Genetically Engineered Triple MAPT-Mutant Human-Induced Pluripotent Stem Cells (N279K, P301L, and E10+16 Mutations) Exhibit Impairments in Mitochondrial Bioenergetics and Dynamics
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Human iPSCs Culture
2.3. ATP Levels
2.4. Determination of MMP
2.5. Profiling Mitochondrial Respiration
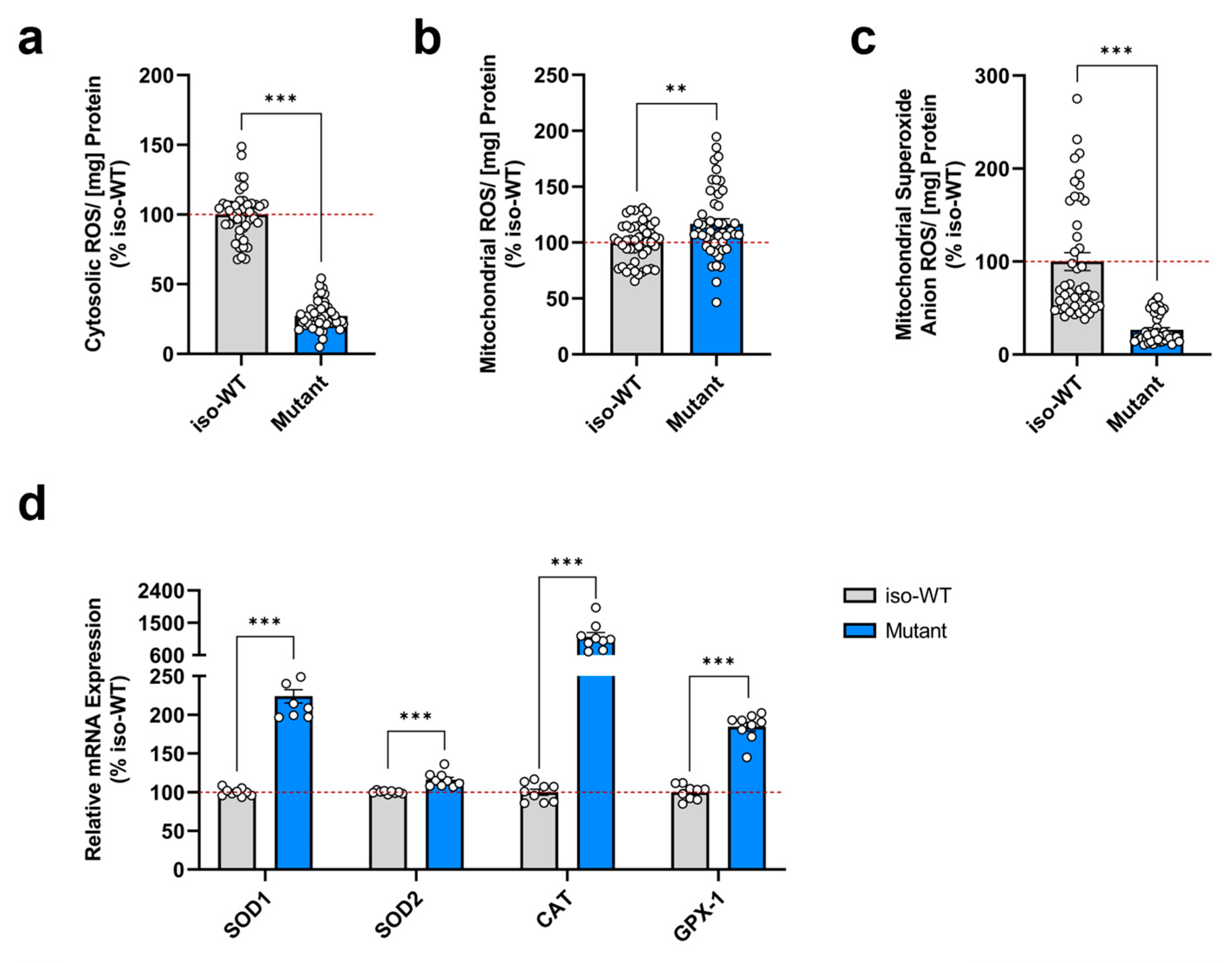
2.6. Detection of ROS Levels
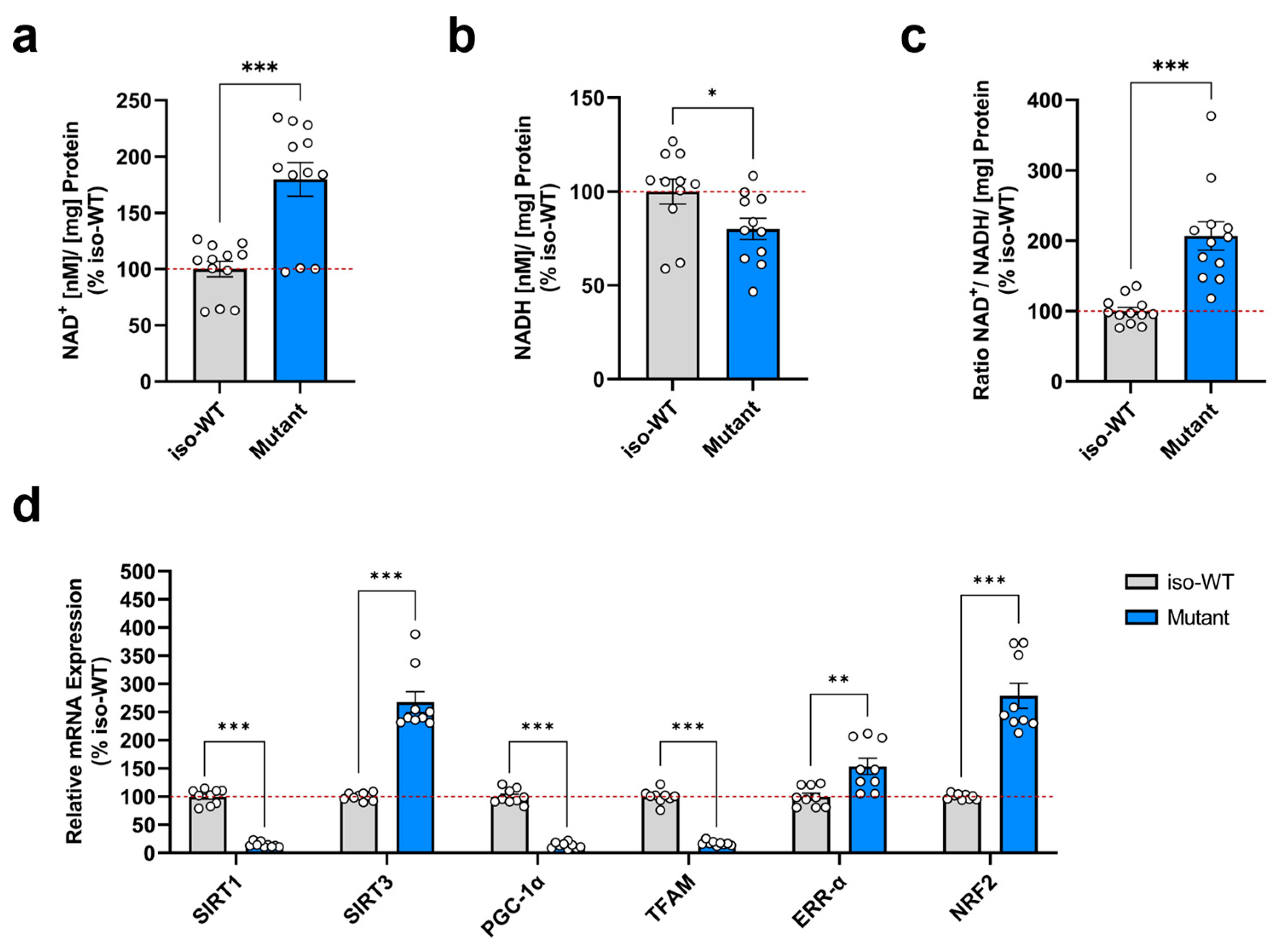
2.7. NAD+ and NADH Quantification
2.8. Protein Content Quantification
2.9. Total RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR
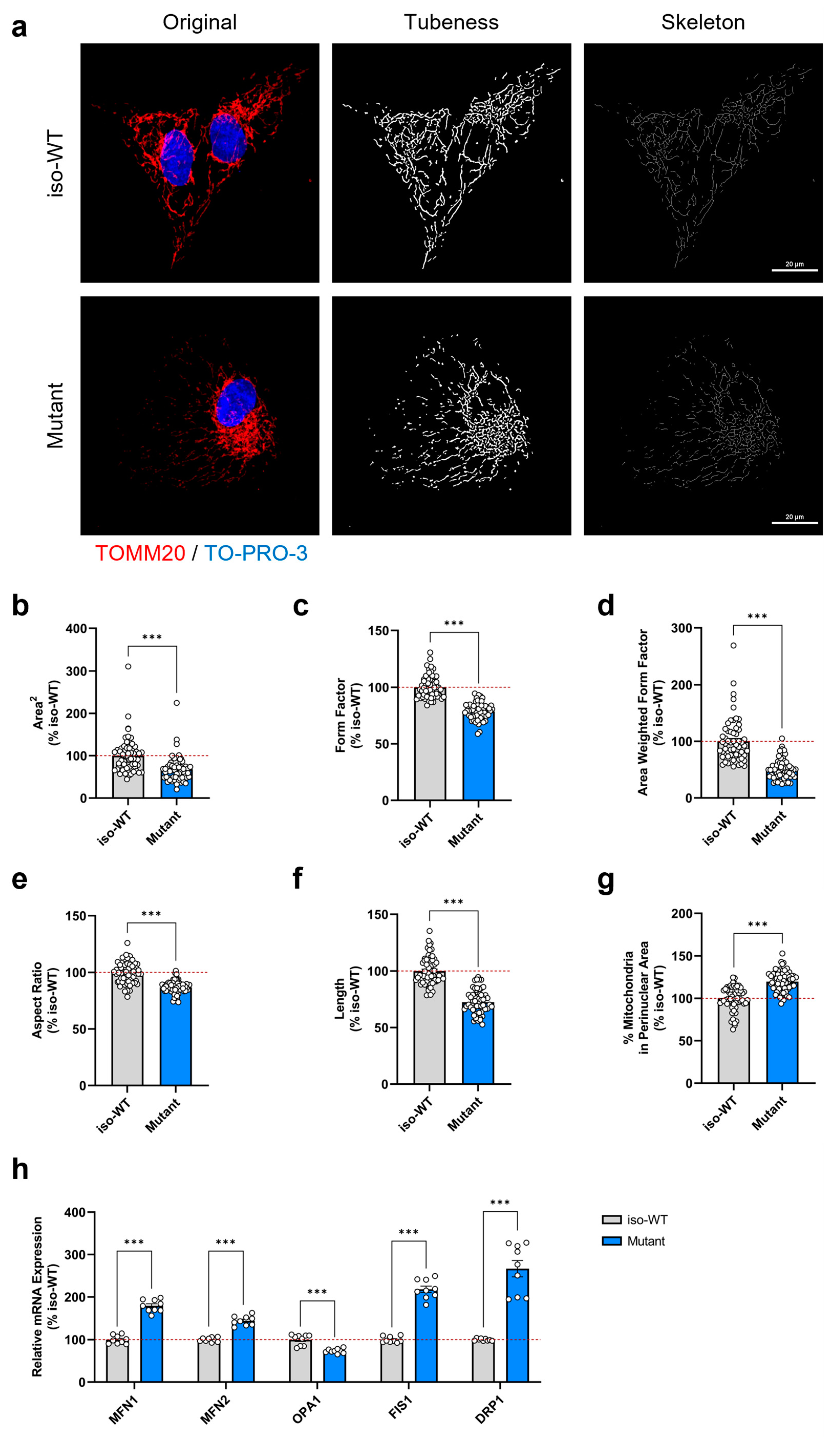
2.10. Mitochondrial Morphology and Distribution
2.11. Statistical Analysis
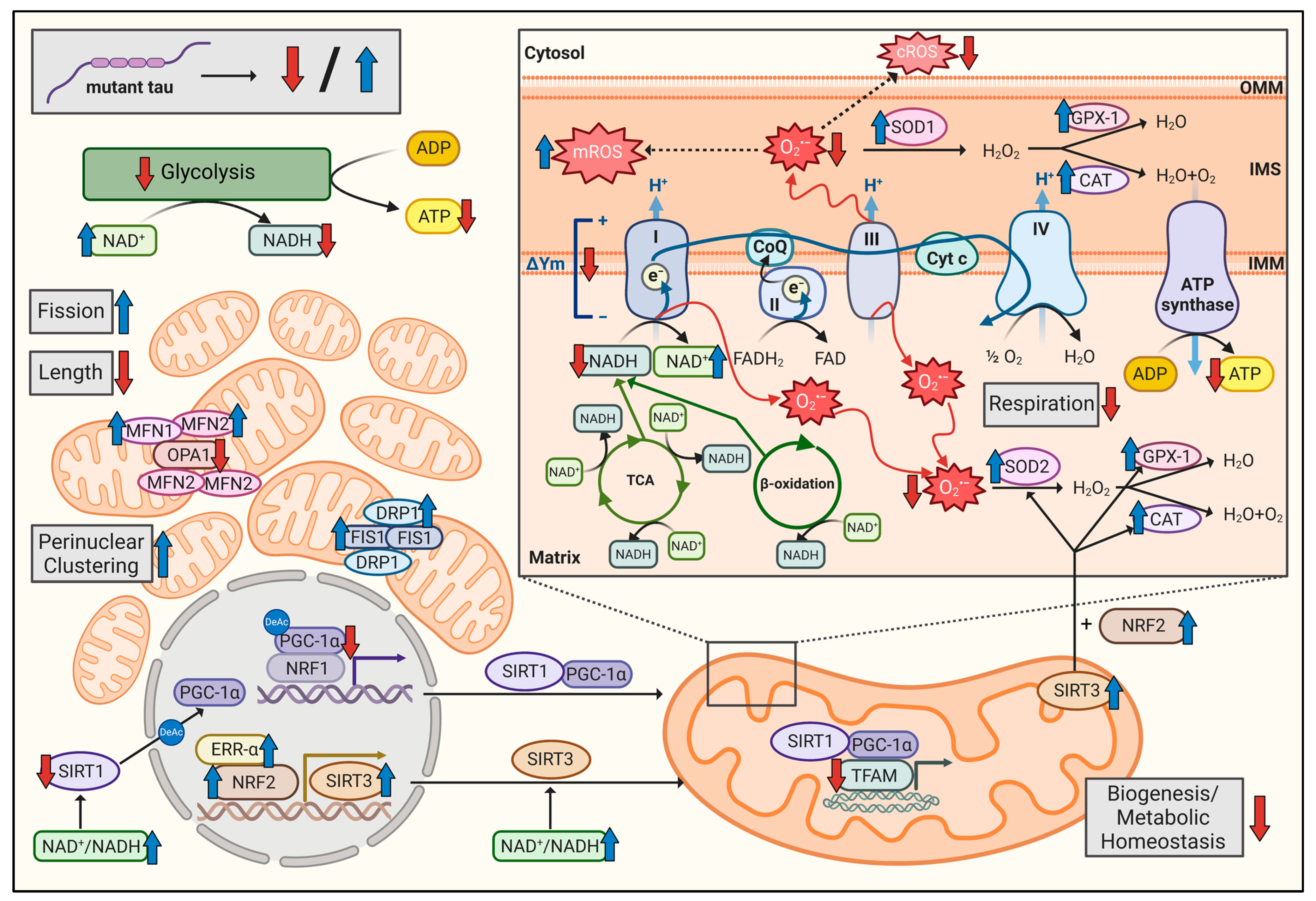
3. Results
3.1. Triple Tau-Mutant iPSCs Display Mitochondrial Bioenergetic Deficits
3.2. Triple Tau Mutations Impair Parameters Linked to the Metabolic Regulation of Mitochondrial Function in iPSCs
3.3. Triple Tau-Mutant iPSCs Exhibit a Disturbed Cellular Redox Homeostasis
3.4. Triple Tau Mutations Modify the Mitochondrial Network Morphology and Distribution in iPSCs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spillantini, M.G.; Goedert, M. Tau pathology and neurodegeneration. Lancet Neurol. 2013, 12, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Goedert, M. Tau protein pathology in neurodegenerative diseases. Trends Neurosci. 1998, 21, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Murrell, J.R.; Goedert, M.; Farlow, M.R.; Klug, A.; Ghetti, B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc. Natl. Acad. Sci. 1998, 95, 7737–7741. [Google Scholar] [CrossRef]
