A Rapid Alkalinization Factor-like Peptide EaF82 Impairs Tapetum Degeneration during Pollen Development through Induced ATP Deficiency
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of Genetic Cassettes
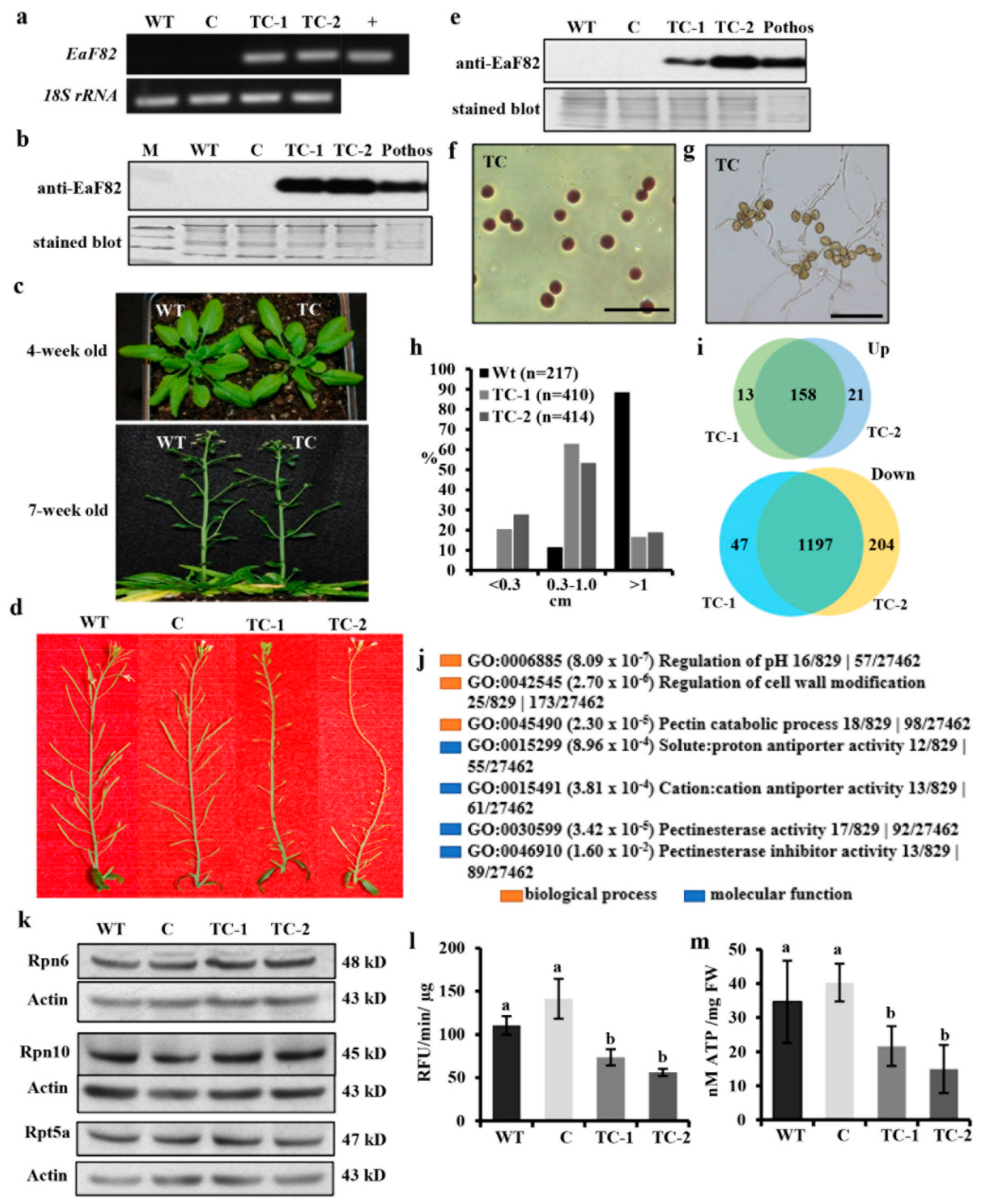
2.2. Created Transgenic Plants
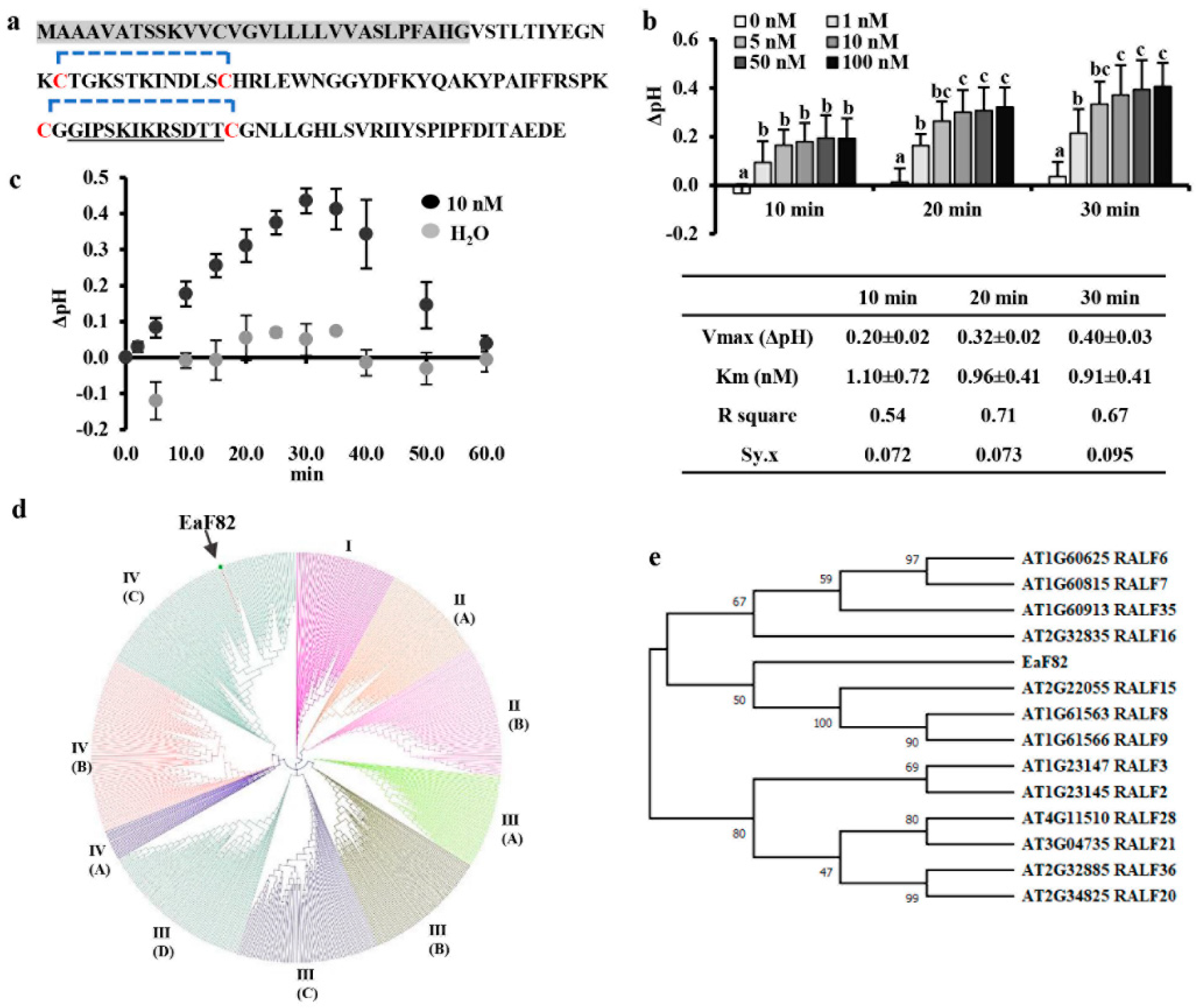
2.3. Alkalinization Assay
2.4. RT-PCR and RT-qPCR
2.5. RNAseq and DEG Analyses
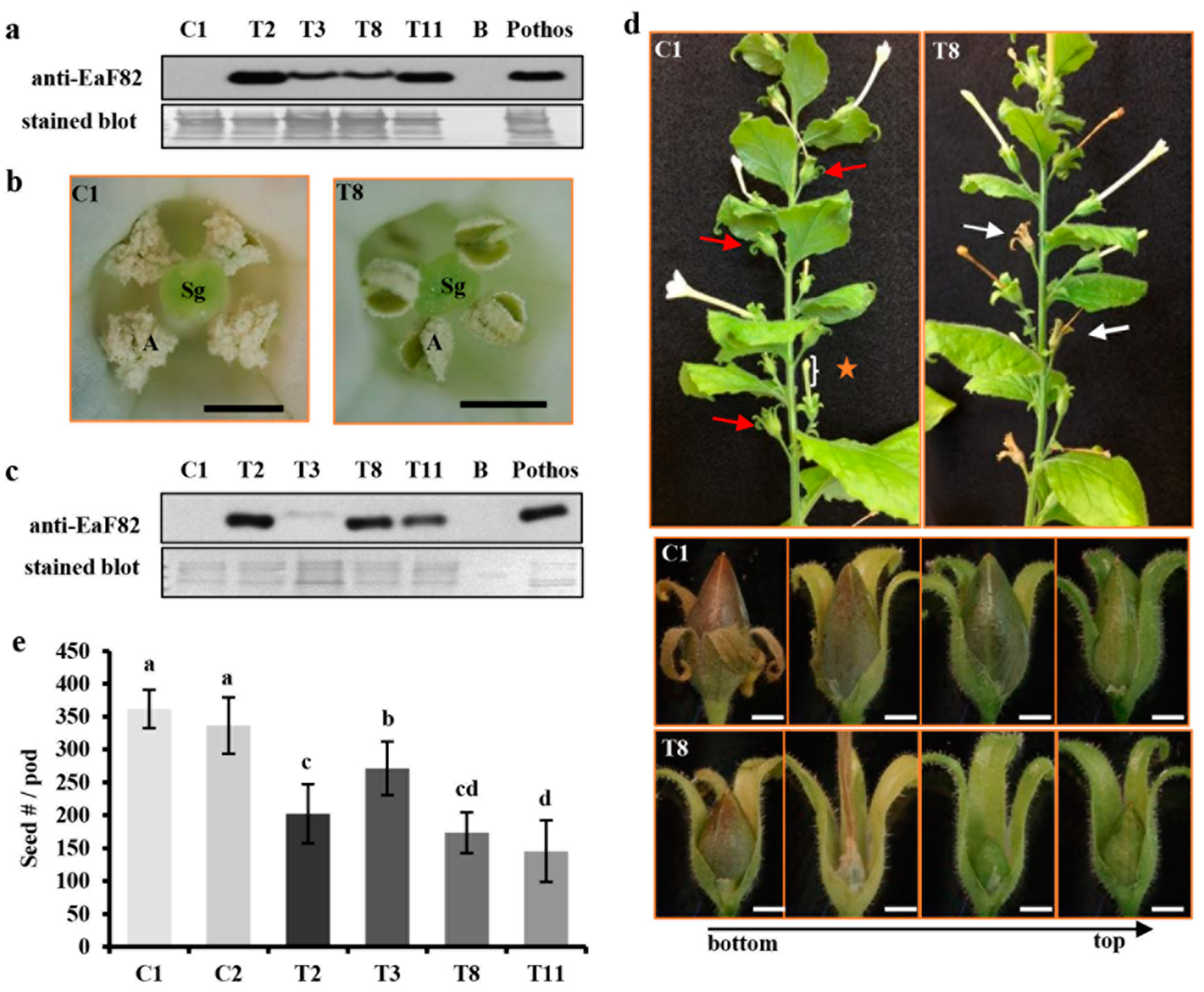
2.6. Protein Isolation, SDS-PAGE and Immunoblotting
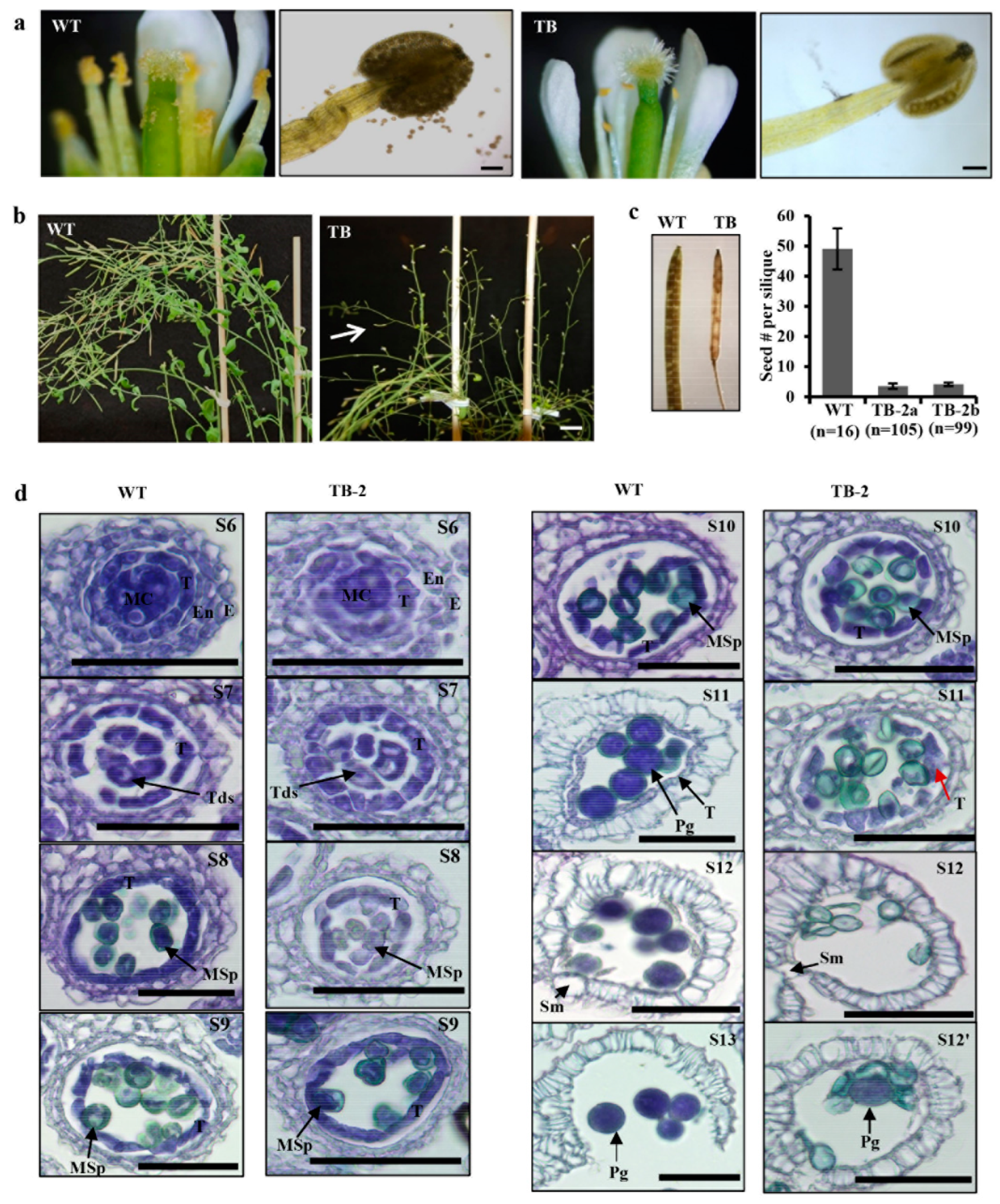
2.7. Histological Analysis and Seed Counting
2.8. ATP Measurement
2.9. Proteasomal Activity Assay
2.10. Yeast Two-Hybrid
2.11. Co-Immunoprecipitation (Co-IP) Analysis
2.12. Peptide Sequence Analysis and Phylogenetic Tree Construction
2.13. Statistical Analysis
3. Results
3.1. EaF82 Is a Clade IV RALF-like Peptide
3.2. EaF82 Is Expressed and Accumulated in Anthers but Not in Pollen
3.3. Overexpressing EaF82 Affected Pollen Development and Seed Setting
3.4. Overexpressing EaF82 Delayed Tapetum Degeneration during Pollen Development
3.5. EaF82 also Affected Pollen Development and Seed-Setting in Tobacco Plants
3.6. Overexpressing EaF82 Induced Transcriptome Changes
3.7. Overexpressing EaF82 Suppressed the Expression of Genes Involved in Cell Wall Modifications and pH Changes
3.8. EaF82 Suppressed the Expression of Genes Involved in Tapetum Degeneration and a Group of AtRALFs
3.9. Overexpressing EaF82 Decreased Proteasome Activity and ATP Levels
3.10. AKIN10 Is an Interacting Partner of EaF82
3.11. Elevated Levels of AKIN10 in EaF82 Transgenic Flowers with ATP Deficiency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pearce, G.; Strydom, D.; Johnson, S.; Ryan, C.A. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 1991, 253, 895–897. [Google Scholar] [CrossRef] [PubMed]
- Olsson, V.; Joos, L.; Zhu, S.; Gevaert, K.; Butenko, M.A.; De Smet, I. Look closely, the beautiful may be small: Precursor-derived peptides in plants. Annu. Rev. Plant Biol. 2019, 70, 153–186. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.; Smith, S.; De Smet, I. Small signaling peptides in Arabidopsis development: How cells communicate over a short distance. Plant Cell 2012, 24, 3198–3217. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Hussain, A.; Mun, B.G.; Imran, Q.M.; Falak, N.; Lee, S.U.; Kim, J.Y.; Hong, J.K.; Loake, G.J.; Ali, A.; et al. Comprehensive analysis of plant rapid alkalization factor (RALF) genes. Plant Physiol. Biochem. 2016, 106, 82–90. [Google Scholar] [CrossRef]
- Campbell, L.; Turner, S.R. A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front. Plant Sci. 2017, 8, 37. [Google Scholar] [CrossRef]
- Blackburn, M.R.; Haruta, M.; Moura, D.S. Twenty years of progress in physiological and biochemical investigation of RALF peptides. Plant Physiol. 2020, 182, 1657–1666. [Google Scholar] [CrossRef]
