Lactiplantibacillus plantarum Lac16 Attenuates Enterohemorrhagic Escherichia coli O157:H7 Infection by Inhibiting Virulence Traits and Improving Intestinal Epithelial Barrier Function
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria
2.2. Antimicrobial Activity
2.3. Aggregation Assay
2.4. Simulated Gastrointestinal Juices Resistance Assay
2.5. Bile Tolerance Assay
2.6. Lactic Acid Production
2.7. Biofilm Formation
2.8. Gene Expression of EHEC
2.9. Caenorhabditis Elegans Experiment
2.10. IPEC-J2 Cell Culture
2.11. Wound-Healing Assay
2.12. CCK-8 Assay and Cell Apoptosis Detection
2.13. LDH and NO Release Analysis, qPCR Assay
2.14. Western Blotting Analysis
2.15. Statistical Analysis
3. Results
3.1. Aggregation Evaluation of Lactic Acid Bacteria
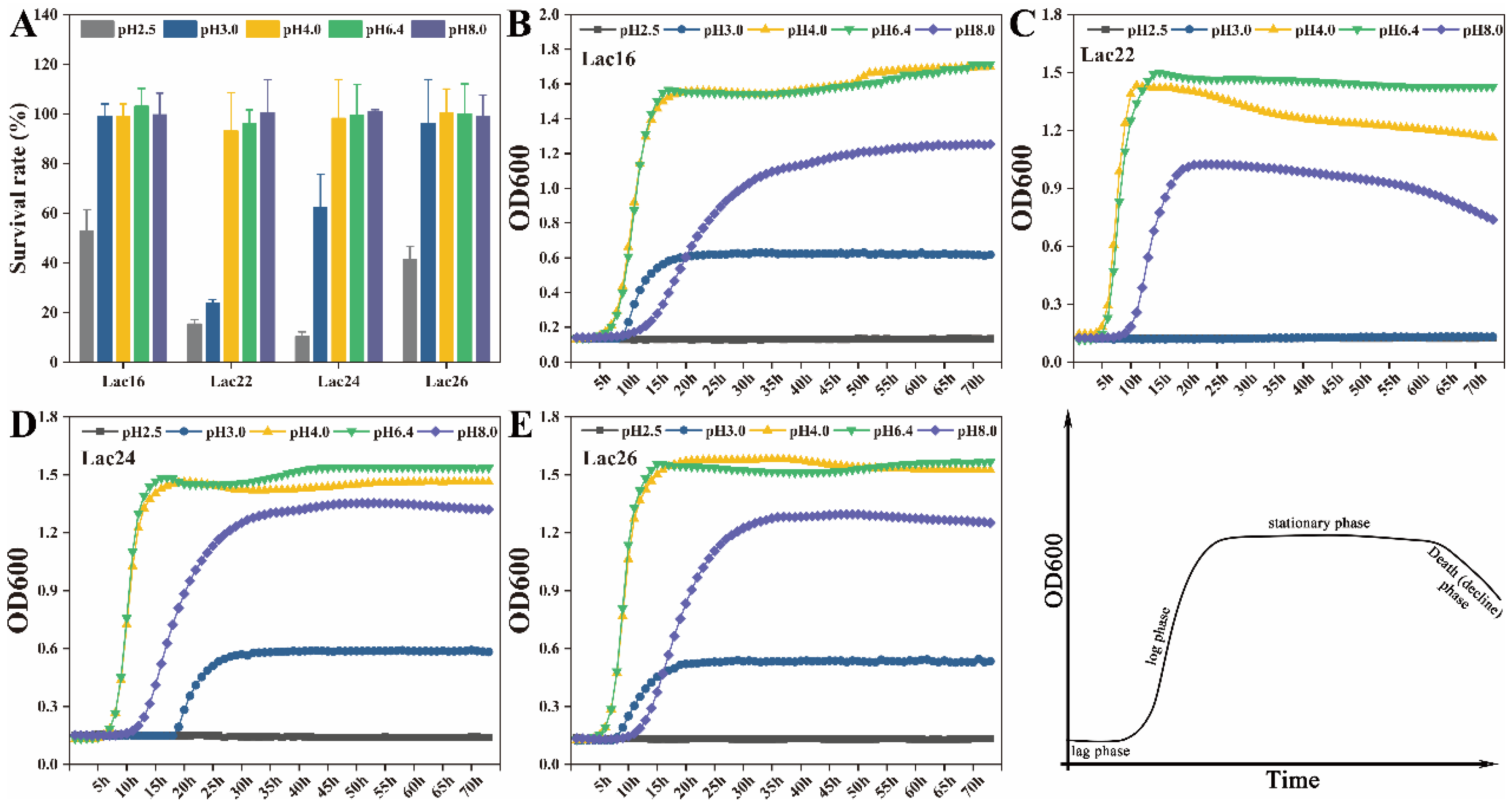
3.2. Tolerance of Lactic Acid Bacteria to Simulated Gastrointestinal Juices and Bile Salts
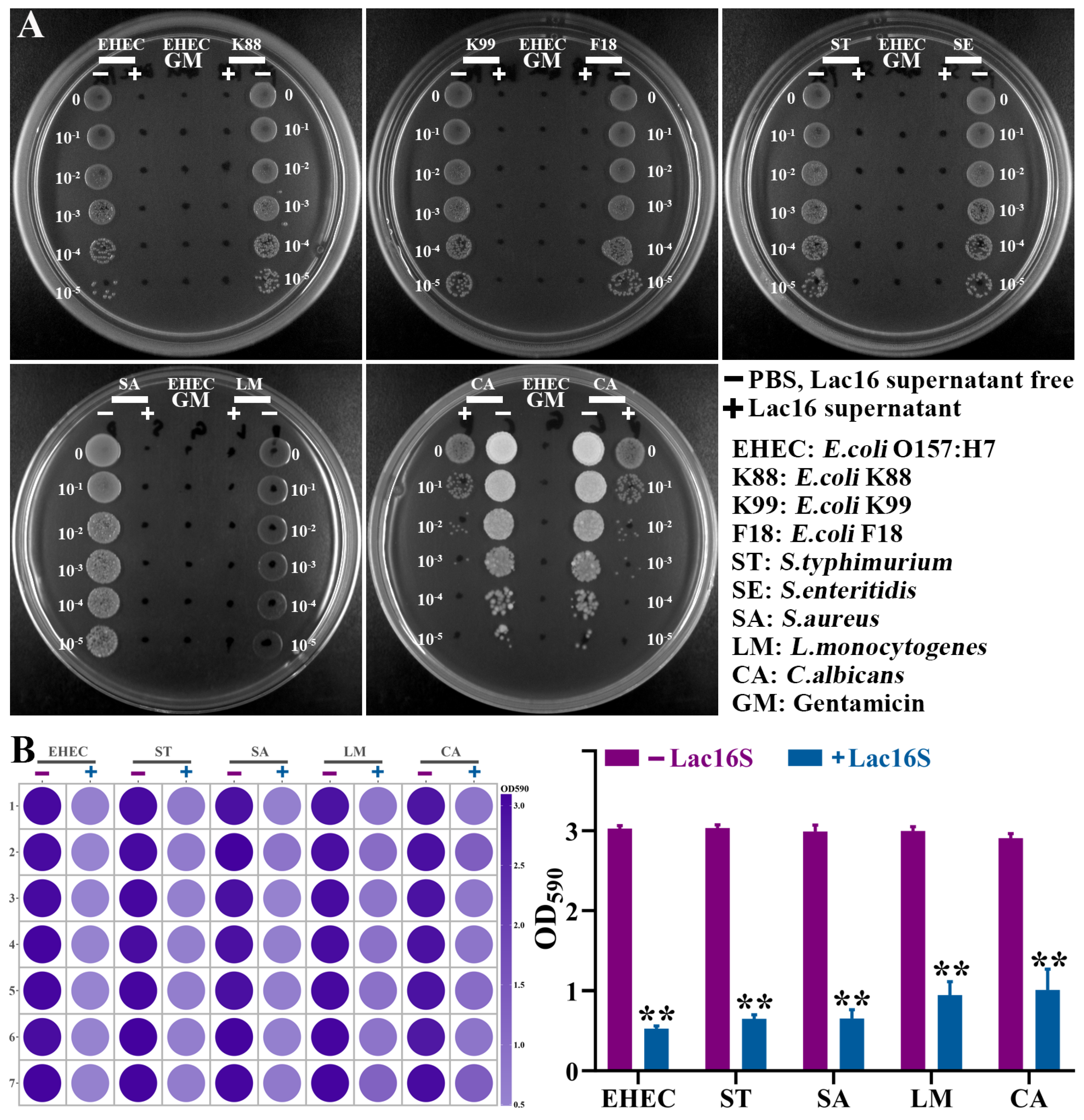
3.3. Antimicrobial Activity of Lactic Acid Bacteria
3.4. Lactic Acid Production of Lactic Acid Bacteria
3.5. Lac16 Inhibited Growth and Biofilm Formation of Zoonotic Pathogens
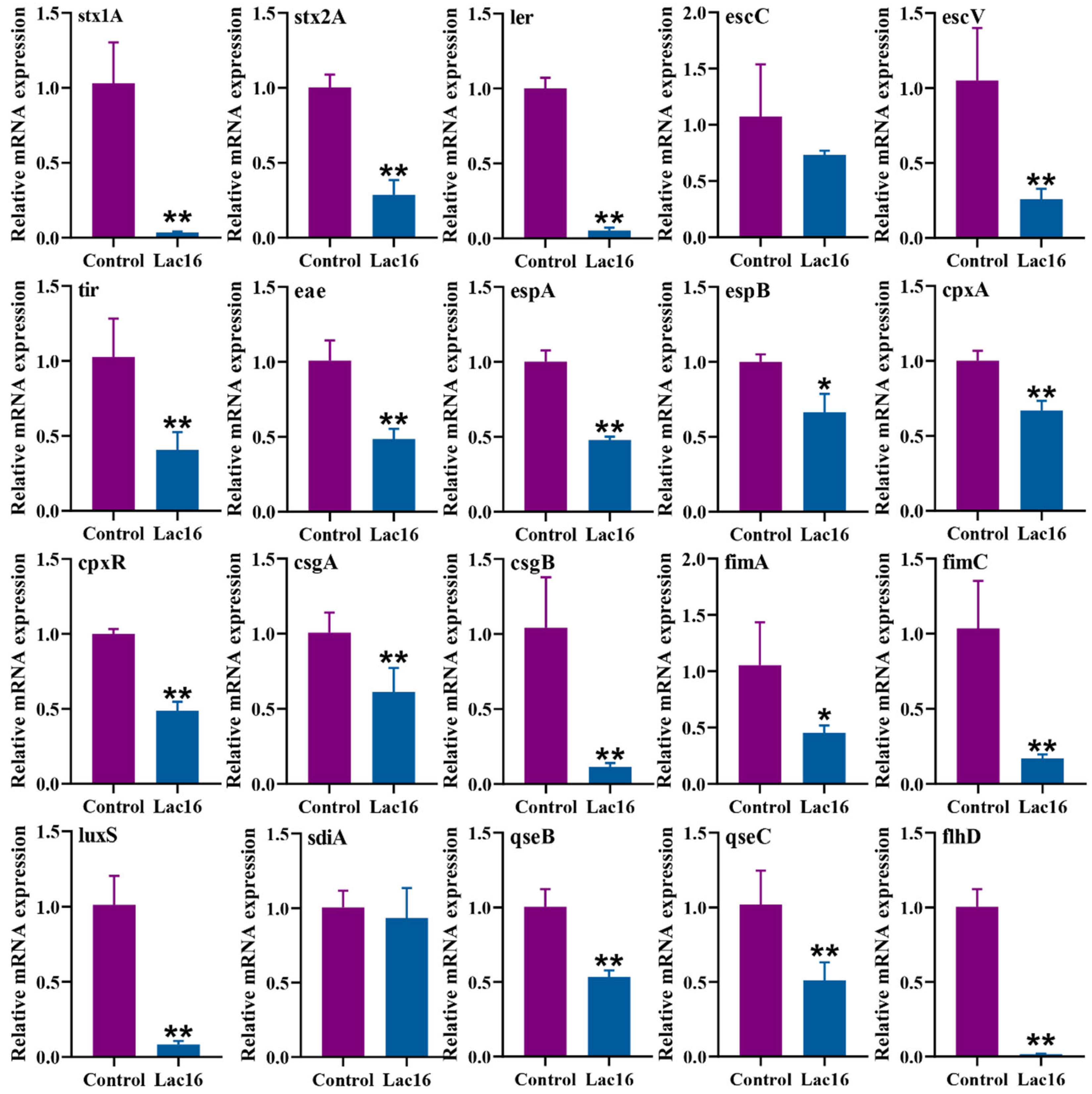
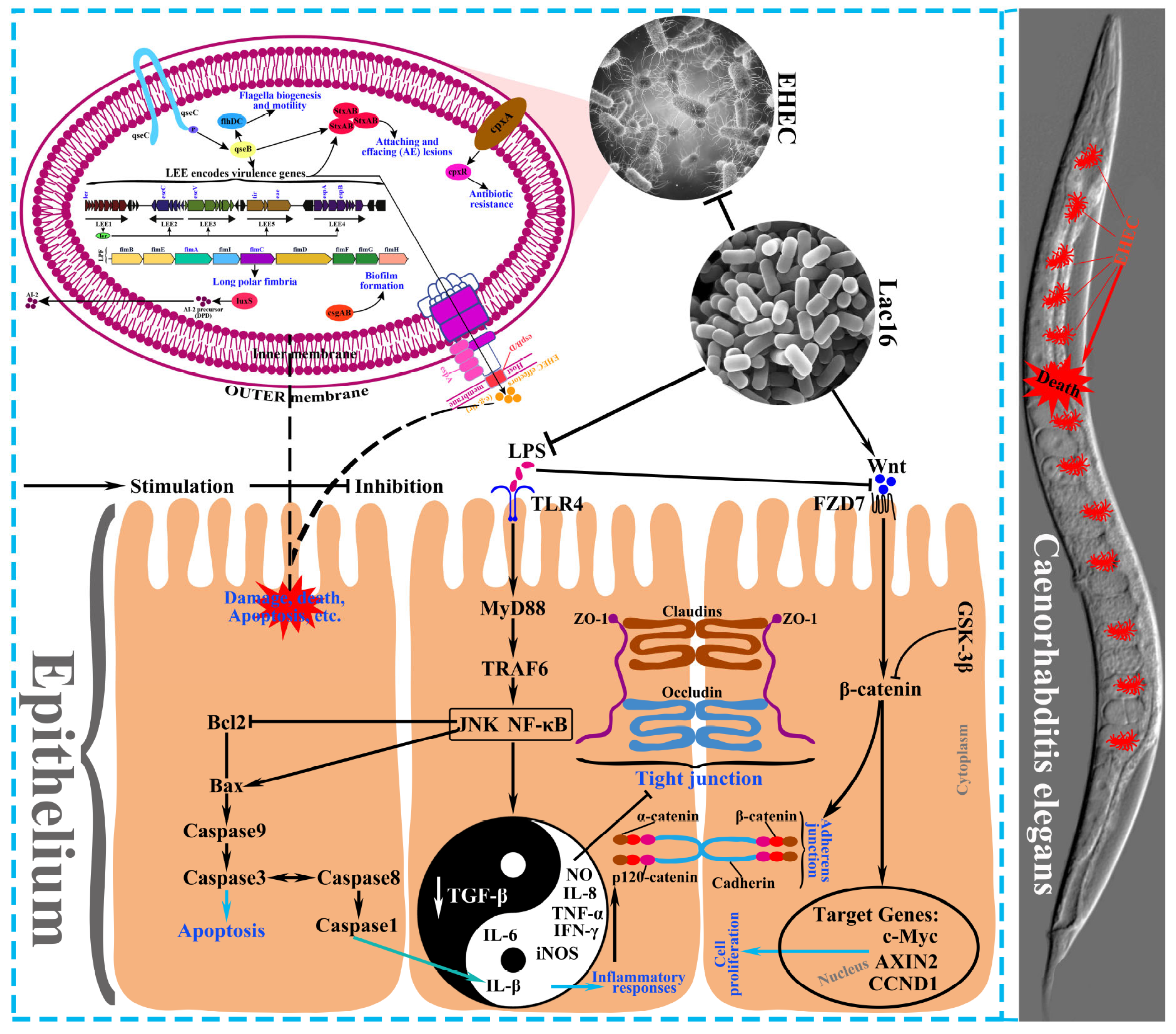
3.6. Lac16 Inhibited the Gene Expression of Key Virulence Traits of EHEC
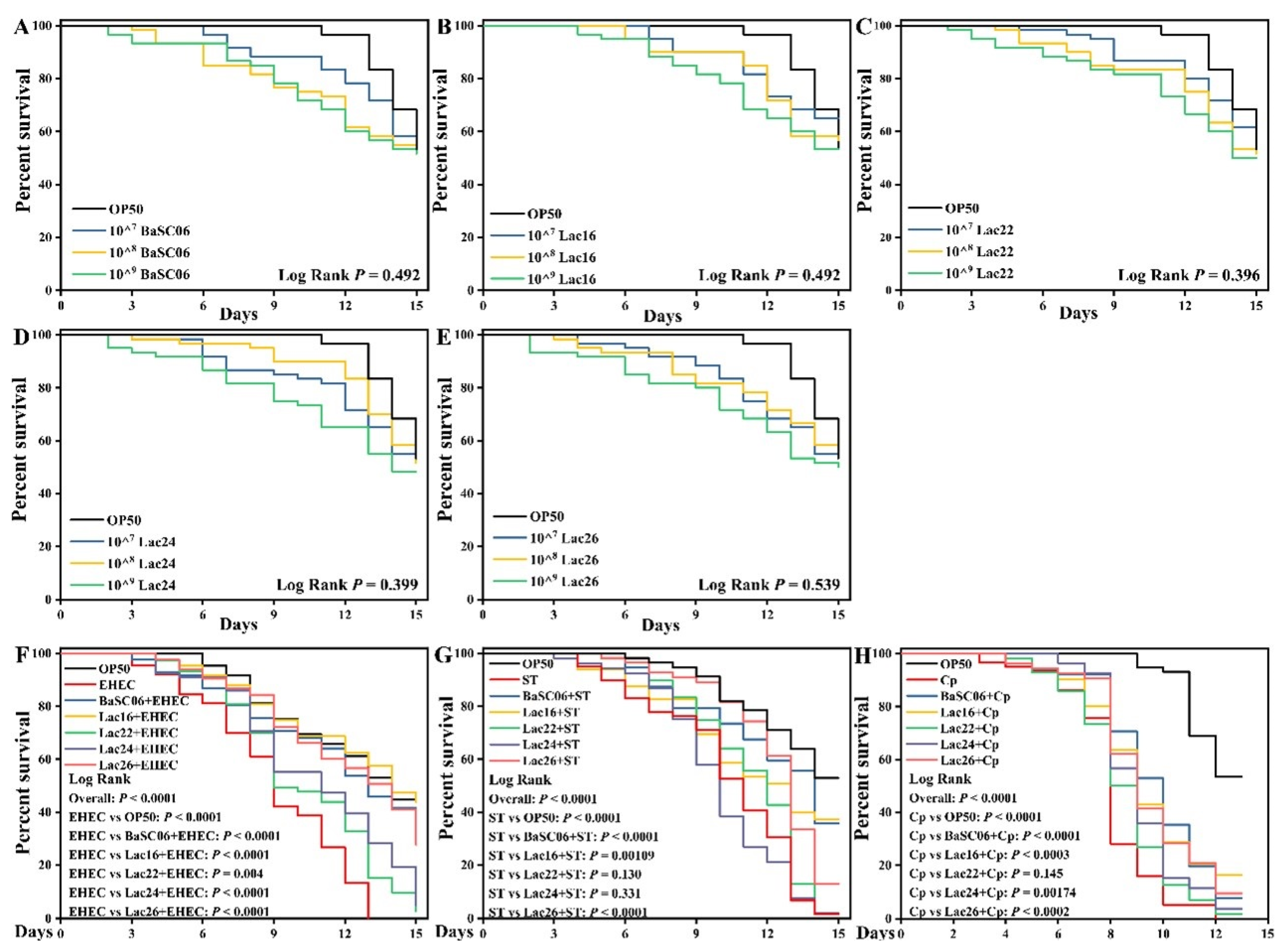
3.7. Lac16 Protected C. elegans against Pathogen Infection
3.8. Lac16 Promoted the Wound Closure of Intestinal Epithelial Cells
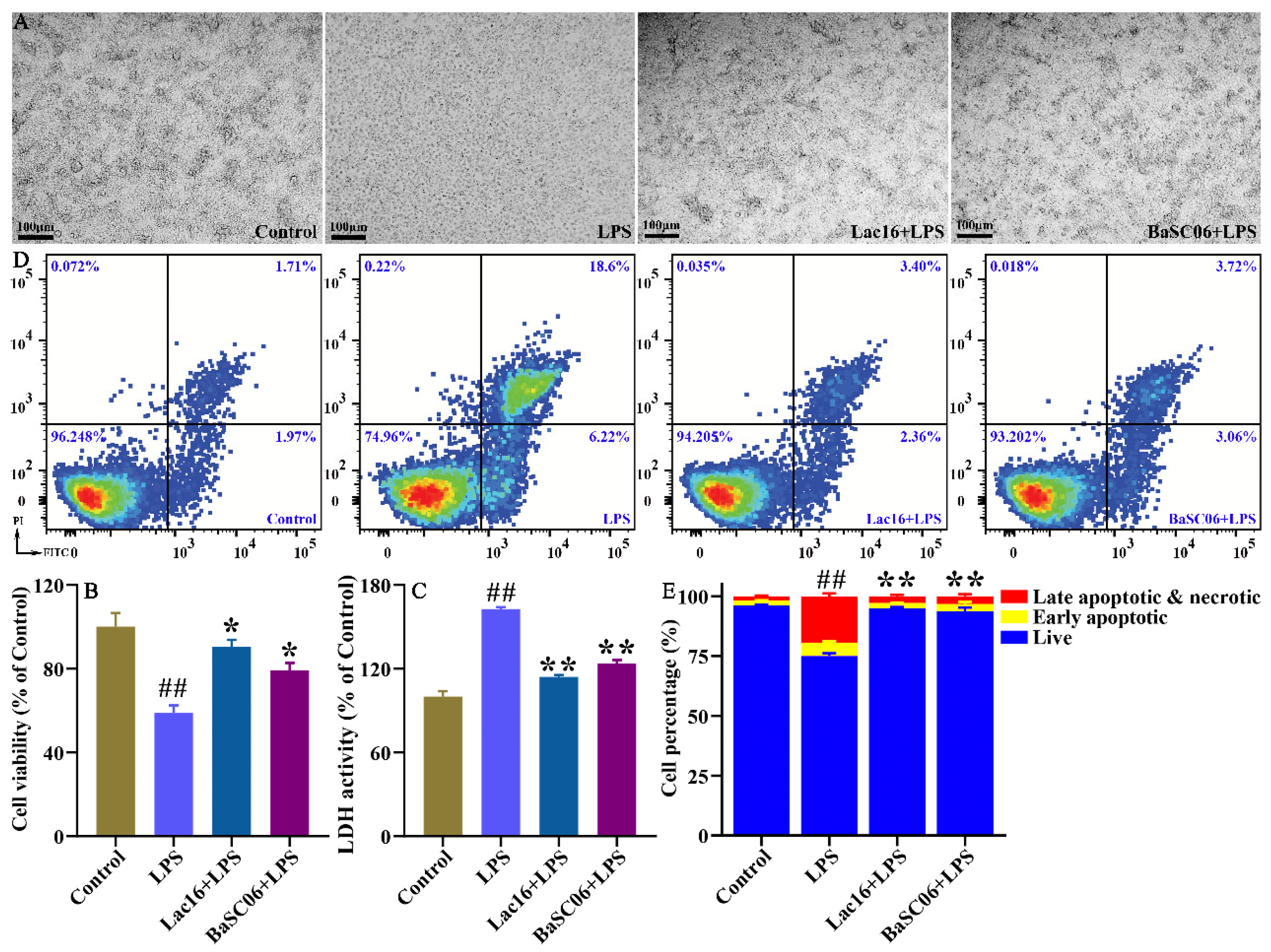
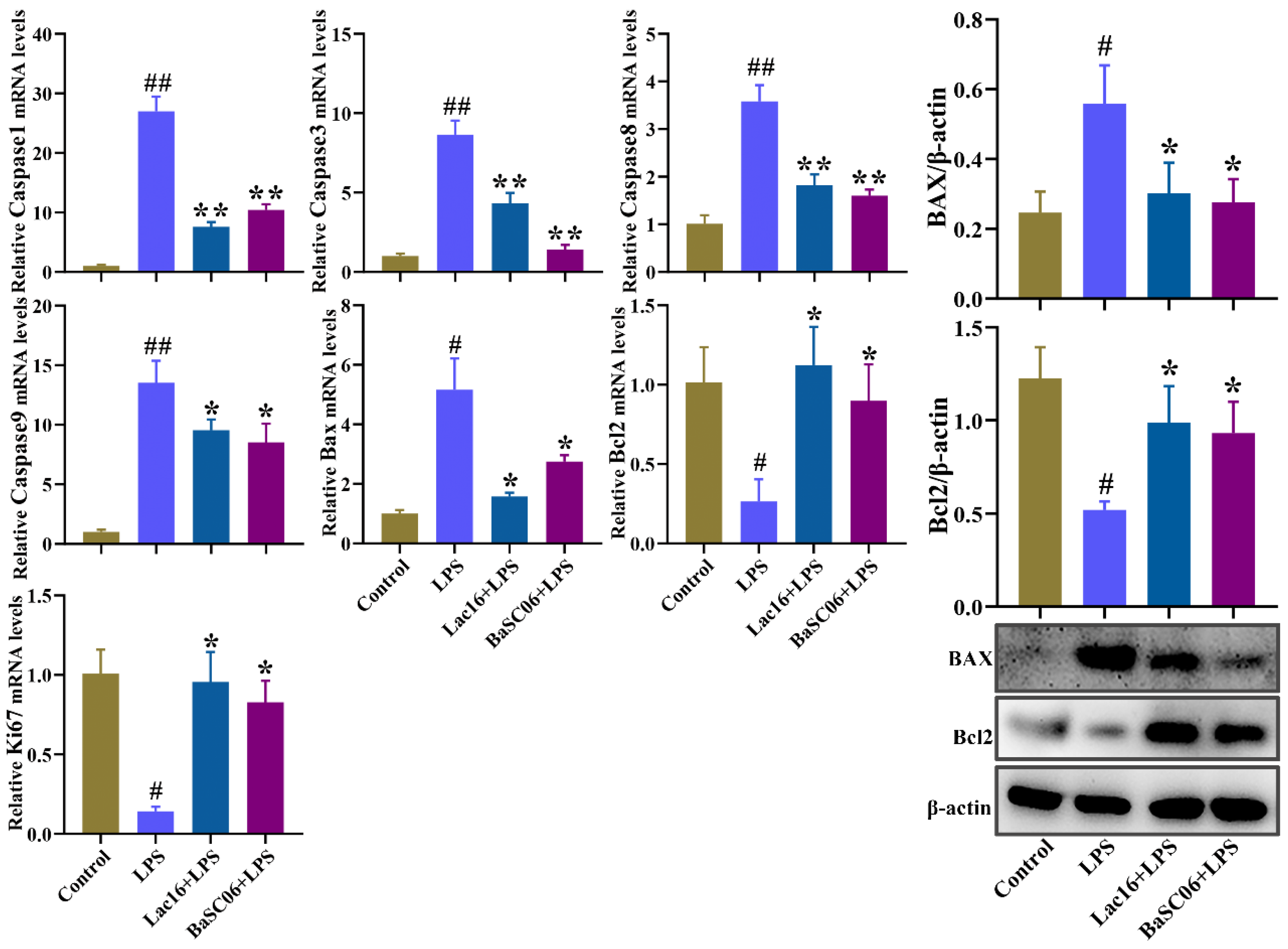
3.9. Lac16 Ameliorated LPS-Induced Apoptosis of Intestinal Epithelial Cells
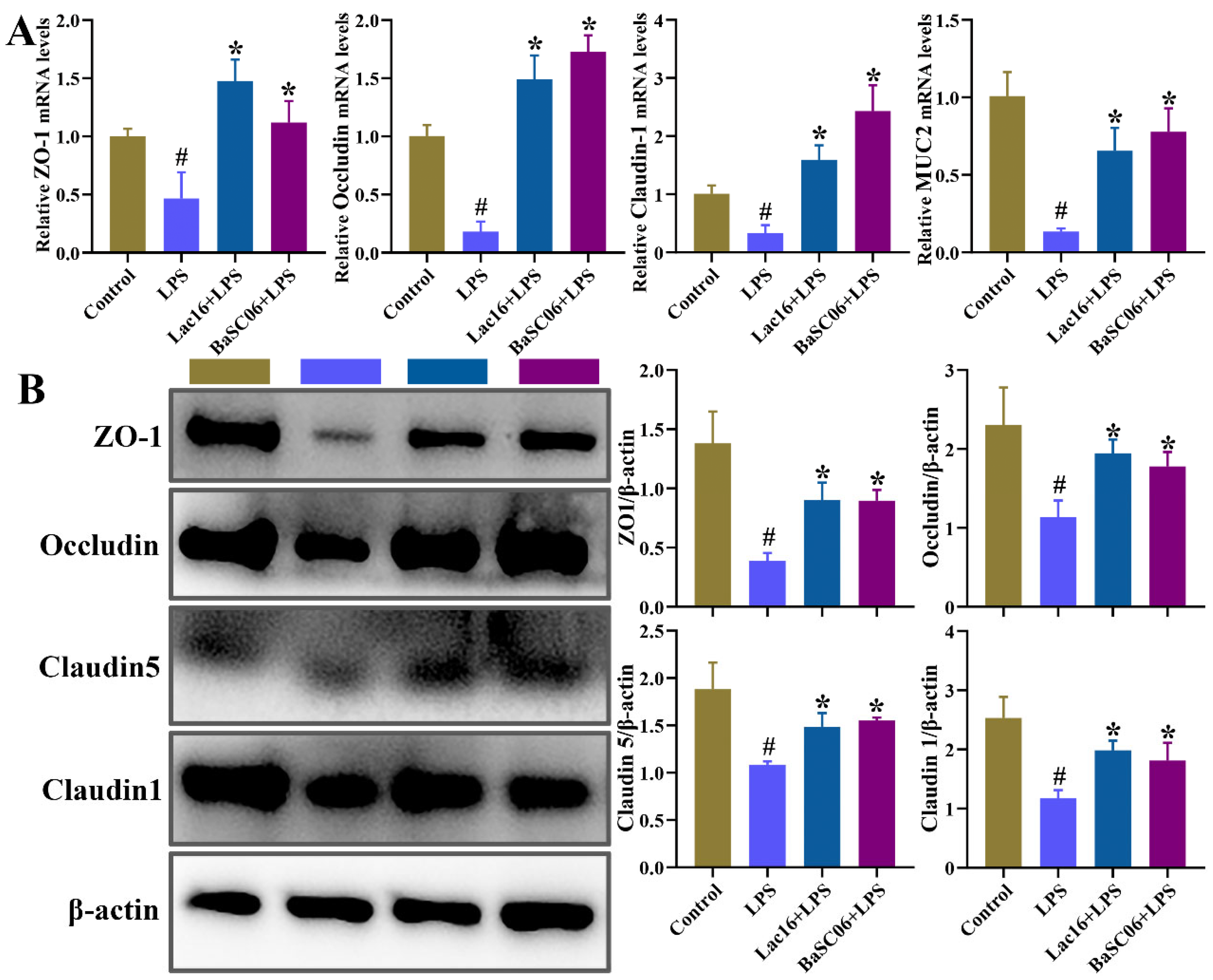
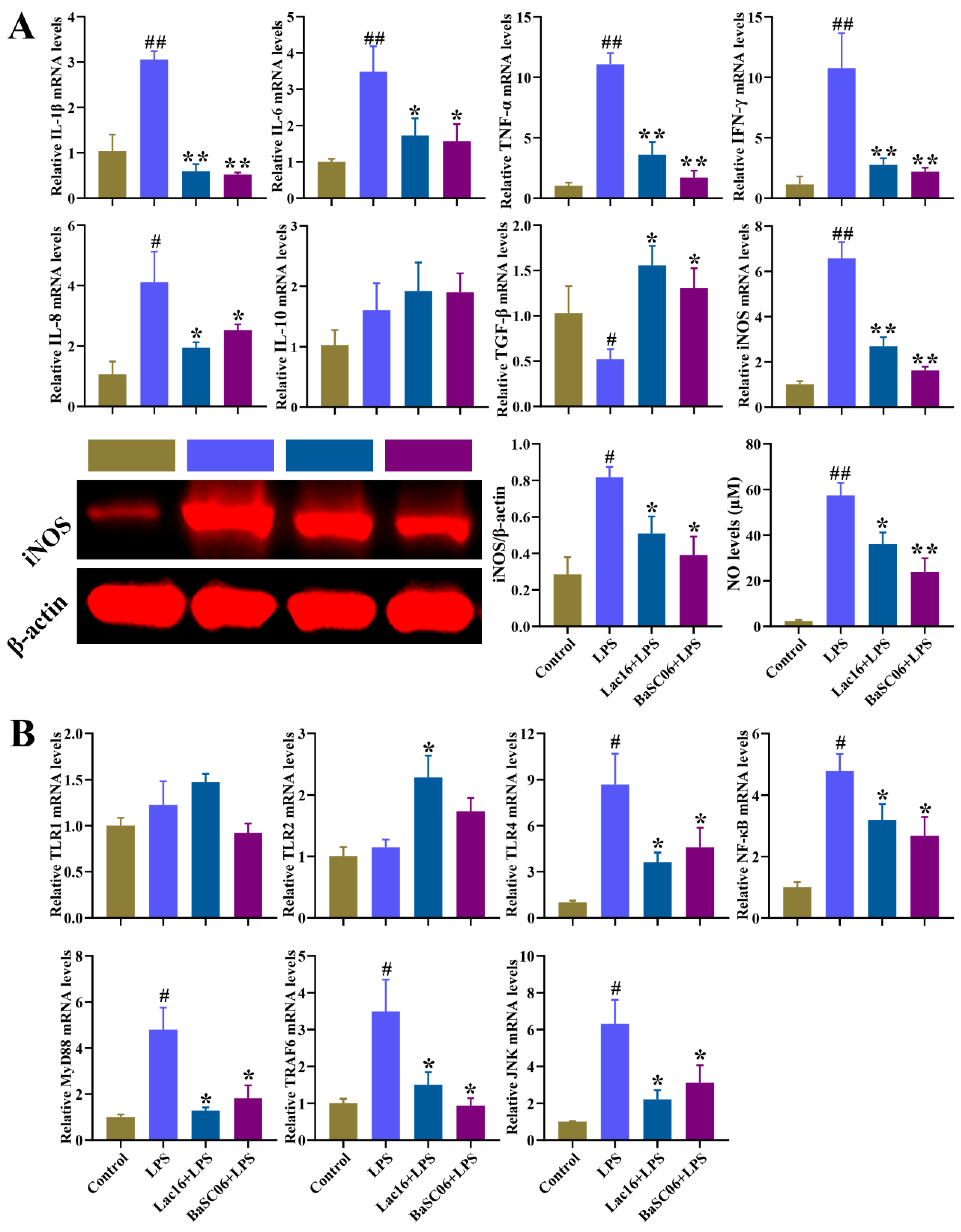
3.10. Lac16 Ameliorated LPS-Induced Intestinal Epithelial Barrier Dysfunction
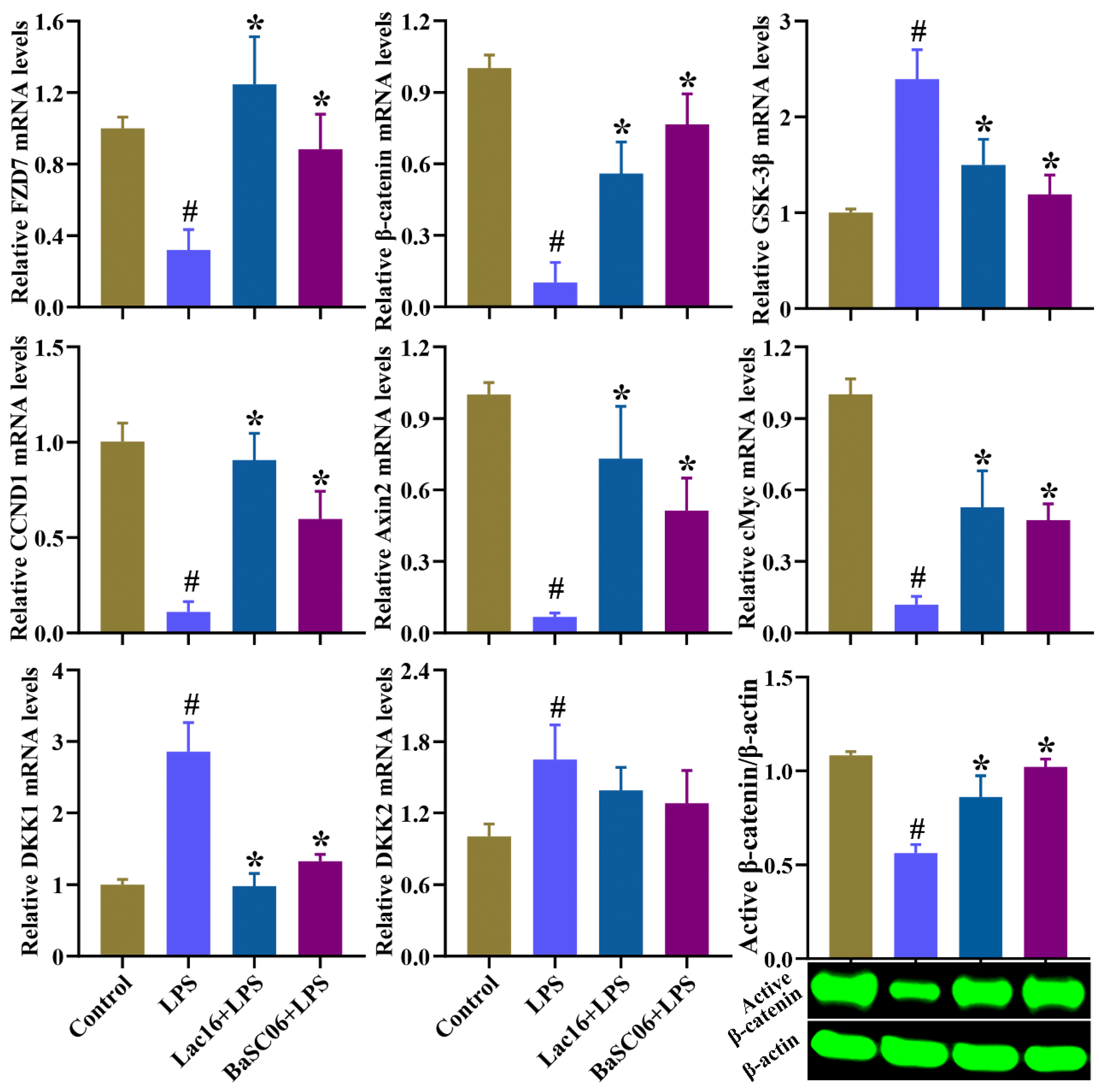
3.11. Lac16 Activated LPS-Induced Inhibition of Wnt/β-Catenin Signaling Pathway of Intestinal Epithelial Cells