- Hutton, M.; Lendon, C.L.; Rizzu, P.; Baker, M.; Froelich, S.; Houlden, H.; Pickering-Brown, S.; Chakraverty, S.; Isaacs, A.; Grover, A. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 1998, 393, 702–705. [Google Scholar] [CrossRef]
- Poorkaj, P.; Bird, T.D.; Wijsman, E.; Nemens, E.; Garruto, R.M.; Anderson, L.; Andreadis, A.; Wiederholt, W.C.; Raskind, M.; Schellenberg, G.D. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann. Neurol. 1998, 43, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Ghetti, B.; Oblak, A.L.; Boeve, B.F.; Johnson, K.A.; Dickerson, B.C.; Goedert, M. Invited review: Frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: A chameleon for neuropathology and neuroimaging. Neuropathol. Appl. Neurobiol. 2015, 41, 24–46. [Google Scholar] [CrossRef]
- von Bergen, M.; Barghorn, S.; Li, L.; Marx, A.; Biernat, J.; Mandelkow, E.-M.; Mandelkow, E. Mutations of tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local β-structure. J. Biol. Chem. 2001, 276, 48165–48174. [Google Scholar] [CrossRef]
- Grover, A.; DeTure, M.; Yen, S.-H.; Hutton, M. Effects on splicing and protein function of three mutations in codon N296 of tau in vitro. Neurosci. Lett. 2002, 323, 33–36. [Google Scholar] [CrossRef]
- Wszolek, Z.K.; Pfeiffer, R.F.; Bhatt, M.H.; Schelper, R.L.; Cordes, M.; Snow, B.J.; Rodnitzky, R.L.; Wolters, E.C.; Arwert, F.; Calne, D.B. Rapidly progressive autosomal dominant parkinsonism and dementia with pallido-ponto-nigral degeneration. Ann. Neurol. 1992, 32, 312–320. [Google Scholar] [CrossRef]
- Arendt, T.; Stieler, J.T.; Holzer, M. Tau and tauopathies. Brain Res. Bull. 2016, 126, 238–292. [Google Scholar] [CrossRef]