- Pearce, G.; Moura, D.S.; Stratmann, J.; Ryan, C.A. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 2001, 98, 12843–12847. [Google Scholar] [CrossRef]
- Tavormina, P.; De Coninck, B.; Nikonorova, N.; De Smet, I.; Cammue, B.P. The plant peptidome: An expanding repertoire of structural features and biological functions. Plant Cell 2015, 27, 2095–2118. [Google Scholar] [CrossRef]
- Haruta, M.; Burch, H.L.; Nelson, R.B.; Barrett-Wilt, G.; Kline, K.G.; Mohsin, S.B.; Young, J.C.; Otegui, M.S.; Sussman, M.R. Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 2010, 285, 17918–17929. [Google Scholar] [CrossRef]
- Covey, P.A.; Subbaiah, C.C.; Parsons, R.L.; Pearce, G.; Lay, F.T.; Anderson, M.A.; Ryan, C.A.; Bedinger, P.A. A pollen-specific RALF from tomato that regulates pollen tube elongation. Plant Physiol. 2010, 153, 703–715. [Google Scholar] [CrossRef]
- Shi, F.; Zhou, X.; Liu, Z.; Feng, H. Rapid alkalinization factor (RALF) genes are related to genic male sterility in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Sci. Hortic. 2017, 225, 480–489. [Google Scholar] [CrossRef]
- Li, D.D.; Xue, J.S.; Zhu, J.; Yang, Z.N. Gene regulatory network for tapetum development in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1559. [Google Scholar] [CrossRef] [PubMed]
- Loraine, A.E.; McCormick, S.; Estrada, A.; Patel, K.; Qin, P. RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol. 2013, 162, 1092–1109. [Google Scholar] [CrossRef]
- Hung, C.Y.; Sun, Y.H.; Chen, J.; Darlington, D.E.; Williams, A.L.; Burkey, K.O.; Xie, J. Identification of a Mg-protoporphyrin IX monomethyl ester cyclase homologue, EaZIP, differentially expressed in variegated Epipremnum aureum ‘Golden Pothos’ is achieved through a unique method of comparative study using tissue regenerated plants. J. Exp. Bot. 2010, 61, 1483–1493. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.Y.; Qiu, J.; Sun, Y.H.; Chen, J.; Kittur, F.S.; Henny, R.J.; Jin, G.; Fan, L.; Xie, J. Gibberellin deficiency is responsible for shy-flowering nature of Epipremnum aureum. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Hung, C.Y.; Umstead, M.L.; Chen, J.; Holliday, B.M.; Kittur, F.S.; Henny, R.J.; Burkey, K.O.; Xie, J. Differential expression of a novel gene EaF82a in green and yellow sectors of variegated Epipremnum aureum leaves is related to uneven distribution of auxin. Physiol. Plant. 2014, 152, 749–762. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, S.; Song, Y.; Men, S.; Wang, J. Gain-of-function analysis of poplar CLE genes in Arabidopsis by exogenous application and over-expression assays. J. Exp. Bot. 2016, 67, 2309–2324. [Google Scholar] [CrossRef]
