Systems Genomics Reveals microRNA Regulation of ICS Response in Childhood Asthma
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohorts: GACRS and CAMP
2.1.1. Discovery Cohort: The Genetics of Asthma in Costa Rica Study (GACRS)
2.1.2. Replication Cohort: The Childhood Asthma Management Program (CAMP)
2.2. CASTER: Cross-Sectional Asthma STEroid Response Measurement
2.3. miRNA Sequencing and Quality Control
2.4. ICS-Response-Associated miRNA Identification
2.5. CAMP Lymphoblastoid Cell Lines (LCL) Transcriptome Data
2.6. Functional Annotation of Replicated miRNAs
2.7. Gene Expression Analysis
3. Results
3.1. Cohort Characteristics
3.1.1. GACRS
| GACRS | CAMP | |||||
|---|---|---|---|---|---|---|
| Characteristics | Poor Responder | Good Responder | p Value | Poor Responder | Good Responder | p Value |
| (n = 379) | (n = 201) | (n = 71) | (n = 116) | |||
| Sex | ||||||
| Male | 156 (41.2%) | 87 (43.3%) | 0.686 | 39 (54.9%) | 70 (60.3%) | 0.565 |
| Female | 223 (58.8%) | 114 (56.7%) | 32 (45.1%) | 46 (39.7%) | ||
| Age (years) | ||||||
| Mean (SD) | 9.43 (1.89) | 8.80 (1.87) | <0.001 | 8.67 (2.18) | 9.16 (2.08) | 0.132 |
| Median [Min, Max] | 9.36 [4.50, 15.2] | 8.53 [6.02, 13.1] | 8.90 [5.18, 13.2] | 8.94 [5.26, 13.1] | ||
| Race/Ethnicity | ||||||
| Non-Hispanic | NA | NA | 50 (70.4%) | 76 (65.5%) | 0.211 | |
| Hispanic White | 379 (100%) | 201 (100%) | 4 (5.6%) | 16 (13.8%) | ||
| Other | NA | NA | 17 (23.9%) | 24 (20.7%) | ||
| Height (cm) | ||||||
| Mean (SD) | 134 (15.2) | 130 (12.3) | <0.001 | 132 (12.9) | 136 (13.7) | 0.0615 |
| Median [Min, Max] | 134 [0, 168] | 128 [103, 164] | 133 [108, 156] | 136 [107, 170] | ||
| Missing | NA | NA | 1 (1.4%) | 0 (0%) | ||
| Weight (kg) | ||||||
| Mean (SD) | 34.5 (11.9) | 32.1 (12.3) | 0.0275 | 32.8 (12.0) | 35.6 (12.5) | 0.133 |
| Median [Min, Max] | 30.9 [15.8, 87.4] | 29.3 [15.0, 94.2] | 30.0 [17.8, 80.5] | 33.0 [17.1, 82.3] | ||
| Missing | 2 (0.5%) | 0 (0%) | NA | NA | ||
| BMI | ||||||
| Mean (SD) | 18.5 (3.81) | 18.4 (4.15) | 0.819 | 18.3 (3.67) | 18.7 (3.67) | 0.442 |
| Median [Min, Max] | 17.8 [8.18, 34.0] | 17.2 [12.8, 41.4] | 17.0 [13.4, 34.3] | 17.5 [14.2, 33.7] | ||
| Missing | 2 (0.5%) | 0 (0%) | 1 (1.4%) | 0 (0%) | ||
| High-Dose Oral Steroid Courses | ||||||
| Mean (SD) | 2.15 (0.388) | 1.41 (0.776) | <0.001 | 2.14 (1.27) | 0.0603 (0.239) | <0.001 |
| Median [Min, Max] | 2.00 [2.00, 5.00] | 2.00 [0, 2.00] | 2.00 [1.00, 6.00] | 0 [0, 1.00] | ||
| ED (emergency department) visits | ||||||
| Mean (SD) | 4.60 (4.19) | 2.75 (3.00) | <0.001 | 0.451 (0.968) | 0.0517 (0.222) | 0.00102 |
| Median [Min, Max] | 4.00 [0, 30.0] | 2.00 [0, 22.0] | 0 [0, 5.00] | 0 [0, 1.00] | ||
| % Predicted Pre-BD FEV1 | ||||||
| Mean (SD) | 91.9 (14.1) | 109 (18.2) | <0.001 | 91.2 (13.0) | 93.8 (13.0) | 0.181 |