3.12. Lac16 Inhibited LPS-Induced Inflammatory Responses of Intestinal Epithelial Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bird, B.H.; Mazet, J.A.K. Detection of Emerging Zoonotic Pathogens: An Integrated One Health Approach. Annu. Rev. Anim. Biosci. 2018, 6, 121–139. [Google Scholar] [CrossRef] [PubMed]
- Atluri, V.L.; Xavier, M.N.; Jong, M.F.D.; den Hartigh, A.B.; Tsolis, R.M. Interactions of the human pathogenic Brucella species with their hosts. Annu. Rev. Microbiol. 2011, 65, 523–541. [Google Scholar] [CrossRef] [PubMed]
- Talukder, B.; Ganguli, N. Climate change-related foodborne zoonotic diseases and pathogens modeling. J. Clim. Chang. Health 2022, 6, 100111. [Google Scholar] [CrossRef]
- Keesing, F.; Ostfeld, R.S. Impacts of biodiversity and biodiversity loss on zoonotic diseases. Proc. Natl. Acad. Sci. USA 2021, 118, e2023540118. [Google Scholar] [CrossRef]
- Marshall, B.M.; Levy, S.B. Food Animals and Antimicrobials: Impacts on Human Health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef]
- Wang, B.; Gong, L.; Zhou, Y.; Tang, L.; Zeng, Z.; Wang, Q.; Zou, P.; Yu, D.; Li, W. Probiotic Paenibacillus polymyxa 10 and Lactobacillus plantarum 16 enhance growth performance of broilers by improving the intestinal health. Anim. Nutr. 2021, 7, 829–840. [Google Scholar] [CrossRef]
- Innes, G.K.; Randad, P.R.; Korinek, A.; Davis, M.F.; Price, L.B.; So, A.D.; Heaney, C.D. External Societal Costs of Antimicrobial Resistance in Humans Attributable to Antimicrobial Use in Livestock. Annu. Rev. Public Health 2020, 41, 141–157. [Google Scholar] [CrossRef]
- Allen, H.K.; Stanton, T.B. Altered egos: Antibiotic effects on food animal microbiomes. Annu. Rev. Microbiol. 2014, 68, 297–315. [Google Scholar] [CrossRef]
- Kelly, T.R.; Machalaba, C.; Karesh, W.B.; Crook, P.Z.; Gilardi, K.; Nziza, J.; Uhart, M.M.; Robles, E.A.; Saylors, K.; Joly, D.O.; et al. Implementing One Health approaches to confront emerging and re-emerging zoonotic disease threats: Lessons from PREDICT. One Health Outlook 2020, 2, 1. [Google Scholar] [CrossRef]