- Musa, T.A.; Hung, C.Y.; Darlington, D.E.; Sane, D.C.; Xie, J. Overexpression of human erythropoietin in tobacco does not affect plant fertility or morphology. Plant Biotechnol. Rep. 2009, 3, 157–165. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Huang, S. Arabidopsis ACT 11 modifies actin turnover to promote pollen germination and maintain the normal rate of tube growth. Plant J. 2015, 83, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, S.; Oppenheimer, D.G. Extragenic suppressors of the Arabidopsis zwi-3 mutation identify new genes that function in trichome branch formation and pollen tube growth. Development 1999, 126, 3079–3088. [Google Scholar] [CrossRef]
- Napolitano, M.J.; Shain, D.H. Distinctions in adenylate metabolism among organisms inhabiting temperature extremes. Extremophiles 2005, 9, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Vallentine, P.; Hung, C.Y.; Xie, J.; Van Hoewyk, D. The ubiquitin–proteasome pathway protects Chlamydomonas reinhardtii against selenite toxicity, but is impaired as reactive oxygen species accumulate. AoB Plants 2014, 6, plu062. [Google Scholar] [CrossRef] [PubMed]
- Vojtek, A.; Hollenberg, S.M. Ras-Raf interaction: Two-hybrid analysis. Methods Enzymol. 1995, 255, 331–342. [Google Scholar] [PubMed]
- Bartel, P.L.; Chein, C.T.; Sternglanz, R.; Fields, S. Using the two hybrid system to detect protein-protein interactions. In Cellular Interactions in Development: A Practical Approach; Hartley, D.A., Ed.; Oxford University Press: Oxford, UK, 1993; pp. 153–179. [Google Scholar]
- Fromont-Racine, M.; Rain, J.C.; Legrain, P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 1997, 16, 277–282. [Google Scholar] [CrossRef]
- Formstecher, E.; Aresta, S.; Collura, V.; Hamburger, A.; Meil, A.; Trehin, A.; Reverdy, C.; Betin, V.; Maire, S.; Brun, C.; et al. Protein interaction mapping: A Drosophila case study. Genome Res. 2005, 15, 376–384. [Google Scholar] [CrossRef]
- Rain, J.C.; Selig, L.; De Reuse, H.; Battaglia, V.; Reverdy, C.; Simon, S.; Lenzen, G.; Petel, F.; Wojcik, J.; Schächter, V.; et al. The protein–protein interaction map of Helicobacter pylori. Nature 2001, 409, 211–215. [Google Scholar] [CrossRef]
- Wojcik, J.; Boneca, I.G.; Legrain, P. Prediction, assessment and validation of protein interaction maps in bacteria. J. Mol. Biol. 2002, 323, 763–770. [Google Scholar] [CrossRef]
- Colland, F.; Jacq, X.; Trouplin, V.; Mougin, C.; Groizeleau, C.; Hamburger, A.; Meil, A.; Wojcik, J.; Legrain, P.; Gauthier, J.M. Functional proteomics mapping of a human signaling pathway. Genome Res. 2004, 14, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Marques, J.R.; Fonseca, R.R.D.; Drury, B.; Melo, A. Amino acid patterns around disulfide bonds. Int. J. Mol. Sci. 2010, 11, 4673–4686. [Google Scholar] [CrossRef]
- Marshall, E.; Costa, L.M.; Gutierrez-Marcos, J. Cysteine-rich peptides (CRPs) mediate diverse aspects of cell–cell communication in plant reproduction and development. J. Exp. Bot. 2011, 62, 1677–1686. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Abarca, A.; Franck, C.M.; Zipfel, C. Family-wide evaluation of RALF peptides in Arabidopsis thaliana. Plant Physiol. 2021, 187, 996–1010. [Google Scholar] [CrossRef]