| Median [Min, Max] | 92.8 [31.8, 135] | 111 [34.6, 154] | 92.0 [62.0, 118] | 95.0 [61.0, 125] | ||
| Airway hyper-responsiveness | ||||||
| Mean (SD) | 0.671 (0.336) | 0.998 (0.532) | <0.001 | −0.260 (1.15) | 0.162 (1.11) | 0.0149 |
| Median [Min, Max] | 0.778 [0.043, 2.2] | 0.778 [0.14, 2.87] | −0.390 [−2.59, 2.48] | 0.140 [−2.94, 2.53] | ||
| Bronchodilator Response as % of baseline FEV1 | ||||||
| Mean (SD) | 6.57 (8.37) | 3.30 (7.33) | <0.001 | 0.130 (0.248) | 0.0764 (0.0731) | 0.0805 |
| Median [Min, Max] | 5.29 [−18.1, 44] | 3.40 [−35.1, 25] | 0.0800 [−0.04, 2.05] | 0.0600 [−0.06, 0.41] | ||
| Log10 IgE | ||||||
| Mean (SD) | 2.61 (0.629) | 2.40 (0.669) | 0.0255 | 2.78 (0.622) | 2.60 (0.669) | 0.0684 |
| Median [Min, Max] | 2.73 [0.260, 3.7] | 2.55 [0.73, 3.67] | 2.89 [1.26, 4.13] | 2.61 [0.30, 4.15] | ||
| Missing | 244 (64.4%) | 118 (58.7%) | NA | NA | ||
| 25 Hydroxyvitamin D (ng/mL) | ||||||
| Mean (SD) | 36.0 (11.0) | 35.0 (9.18) | 0.484 | 39.9 (15.7) | 40.2 (15.2) | 0.902 |
| Median [Min, Max] | 34.7 [12.5, 71.5] | 34.4 [18.4, 63.1] | 37.8 [14.6, 80.0] | 39.5 [9.00, 75.9] | ||
| Missing | 244 (64.4%) | 118 (58.7%) | NA | NA | ||
| Blood Eosinophils (Log10) | ||||||
| Mean (SD) | 2.63 (0.439) | 2.52 (0.394) | 0.00373 | 2.57 (0.462) | 2.52 (0.427) | 0.486 |
| Median [Min, Max] | 2.73 [−1.0, 3.30] | 2.57 [1.0, 3.27] | 2.67 [0, 3.40] | 2.61 [0, 3.24] | ||
| Missing | 13 (3.4%) | 6 (3.0%) | 1 (1.4%) | 2 (1.7%) | ||
| Smoking | ||||||
| No | 281 (74.1%) | 152 (75.6%) | 0.59 | 46 (64.8%) | 67 (57.8%) | 0.508 |
| Yes | 97 (25.6%) | 46 (22.9%) | 25 (35.2%) | 47 (40.5%) | ||
| Missing | 1 (0.3%) | 3 (1.5%) | 0 (0%) | 2 (1.7%) | ||
| Asthma Severity | ||||||
| Mild | NA | NA | 29 (40.8%) | 51 (44.0%) | 0.79 | |
| Moderate | NA | NA | 42 (59.2%) | 65 (56.0%) | ||
| Genetic Ancestry Proportion | (n = 366) | (n = 194) | (n = 48) | (n = 83) | ||
| Sub Saharan African | 0.047 | 0.045 | 0.29 | 0.167 | 0.179 | 0.84 |
| Central and South Asia | 0.014 | 0.014 | 0.40 | 0.009 | 0.01 | 0.33 |
| East Asia | 0.003 | 0.003 | 0.70 | 0.002 | 0.003 | 0.18 |
| Europe | 0.533 | 0.539 | 0.26 | 0.772 | 0.734 | 0.52 |
| Native America | 0.320 | 0.317 | 0.61 | 0.030 | 0.045 | 0.35 |
| Oceania | 0 | 0 | 0.06 | 0 | 0 | 0.7 |
| Middle East | 0.082 | 0.080 | 0.3 | 0.019 | 0.027 | 0.15 |
3.1.2. CAMP
3.2. Identification of miRNAs Associated with ICS Response
3.3. Target Identification and Functional Enrichment Analysis of Replicated miRNAs
3.4. In Vitro Steroid Response Lymphoblastoid Gene Expression
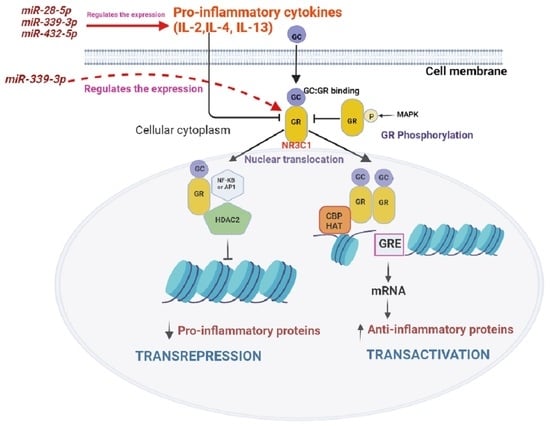
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- 2019 National Health Interview Survey Data; U.S. Department of Health & Human, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2020. Available online: https://www.cdc.gov/asthma/nhis/2019/data.htm (accessed on 29 April 2023).