- Noyes, N.R.; Slizovskiy, I.B.; Singer, R.S. Beyond Antimicrobial Use: A Framework for Prioritizing Antimicrobial Resistance Interventions. Annu. Rev. Anim. Biosci. 2021, 9, 313–332. [Google Scholar] [CrossRef]
- Teillant, A.; Brower, C.H.; Laxminarayan, R. Economics of Antibiotic Growth Promoters in Livestock. Annu. Rev. Resour. Econ. 2015, 7, 349–374. [Google Scholar] [CrossRef]
- Buntyn, J.O.; Schmidt, T.B.; Nisbet, D.J.; Callaway, T. The Role of Direct-Fed Microbials in Conventional Livestock Production. Annu. Rev. Anim. Biosci. 2016, 4, 335–355. [Google Scholar] [CrossRef] [PubMed]
- Odenwald, M.A.; Turner, J.R. The intestinal epithelial barrier: A therapeutic target? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Marchiando, A.M.; Graham, W.V.; Turner, J.R. Epithelial barriers in homeostasis and disease. Annu. Rev. Pathol. 2010, 5, 119–144. [Google Scholar] [CrossRef]
- Kayama, H.; Okumura, R.; Takeda, K. Interaction Between the Microbiota, Epithelia, and Immune Cells in the Intestine. Annu. Rev. Immunol. 2020, 38, 23–48. [Google Scholar] [CrossRef]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Turner, J.R. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009, 9, 799–809. [Google Scholar] [CrossRef]
- Bhunia, A.K. Foodborne Microbial Pathogens: Mechanisms and Pathogenesis; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Rogers, A.P.; Mileto, S.J.; Lyras, D. Impact of enteric bacterial infections at and beyond the epithelial barrier. Nat. Rev. Microbiol. 2022, 21, 260–274. [Google Scholar] [CrossRef]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Núñez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef]
- Nobile, C.J.; Johnson, A.D. Candida albicans Biofilms and Human Disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef]