- De Smet, I.; Jürgens, G. Patterning the axis in plants–auxin in control. Curr. Opin. Genet. Dev. 2007, 17, 337–343. [Google Scholar] [CrossRef]
- Yao, X.; Tian, L.; Yang, J.; Zhao, Y.N.; Zhu, Y.X.; Dai, X.; Zhao, Y.; Yang, Z.N. Auxin production in diploid microsporocytes is necessary and sufficient for early stages of pollen development. PLoS Genet. 2018, 14, e1007397. [Google Scholar] [CrossRef]
- Smyth, D.R.; Bowman, J.L.; Meyerowitz, E.M. Early flower development in Arabidopsis. Plant Cell 1990, 2, 755–767. [Google Scholar]
- Sanders, P.M.; Bui, A.Q.; Weterings, K.; McIntire, K.N.; Hsu, Y.C.; Lee, P.Y.; Truong, M.T.; Beals, T.P.; Goldberg, R.B. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999, 11, 297–322. [Google Scholar] [CrossRef]
- Parenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Barnes, W.J.; Anderson, C.T. Release, recycle, rebuild: Cell-wall remodeling, autodegradation, and sugar salvage for new wall biosynthesis during plant development. Mol. Plant 2018, 11, 31–46. [Google Scholar] [CrossRef]
- Pelloux, J.; Rusterucci, C.; Mellerowicz, E.J. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007, 12, 267–277. [Google Scholar] [CrossRef]
- Pina, C.; Pinto, F.; Feijó, J.A.; Becker, J.D. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 2005, 138, 744–756. [Google Scholar] [CrossRef] [PubMed]
- Pinzón-Latorre, D.; Deyholos, M.K. Characterization and transcript profiling of the pectin methylesterase (PME) and pectin methylesterase inhibitor (PMEI) gene families in flax (Linum usitatissimum). BMC Genom. 2013, 14, 742. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.W.; Chen, M.H.; Zaltsman, A.; Citovsky, V. Pollen-specific pectin methylesterase involved in pollen tube growth. Dev. Biol. 2006, 294, 83–91. [Google Scholar] [CrossRef]
- Phan, H.A.; Iacuone, S.; Li, S.F.; Parish, R.W. The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 2011, 23, 2209–2224. [Google Scholar] [CrossRef]
- Jia, Q.S.; Zhu, J.; Xu, X.F.; Lou, Y.; Zhang, Z.L.; Zhang, Z.P.; Yang, Z.N. Arabidopsis AT-hook protein TEK positively regulates the expression of arabinogalactan proteins for Nexine formation. Mol. Plant 2015, 8, 251–260. [Google Scholar] [CrossRef]
- Coimbra, S.; Costa, M.; Jones, B.; Mendes, M.A.; Pereira, L.G. Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. J. Exp. Bot. 2009, 60, 3133–3142. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, R.B.; Beals, T.P.; Sanders, P.M. Anther development: Basic principles and practical applications. Plant Cell 1993, 5, 1217–1229. [Google Scholar] [PubMed]
- Scott, R.J.; Spielman, M.; Dickinson, H.G. Stamen structure and function. Plant Cell 2004, 16, S46–S60. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Liu, B. Tapetum-dependent male meiosis progression in plants: Increasing evidence emerges. Front. Plant Sci. 2019, 10, 1667. [Google Scholar] [CrossRef]