- Rossi, G.; Tagliavini, F. Frontotemporal lobar degeneration: Old knowledge and new insight into the pathogenetic mechanisms of tau mutations. Front. Aging Neurosci. 2015, 7, 192. [Google Scholar] [CrossRef]
- Li, L.; Chao, J.; Shi, Y. Modeling neurological diseases using iPSC-derived neural cells: iPSC modeling of neurological diseases. Cell Tissue Res. 2018, 371, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Nakao, S.; Ueyama, T.; Harada, Y.; Kawamura, T. Metabolic remodeling during somatic cell reprogramming to induced pluripotent stem cells: Involvement of hypoxia-inducible factor 1. Inflamm. Regen. 2020, 40, 8. [Google Scholar] [CrossRef]
- Li, Q.V.; Rosen, B.P.; Huangfu, D. Decoding pluripotency: Genetic screens to interrogate the acquisition, maintenance, and exit of pluripotency. WIREs Syst. Biol. Med. 2020, 12, e1464. [Google Scholar] [CrossRef] [PubMed]
- Lines, G.; Casey, J.M.; Preza, E.; Wray, S. Modelling frontotemporal dementia using patient-derived induced pluripotent stem cells. Mol. Cell Neurosci. 2020, 109, 103553. [Google Scholar] [CrossRef]
- Kühn, R.; Mahajan, A.; Canoll, P.; Hargus, G. Human Induced Pluripotent Stem Cell Models of Frontotemporal Dementia with Tau Pathology. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, K.; Grimm, A.; Kazmierczak, A.; Strosznajder, J.B.; Götz, J.; Eckert, A. Insights into mitochondrial dysfunction: Aging, amyloid-β, and tau-A deleterious trio. Antioxid. Redox Signal. 2012, 16, 1456–1466. [Google Scholar] [CrossRef]
- Pérez, M.J.; Jara, C.; Quintanilla, R.A. Contribution of Tau Pathology to Mitochondrial Impairment in Neurodegeneration. Front. Neurosci. 2018, 12, 441. [Google Scholar] [CrossRef] [PubMed]