- Wang, B.; Ye, X.; Zhou, Y.; Zhao, P.; Mao, Y. Glycyrrhizin Attenuates Salmonella Typhimurium-Induced Tissue Injury, Inflammatory Response, and Intestinal Dysbiosis in C57BL/6 Mice. Front. Vet. Sci. 2021, 8, 648698. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, B.; Delgado, S.; Blanco-Miguez, A.; Lourenço, A.; Gueimonde, M.; Margolles, A. Probiotics, gut microbiota, and their influence on host health and disease. Mol. Nutr. Food Res. 2017, 61, 1600240. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.L.Y.; Forsythe, S.J.; El-Nezami, H. Probiotics interaction with foodborne pathogens: A potential alternative to antibiotics and future challenges. Crit. Rev. Food Sci. Nutr. 2019, 59, 3320–3333. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef]
- Li, Y.; Liu, T.; Zhao, M.; Zhong, H.; Luo, W.; Feng, F. In vitro and in vivo investigations of probiotic properties of lactic acid bacteria isolated from Chinese traditional sourdough. Appl. Microbiol. Biotechnol. 2019, 103, 1893–1903. [Google Scholar] [CrossRef]
- Hu, P.L.; Yuan, Y.H.; Yue, T.L.; Guo, C.-F. A new method for the in vitro determination of the bile tolerance of potentially probiotic lactobacilli. Appl. Microbiol. Biot. 2018, 102, 1903–1910. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Girard, L.R.; Fiedler, T.J.; Harris, T.W.; Carvalho, F.; Antoshechkin, I.; Han, M.; Sternberg, P.W.; Stein, L.D.; Chalfie, M. WormBook: The online review of Caenorhabditis elegans biology. Nucleic Acids Res. 2007, 35, D472–D475. [Google Scholar] [CrossRef]
- Rangan, K.J.; Pedicord, V.A.; Wang, Y.C.; Kim, B.; Lu, Y.; Shaham, S.; Mucida, D.; Hang, H.C. A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science 2016, 353, 1434–1437. [Google Scholar] [CrossRef]
- Xiao, Z.; Liu, L.; Tao, W.; Pei, X.; Wang, G.; Wang, M. Clostridium Tyrobutyricum Protect Intestinal Barrier Function from LPS-Induced Apoptosis via P38/JNK Signaling Pathway in IPEC-J2 Cells. Cell. Physiol. Biochem. 2018, 46, 1779–1792. [Google Scholar] [CrossRef]
