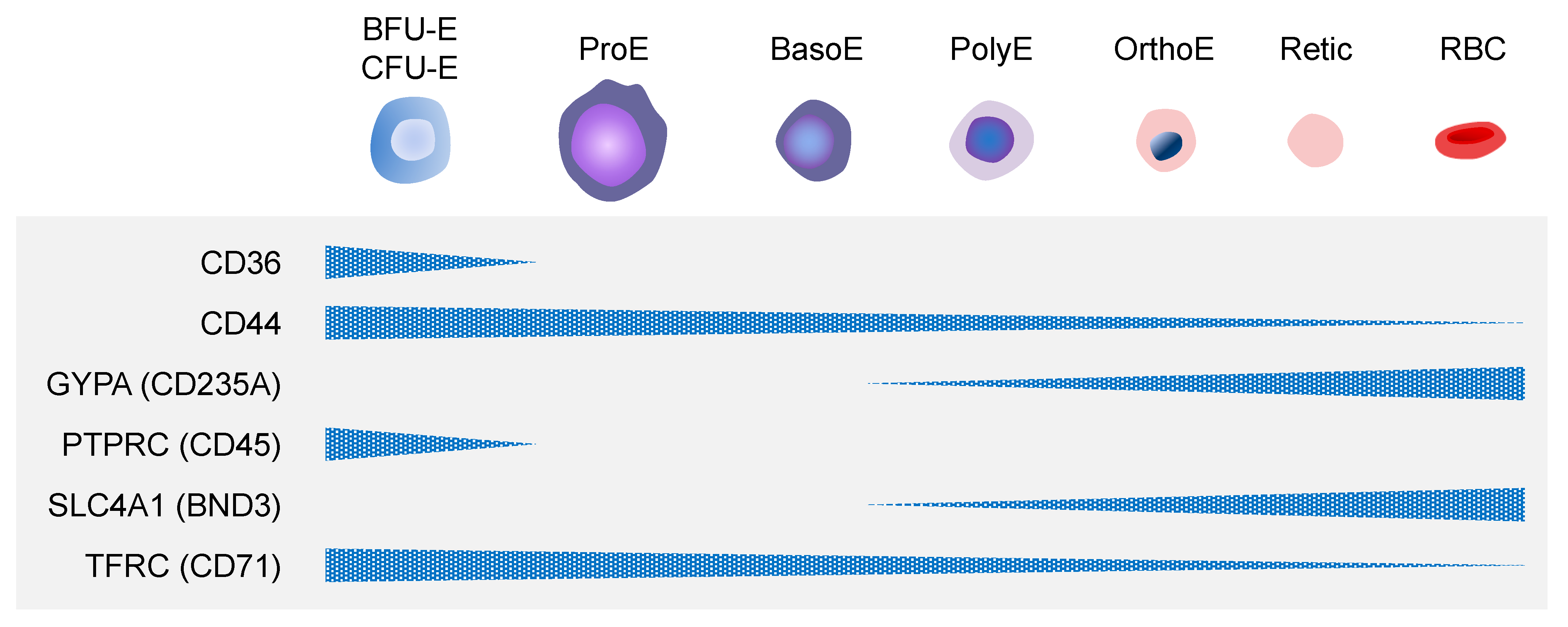
2. Erythropoiesis
2.1. Primitive Erythropoiesis
2.2. Definitive Erythropoiesis
3. Mechanisms of Erythropoiesis
3.1. Paracrine Mechanisms of Erythropoiesis
3.2. Erythroblastic Islands
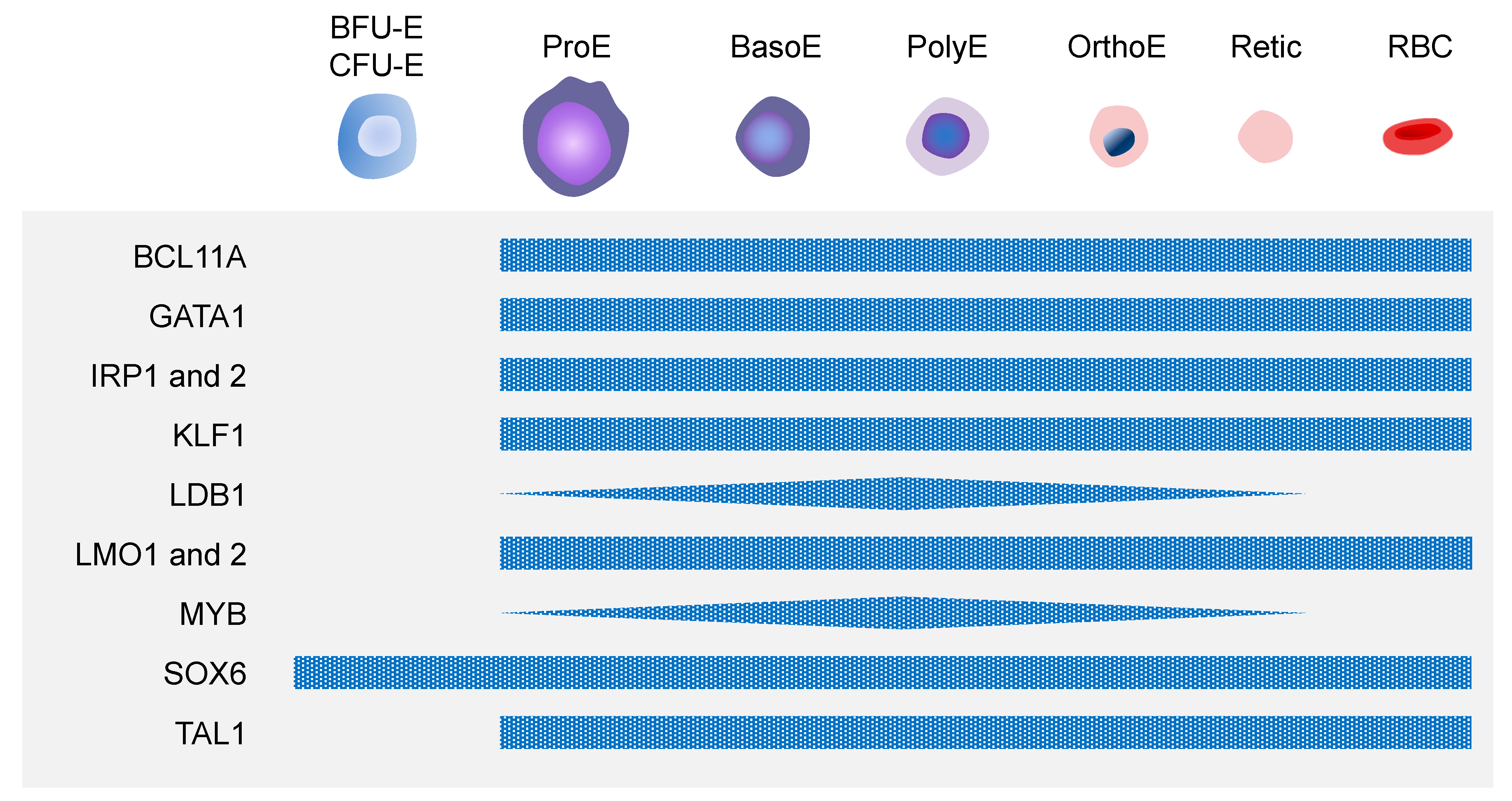
3.3. Molecular Mechanisms of Erythropoiesis
4. Human PSC-Derived Erythroid Lineage Cells
4.1. Generation of RBCs from CD34+ HSCs and/or HSPCs
4.2. Generation of RBCs from hPSCs
5. Current Challenges and Future Directions Using hiPSC-Derived Erythroid Lineage Cells
5.1. Heme Synthesis during Erythropoiesis
5.2. β-Globin Expression in Erythrocytes
5.3. Iron Supplementation for Erythrocytes
5.4. Enucleation of OrthoE
5.5. Methods for Enucleation of hPSC-Derived OrthoE
5.6. Future Directions of Clinical Applications Using hiPSC-Derived Erythrocytes
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Abbreviation | Full name |
| ACVR1B | Activin A receptor type 1B |
| BCL-xL | B-cell lymphoma-extra large |
| BCL11A | BCL11 transcription factor A |
| BMP4, 11 | Bone morphogenetic protein 4 and 11 |
| CEBPA | CCAAT enhancer binding protein alpha |
| EPO, EPOR | Erythropoietin, erythropoietin receptor |
| ERK | Extracellular regulated MAP kinase |
| FOXO3 | Forkhead box O3 |
| GATA1 | GATA binding protein 1 |
| GYPA (CD235A; GPA) | Glycophorin A |
| IGF1, 2 | Insulin like growth factor 1 and 2 |
| IRE | Iron responsive elements |
| IRF2, 6 | Interferon regulatory factor 2 and 6 |
| JAK2 | Janus kinase 2 |
| KIT | KIT proto-oncogene, receptor tyrosine kinase |
| KLF1 | LIM domain binding 1 |
| LDB1 | LIM domain binding 1 |
| LMO1, 2 | Krüppel-Like Factor 1 |
| MEK | MAP kinase-ERK kinase |
| MYB | MYB proto-oncogene, transcription factor |
| PI3K | Phosphatidylinositol 3-kinase |
| RAS | Rat sarcoma |
| SCF | Stem cell factor |
| SCL | Stem cell leukemia protein |
| SLC4A1 (BND3), 11A2 (DMT1), 40A1 (FPN1) | Solute carrier family 4 member 1, 11 member 2, and 40 member 1 |
| SMAD1 | SMAD family member 1 |
| SOX6 | SRY-box transcription factor 6 |
| SPI1 (PU.1) | Spi-1 proto-oncogene |
| SPN (CD43) | Sialophorin |
| STAT5 | Signal transducer and activator of transcription 5 |
| TAL1 | TAL bHLH transcription factor 1, erythroid differentiation factor |
| TCF7L2 | Transcription factor 7 like 2 |
| TFRC (CD71) | Transferrin receptor |
| TGFβ | Transforming growth factor B |
| TNFα | Tumor necrosis factor A |
| VEGF | Vascular endothelial growth factor |
| WNT3A | Wnt family member 3A |
| ZFPM1 (FOG1) | Zinc finger protein, FOG family member 1 |
References
- Greinacher, A.; Fendrich, K.; Hoffmann, W. Demographic Changes: The Impact for Safe Blood Supply. Transfus. Med. Hemother. 2010, 37, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Douay, L.; Andreu, G. Ex vivo production of human red blood cells from hematopoietic stem cells: What is the future in transfusion? Transfus. Med. Rev. 2007, 21, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Migliaccio, A.R.; Whitsett, C.; Migliaccio, G. Erythroid cells in vitro: From developmental biology to blood transfusion products. Curr. Opin. Hematol. 2009, 16, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef]
- Dzierzak, E.; Philipsen, S. Erythropoiesis: Development and differentiation. Cold Spring Harb. Perspect. Med. 2013, 3, a011601. [Google Scholar] [CrossRef]
- Park, Y.J.; Jeon, S.H.; Kim, H.K.; Suh, E.J.; Choi, S.J.; Kim, S.; Kim, H.O. Human induced pluripotent stem cell line banking for the production of rare blood type erythrocytes. J. Transl. Med. 2020, 18, 236. [Google Scholar] [CrossRef]
- Lim, Z.R.; Vassilev, S.; Leong, Y.W.; Hang, J.W.; Renia, L.; Malleret, B.; Oh, S.K. Industrially Compatible Transfusable iPSC-Derived RBCs: Progress, Challenges and Prospective Solutions. Int. J. Mol. Sci. 2021, 22, 9808. [Google Scholar] [CrossRef]
- Caulier, A.L.; Sankaran, V.G. Molecular and cellular mechanisms that regulate human erythropoiesis. Blood 2022, 139, 2450–2459. [Google Scholar] [CrossRef]
- Palis, J. Primitive and definitive erythropoiesis in mammals. Front. Physiol. 2014, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Barminko, J.; Reinholt, B.; Baron, M.H. Development and differentiation of the erythroid lineage in mammals. Dev. Comp. Immunol. 2016, 58, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lewis, K. Erythroid Lineage Cells in the Liver: Novel Immune Regulators and Beyond. J. Clin. Transl. Hepatol. 2020, 8, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Liu, J.; Heck, S.; Chasis, J.A.; An, X.; Mohandas, N. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc. Natl. Acad. Sci. USA 2009, 106, 17413–17418. [Google Scholar] [CrossRef] [PubMed]
- Palis, J.; Yoder, M.C. Yolk-sac hematopoiesis: The first blood cells of mouse and man. Exp. Hematol. 2001, 29, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Palis, J.; Robertson, S.; Kennedy, M.; Wall, C.; Keller, G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 1999, 126, 5073–5084. [Google Scholar] [CrossRef]
- Kingsley, P.D.; Malik, J.; Fantauzzo, K.A.; Palis, J. Yolk sac-derived primitive erythroblasts enucleate during mammalian embryogenesis. Blood 2004, 104, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, P.D.; Malik, J.; Emerson, R.L.; Bushnell, T.P.; McGrath, K.E.; Bloedorn, L.A.; Bulger, M.; Palis, J. “Maturational” globin switching in primary primitive erythroid cells. Blood 2006, 107, 1665–1672. [Google Scholar] [CrossRef]
- van Deurs, B.; Behnke, O. The microtubule marginal band of mammalian red blood cells. Z. Anat. Entwickl. 1973, 143, 43–47. [Google Scholar] [CrossRef]
- Sangiorgi, F.; Woods, C.M.; Lazarides, E. Vimentin downregulation is an inherent feature of murine erythropoiesis and occurs independently of lineage. Development 1990, 110, 85–96. [Google Scholar] [CrossRef]
- Ema, H.; Nakauchi, H. Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 2000, 95, 2284–2288. [Google Scholar] [CrossRef] [PubMed]