- Szabo, L.; Eckert, A.; Grimm, A. Insights into Disease-Associated Tau Impact on Mitochondria. Int. J. Mol. Sci. 2020, 21, 6344. [Google Scholar] [CrossRef]
- Khacho, M.; Harris, R.; Slack, R.S. Mitochondria as central regulators of neural stem cell fate and cognitive function. Nat. Rev. Neurosci. 2019, 20, 34–48. [Google Scholar] [CrossRef]
- Qi, Y.; Ye, Y.; Wang, R.; Yu, S.; Zhang, Y.; Lv, J.; Jin, W.; Xia, S.; Jiang, W.; Li, Y.; et al. Mitochondrial dysfunction by TFAM depletion disrupts self-renewal and lineage differentiation of human PSCs by affecting cell proliferation and YAP response. Redox Biol. 2022, 50, 102248. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; Cheng, C.; Mair, W.; Almeida, S.; Fong, H.; Biswas, M.H.U.; Zhang, Z.; Huang, Y.; Temple, S.; Coppola, G.; et al. Human iPSC-Derived Neuronal Model of Tau-A152T Frontotemporal Dementia Reveals Tau-Mediated Mechanisms of Neuronal Vulnerability. Stem Cell Rep. 2016, 7, 325–340. [Google Scholar] [CrossRef]
- García-León, J.A.; Cabrera-Socorro, A.; Eggermont, K.; Swijsen, A.; Terryn, J.; Fazal, R.; Nami, F.; Ordovás, L.; Quiles, A.; Lluis, F.; et al. Generation of a human induced pluripotent stem cell–based model for tauopathies combining three microtubule-associated protein TAU mutations which displays several phenotypes linked to neurodegeneration. Alzheimer’s Dement. 2018, 14, 1261–1280. [Google Scholar] [CrossRef]
- Merrill, R.A.; Flippo, K.H.; Strack, S. Measuring Mitochondrial Shape with ImageJ. In Techniques to Investigate Mitochondrial Function in Neurons; Strack, S., Usachev, Y.M., Eds.; Springer: New York, NY, USA, 2017; pp. 31–48. [Google Scholar]