- Tantisira, K.G.; Lake, S.; Silverman, E.S.; Palmer, L.J.; Lazarus, R.; Silverman, E.K.; Liggett, S.B.; Gelfand, E.W.; Rosenwasser, L.J.; Richter, B.; et al. Corticosteroid pharmacogenetics: Association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum. Mol. Genet. 2004, 13, 1353–1359. [Google Scholar] [CrossRef]
- Bengtson, L.G.S.; Yu, Y.; Wang, W.; Cao, F.; Hulbert, E.M.; Wolbeck, R.; Elliott, C.A.; Buikema, A.R. Inhaled Corticosteroid-Containing Treatment Escalation and Outcomes for Patients with Asthma in a U.S. Health Care Organization. J. Manag. Care Spec. Pharm. 2017, 23, 1149–1159. [Google Scholar] [CrossRef]
- Chan, M.T.S.; Leung, D.Y.M.; Szefler, S.J.; Spahn, J.D. Difficult-to-control asthma: Clinical characteristics of steroid-insensitive asthma. J. Allergy Clin. Immunol. 1998, 101, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Sohel, M.H. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef]
- Wang, L.; Xiong, Y.; Fu, B.; Guo, D.; Zaky, M.Y.; Lin, X.; Wu, H. MicroRNAs as immune regulators and biomarkers in tuberculosis. Front. Immunol. 2022, 13, 1027472. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Zhang, Z.; Xiao, Z.; Li, X.; Luo, S.; Zhou, Z. Circular RNAs in diabetes and its complications: Current knowledge and future prospects. Front. Genet. 2022, 13, 1006307. [Google Scholar] [CrossRef]
- Sharma, R.; Tiwari, A.; McGeachie, M.J. Recent miRNA Research in Asthma. Curr. Allergy Asthma Rep. 2022, 22, 231–258. [Google Scholar] [CrossRef]
- Clemmer, G.L.; Wu, A.C.; Rosner, B.; McGeachie, M.J.; Litonjua, A.A.; Tantisira, K.G.; Weiss, S.T. Measuring the corticosteroid responsiveness endophenotype in asthmatic patients. J. Allergy Clin. Immunol. 2015, 136, 274–281.e8. [Google Scholar] [CrossRef]
- Kho, A.T.; Sordillo, J.; Wu, A.C.; Cho, M.H.; Sharma, S.; Tiwari, A.; Lasky-Su, J.; Weiss, S.T.; Tantisira, K.G.; McGeachie, M.J. Caster: Cross-sectional asthma steroid response measurement. J. Pers. Med. 2020, 10, 95. [Google Scholar] [CrossRef]