- Kumaravelu, P.; Hook, L.; Morrison, A.M.; Ure, J.; Zhao, S.; Zuyev, S.; Ansell, J.; Medvinsky, A. Quantitative developmental anatomy of definitive haematopoietic stem cells/long-term repopulating units (HSC/RUs): Role of the aorta-gonad-mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Development 2002, 129, 4891–4899. [Google Scholar] [CrossRef] [PubMed]
- Mikkola, H.K.; Orkin, S.H. The journey of developing hematopoietic stem cells. Development 2006, 133, 3733–3744. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Cai, J.Y.; Ma, J.; Huang, Z.; Guo, M.X.; Fu, L.Z.; Shi, Y.B.; Li, W.X. Global expression profiling reveals genetic programs underlying the developmental divergence between mouse and human embryogenesis. BMC Genom. 2013, 14, 568. [Google Scholar] [CrossRef]
- Popescu, D.M.; Botting, R.A.; Stephenson, E.; Green, K.; Webb, S.; Jardine, L.; Calderbank, E.F.; Polanski, K.; Goh, I.; Efremova, M.; et al. Decoding human fetal liver haematopoiesis. Nature 2019, 574, 365–371. [Google Scholar] [CrossRef]
- Shemin, D.; Rittenberg, D. The life span of the human red blood cell. J. Biol. Chem. 1946, 166, 627–636. [Google Scholar] [CrossRef]
- Kuhrt, D.; Wojchowski, D.M. Emerging EPO and EPO receptor regulators and signal transducers. Blood 2015, 125, 3536–3541. [Google Scholar] [CrossRef]
- Wu, H.; Liu, X.; Jaenisch, R.; Lodish, H.F. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell 1995, 83, 59–67. [Google Scholar] [CrossRef]
- Singbrant, S.; Russell, M.R.; Jovic, T.; Liddicoat, B.; Izon, D.J.; Purton, L.E.; Sims, N.A.; Martin, T.J.; Sankaran, V.G.; Walkley, C.R. Erythropoietin couples erythropoiesis, B-lymphopoiesis, and bone homeostasis within the bone marrow microenvironment. Blood 2011, 117, 5631–5642. [Google Scholar] [CrossRef]
- Malik, J.; Kim, A.R.; Tyre, K.A.; Cherukuri, A.R.; Palis, J. Erythropoietin critically regulates the terminal maturation of murine and human primitive erythroblasts. Haematologica 2013, 98, 1778–1787. [Google Scholar] [CrossRef]
- Nocka, K.; Majumder, S.; Chabot, B.; Ray, P.; Cervone, M.; Bernstein, A.; Besmer, P. Expression of c-kit gene products in known cellular targets of W mutations in normal and W mutant mice—Evidence for an impaired c-kit kinase in mutant mice. Genes Dev. 1989, 3, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Klingmuller, U.; Besmer, P.; Lodish, H.F. Interaction of the erythropoietin and stem-cell-factor receptors. Nature 1995, 377, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Papayannopoulou, T.; Brice, M.; Blau, C.A. Kit ligand in synergy with interleukin-3 amplifies the erythropoietin-independent, globin-synthesizing progeny of normal human burst-forming units-erythroid in suspension cultures: Physiologic implications. Blood 1993, 81, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tang, Z.Y.; Ka, W.; Sun, D.; Yao, W.; Wen, Z.; Chien, S. Synergistic effect of cytokines EPO, IL-3 and SCF on the proliferation, differentiation and apoptosis of erythroid progenitor cells. Clin. Hemorheol. Microcirc. 2007, 37, 291–299. [Google Scholar]
- Dussiot, M.; Maciel, T.T.; Fricot, A.; Chartier, C.; Negre, O.; Veiga, J.; Grapton, D.; Paubelle, E.; Payen, E.; Beuzard, Y.; et al. An activin receptor IIA ligand trap corrects ineffective erythropoiesis in beta-thalassemia. Nat. Med. 2014, 20, 398–407. [Google Scholar] [CrossRef]
- Suragani, R.N.; Cadena, S.M.; Cawley, S.M.; Sako, D.; Mitchell, D.; Li, R.; Davies, M.V.; Alexander, M.J.; Devine, M.; Loveday, K.S.; et al. Transforming growth factor-beta superfamily ligand trap ACE-536 corrects anemia by promoting late-stage erythropoiesis. Nat. Med. 2014, 20, 408–414. [Google Scholar] [CrossRef]
- Mohandas, N.; Prenant, M. Three-dimensional model of bone marrow. Blood 1978, 51, 633–643. [Google Scholar] [CrossRef]
- Sonoda, Y.; Sasaki, K. Hepatic extramedullary hematopoiesis and macrophages in the adult mouse: Histometrical and immunohistochemical studies. Cells Tissues Organs 2012, 196, 555–564. [Google Scholar] [CrossRef]
- May, A.; Forrester, L.M. The erythroblastic island niche: Modeling in health, stress, and disease. Exp. Hematol. 2020, 91, 10–21. [Google Scholar] [CrossRef]
- Paulson, R.F.; Hariharan, S.; Little, J.A. Stress erythropoiesis: Definitions and models for its study. Exp. Hematol. 2020, 89, 43–54. [Google Scholar] [CrossRef]
- Bessis, M. [Erythroblastic island, functional unity of bone marrow]. Rev. Hematol. 1958, 13, 8–11. [Google Scholar]
- Rhodes, M.M.; Kopsombut, P.; Bondurant, M.C.; Price, J.O.; Koury, M.J. Adherence to macrophages in erythroblastic islands enhances erythroblast proliferation and increases erythrocyte production by a different mechanism than erythropoietin. Blood 2008, 111, 1700–1708. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Kawane, K.; Koike, M.; Mori, Y.; Uchiyama, Y.; Nagata, S. Phosphatidylserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature 2005, 437, 754–758. [Google Scholar] [CrossRef] [PubMed]
- Yeo, J.H.; Lam, Y.W.; Fraser, S.T. Cellular dynamics of mammalian red blood cell production in the erythroblastic island niche. Biophys. Rev. 2019, 11, 873–894. [Google Scholar] [CrossRef] [PubMed]
- Porcher, C.; Chagraoui, H.; Kristiansen, M.S. SCL/TAL1: A multifaceted regulator from blood development to disease. Blood 2017, 129, 2051–2060. [Google Scholar] [CrossRef]
- Xu, J.; Shao, Z.; Glass, K.; Bauer, D.E.; Pinello, L.; Van Handel, B.; Hou, S.; Stamatoyannopoulos, J.A.; Mikkola, H.K.; Yuan, G.C.; et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev. Cell. 2012, 23, 796–811. [Google Scholar] [CrossRef]
- Sankaran, V.G.; Ghazvinian, R.; Do, R.; Thiru, P.; Vergilio, J.A.; Beggs, A.H.; Sieff, C.A.; Orkin, S.H.; Nathan, D.G.; Lander, E.S.; et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J. Clin. Investig. 2012, 122, 2439–2443. [Google Scholar] [CrossRef]