- Schulz, K.L.; Eckert, A.; Rhein, V.; Mai, S.; Haase, W.; Reichert, A.S.; Jendrach, M.; Müller, W.E.; Leuner, K. A New Link to Mitochondrial Impairment in Tauopathies. Mol. Neurobiol. 2012, 46, 205–216. [Google Scholar] [CrossRef]
- Grimm, A.; Biliouris, E.E.; Lang, U.E.; Götz, J.; Mensah-Nyagan, A.G.; Eckert, A. Sex hormone-related neurosteroids differentially rescue bioenergetic deficits induced by amyloid-β or hyperphosphorylated tau protein. Cell. Mol. Life Sci. 2016, 73, 201–215. [Google Scholar] [CrossRef]
- Grimm, A.; Lejri, I.; Hallé, F.; Schmitt, M.; Götz, J.; Bihel, F.; Eckert, A. Mitochondria modulatory effects of new TSPO ligands in a cellular model of tauopathies. J. Neuroendocrinol. 2020, 32, e12796. [Google Scholar] [CrossRef] [PubMed]
- Connolly, N.M.C.; Theurey, P.; Adam-Vizi, V.; Bazan, N.G.; Bernardi, P.; Bolaños, J.P.; Culmsee, C.; Dawson, V.L.; Deshmukh, M.; Duchen, M.R.; et al. Guidelines on experimental methods to assess mitochondrial dysfunction in cellular models of neurodegenerative diseases. Cell Death Differ. 2018, 25, 542–572. [Google Scholar] [CrossRef]
- Nsiah-Sefaa, A.; McKenzie, M. Combined defects in oxidative phosphorylation and fatty acid β-oxidation in mitochondrial disease. Biosci. Rep. 2016, 36. [Google Scholar] [CrossRef]
- Zapata-Pérez, R.; Wanders, R.J.A.; van Karnebeek, C.D.M.; Houtkooper, R.H. NAD(+) homeostasis in human health and disease. EMBO Mol. Med. 2021, 13, e13943. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Zhang, L.; Gao, W.; Huang, C.; Huber, P.E.; Zhou, X.; Li, C.; Shen, G.; Zou, B. NAD+ metabolism: Pathophysiologic mechanisms and therapeutic potential. Signal Transduct. Target. Ther. 2020, 5, 227. [Google Scholar] [CrossRef] [PubMed]