- Parish, R.W.; Li, S.F. Death of a tapetum: A programme of developmental altruism. Plant Sci. 2010, 178, 73–89. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, D.; Lv, X.; Wang, Y.; Xun, Z.; Liu, Z.; Li, F.; Lu, H. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 2014, 26, 2939–2961. [Google Scholar] [CrossRef]
- Zhang, D.S.; Liang, W.Q.; Yuan, Z.; Li, N.; Shi, J.; Wang, J.; Liu, Y.M.; Yu, W.J.; Zhang, D.B. Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol. Plant 2008, 1, 599–610. [Google Scholar] [CrossRef]
- Kawanabe, T.; Ariizumi, T.; Kawai-Yamada, M.; Uchimiya, H.; Toriyama, K. Abolition of the tapetum suicide program ruins microsporogenesis. Plant Cell Physiol. 2006, 47, 784–787. [Google Scholar] [CrossRef]
- Buono, R.A.; Hudecek, R.; Nowack, M.K. Plant proteases during developmental programmed cell death. J. Exp. Bot. 2019, 70, 2097–2112. [Google Scholar] [CrossRef]
- Gallois, J.L.; Guyon-Debast, A.; Lécureuil, A.; Vezon, D.; Carpentier, V.; Bonhomme, S.; Guerche, P. The Arabidopsis proteasome RPT5 subunits are essential for gametophyte development and show accession-dependent redundancy. Plant Cell 2009, 21, 442–459. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, C.L.; Deng, S.R.; Lu, C.F.; Shen, X.; Zhou, X.Y.; Zheng, X.J.; Hu, Z.M.; Chen, S.L. An ATP signalling pathway in plant cells: Extracellular ATP triggers programmed cell death in Populus euphratica. Plant Cell Environ. 2012, 35, 893–916. [Google Scholar] [CrossRef] [PubMed]
- Van Aken, O.; Van Breusegem, F. Licensed to kill: Mitochondria, chloroplasts, and cell death. Trends Plant Sci. 2015, 20, 754–766. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Vierstra, R.D. Dynamic regulation of the 26S proteasome: From synthesis to degradation. Front. Mol. Biosci. 2019, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Brookes, P.S.; Yoon, Y.; Robotham, J.L.; Anders, M.W.; Sheu, S.S. Calcium, ATP, and ROS: A mitochondrial love-hate triangle. Am. J. Physiol. Cell Physiol. 2004, 287, C817–C833. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Gilroy, S.; Jones, A.M.; Stacey, G. Extracellular ATP signaling in plants. Trends Cell Biol. 2010, 20, 601–608. [Google Scholar] [CrossRef]
- Berthomé, R.; Thomasset, M.; Maene, M.; Bourgeois, N.; Froger, N.; Budar, F. pur4 mutations are lethal to the male, but not the female, gametophyte and affect sporophyte development in Arabidopsis. Plant Physiol. 2008, 147, 650–660. [Google Scholar] [CrossRef]
- Li, W.Q.; Zhang, X.Q.; Xia, C.; Deng, Y.; Ye, D. MALE GAMETOPHYTE DEFECTIVE 1, encoding the FAd subunit of mitochondrial F1F0-ATP synthase, is essential for pollen formation in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 923–935. [Google Scholar] [CrossRef]
- Liang, C.; Zhang, Y.; Cheng, S.; Osorio, S.; Sun, Y.; Fernie, A.R.; Cheung, C.Y.; Lim, B.L. Impacts of high ATP supply from chloroplasts and mitochondria on the leaf metabolism of Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 922. [Google Scholar] [CrossRef]