- Kho, A.T.; McGeachie, M.J.; Li, J.; Chase, R.P.; Amr, S.S.; Hastie, A.T.; Hawkins, G.A.; Li, X.; Chupp, G.L.; Meyers, D.A.; et al. Lung function, airway and peripheral basophils and eosinophils are associated with molecular pharmacogenomic endotypes of steroid response in severe asthma. Thorax 2022, 77, 452–460. [Google Scholar] [CrossRef]
- Li, J.; Panganiban, R.; Kho, A.T.; McGeachie, M.J.; Farnam, L.; Chase, R.P.; Weiss, S.T.; Lu, Q.; Tantisira, K.G. Circulating micrornas and treatment response in childhood asthma. Am. J. Respir. Crit. Care Med. 2020, 202, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Tantisira, K.G.; Lasky-Su, J.; Harada, M.; Murphy, A.; Litonjua, A.A.; Himes, B.E.; Lange, C.; Lazarus, R.; Sylvia, J.; Klanderman, B.; et al. Genomewide Association between GLCCI1 and Response to Glucocorticoid Therapy in Asthma. N. Engl. J. Med. 2011, 365, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Rogers, A.J.; Damask, A.; Raby, B.A.; Klanderman, B.J.; Duan, Q.L.; Tyagi, S.; Niu, S.; Anderson, C.; Cahir-Mcfarland, E.; et al. Pharmacogenomics: Novel loci identification via integrating gene differential analysis and eQTL analysis. Hum. Mol. Genet. 2014, 23, 5017–5024. [Google Scholar] [CrossRef]
- McGeachie, M.J.; Clemmer, G.L.; Hayete, B.; Xing, H.; Runge, K.; Wu, A.C.; Jiang, X.; Lu, Q.; Church, B.; Khalil, I.; et al. Systems biology and in vitro validation identifies family with sequence similarity 129 member A (FAM129A) as an asthma steroid response modulator. J. Allergy Clin. Immunol. 2018, 142, 1479–1488.e12. [Google Scholar] [CrossRef]
- Kachroo, P.; Sordillo, J.E.; Lutz, S.M.; Weiss, S.T.; Kelly, R.S.; McGeachie, M.J.; Wu, A.C.; Lasky-Su, J.A. Pharmaco-Metabolomics of Inhaled Corticosteroid Response in Individuals with Asthma. J. Pers. Med. 2021, 11, 1148. [Google Scholar] [CrossRef]
- Hunninghake, G.M.; Soto-Quiros, M.E.; Avila, L.; Ly, N.P.; Liang, C.; Sylvia, J.S.; Klanderman, B.J.; Silverman, E.K.; Celedón, J.C. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J. Allergy Clin. Immunol. 2007, 119, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, G.G.; DuHamel, T.R.; Wighton, T.G.; Chinn, T.; Bierman, C.W.; Furukawa, C.T.; Altman, L.C.; Virant, F.S.; Williams, P.V.; Minotti, D.A.; et al. The Childhood Asthma Management Program (CAMP): Design, Rationale, and Methods. Control. Clin. Trials 1999, 20, 91–120. [Google Scholar] [CrossRef]