- Clement, J.W.M.; Poljak, A.; Sachdev, P.; Braidy, N. The Plasma NAD+ Metabolome Is Dysregulated in “Normal” Aging. Rejuvenation Res. 2019, 22, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.S.; Adriaanse, B.A.; Greig, N.H.; Mattson, M.P.; Cader, M.Z.; Bohr, V.A.; Fang, E.F. Mitophagy and Alzheimer’s Disease: Cellular and Molecular Mechanisms. Trends Neurosci. 2017, 40, 151–166. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Cruzat, V.F.; Newsholme, P.; Cheng, J.; Chen, Y.; Lu, Y. Regulation of SIRT1 in aging: Roles in mitochondrial function and biogenesis. Mech. Ageing Dev. 2016, 155, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Aquilano, K.; Vigilanza, P.; Baldelli, S.; Pagliei, B.; Rotilio, G.; Ciriolo, M.R. Peroxisome Proliferator-activated Receptor γ Co-activator 1α (PGC-1α) and Sirtuin 1 (SIRT1) Reside in Mitochondria: Possible Direct Function in Mitochondrial Biogenesis *. J. Biol. Chem. 2010, 285, 21590–21599. [Google Scholar] [CrossRef]
- Luo, C.; Ding, W.; Yang, C.; Zhang, W.; Liu, X.; Deng, H. Nicotinamide Mononucleotide Administration Restores Redox Homeostasis via the Sirt3–Nrf2 Axis and Protects Aged Mice from Oxidative Stress-Induced Liver Injury. J. Proteome Res. 2022, 21, 1759–1770. [Google Scholar] [CrossRef] [PubMed]
- Sidorova-Darmos, E.; Sommer, R.; Eubanks, J.H. The Role of SIRT3 in the Brain under Physiological and Pathological Conditions. Front. Cell Neurosci. 2018, 12, 196. [Google Scholar] [CrossRef]
- Grimm, A.; Eckert, A. Brain aging and neurodegeneration: From a mitochondrial point of view. J. Neurochem. 2017, 143, 418–431. [Google Scholar] [CrossRef]