- Geens, M.M.; Niewold, T.A. Preliminary Characterization of the Transcriptional Response of the Porcine Intestinal Cell Line IPEC-J2 to Enterotoxigenic Escherichia coli, Escherichia coli, and E. coli Lipopolysaccharide. Comp. Funct. Genomics 2010, 2010, 469583. [Google Scholar] [CrossRef] [PubMed]
- Kunyeit, L.; Kurrey, N.K.; Anu-Appaiah, K.A.; Rao, R.P. Probiotic Yeasts Inhibit Virulence of Non-albicans Candida Species. mBio 2019, 10, e02307-19. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.P.; Wang, Y.; Zou, H.; Wang, B.; Sun, Q.; Fu, A.; Wang, Y.; Wang, Y.; Xu, X.; Li, W. Probiotic Bacillus amyloliquefaciens SC06 Induces Autophagy to Protect against Pathogens in Macrophages. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Fu, A.K.; Mo, Q.F.; Wu, Y.P.; Wang, B.; Liu, R.; Tang, L.; Zeng, Z.; Zhang, X.; Li, W. Protective effect of Bacillus amyloliquefaciens against Salmonella via polarizing macrophages to M1 phenotype directly and to M2 depended on microbiota. Food Funct. 2019, 10, 7653–7666. [Google Scholar] [CrossRef]
- Ekmekci, H.; Aslim, B.; Ozturk, S. Characterization of vaginal lactobacilli coaggregation ability with Escherichia coli. Microbiol. Immunol. 2009, 53, 59–65. [Google Scholar] [CrossRef]
- Trunk, T.; Khalil, H.S.; Leo, J.C. Bacterial autoaggregation. AIMS Microbiol. 2018, 4, 140. [Google Scholar] [CrossRef]
- Rickard, A.H.; Gilbert, P.; High, N.J.; Kolenbrander, P.E.; Handley, P.S. Bacterial coaggregation: An integral process in the development of multi-species biofilms. Trends Microbiol. 2003, 11, 94–100. [Google Scholar] [CrossRef]
- Trivedi, D.; Jena, P.K.; Patel, J.K.; Seshadri, S. Partial Purification and Characterization of a Bacteriocin DT24 Produced by Probiotic Vaginal Lactobacillus brevis DT24 and Determination of its Anti-Uropathogenic Escherichia coli Potential. Probiotics Antimicrob. Proteins 2013, 5, 142–151. [Google Scholar] [CrossRef]