- He, T.; Hildhood, C.; Sthma, A.; Anagement, M.; Rogram, P.; Esearch, R.; Roup, G. Long-term effects of budesonide or nedocromil in children with asthma. N. Engl. J. Med. 2000, 343, 1054–1063. [Google Scholar] [CrossRef]
- Tiwari, A.; Li, J.; Kho, A.T.; Sun, M.; Lu, Q.; Weiss, S.T.; Tantisira, K.G.; McGeachie, M.J. COPD-associated miR-145-5p is downregulated in early-decline FEV1 trajectories in childhood asthma. J. Allergy Clin. Immunol. 2021, 147, 2181–2190. [Google Scholar] [CrossRef]
- Rozowsky, J.; Kitchen, R.R.; Park, J.J.; Galeev, T.R.; Diao, J.; Warrell, J.; Thistlethwaite, W.; Subramanian, S.L.; Milosavljevic, A.; Gerstein, M. exceRpt: A Comprehensive Analytic Platform for Extracellular RNA Profiling. Cell Syst. 2019, 8, 352–357.e3. [Google Scholar] [CrossRef]
- Reese, S.E.; Archer, K.J.; Therneau, T.M.; Atkinson, E.J.; Vachon, C.M.; de Andrade, M.; Kocher, J.-P.A.; Eckel-Passow, J.E. A new statistic for identifying batch effects in high-throughput genomic data that uses guided principal component analysis. Bioinformatics 2013, 29, 2877–2883. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ru, Y.; Kechris, K.J.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.A.; Bowler, R.; Mahaffey, S.; Rossi, S.; Calin, G.A.; Bemis, L.; et al. The multiMiR R package and database: Integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.; Mächler, M.; Zurich, E.; Bolker, B.M.; Walker, S.C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Li, J.; Zhou, D.; Qiu, W.; Shi, Y.; Yang, J.-J.; Chen, S.; Wang, Q.; Pan, H. Application of Weighted Gene Co-expression Network Analysis for Data from Paired Design. Sci. Rep. 2018, 8, 622. [Google Scholar] [CrossRef]
- Koturbash, I.; Tolleson, W.H.; Guo, L.; Yu, D.; Chen, S.; Hong, H.; Mattes, W.; Ning, B. microRNAs as pharmacogenomic biomarkers for drug efficacy and drug safety assessment. Biomark. Med. 2015, 9, 1153–1176. [Google Scholar] [CrossRef]