- Borg, J.; Patrinos, G.P.; Felice, A.E.; Philipsen, S. Erythroid phenotypes associated with KLF1 mutations. Haematologica 2011, 96, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Drissen, R.; von Lindern, M.; Kolbus, A.; Driegen, S.; Steinlein, P.; Beug, H.; Grosveld, F.; Philipsen, S. The erythroid phenotype of EKLF-null mice: Defects in hemoglobin metabolism and membrane stability. Mol. Cell. Biol. 2005, 25, 5205–5214. [Google Scholar] [CrossRef]
- Kitajima, K.; Zheng, J.; Yen, H.; Sugiyama, D.; Nakano, T. Multipotential differentiation ability of GATA-1-null erythroid-committed cells. Genes Dev. 2006, 20, 654–659. [Google Scholar] [CrossRef]
- Hodge, D.; Coghill, E.; Keys, J.; Maguire, T.; Hartmann, B.; McDowall, A.; Weiss, M.; Grimmond, S.; Perkins, A. A global role for EKLF in definitive and primitive erythropoiesis. Blood 2006, 107, 3359–3370. [Google Scholar] [CrossRef]
- Gutierrez, L.; Tsukamoto, S.; Suzuki, M.; Yamamoto-Mukai, H.; Yamamoto, M.; Philipsen, S.; Ohneda, K. Ablation of Gata1 in adult mice results in aplastic crisis, revealing its essential role in steady-state and stress erythropoiesis. Blood 2008, 111, 4375–4385. [Google Scholar] [CrossRef]
- Wontakal, S.N.; Guo, X.; Smith, C.; MacCarthy, T.; Bresnick, E.H.; Bergman, A.; Snyder, M.P.; Weissman, S.M.; Zheng, D.; Skoultchi, A.I. A core erythroid transcriptional network is repressed by a master regulator of myelo-lymphoid differentiation. Proc. Natl. Acad. Sci. USA 2012, 109, 3832–3837. [Google Scholar] [CrossRef]
- Sankaran, V.G.; Menne, T.F.; Xu, J.; Akie, T.E.; Lettre, G.; Van Handel, B.; Mikkola, H.K.; Hirschhorn, J.N.; Cantor, A.B.; Orkin, S.H. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 2008, 322, 1839–1842. [Google Scholar] [CrossRef] [PubMed]
- Stadhouders, R.; Aktuna, S.; Thongjuea, S.; Aghajanirefah, A.; Pourfarzad, F.; van Ijcken, W.; Lenhard, B.; Rooks, H.; Best, S.; Menzel, S.; et al. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J. Clin. Investig. 2014, 124, 1699–1710. [Google Scholar] [CrossRef] [PubMed]
- Cantu, C.; Ierardi, R.; Alborelli, I.; Fugazza, C.; Cassinelli, L.; Piconese, S.; Bose, F.; Ottolenghi, S.; Ferrari, G.; Ronchi, A. Sox6 enhances erythroid differentiation in human erythroid progenitors. Blood 2011, 117, 3669–3679. [Google Scholar] [CrossRef]
- Trompouki, E.; Bowman, T.V.; Lawton, L.N.; Fan, Z.P.; Wu, D.C.; DiBiase, A.; Martin, C.S.; Cech, J.N.; Sessa, A.K.; Leblanc, J.L.; et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 2011, 147, 577–589. [Google Scholar] [CrossRef]
- Boria, I.; Garelli, E.; Gazda, H.T.; Aspesi, A.; Quarello, P.; Pavesi, E.; Ferrante, D.; Meerpohl, J.J.; Kartal, M.; Da Costa, L.; et al. The ribosomal basis of Diamond-Blackfan Anemia: Mutation and database update. Hum. Mutat. 2010, 31, 1269–1279. [Google Scholar] [CrossRef]
- Ludwig, L.S.; Gazda, H.T.; Eng, J.C.; Eichhorn, S.W.; Thiru, P.; Ghazvinian, R.; George, T.I.; Gotlib, J.R.; Beggs, A.H.; Sieff, C.A.; et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat. Med. 2014, 20, 748–753. [Google Scholar] [CrossRef]
- Hentze, M.W.; Kuhn, L.C. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. USA 1996, 93, 8175–8182. [Google Scholar] [CrossRef]
- Fibach, E.; Manor, D.; Oppenheim, A.; Rachmilewitz, E.A. Proliferation and maturation of human erythroid progenitors in liquid culture. Blood 1989, 73, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Wada, H.; Suda, T.; Miura, Y.; Kajii, E.; Ikemoto, S.; Yawata, Y. Expression of major blood group antigens on human erythroid cells in a two phase liquid culture system. Blood 1990, 75, 505–511. [Google Scholar] [CrossRef]
- Fibach, E.; Rachmilewitz, E.A. A two-step liquid culture—A novel culture procedure for studying erythroid cell development. Haematologia 1991, 24, 211–220. [Google Scholar] [PubMed]
- Wright, D.E.; Wagers, A.J.; Gulati, A.P.; Johnson, F.L.; Weissman, I.L. Physiological migration of hematopoietic stem and progenitor cells. Science 2001, 294, 1933–1936. [Google Scholar] [CrossRef] [PubMed]
- Massberg, S.; Schaerli, P.; Knezevic-Maramica, I.; Kollnberger, M.; Tubo, N.; Moseman, E.A.; Huff, I.V.; Junt, T.; Wagers, A.J.; Mazo, I.B.; et al. Immunosurveillance by hematopoietic progenitor cells trafficking through blood, lymph, and peripheral tissues. Cell 2007, 131, 994–1008. [Google Scholar] [CrossRef]
- Singh, V.K.; Saini, A.; Tsuji, K.; Sharma, P.B.; Chandra, R. Manufacturing blood ex vivo: A futuristic approach to deal with the supply and safety concerns. Front. Cell. Dev. Biol. 2014, 2, 26. [Google Scholar] [CrossRef]
- Neildez-Nguyen, T.M.; Wajcman, H.; Marden, M.C.; Bensidhoum, M.; Moncollin, V.; Giarratana, M.C.; Kobari, L.; Thierry, D.; Douay, L. Human erythroid cells produced ex vivo at large scale differentiate into red blood cells in vivo. Nat. Biotechnol. 2002, 20, 467–472. [Google Scholar] [CrossRef]
- Giarratana, M.C.; Kobari, L.; Lapillonne, H.; Chalmers, D.; Kiger, L.; Cynober, T.; Marden, M.C.; Wajcman, H.; Douay, L. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat. Biotechnol. 2005, 23, 69–74. [Google Scholar] [CrossRef]
- Miharada, K.; Hiroyama, T.; Sudo, K.; Nagasawa, T.; Nakamura, Y. Efficient enucleation of erythroblasts differentiated in vitro from hematopoietic stem and progenitor cells. Nat. Biotechnol. 2006, 24, 1255–1256. [Google Scholar] [CrossRef]
- Ohneda, O.; Bautch, V.L. Murine endothelial cells support fetal liver erythropoiesis and myelopoiesis via distinct interactions. Br. J. Haematol. 1997, 98, 798–808. [Google Scholar] [CrossRef]
- Hanspal, M.; Smockova, Y.; Uong, Q. Molecular identification and functional characterization of a novel protein that mediates the attachment of erythroblasts to macrophages. Blood 1998, 92, 2940–2950. [Google Scholar] [CrossRef]