- Kaundal, A.; Ramu, V.S.; Oh, S.; Lee, S.; Pant, B.; Lee, H.K.; Rojas, C.M.; Senthil-Kumar, M.; Mysore, K.S. GENERAL CONTROL NONREPRESSIBLE4 degrades 14-3-3 and the RIN4 complex to regulate stomatal aperture with implications on nonhost disease resistance and drought tolerance. Plant Cell 2017, 29, 2233–2248. [Google Scholar] [CrossRef]
- Phee, B.K.; Kim, J.I.; Shin, D.H.; Yoo, J.; Park, K.J.; Han, Y.J.; Kwon, Y.K.; Cho, M.H.; Jeon, J.S.; Bhoo, S.H.; et al. A novel protein phosphatase indirectly regulates phytochrome-interacting factor 3 via phytochrome. Biochem. J. 2008, 415, 247–255. [Google Scholar] [CrossRef]
- Nukarinen, E.; Nägele, T.; Pedrotti, L.; Wurzinger, B.; Mair, A.; Landgraf, R.; Börnke, F.; Hanson, J.; Teige, M.; Baena-Gonzalez, E.; et al. Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 2016, 6, 31697. [Google Scholar] [CrossRef]
- Hardie, D.G.; Schaffer, B.E.; Brunet, A. AMPK: An energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol. 2016, 26, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Wen, T.N.; Wang, Y.T.; Shih, M.C. Quantitative phosphoproteomics of protein kinase SnRK1 regulated protein phosphorylation in Arabidopsis under submergence. J. Exp. Bot. 2016, 67, 2745–2760. [Google Scholar] [CrossRef] [PubMed]
- Tsai, A.Y.L.; Gazzarrini, S. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 2012, 69, 809–821. [Google Scholar] [CrossRef]
- Jeong, E.Y.; Seo, P.J.; Woo, J.C.; Park, C.M. AKIN10 delays flowering by inactivating IDD8 transcription factor through protein phosphorylation in Arabidopsis. BMC Plant Bio. 2015, 15, 110. [Google Scholar] [CrossRef] [PubMed]
- Farrás, R.; Ferrando, A.; Jásik, J.; Kleinow, T.; Okrész, L.; Tiburcio, A.; Salchert, K.; del Pozo, C.; Schell, J.; Koncz, C. SKP1–SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 2001, 20, 2742–2756. [Google Scholar] [CrossRef] [PubMed]
- Broeckx, T.; Hulsmans, S.; Rolland, F. The plant energy sensor: Evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J. Exp. Bot. 2016, 67, 6215–6252. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef]
- Gómez, J.F.; Talle, B.; Wilson, Z.A. Anther and pollen development: A conserved developmental pathway. J. Integr. Plant Biol. 2015, 57, 876–891. [Google Scholar] [CrossRef]
- Truskina, J.; Brück, S.; Stintzi, A.; Boeuf, S.; Doll, N.M.; Fujita, S.; Geldner, N.; Schaller, A.; Ingram, G.C. A peptide-mediated, multilateral molecular dialogue for the coordination of pollen wall formation. Proc. Natl. Acad. Sci. USA 2022, 119, e2201446119. [Google Scholar] [CrossRef]
- Cardarelli, M.; Cecchetti, V. Auxin polar transport in stamen formation and development: How many actors? Front. Plant Sci. 2014, 5, 333. [Google Scholar] [CrossRef] [PubMed]
- Bitrián, M.; Roodbarkelari, F.; Horváth, M.; Koncz, C. BAC-recombineering for studying plant gene regulation: Developmental control and cellular localization of SnRK1 kinase subunits. Plant J. 2011, 65, 829–842. [Google Scholar] [CrossRef]