- Tsitsiou, E.; Williams, A.E.; Moschos, S.A.; Patel, K.; Rossios, C.; Jiang, X.; Adams, O.-D.; Macedo, P.; Booton, R.; Gibeon, D.; et al. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J. Allergy Clin. Immunol. 2012, 129, 95–103. [Google Scholar] [CrossRef]
- Gurgone, D.; McShane, L.; McSharry, C.; Guzik, T.J.; Maffia, P. Cytokines at the Interplay Between Asthma and Atherosclerosis? Front. Pharmacol. 2020, 11, 166. [Google Scholar] [CrossRef] [PubMed]
- Kanagalingam, T.; Solomon, L.; Vijeyakumaran, M.; Palikhe, N.S.; Vliagoftis, H.; Cameron, L. IL-2 modulates Th2 cell responses to glucocorticosteroid: A cause of persistent type 2 inflammation? Immunity Inflamm. Dis. 2019, 7, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Cain, D.W.; Cidlowski, J.A. Immune regulation by glucocorticoids. Nat. Rev. Immunol. 2017, 17, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Faiz, A.; Steiling, K.; Roffel, M.P.; Postma, D.S.; Spira, A.; Lenburg, M.E.; Borggrewe, M.; Eijgenraam, T.R.; Jonker, M.R.; Koppelman, G.H.; et al. Effect of long-term corticosteroid treatment on microRNA and gene-expression profiles in COPD. Eur. Respir. J. 2019, 53, 1801202. [Google Scholar] [CrossRef] [PubMed]
- Cornell, L.; Wander, S.A.; Visal, T.; Wagle, N.; Shapiro, G.I. MicroRNA-Mediated Suppression of the TGF-β Pathway Confers Transmissible and Reversible CDK4/6 Inhibitor Resistance. Cell Rep. 2019, 26, 2667–2680.e7. [Google Scholar] [CrossRef]
- Leung, D.Y.; Martin, R.J.; Szefler, S.J.; Sher, E.R.; Ying, S.; Kay, A.B.; Hamid, Q. Dysregulation of interleukin 4, interleukin 5, and interferon gamma gene expression in steroid-resistant asthma. J. Exp. Med. 1995, 181, 33–40. [Google Scholar] [CrossRef]
- Peters, M.C.; Wenzel, S.E. Intersection of biology and therapeutics: Type 2 targeted therapeutics for adult asthma. Lancet 2020, 395, 371–383. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, J.S.; Kim, S.R.; Lee, Y.C. Defining Bronchial Asthma with Phosphoinositide 3-Kinase Delta Activation: Towards Endotype-Driven Management. Int. J. Mol. Sci. 2019, 20, 3525. [Google Scholar] [CrossRef]
- Bi, J.; Min, Z.; Yuan, H.; Jiang, Z.; Mao, R.; Zhu, T.; Liu, C.; Zeng, Y.; Song, J.; Du, C.; et al. PI3K inhibitor treatment ameliorates the glucocorticoid insensitivity of PBMCs in severe asthma. Clin. Transl. Med. 2020, 9, e22. [Google Scholar] [CrossRef]