- Chasis, J.A.; Mohandas, N. Erythroblastic islands: Niches for erythropoiesis. Blood 2008, 112, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Fujimi, A.; Matsunaga, T.; Kobune, M.; Kawano, Y.; Nagaya, T.; Tanaka, I.; Iyama, S.; Hayashi, T.; Sato, T.; Miyanishi, K.; et al. Ex vivo large-scale generation of human red blood cells from cord blood CD34+ cells by co-culturing with macrophages. Int. J. Hematol. 2008, 87, 339–350. [Google Scholar] [CrossRef]
- Hanspal, M.; Hanspal, J.S. The association of erythroblasts with macrophages promotes erythroid proliferation and maturation: A 30-kD heparin-binding protein is involved in this contact. Blood 1994, 84, 3494–3504. [Google Scholar] [CrossRef]
- Iavarone, A.; King, E.R.; Dai, X.M.; Leone, G.; Stanley, E.R.; Lasorella, A. Retinoblastoma promotes definitive erythropoiesis by repressing Id2 in fetal liver macrophages. Nature 2004, 432, 1040–1045. [Google Scholar] [CrossRef] [PubMed]
- Webb, S. Banking on cord blood stem cells. Nat. Biotechnol. 2013, 31, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Olivier, E.N.; Qiu, C.; Velho, M.; Hirsch, R.E.; Bouhassira, E.E. Large-scale production of embryonic red blood cells from human embryonic stem cells. Exp. Hematol. 2006, 34, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Wang, D.; Hanada, S.; Ebihara, Y.; Kawasaki, H.; Zaike, Y.; Heike, T.; Nakahata, T.; Tsuji, K. Novel method for efficient production of multipotential hematopoietic progenitors from human embryonic stem cells. Int. J. Hematol. 2007, 85, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Ebihara, Y.; Umeda, K.; Sakai, H.; Hanada, S.; Zhang, H.; Zaike, Y.; Tsuchida, E.; Nakahata, T.; Nakauchi, H.; et al. Generation of functional erythrocytes from human embryonic stem cell-derived definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 2008, 105, 13087–13092. [Google Scholar] [CrossRef]
- Dias, J.; Gumenyuk, M.; Kang, H.; Vodyanik, M.; Yu, J.; Thomson, J.A.; Slukvin, I.I. Generation of red blood cells from human induced pluripotent stem cells. Stem Cells Dev. 2011, 20, 1639–1647. [Google Scholar] [CrossRef]
- Haro-Mora, J.J.; Uchida, N.; Demirci, S.; Wang, Q.; Zou, J.; Tisdale, J.F. Biallelic correction of sickle cell disease-derived induced pluripotent stem cells (iPSCs) confirmed at the protein level through serum-free iPS-sac/erythroid differentiation. Stem Cells Transl. Med. 2020, 9, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Uchida, N.; Haro-Mora, J.J.; Fujita, A.; Lee, D.Y.; Winkler, T.; Hsieh, M.M.; Tisdale, J.F. Efficient Generation of beta-Globin-Expressing Erythroid Cells Using Stromal Cell-Derived Induced Pluripotent Stem Cells from Patients with Sickle Cell Disease. Stem Cells 2017, 35, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Cerdan, C.; Rouleau, A.; Bhatia, M. VEGF-A165 augments erythropoietic development from human embryonic stem cells. Blood 2004, 103, 2504–2512. [Google Scholar] [CrossRef]
- Chang, K.H.; Nelson, A.M.; Cao, H.; Wang, L.; Nakamoto, B.; Ware, C.B.; Papayannopoulou, T. Definitive-like erythroid cells derived from human embryonic stem cells coexpress high levels of embryonic and fetal globins with little or no adult globin. Blood 2006, 108, 1515–1523. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.J.; Feng, Q.; Park, J.S.; Vida, L.; Lee, B.S.; Strausbauch, M.; Wettstein, P.J.; Honig, G.R.; Lanza, R. Biologic properties and enucleation of red blood cells from human embryonic stem cells. Blood 2008, 112, 4475–4484. [Google Scholar] [CrossRef]
- Lapillonne, H.; Kobari, L.; Mazurier, C.; Tropel, P.; Giarratana, M.C.; Zanella-Cleon, I.; Kiger, L.; Wattenhofer-Donze, M.; Puccio, H.; Hebert, N.; et al. Red blood cell generation from human induced pluripotent stem cells: Perspectives for transfusion medicine. Haematologica 2010, 95, 1651–1659. [Google Scholar] [CrossRef]
- Dorn, I.; Klich, K.; Arauzo-Bravo, M.J.; Radstaak, M.; Santourlidis, S.; Ghanjati, F.; Radke, T.F.; Psathaki, O.E.; Hargus, G.; Kramer, J.; et al. Erythroid differentiation of human induced pluripotent stem cells is independent of donor cell type of origin. Haematologica 2015, 100, 32–41. [Google Scholar] [CrossRef]
- Olivier, E.N.; Marenah, L.; McCahill, A.; Condie, A.; Cowan, S.; Mountford, J.C. High-Efficiency Serum-Free Feeder-Free Erythroid Differentiation of Human Pluripotent Stem Cells Using Small Molecules. Stem Cells Transl. Med. 2016, 5, 1394–1405. [Google Scholar] [CrossRef]
- Kessel, K.U.; Bluemke, A.; Scholer, H.R.; Zaehres, H.; Schlenke, P.; Dorn, I. Emergence of CD43-Expressing Hematopoietic Progenitors from Human Induced Pluripotent Stem Cells. Transfus. Med. Hemother. 2017, 44, 143–150. [Google Scholar] [CrossRef]
- Bernecker, C.; Ackermann, M.; Lachmann, N.; Rohrhofer, L.; Zaehres, H.; Arauzo-Bravo, M.J.; van den Akker, E.; Schlenke, P.; Dorn, I. Enhanced Ex Vivo Generation of Erythroid Cells from Human Induced Pluripotent Stem Cells in a Simplified Cell Culture System with Low Cytokine Support. Stem Cells Dev. 2019, 28, 1540–1551. [Google Scholar] [CrossRef]
- Roh, J.; Kim, S.; Cheong, J.W.; Jeon, S.H.; Kim, H.K.; Kim, M.J.; Kim, H.O. Erythroid Differentiation of Induced Pluripotent Stem Cells Co-cultured with OP9 Cells for Diagnostic Purposes. Ann. Lab. Med. 2022, 42, 457–466. [Google Scholar] [CrossRef]
- Smith, B.W.; Rozelle, S.S.; Leung, A.; Ubellacker, J.; Parks, A.; Nah, S.K.; French, D.; Gadue, P.; Monti, S.; Chui, D.H.; et al. The aryl hydrocarbon receptor directs hematopoietic progenitor cell expansion and differentiation. Blood 2013, 122, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Olivier, E.N.; Zhang, S.; Yan, Z.; Suzuka, S.; Roberts, K.; Wang, K.; Bouhassira, E.E. PSC-RED and MNC-RED: Albumin-free and low-transferrin robust erythroid differentiation protocols to produce human enucleated red blood cells. Exp. Hematol. 2019, 75, 31–52.e15. [Google Scholar] [CrossRef]