- Williams, S.P.; Rangarajan, P.; Donahue, J.L.; Hess, J.E.; Gillaspy, G.E. Regulation of sucrose non-fermenting related kinase 1 genes in Arabidopsis thaliana. Front. Plant Sci. 2014, 5, 324. [Google Scholar] [CrossRef]
- Ramon, M.; Dang, T.V.T.; Broeckx, T.; Hulsmans, S.; Crepin, N.; Sheen, J.; Rolland, F. Default activation and nuclear translocation of the plant cellular energy sensor SnRK1 regulate metabolic stress responses and development. Plant Cell 2019, 31, 1614–1632. [Google Scholar] [CrossRef] [PubMed]
- Emptage, R.P.; Lemmon, M.A.; Ferguson, K.M. Molecular determinants of KA1 domain-mediated autoinhibition and phospholipid activation of MARK1 kinase. Biochem. J. 2017, 474, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.T.; Zhang, Q.; Xie, J.H.; Hung, C.-Y.; Cui, J.; Henny, R.J.; Chen, J. Plant regeneration via direct somatic embryogenesis from leaf and petiole explants of Epipremnum aureum ‘Marble Queen’ and characterization of selected variants. Acta Physiol. Plant. 2012, 34, 1461–1469. [Google Scholar] [CrossRef]


| Gene_ID a | Gene Description b | TC-1 (n = 3) | TC-2 (n = 3) | ||
|---|---|---|---|---|---|
| RNAseq (FC) | RT-qPCR (FC ± SD) | RNAseq (FC) | RT-qPCR (FC ± SD) | ||
| Flowering and pollen development related | |||||
| AT2G45660 | SOC1/AGL20 (AGAMOUS-like) | 2.52 | 2.15 ± 0.27 | 2.55 | 2.57 ± 0.13 |
| AT4G24540 | AGL24 (AGAMOUS-like) | 3.02 | 2.96 ± 0.35 | 3.27 | 3.80 ± 0.86 |
| AT5G62165 | AGL42 (AGAMOUS-like) | 2.10 | 2.13 ± 0.05 | 2.10 | 2.35 ± 0.18 |
| AT4G35900 | FD (Basic-leucine zipper transcription factor) | 3.26 | 1.80 ± 0.16 | 3.84 | 2.24 ± 0.07 |
| AT1G19890 | MGH3 (male-gamete-specific histone H3) | −2.93 | −4.34 ± 1.32 | −4.91 | −4.34 ± 1.65 |
| AT1G19960 | transcription factor | −5.10 | −6.60 ± 0.87 | −7.50 | −8.55 ± 2.86 |
| AT1G21000 | PLATZ family transcription factor | −4.16 | −4.13 ± 1.52 | −4.42 | −3.86 ± 1.08 |
| AT1G35490 | bZIP family transcription factor | −5.35 | −7.04 ± 3.20 | −7.11 | −7.31 ± 2.50 |
| AT2G36080 | ABS2/NGAL1 (AP2/B3-like transcription factor) | −4.40 | −5.75 ± 1.67 | −4.58 | −5.69 ± 1.69 |
| AT1G24520 | BCP1 (Brassica campestris homolog pollen protein 1) | −6.65 | −3.15 ± 1.65 | −14.91 | −98.37 ± 5.98 |
| AT5G17480 | PC1/APC1/CML29 (pollen calcium-binding protein 1) | −2.44 | −4.17 ± 1.85 | −2.66 | −3.99 ± 0.64 |
| AT4G10603 | SLR1-BP (S locus-related glycoprotein 1 binding pollen coat protein) | −2.29 | −3.15 ± 0.55 | −2.64 | −2.57 ± 0.57 |
| AT1G29140 | Pollen Ole e 1 allergen and extensin family protein | −3.75 | −5.85 ± 1.94 | −4.76 | −6.05 ± 3.63 |
| AT5G45880 | Pollen Ole e 1 allergen and extensin family protein | −4.09 | −6.26 ± 2.00 | −6.70 | −5.81 ± 0.82 |
| H+-ATPase | |||||
| AT5G57350 | HA3 | −2.12 | −2.83 ± 0.74 | −2.28 | −2.16 ± 0.46 |
| AT2G07560 | HA6 | −2.95 | −4.31 ± 1.35 | −3.48 | −3.87 ± 1.00 |
| AT3G42640 | HA8 | −2.19 | −2.50 ± 0.95 | −2.74 | −2.54 ± 1.49 |
| AT1G80660 | HA9 | −3.84 | −4.92 ± 1.00 | −4.52 | −5.08 ± 1.54 |