- Gómez, N.C.; Ramiro, J.M.; Quecan, B.X.; Franco, B.D.G.D.M. Use of potential probiotic lactic acid bacteria (LAB) biofilms for the control of Listeria monocytogenes, Salmonella Typhimurium, and Escherichia coli O157: H7 biofilms formation. Front. Microbiol. 2016, 7, 863. [Google Scholar] [CrossRef]
- Kalia, V.C.; Shim, W.Y.; Patel, S.K.S.; Gong, C.; Lee, J.-K. Recent developments in antimicrobial growth promoters in chicken health: Opportunities and challenges. Sci. Total Environ. 2022, 834, 155300. [Google Scholar] [CrossRef]
- Crabbe, A.; Jensen, P.O.; Bjarnsholt, T.; Coenye, T. Antimicrobial Tolerance and Metabolic Adaptations in Microbial Biofilms. Trends Microbiol. 2019, 27, 850–863. [Google Scholar] [CrossRef] [PubMed]
- Motta, J.P.; Wallace, J.L.; Buret, A.G.; Deraison, C.; Vergnolle, N. Gastrointestinal biofilms in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 314–334. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zou, H.; Li, J.; Song, T.; Lv, W.; Wang, W.; Wang, Z.; Tao, S. Impact of quorum sensing signaling molecules in gram-negative bacteria on host cells: Current understanding and future perspectives. Gut Microbes 2022, 14, 2039048. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Baruah, A.; Tomioka, M.; Iino, Y.; Kalita, M.C.; Khan, M. Caenorhabditis elegans: A model to understand host-microbe interactions. Cell. Mol. Life Sci. 2020, 77, 1229–1249. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.K.; Podia, M.; Raut, S.; Singh, S.; Pinnaka, A.K.; Khatri, N. Insight Into the Beneficial Role of Lactiplantibacillus plantarum Supernatant Against Bacterial Infections, Oxidative Stress, and Wound Healing in A549 Cells and BALB/c Mice. Front. Pharmacol. 2021, 12, 2817. [Google Scholar] [CrossRef] [PubMed]
- Rim, E.Y.; Clevers, H.; Nusse, R. The Wnt Pathway: From Signaling Mechanisms to Synthetic Modulators. Annu. Rev. Biochem. 2022, 91, 571–598. [Google Scholar] [CrossRef]
- Niehrs, C. The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell. Biol. 2012, 13, 767–779. [Google Scholar] [CrossRef]