- Poldee, S.; Metheetrairut, C.; Nugoolsuksiri, S.; Frayne, J.; Trakarnsanga, K. Optimization of an erythroid culture system to reduce the cost of in vitro production of red blood cells. MethodsX 2018, 5, 1626–1632. [Google Scholar] [CrossRef]
- Netsrithong, R.; Suwanpitak, S.; Boonkaew, B.; Trakarnsanga, K.; Chang, L.J.; Tipgomut, C.; Vatanashevanopakorn, C.; Pattanapanyasat, K.; Wattanapanitch, M. Multilineage differentiation potential of hematoendothelial progenitors derived from human induced pluripotent stem cells. Stem Cell Res. Ther. 2020, 11, 481. [Google Scholar] [CrossRef] [PubMed]
- Niwa, A.; Heike, T.; Umeda, K.; Oshima, K.; Kato, I.; Sakai, H.; Suemori, H.; Nakahata, T.; Saito, M.K. A novel serum-free monolayer culture for orderly hematopoietic differentiation of human pluripotent cells via mesodermal progenitors. PLoS ONE 2011, 6, e22261. [Google Scholar] [CrossRef]
- Chou, S.T.; Byrska-Bishop, M.; Tober, J.M.; Yao, Y.; Vandorn, D.; Opalinska, J.B.; Mills, J.A.; Choi, J.K.; Speck, N.A.; Gadue, P.; et al. Trisomy 21-associated defects in human primitive hematopoiesis revealed through induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2012, 109, 17573–17578. [Google Scholar] [CrossRef] [PubMed]
- Tursky, M.L.; Loi, T.H.; Artuz, C.M.; Alateeq, S.; Wolvetang, E.J.; Tao, H.; Ma, D.D.; Molloy, T.J. Direct Comparison of Four Hematopoietic Differentiation Methods from Human Induced Pluripotent Stem Cells. Stem Cell Rep. 2020, 15, 735–748. [Google Scholar] [CrossRef]
- Leary, A.G.; Ikebuchi, K.; Hirai, Y.; Wong, G.G.; Yang, Y.C.; Clark, S.C.; Ogawa, M. Synergism between interleukin-6 and interleukin-3 in supporting proliferation of human hematopoietic stem cells: Comparison with interleukin-1 alpha. Blood 1988, 71, 1759–1763. [Google Scholar] [CrossRef]
- Ulich, T.R.; del Castillo, J.; Yin, S.M.; Egrie, J.C. The erythropoietic effects of interleukin 6 and erythropoietin in vivo. Exp. Hematol. 1991, 19, 29–34. [Google Scholar]
- Kieran, M.W.; Perkins, A.C.; Orkin, S.H.; Zon, L.I. Thrombopoietin rescues in vitro erythroid colony formation from mouse embryos lacking the erythropoietin receptor. Proc. Natl. Acad. Sci. USA 1996, 93, 9126–9131. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Klingmuller, U.; Acurio, A.; Hsiao, J.G.; Lodish, H.F. Functional interaction of erythropoietin and stem cell factor receptors is essential for erythroid colony formation. Proc. Natl. Acad. Sci. USA 1997, 94, 1806–1810. [Google Scholar] [CrossRef] [PubMed]
- Lyman, S.D.; Jacobsen, S.E. c-kit ligand and Flt3 ligand: Stem/progenitor cell factors with overlapping yet distinct activities. Blood 1998, 91, 1101–1134. [Google Scholar] [CrossRef]
- Wannatung, T.; Lithanatudom, P.; Leecharoenkiat, A.; Svasti, S.; Fucharoen, S.; Smith, D.R. Increased erythropoiesis of beta-thalassaemia/Hb E proerythroblasts is mediated by high basal levels of ERK1/2 activation. Br. J. Haematol. 2009, 146, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.F.; Li, N.; Lee, H.J.; Adamo, L.; Evans, S.M.; Willey, H.E.; Arora, N.; Torisawa, Y.S.; Vickers, D.A.; Morris, S.A.; et al. Biomechanical forces promote blood development through prostaglandin E2 and the cAMP-PKA signaling axis. J. Exp. Med. 2015, 212, 665–680. [Google Scholar] [CrossRef]
- Bratt-Leal, A.M.; Carpenedo, R.L.; McDevitt, T.C. Engineering the embryoid body microenvironment to direct embryonic stem cell differentiation. Biotechnol. Prog. 2009, 25, 43–51. [Google Scholar] [CrossRef]
- McGrath, K.E.; Frame, J.M.; Fromm, G.J.; Koniski, A.D.; Kingsley, P.D.; Little, J.; Bulger, M.; Palis, J. A transient definitive erythroid lineage with unique regulation of the beta-globin locus in the mammalian embryo. Blood 2011, 117, 4600–4608. [Google Scholar] [CrossRef]
- Friedman, J.M. Structure, dynamics, and reactivity in hemoglobin. Science 1985, 228, 1273–1280. [Google Scholar] [CrossRef]
- Gell, D.A. Structure and function of haemoglobins. Blood Cells Mol. Dis. 2018, 70, 13–42. [Google Scholar] [CrossRef]
- Wood, W.G.; Weatherall, D.J. Haemoglobin synthesis during human foetal development. Nature 1973, 244, 162–165. [Google Scholar] [CrossRef]
- Qiu, C.; Hanson, E.; Olivier, E.; Inada, M.; Kaufman, D.S.; Gupta, S.; Bouhassira, E.E. Differentiation of human embryonic stem cells into hematopoietic cells by coculture with human fetal liver cells recapitulates the globin switch that occurs early in development. Exp. Hematol. 2005, 33, 1450–1458. [Google Scholar] [CrossRef]
- Umeda, K.; Heike, T.; Nakata-Hizume, M.; Niwa, A.; Arai, M.; Shinoda, G.; Ma, F.; Suemori, H.; Luo, H.Y.; Chui, D.H.; et al. Sequential analysis of alpha- and beta-globin gene expression during erythropoietic differentiation from primate embryonic stem cells. Stem Cells 2006, 24, 2627–2636. [Google Scholar] [CrossRef] [PubMed]
- Peschle, C.; Mavilio, F.; Care, A.; Migliaccio, G.; Migliaccio, A.R.; Salvo, G.; Samoggia, P.; Petti, S.; Guerriero, R.; Marinucci, M.; et al. Haemoglobin switching in human embryos: Asynchrony of zeta → alpha and epsilon → gamma-globin switches in primitive and definite erythropoietic lineage. Nature 1985, 313, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Olivier, E.N.; Velho, M.; Bouhassira, E.E. Globin switches in yolk sac-like primitive and fetal-like definitive red blood cells produced from human embryonic stem cells. Blood 2008, 111, 2400–2408. [Google Scholar] [CrossRef]
- Ochi, K.; Takayama, N.; Hirose, S.; Nakahata, T.; Nakauchi, H.; Eto, K. Multicolor staining of globin subtypes reveals impaired globin switching during erythropoiesis in human pluripotent stem cells. Stem Cells Transl. Med. 2014, 3, 792–800. [Google Scholar] [CrossRef]
- Pevny, L.; Simon, M.C.; Robertson, E.; Klein, W.H.; Tsai, S.F.; D’Agati, V.; Orkin, S.H.; Costantini, F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 1991, 349, 257–260. [Google Scholar] [CrossRef]