- Zhang, Y.; Leung, D.Y.M.; Nordeen, S.K.; Goleva, E. Estrogen inhibits glucocorticoid action via protein phosphatase 5 (PP5)-mediated glucocorticoid receptor dephosphorylation. J. Biol. Chem. 2009, 284, 24542–24552. [Google Scholar] [CrossRef]
- Mittelstadt, P.R.; Ashwell, J.D. Inhibition of AP-1 by the glucocorticoid-inducible protein GILZ. J. Biol. Chem. 2001, 276, 29603–29610. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. Corticosteroid effects on cell signalling. Eur. Respir. J. 2006, 27, 413–426. [Google Scholar] [CrossRef]
- Hart, L.; Lim, S.; Adcock, I.; Barnes, P.J.; Chung, K.F. Effects of inhaled corticosteroid therapy on expression and DNA-binding activity of nuclear factor kappaB in asthma. Am. J. Respir. Crit. Care Med. 2000, 161, 224–231. [Google Scholar] [CrossRef]
- Zhang, Y.; Jing, Y.; Qiao, J.; Luan, B.; Wang, X.; Wang, L.; Song, Z. Activation of the mTOR signaling pathway is required for asthma onset. Sci. Rep. 2017, 7, 4532. [Google Scholar] [CrossRef] [PubMed]
- Mitani, A.; Ito, K.; Vuppusetty, C.; Barnes, P.J.; Mercado, N. Restoration of Corticosteroid Sensitivity in Chronic Obstructive Pulmonary Disease by Inhibition of Mammalian Target of Rapamycin. Am. J. Respir. Crit. Care Med. 2016, 193, 143–153. [Google Scholar] [CrossRef]
- Barnes, P.J.; Adcock, I.M.; Ito, K. Histone acetylation and deacetylation: Importance in inflammatory lung diseases. Eur. Respir. J. 2005, 25, 552–563. [Google Scholar] [CrossRef]
- Gunawardhana, L.P.; Gibson, P.G.; Simpson, J.L.; Powell, H.; Baines, K.J. Activity and expression of histone acetylases and deacetylases in inflammatory phenotypes of asthma. Clin. Exp. Allergy 2014, 44, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Reddel, H.K.; Bacharier, L.B.; Bateman, E.D.; Brightling, C.E.; Brusselle, G.G.; Buhl, R.; Cruz, A.A.; Duijts, L.; Drazen, J.M.; FitzGerald, J.M.; et al. Global Initiative for Asthma Strategy 2021: Executive Summary and Rationale for Key Changes. Am. J. Respir. Crit. Care Med. 2022, 205, 17–35. [Google Scholar] [CrossRef]