| AT3G08560 | VHA-E2 | −2.50 | −3.41 ± 0.21 | −2.90 | −2.94 ± 1.14 |
| Protein kinases | |||||
| AT2G07040 | PRK2A (Leucine-rich repeat protein kinase) | −5.07 | −8.55 ± 4.81 | −7.81 | −8.47 ± 3.81 |
| AT2G18470 | PERK4 (Proline-rich protein kinase) | −3.24 | −4.52 ± 1.24 | −4.07 | −4.18 ± 0.51 |
| AT2G21480 | BUPS2 (Malectin/receptor-like) | −3.96 | −6.52 ± 4.87 | −5.45 | −7.75 ± 2.38 |
| AT4G39110 | BUPS1 (Malectin/receptor-like) | −3.14 | −1.99 ± 0.49 | −4.65 | −2.10 ± 0.84 |
| AT5G28680 | ANX2 (Malectin/receptor-like) | −5.86 | −8.16 ± 6.36 | −12.79 | −25.23 ± 13.51 |
| Gene_ID a | Gene Description (Clade) b | TC-1 (n = 3) | TC-2 (n = 3) | Predicted pI and MW c | |||
|---|---|---|---|---|---|---|---|
| RNAseq (FC) | RT-qPCR (FC ± SD) | RNAseq (FC) | RT-qPCR (FC ± SD) | pI | MW (kD) | ||
| AT1G28270 | RALF4 (III-B) | −4.27 | −7.00 ± 2.79 | −5.96 | −8.37 ± 4.59 | 9.76 | 8.7 |
| AT1G61563 | RALF8 (IV-C) | −3.67 | −5.26 ± 1.60 | −5.35 | −5.66 ± 1.00 | 9.30 | 6.6 |
| AT1G61566 | RALF9 (IV-C) | −3.51 | −4.77 ± 1.26 | −5.27 | −5.74 ± 1.38 | 9.27 | 6.6 |
| AT2G22055 | RALF15 (IV-C) | −5.73 | −2.48 ± 0.39 | −23.41 | −16.92 ± 2.40 | 9.76 | 6.5 |
| AT2G33775 | RALF19 (III-B) | −5.26 | −9.10 ± 5.00 | −9.69 | −8.14 ± 3.43 | 10.57 | 7.5 |
| AT3G25165 | RALF25 (IV-B) | −8.24 | −2.48 ± 0.67 | −23.55 | −82.52 ± 18.38 | 10.08 | 7.0 |
| AT4G14020 | RALF family protein (IV-A) | −3.39 | −4.29 ± 1.30 | −3.78 | −4.02 ± 1.27 | 10.17 | 6.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hung, C.-Y.; Kittur, F.S.; Wharton, K.N.; Umstead, M.L.; Burwell, D.B.; Thomas, M.; Qi, Q.; Zhang, J.; Oldham, C.E.; Burkey, K.O.; et al. A Rapid Alkalinization Factor-like Peptide EaF82 Impairs Tapetum Degeneration during Pollen Development through Induced ATP Deficiency. Cells 2023, 12, 1542. https://doi.org/10.3390/cells12111542
Hung C-Y, Kittur FS, Wharton KN, Umstead ML, Burwell DB, Thomas M, Qi Q, Zhang J, Oldham CE, Burkey KO, et al. A Rapid Alkalinization Factor-like Peptide EaF82 Impairs Tapetum Degeneration during Pollen Development through Induced ATP Deficiency. Cells. 2023; 12(11):1542. https://doi.org/10.3390/cells12111542
Chicago/Turabian StyleHung, Chiu-Yueh, Farooqahmed S. Kittur, Keely N. Wharton, Makendra L. Umstead, D’Shawna B. Burwell, Martinique Thomas, Qi Qi, Jianhui Zhang, Carla E. Oldham, Kent O. Burkey, and et al. 2023. "A Rapid Alkalinization Factor-like Peptide EaF82 Impairs Tapetum Degeneration during Pollen Development through Induced ATP Deficiency" Cells 12, no. 11: 1542. https://doi.org/10.3390/cells12111542
APA StyleHung, C.-Y., Kittur, F. S., Wharton, K. N., Umstead, M. L., Burwell, D. B., Thomas, M., Qi, Q., Zhang, J., Oldham, C. E., Burkey, K. O., Chen, J., & Xie, J. (2023). A Rapid Alkalinization Factor-like Peptide EaF82 Impairs Tapetum Degeneration during Pollen Development through Induced ATP Deficiency. Cells, 12(11), 1542. https://doi.org/10.3390/cells12111542