- Basu, P.; Lung, T.K.; Lemsaddek, W.; Sargent, T.G.; Williams, D.C., Jr.; Basu, M.; Redmond, L.C.; Lingrel, J.B.; Haar, J.L.; Lloyd, J.A. EKLF and KLF2 have compensatory roles in embryonic beta-globin gene expression and primitive erythropoiesis. Blood 2007, 110, 3417–3425. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.T.; Ma, R.; Axton, R.A.; Jackson, M.; Taylor, A.H.; Fidanza, A.; Marenah, L.; Frayne, J.; Mountford, J.C.; Forrester, L.M. Activation of KLF1 Enhances the Differentiation and Maturation of Red Blood Cells from Human Pluripotent Stem Cells. Stem Cells 2017, 35, 886–897. [Google Scholar] [CrossRef]
- Song, S.H.; Hou, C.; Dean, A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol. Cell 2007, 28, 810–822. [Google Scholar] [CrossRef]
- Honig, G.R.; Lu, S.J.; Feng, Q.; Vida, L.N.; Lee, B.S.; Lanza, R. Alpha-Thalassemia-like globin gene expression by primitive erythrocytes derived from human embryonic stem cells. Hemoglobin 2010, 34, 145–150. [Google Scholar] [CrossRef]
- Fujita, A.; Uchida, N.; Haro-Mora, J.J.; Winkler, T.; Tisdale, J. Beta-Globin-Expressing Definitive Erythroid Progenitor Cells Generated from Embryonic and Induced Pluripotent Stem Cell-Derived Sacs. Stem Cells 2016, 34, 1541–1552. [Google Scholar] [CrossRef] [PubMed]
- Sassa, S.; Nagai, T. The role of heme in gene expression. Int. J. Hematol. 1996, 63, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.S.; Xu, J.; Wardan, H.; McColl, B.; Orkin, S.; Vadolas, J. Generation of a genomic reporter assay system for analysis of gamma- and beta-globin gene regulation. FASEB J. 2012, 26, 1736–1744. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Yamamoto, M.; Engel, J.D. Fetal globin gene repressors as drug targets for molecular therapies to treat the beta-globinopathies. Mol. Cell. Biol. 2014, 34, 3560–3569. [Google Scholar] [CrossRef]
- Yu, L.; Myers, G.; Engel, J.D. Small molecule therapeutics to treat the beta-globinopathies. Curr. Opin. Hematol. 2020, 27, 129–140. [Google Scholar] [CrossRef]
- Soboleva, S.; Kurita, R.; Ek, F.; Akerstrand, H.; Silverio-Alves, R.; Olsson, R.; Nakamura, Y.; Miharada, K. Identification of potential chemical compounds enhancing generation of enucleated cells from immortalized human erythroid cell lines. Commun. Biol. 2021, 4, 677. [Google Scholar] [CrossRef]
- Xu, J.; Sankaran, V.G.; Ni, M.; Menne, T.F.; Puram, R.V.; Kim, W.; Orkin, S.H. Transcriptional silencing of gamma-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 2010, 24, 783–798. [Google Scholar] [CrossRef]
- Trakarnsanga, K.; Wilson, M.C.; Lau, W.; Singleton, B.K.; Parsons, S.F.; Sakuntanaga, P.; Kurita, R.; Nakamura, Y.; Anstee, D.J.; Frayne, J. Induction of adult levels of beta-globin in human erythroid cells that intrinsically express embryonic or fetal globin by transduction with KLF1 and BCL11A-XL. Haematologica 2014, 99, 1677–1685. [Google Scholar] [CrossRef]
- Bauer, D.E.; Orkin, S.H. Hemoglobin switching’s surprise: The versatile transcription factor BCL11A is a master repressor of fetal hemoglobin. Curr. Opin. Genet. Dev. 2015, 33, 62–70. [Google Scholar] [CrossRef]
- Liu, N.; Hargreaves, V.V.; Zhu, Q.; Kurland, J.V.; Hong, J.; Kim, W.; Sher, F.; Macias-Trevino, C.; Rogers, J.M.; Kurita, R.; et al. Direct Promoter Repression by BCL11A Controls the Fetal to Adult Hemoglobin Switch. Cell 2018, 173, 430–442.e417. [Google Scholar] [CrossRef]
- Chang, K.H.; Huang, A.; Hirata, R.K.; Wang, P.R.; Russell, D.W.; Papayannopoulou, T. Globin phenotype of erythroid cells derived from human induced pluripotent stem cells. Blood 2010, 115, 2553–2554. [Google Scholar] [CrossRef]
- Ng, E.S.; Azzola, L.; Bruveris, F.F.; Calvanese, V.; Phipson, B.; Vlahos, K.; Hirst, C.; Jokubaitis, V.J.; Yu, Q.C.; Maksimovic, J.; et al. Differentiation of human embryonic stem cells to HOXA+ hemogenic vasculature that resembles the aorta-gonad-mesonephros. Nat. Biotechnol. 2016, 34, 1168–1179. [Google Scholar] [CrossRef] [PubMed]
- Razaq, M.A.; Taylor, S.; Roberts, D.J.; Carpenter, L. A molecular roadmap of definitive erythropoiesis from human induced pluripotent stem cells. Br. J. Haematol. 2017, 176, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Vanuytsel, K.; Matte, T.; Leung, A.; Naing, Z.H.; Morrison, T.; Chui, D.H.K.; Steinberg, M.H.; Murphy, G.J. Induced pluripotent stem cell-based mapping of beta-globin expression throughout human erythropoietic development. Blood Adv. 2018, 2, 1998–2011. [Google Scholar] [CrossRef]
- Trakarnsanga, K.; Wilson, M.C.; Heesom, K.J.; Andrienko, T.N.; Srisawat, C.; Frayne, J. Secretory factors from OP9 stromal cells delay differentiation and increase the expansion potential of adult erythroid cells in vitro. Sci. Rep. 2018, 8, 1983. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Liu, K.; Sun, C.W.; Pawlik, K.M.; Townes, T.M. KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat. Genet. 2010, 42, 742–744. [Google Scholar] [CrossRef]
- Hanna, J.; Wernig, M.; Markoulaki, S.; Sun, C.W.; Meissner, A.; Cassady, J.P.; Beard, C.; Brambrink, T.; Wu, L.C.; Townes, T.M.; et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 2007, 318, 1920–1923. [Google Scholar] [CrossRef]
- Abboud, S.; Haile, D.J. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J. Biol. Chem. 2000, 275, 19906–19912. [Google Scholar] [CrossRef]
- Nemeth, E.; Tuttle, M.S.; Powelson, J.; Vaughn, M.B.; Donovan, A.; Ward, D.M.; Ganz, T.; Kaplan, J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 2004, 306, 2090–2093. [Google Scholar] [CrossRef]
- Rouault, T.A. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2006, 2, 406–414. [Google Scholar] [CrossRef]