- Beumer, J.; Clevers, H. Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol. 2021, 22, 39–53. [Google Scholar] [CrossRef]
- Barker, N. Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014, 15, 19–33. [Google Scholar] [CrossRef]
- Wang, Z.; Havasi, A.; Gall, J.M.; Mao, H.; Schwartz, J.H.; Borkan, S.C. Beta-catenin promotes survival of renal epithelial cells by inhibiting Bax. J. Am. Soc. Nephrol. 2009, 20, 1919–1928. [Google Scholar] [CrossRef] [PubMed]
- You, L.; He, B.; Uematsu, K.; Xu, Z.; Mazieres, J.; Lee, A.; McCormick, F.; Jablons, D.M. Inhibition of Wnt-1 signaling induces apoptosis in beta-catenin-deficient mesothelioma cells. Cancer Res. 2004, 64, 3474–3478. [Google Scholar] [CrossRef] [PubMed]
- Sodhi, C.P.; Shi, X.H.; Richardson, W.M.; Grant, Z.S.; Shapiro, R.A.; Prindle, T., Jr.; Branca, M.; Russo, A.; Gribar, S.C.; Ma, C.; et al. Toll-like receptor-4 inhibits enterocyte proliferation via impaired beta-catenin signaling in necrotizing enterocolitis. Gastroenterology 2010, 138, 185–196. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Min, Y.J. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu. Rev. Biochem. 2007, 76, 447–480. [Google Scholar] [CrossRef]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef]
- Wang, B.; Hussain, A.; Zhou, Y.; Zeng, Z.; Wang, Q.; Zou, P.; Gong, L.; Zhao, P.; Li, W. Saccharomyces boulardii attenuates inflammatory response induced by Clostridium perfringens via TLR4/TLR15-MyD8 pathway in HD11 avian macrophages. Poult. Sci. 2020, 99, 5356–5365. [Google Scholar] [CrossRef]
- Martinez, F.O.; Helming, L.; Gordon, S. Alternative activation of macrophages: An immunologic functional perspective. Annu. Rev. Immunol. 2009, 27, 451–483. [Google Scholar] [CrossRef]




| Strains | Lac16 | Lac22 | Lac24 | Lac26 | |
|---|---|---|---|---|---|
| Oxgall (%) | |||||
| Time to reach OD620 of 0.3 units (h) | |||||
| 0% | 9.06 ± 0.08 | 6.50 ± 0.14 | 7.83 ± 0.13 | 8.94 ± 0.08 | |