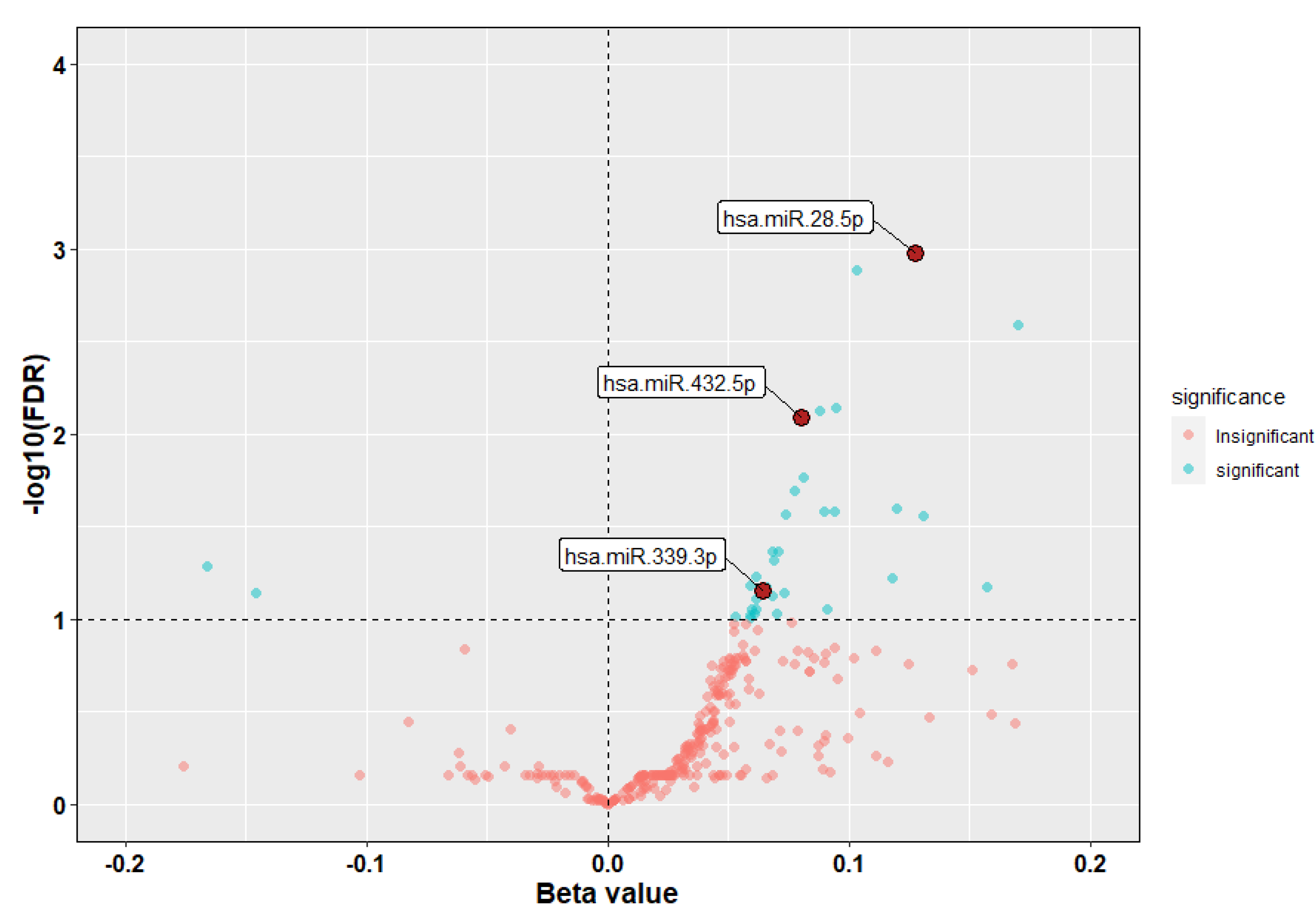
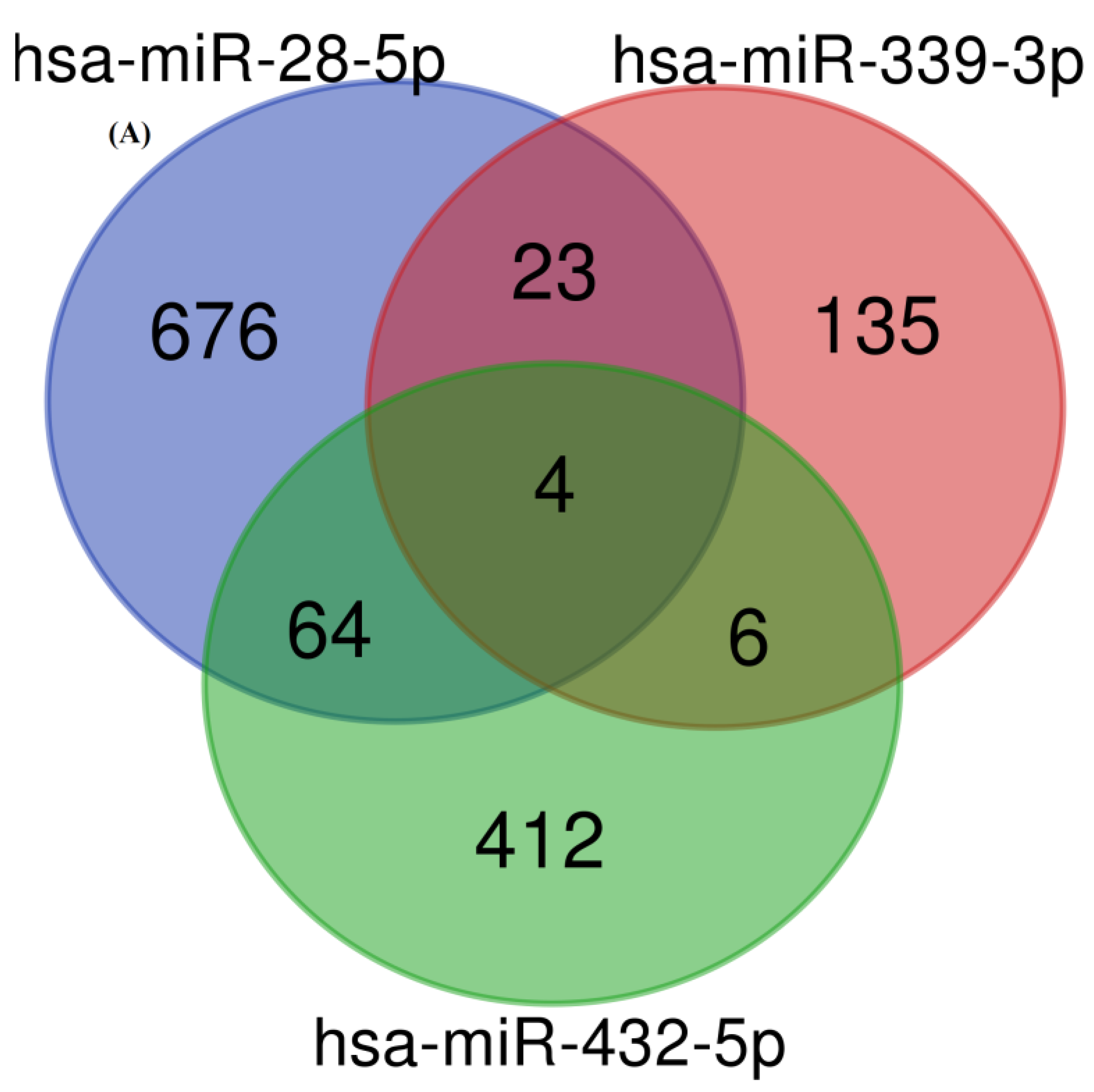
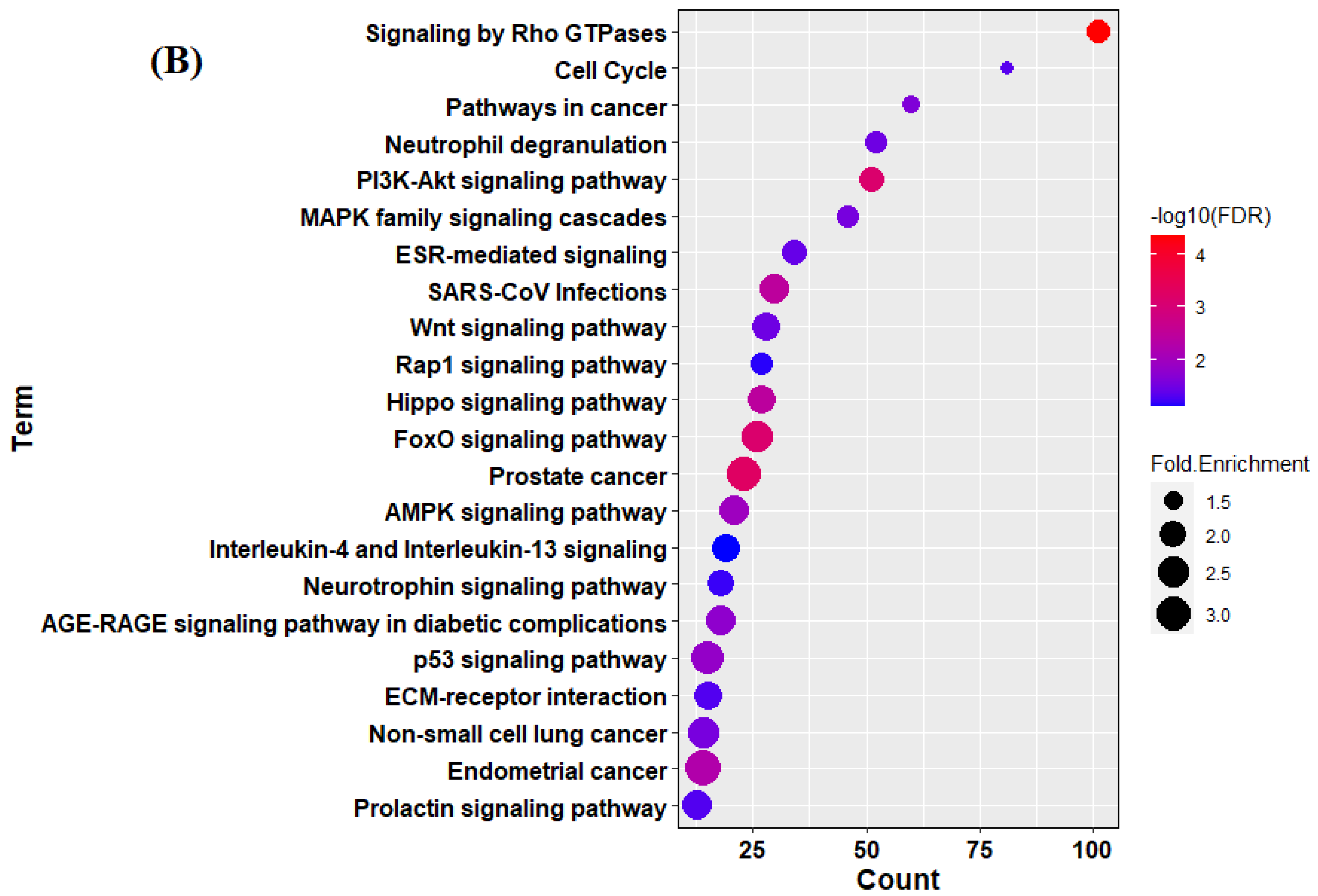
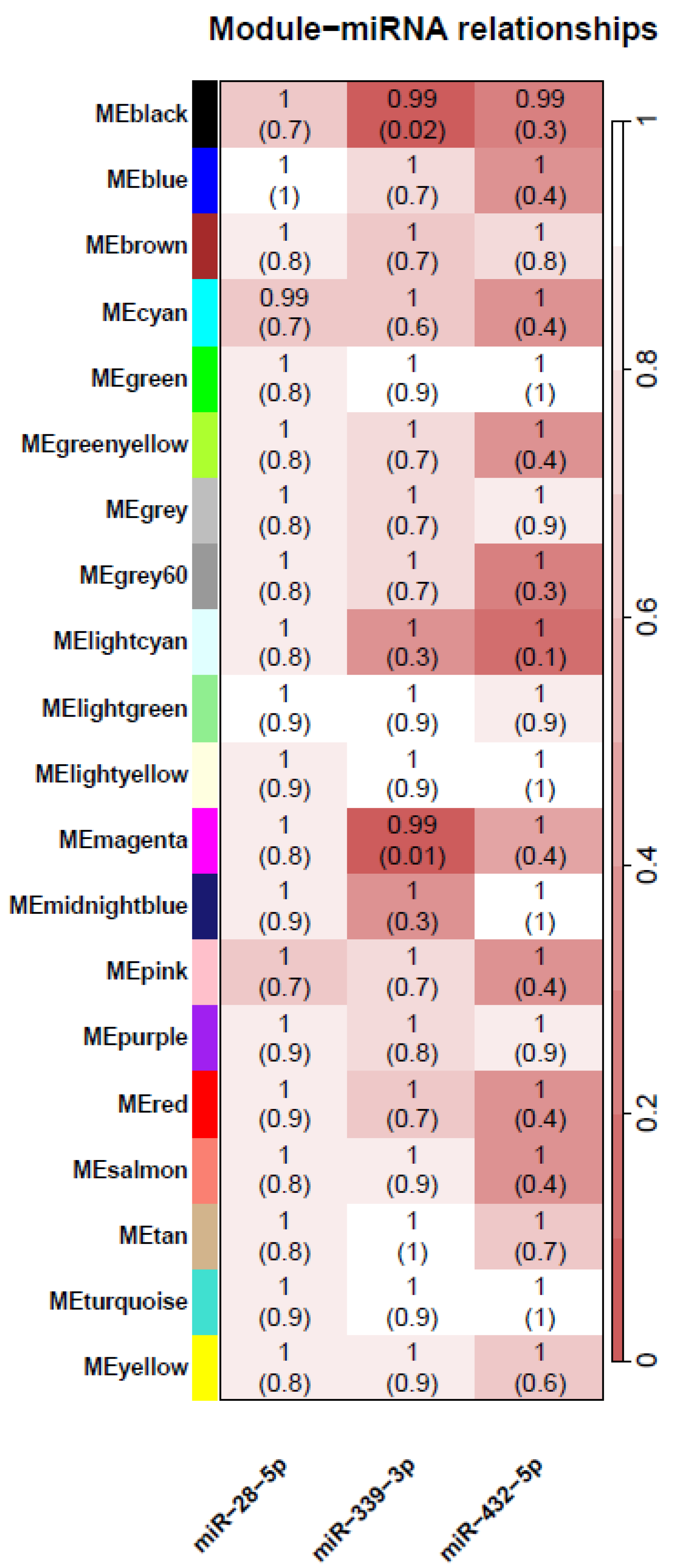
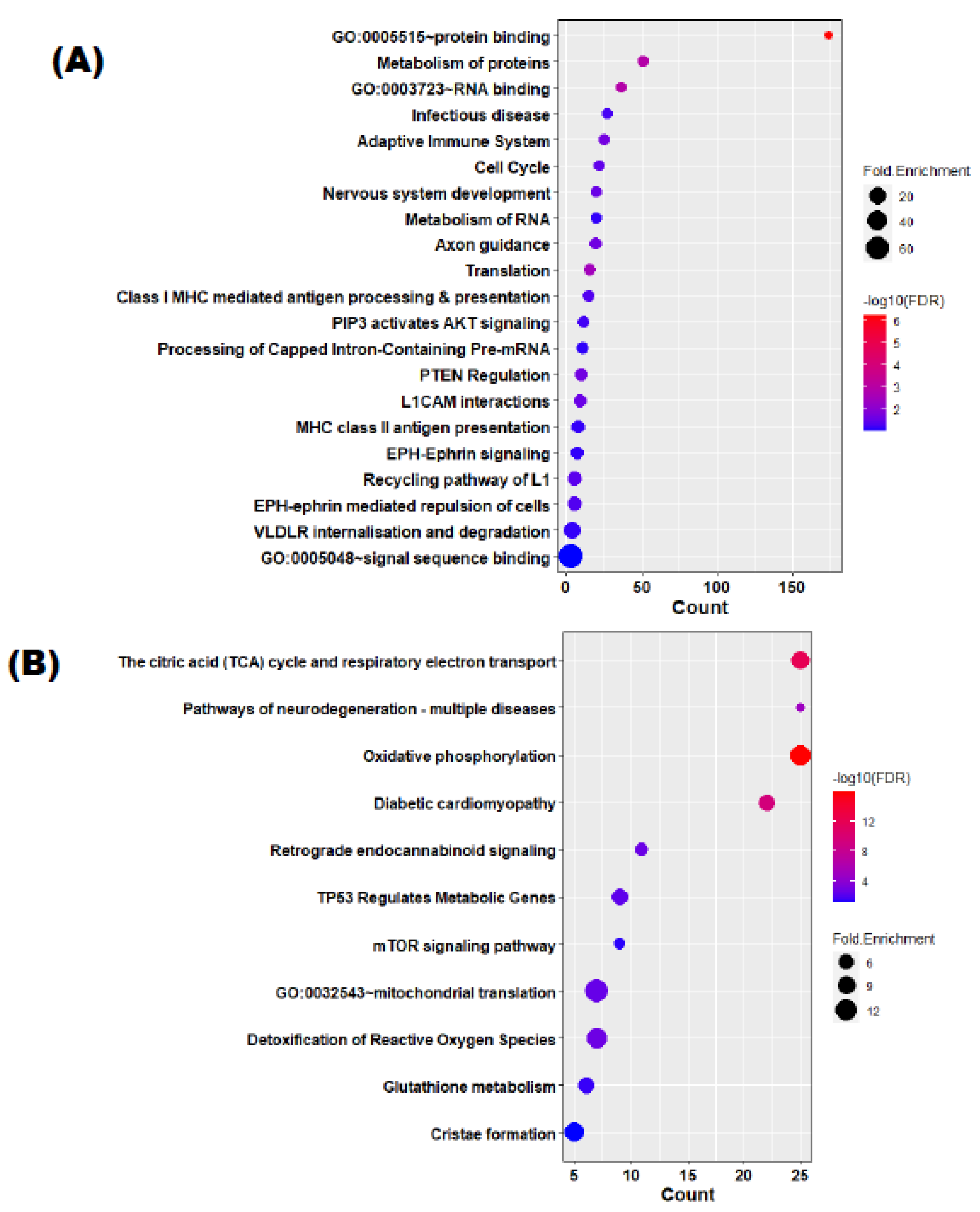
| (A) | ||||||
| Logistic Regression GACRS (Discovery Cohort) | ||||||
| Variable | Beta | Z | p Value | p Adjust | OR | OR (95% CI) |
| hsa-miR-339-3p | 0.064 | 2.238 | 0.025 | 0.070 | 1.066 | 1.008–1.128 |
| hsa-miR-28-5p | 0.128 | 3.581 | 0.000 | 0.001 | 1.136 | 1.059–1.218 |
| hsa-miR-432-5p | 0.080 | 2.988 | 0.003 | 0.008 | 1.084 | 1.028–1.142 |
| (B) | ||||||
| Logistic Regression CAMP (Replication Cohort) | ||||||
| Variable | Beta | Z | p Value | OR | OR (95% CI) | |
| hsa-miR-339-3p | 0.199 | 2.620 | 0.009 | 1.221 | 1.052–1.417 | |
| hsa-miR-28-5p | 0.172 | 2.408 | 0.016 | 1.188 | 1.033–1.366 | |
| hsa-miR-432-5p | 0.128 | 1.898 | 0.058 | 1.137 | 0.996–1.298 | |
| microRNAs Name | Number of Target Genes |
|---|---|
| hsa-miR-28-5p | 767 |
| hsa-miR-339-3p | 168 |
| hsa-miR-432-5p | 486 |
| Overall number of unique target genes | 1320 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, R.; Tiwari, A.; Kho, A.T.; Celedón, J.C.; Weiss, S.T.; Tantisira, K.G.; McGeachie, M.J. Systems Genomics Reveals microRNA Regulation of ICS Response in Childhood Asthma. Cells 2023, 12, 1505. https://doi.org/10.3390/cells12111505
Sharma R, Tiwari A, Kho AT, Celedón JC, Weiss ST, Tantisira KG, McGeachie MJ. Systems Genomics Reveals microRNA Regulation of ICS Response in Childhood Asthma. Cells. 2023; 12(11):1505. https://doi.org/10.3390/cells12111505
Chicago/Turabian StyleSharma, Rinku, Anshul Tiwari, Alvin T. Kho, Juan C. Celedón, Scott T. Weiss, Kelan G. Tantisira, and Michael J. McGeachie. 2023. "Systems Genomics Reveals microRNA Regulation of ICS Response in Childhood Asthma" Cells 12, no. 11: 1505. https://doi.org/10.3390/cells12111505
APA StyleSharma, R., Tiwari, A., Kho, A. T., Celedón, J. C., Weiss, S. T., Tantisira, K. G., & McGeachie, M. J. (2023). Systems Genomics Reveals microRNA Regulation of ICS Response in Childhood Asthma. Cells, 12(11), 1505. https://doi.org/10.3390/cells12111505