- Crooks, D.R.; Ghosh, M.C.; Haller, R.G.; Tong, W.H.; Rouault, T.A. Posttranslational stability of the heme biosynthetic enzyme ferrochelatase is dependent on iron availability and intact iron-sulfur cluster assembly machinery. Blood 2010, 115, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.C.; Zhang, D.L.; Jeong, S.Y.; Kovtunovych, G.; Ollivierre-Wilson, H.; Noguchi, A.; Tu, T.; Senecal, T.; Robinson, G.; Crooks, D.R.; et al. Deletion of iron regulatory protein 1 causes polycythemia and pulmonary hypertension in mice through translational derepression of HIF2alpha. Cell Metab. 2013, 17, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, N.; Pantopoulos, K. IRP1 regulates erythropoiesis and systemic iron homeostasis by controlling HIF2alpha mRNA translation. Blood 2013, 122, 1658–1668. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.L.; Ghosh, M.C.; Rouault, T.A. The physiological functions of iron regulatory proteins in iron homeostasis—An update. Front. Pharmacol. 2014, 5, 124. [Google Scholar] [CrossRef]
- Meyron-Holtz, E.G.; Ghosh, M.C.; Iwai, K.; LaVaute, T.; Brazzolotto, X.; Berger, U.V.; Land, W.; Ollivierre-Wilson, H.; Grinberg, A.; Love, P.; et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004, 23, 386–395. [Google Scholar] [CrossRef]
- Smith, S.R.; Ghosh, M.C.; Ollivierre-Wilson, H.; Hang Tong, W.; Rouault, T.A. Complete loss of iron regulatory proteins 1 and 2 prevents viability of murine zygotes beyond the blastocyst stage of embryonic development. Blood Cells Mol. Dis. 2006, 36, 283–287. [Google Scholar] [CrossRef]
- Cooperman, S.S.; Meyron-Holtz, E.G.; Olivierre-Wilson, H.; Ghosh, M.C.; McConnell, J.P.; Rouault, T.A. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood 2005, 106, 1084–1091. [Google Scholar] [CrossRef]
- Soboleva, S.; Miharada, K. Induction of enucleation in primary and immortalized erythroid cells. Int. J. Hematol. 2022, 116, 192–198. [Google Scholar] [CrossRef]
- Kawabata, H. Transferrin and transferrin receptors update. Free Radic. Biol. Med. 2019, 133, 46–54. [Google Scholar] [CrossRef]
- Canonne-Hergaux, F.; Zhang, A.S.; Ponka, P.; Gros, P. Characterization of the iron transporter DMT1 (NRAMP2/DCT1) in red blood cells of normal and anemic mk/mk mice. Blood 2001, 98, 3823–3830. [Google Scholar] [CrossRef]
- Meng, F.; Fleming, B.A.; Jia, X.; Rousek, A.A.; Mulvey, M.A.; Ward, D.M. Lysosomal iron recycling in mouse macrophages is dependent upon both LcytB and Steap3 reductases. Blood Adv. 2022, 6, 1692–1707. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.; Firpo, M.; Choi, K.; Wall, C.; Robertson, S.; Kabrun, N.; Keller, G. A common precursor for primitive erythropoiesis and definitive haematopoiesis. Nature 1997, 386, 488–493. [Google Scholar] [CrossRef] [PubMed]
- McGrath, K.E.; Kingsley, P.D.; Koniski, A.D.; Porter, R.L.; Bushnell, T.P.; Palis, J. Enucleation of primitive erythroid cells generates a transient population of “pyrenocytes” in the mammalian fetus. Blood 2008, 111, 2409–2417. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, M.; Forouzesh, M.; Raoufi, S.; Ramazii, M.; Ghaedrahmati, F.; Farzaneh, M. Differentiation of human induced pluripotent stem cells into erythroid cells. Stem Cell. Res. Ther. 2020, 11, 483. [Google Scholar] [CrossRef]
- Moras, M.; Lefevre, S.D.; Ostuni, M.A. From Erythroblasts to Mature Red Blood Cells: Organelle Clearance in Mammals. Front. Physiol. 2017, 8, 1076. [Google Scholar] [CrossRef]
- Simpson, C.F.; Kling, J.M. The mechanism of denucleation in circulating erythroblasts. J. Cell. Biol. 1967, 35, 237–245. [Google Scholar] [CrossRef]
- Tallack, M.R.; Keys, J.R.; Humbert, P.O.; Perkins, A.C. EKLF/KLF1 controls cell cycle entry via direct regulation of E2f2. J. Biol. Chem. 2009, 284, 20966–20974. [Google Scholar] [CrossRef]
- Gnanapragasam, M.N.; McGrath, K.E.; Catherman, S.; Xue, L.; Palis, J.; Bieker, J.J. EKLF/KLF1-regulated cell cycle exit is essential for erythroblast enucleation. Blood 2016, 128, 1631–1641. [Google Scholar] [CrossRef]
- Swartz, K.L.; Wood, S.N.; Murthy, T.; Ramirez, O.; Qin, G.; Pillai, M.M.; Rao, S.; Minella, A.C. E2F-2 Promotes Nuclear Condensation and Enucleation of Terminally Differentiated Erythroblasts. Mol. Cell. Biol. 2017, 37, e00274-16. [Google Scholar] [CrossRef]
- Jayapal, S.R.; Lee, K.L.; Ji, P.; Kaldis, P.; Lim, B.; Lodish, H.F. Down-regulation of Myc is essential for terminal erythroid maturation. J. Biol. Chem. 2010, 285, 40252–40265. [Google Scholar] [CrossRef]
- Zhao, B.; Mei, Y.; Schipma, M.J.; Roth, E.W.; Bleher, R.; Rappoport, J.Z.; Wickrema, A.; Yang, J.; Ji, P. Nuclear Condensation during Mouse Erythropoiesis Requires Caspase-3-Mediated Nuclear Opening. Dev. Cell 2016, 36, 498–510. [Google Scholar] [CrossRef]
- Ji, P.; Yeh, V.; Ramirez, T.; Murata-Hori, M.; Lodish, H.F. Histone deacetylase 2 is required for chromatin condensation and subsequent enucleation of cultured mouse fetal erythroblasts. Haematologica 2010, 95, 2013–2021. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Flygare, J.; Wong, P.; Lim, B.; Lodish, H.F. miR-191 regulates mouse erythroblast enucleation by down-regulating Riok3 and Mxi1. Genes Dev. 2011, 25, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Jayapal, S.R.; Lodish, H.F. Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat. Cell. Biol. 2008, 10, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Keerthivasan, G.; Small, S.; Liu, H.; Wickrema, A.; Crispino, J.D. Vesicle trafficking plays a novel role in erythroblast enucleation. Blood 2010, 116, 3331–3340. [Google Scholar] [CrossRef] [PubMed]
- Keerthivasan, G.; Liu, H.; Gump, J.M.; Dowdy, S.F.; Wickrema, A.; Crispino, J.D. A novel role for survivin in erythroblast enucleation. Haematologica 2012, 97, 1471–1479. [Google Scholar] [CrossRef]
- Timmins, N.E.; Athanasas, S.; Gunther, M.; Buntine, P.; Nielsen, L.K. Ultra-high-yield manufacture of red blood cells from hematopoietic stem cells. Tissue Eng. C Methods 2011, 17, 1131–1137. [Google Scholar] [CrossRef]