| 0.10% | 6.56 ± 0.08 | 6.78 ± 0.21 | 6.67 ± 0.13 | 9.17 ± 0.13 | |
| 0.20% | 5.00 ± 0.14 | 6.56 ± 0.08 | 6.39 ± 0.21 | 9.56 ± 0.08 | |
| 0.30% | 3.22 ± 0.08 | 4.22 ± 0.08 | 5.28 ± 0.16 | 3.89 ± 0.21 | |
| 0.40% | 1.94 ± 0.08 | 3.22 ± 0.08 | 4.28 ± 0.21 | 2.50 ± 0.14 | |
| Lag time (h) | |||||
| 0.10% | −2.50 ± 0.08 c | 0.28 ± 0.21 a | −1.17 ± 0.13 b | 0.22 ± 0.13 a | |
| 0.20% | −4.06 ± 0.14 d | 0.06 ± 0.08 b | −1.44 ± 0.21 c | 0.61 ± 0.08 a | |
| 0.30% | −5.83 ± 0.08 c | −2.28 ± 0.08 a | −2.55 ± 0.16 a | −5.05 ± 0.21 b | |
| 0.40% | −7.11 ± 0.08 c | −3.28 ± 0.08 a | −3.56 ± 0.21 a | −6.44 ± 0.14 b | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Zhou, Y.; Wang, Q.; Xu, S.; Wang, F.; Yue, M.; Zeng, Z.; Li, W. Lactiplantibacillus plantarum Lac16 Attenuates Enterohemorrhagic Escherichia coli O157:H7 Infection by Inhibiting Virulence Traits and Improving Intestinal Epithelial Barrier Function. Cells 2023, 12, 1438. https://doi.org/10.3390/cells12101438
Wang B, Zhou Y, Wang Q, Xu S, Wang F, Yue M, Zeng Z, Li W. Lactiplantibacillus plantarum Lac16 Attenuates Enterohemorrhagic Escherichia coli O157:H7 Infection by Inhibiting Virulence Traits and Improving Intestinal Epithelial Barrier Function. Cells. 2023; 12(10):1438. https://doi.org/10.3390/cells12101438
Chicago/Turabian StyleWang, Baikui, Yuanhao Zhou, Qi Wang, Shujie Xu, Fei Wang, Min Yue, Zhonghua Zeng, and Weifen Li. 2023. "Lactiplantibacillus plantarum Lac16 Attenuates Enterohemorrhagic Escherichia coli O157:H7 Infection by Inhibiting Virulence Traits and Improving Intestinal Epithelial Barrier Function" Cells 12, no. 10: 1438. https://doi.org/10.3390/cells12101438
APA StyleWang, B., Zhou, Y., Wang, Q., Xu, S., Wang, F., Yue, M., Zeng, Z., & Li, W. (2023). Lactiplantibacillus plantarum Lac16 Attenuates Enterohemorrhagic Escherichia coli O157:H7 Infection by Inhibiting Virulence Traits and Improving Intestinal Epithelial Barrier Function. Cells, 12(10), 1438. https://doi.org/10.3390/cells12101438