- Bernecker, C.; Kofeler, H.; Pabst, G.; Trotzmuller, M.; Kolb, D.; Strohmayer, K.; Trajanoski, S.; Holzapfel, G.A.; Schlenke, P.; Dorn, I. Cholesterol Deficiency Causes Impaired Osmotic Stability of Cultured Red Blood Cells. Front. Physiol. 2019, 10, 1529. [Google Scholar] [CrossRef]

| Culture System | Source Cell Type | Differentiated Cell Type(s) | Factor(s) | Culture Duration | Molecular Characterization | Functional Assessment | Note | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| Feeder cell | MS5 | hESC | Early erythroblast | Insulin, transferrin, IL3, BMP4, FLT3 ligand, SCF, EPO, IGF1, hemin | 24 days (+15 days for HSC differentiation) | CD34+GYPA+TFRC+, orthochromatic, embryonic and fetal globin | Hemoglobin production | Large-scale production of RBCs | [77] |
| mFLSC | hESC | RBC | SCF, IL3, IL6, EPO, TPO, CSF3 | 18 days | GYPA+TFRC+, clonogenic, embryonic, fetal, and adult globin | Oxygen dissociation curve | Adapted from [78] | [79] | |
| MS5 | hiPSC, hESC | RBC | Dex, insulin, SCF, EPO, TPO, IL3, IL6 | 40–45 days (+7–8 days for HSC differentiation) | CD34+SPN+GYPA+TFRC+, embryonic and fetal globin | N/A | [80] | ||
| OP9 | hiPSC, hESC | RBC | SCF, FLT3 ligand, EPO, TPO, IL3, BMP4; VEGF | 15 days (+15 days for sac differentiation) | CD34+ PTPRC+GYPA+, clonogenic, embryonic, fetal, and adult globin | N/A | Gene correction in patient-iPSC from [81] | [82] | |
| VEGF, BMP4, SCF, FLT3 ligand, IL3, IL6, CSF3, EPO | hESC | Erythroid precursor, early erythroblast | VEGF, BMP4, SCF, FLT3 ligand, IL3, IL6, CSF3 | 15 days (+15 days for EB formation) | CD34+PTPRC+KDR+GYPA+, embryonic globin, clonogenic, self-renewal | N/A | Importance of VEGF | [83] | |
| EB | Transferrin, ascorbic acid, FGF2, VEGF | hESC | RBC | Dex, EPO, TPO, SCF, FLT3 ligand, IL3, IL6, CSF3, transferrin, FGF2, VEGF | 13–29 days (+14 days for EB formation) | CD34+GYPA+, clonogenic, embryonic and fetal globin | N/A | [84] | |
| BMP4, VEGF, FGF2, SCF, TPO, FLT3 ligand; FGF2, tPTD-HoxB4 | hESC | RBC | EPO, SCF | 21 days (+3–5 days for EB formation; +10 days for blast colony formation and expansion) | TFRC+GYPA+CD47+, embryonic and fetal globin | Oxygen dissociation curve | Blast colony formation; enucleation on days 36–42 | [85] | |
| SCF, TPO, FLT3 ligand, BMP4, VEGF, IL3, IL6, EPO | hiPSC, hESC | RBC | Insulin, heparin, SCF, IL3, EPO, human plasma | 25 days (+20 days for EB formation) | CD34+PTPRC+TFRC+GYPA+, enucleated/orthochromatic, fetal globin | Hemoglobin production, hemoglobin allosteric transition | Adapted from [67,68] | [86] | |
| SCF, TPO, FLT3 ligand, IL3, IL6, VEGF, BMP4, EPO | hiPSC, hESC | RBC | Human plasma, insulin, transferrin, SCF, IL3 | 25 days (+20 days for EB formation) | Enucleated, embryonic and fetal globin | N/A | [87] | ||
| BMP4, VEGF, activin A, WNT3A, GSK3βi VIII, FGF1, SCF, β-estradiol | hiPSC, hESC | RBC | BMP4, VEGF, FGF1, IGF2, TPO, heparin, IBMX, β-estradiol, hydrocortisone, FLT3 ligand, IL3, IL11, IGF1 | 24–31 days (+2 days for EB formation) | CD36+GYPA+, enucleated, embryonic and fetal globin | N/A | cGMP-compatible | [88] | |
| BMP4, VEGF, FLT3 ligand, IL3, IL6, SCF, TOP, EPO | hiPSC | RBC | IL3, SCF, EPO | 18 days (+21 days for EB formation) | CD36+SPN+GYPA+, clonogenic, enucleated, fetal and adult globin | N/A | [89,90] | ||
| ROCKi, BMP4, VEGF, WNT3A, FGF1, SCF, activin A, GSK3βi VIII, β-estradiol; FGF2 | hiPSC, hESC | RBC | BMP4, SCF, VEGF, IGF2, FGF1, TPO, heparin, EPO, IBMX, β-estradiol, hydrocortisone, IL3, ferric nitrate, poloxamer 188; FGF2, human plasma | 24 days (+3 days for EB formation) | SPN+TFRC+GYPA+, enucleated/orthochromatic, fetal globin | Oxygen dissociation curve | Further modified by [8] | [91] | |
| Monolayer | Matrigel-coated, FGF2, ROCKi | hiPSC | RBC | Human plasma-mimetic, FICZ, EPO, BMP4, VEGF, WNT3A, FGF2, SCF, FLT3 ligand, TPO, IL6, ascorbic acid | 60 days (+10–15 days for HSC differentiation) | GYPA+, embryonic and fetal globin | Hypoxia | Importance of aryl hydrocarbon receptor | [92] |
| Vitronectin-coated, FGF2, BMP4, SCF, VEGF, WNT3A, WNT5A, activin A, GSK3βi VIII, β-estradiol | hiPSC | RBC | BMP4, SCF, FGF2, TPO, VEGF, IGF2, β-estradiol, SB431542, heparin, IBMX, EPO, UM171, Dex, RU486, Optiferrin | 32–46 days (+7 days for HSC differentiation) | CD36+SPN+ TFRC+GYPA+, enucleated/orthochromatic, embryonic, fetal, and adult globin | N/A | Adapted from [88] | [93] | |
| Matrigel-coated, ROCKi | hiPSC | RBC | Ascorbic acid, FGF2, CHIR990921, VEGF, SCF, SB431542, transferrin, IL3, EPO | 20 days (+8–12 days of HSPC differentiation) | CD34+GYPA+SLC4A1+, clonogenic, embryonic, fetal, and adult globin | N/A | Adapted from [94] | [95] | |
| ECM-coated | hiPSC | RBC | Monothioglycerol, SCF, ascorbic acid, FGF2, WNT3A, IL3, BMP4, EPO, VEGF, TPO, FLT3 ligand, FICZ, IGF1, Dex | 16–17 days (+16 days for HSC differentiation) | CD34+PTPRC+GYPA+, clonogenic, embryonic and fetal globin | N/A | Direct comparison of four methods from [86,92,96,97] | [98] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-J.; Jung, C.; Oh, J.E.; Kim, S.; Lee, S.; Lee, J.Y.; Yoon, Y.-s. Generation of Red Blood Cells from Human Pluripotent Stem Cells—An Update. Cells 2023, 12, 1554. https://doi.org/10.3390/cells12111554
Lee S-J, Jung C, Oh JE, Kim S, Lee S, Lee JY, Yoon Y-s. Generation of Red Blood Cells from Human Pluripotent Stem Cells—An Update. Cells. 2023; 12(11):1554. https://doi.org/10.3390/cells12111554
Chicago/Turabian StyleLee, Shin-Jeong, Cholomi Jung, Jee Eun Oh, Sangsung Kim, Sangho Lee, Ji Yoon Lee, and Young-sup Yoon. 2023. "Generation of Red Blood Cells from Human Pluripotent Stem Cells—An Update" Cells 12, no. 11: 1554. https://doi.org/10.3390/cells12111554
APA StyleLee, S.-J., Jung, C., Oh, J. E., Kim, S., Lee, S., Lee, J. Y., & Yoon, Y.-s. (2023). Generation of Red Blood Cells from Human Pluripotent Stem Cells—An Update. Cells, 12(11), 1554. https://doi.org/10.3390/cells12111554