- Lejri, I.; Agapouda, A.; Grimm, A.; Eckert, A. Mitochondria- and Oxidative Stress-Targeting Substances in Cognitive Decline-Related Disorders: From Molecular Mechanisms to Clinical Evidence. Oxid. Med. Cell Longev. 2019, 2019, 9695412. [Google Scholar] [CrossRef]
- Tönnies, E.; Trushina, E. Oxidative Stress, Synaptic Dysfunction, and Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 57, 1105–1121. [Google Scholar] [CrossRef]
- Lee, K.H.; Cha, M.; Lee, B.H. Neuroprotective Effect of Antioxidants in the Brain. Int. J. Mol. Sci. 2020, 21, 7152. [Google Scholar] [CrossRef] [PubMed]
- Benard, G.; Bellance, N.; James, D.; Parrone, P.; Fernandez, H.; Letellier, T.; Rossignol, R. Mitochondrial bioenergetics and structural network organization. J. Cell Sci. 2007, 120, 838–848. [Google Scholar] [CrossRef]
- Prieto, J.; Ponsoda, X.; Izpisua Belmonte, J.C.; Torres, J. Mitochondrial dynamics and metabolism in induced pluripotency. Exp. Gerontol. 2020, 133, 110870. [Google Scholar] [CrossRef]
- Galloway, C.A.; Lee, H.; Yoon, Y. Mitochondrial morphology—Emerging role in bioenergetics. Free. Radic. Biol. Med. 2012, 53, 2218–2228. [Google Scholar] [CrossRef]
- Wade, S.; Khacho, M. Mitochondria: Mitochondrial dynamics in the regulation of stem cells. Int. J. Biochem. Cell Biol. 2022, 144, 106158. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.Y.; Weick, J.P.; Yu, J.; Ma, L.X.; Zhang, X.Q.; Thomson, J.A.; Zhang, S.C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA 2010, 107, 4335–4340. [Google Scholar] [CrossRef]
- Yellen, G. Fueling thought: Management of glycolysis and oxidative phosphorylation in neuronal metabolism. J. Cell Biol. 2018, 217, 2235–2246. [Google Scholar] [CrossRef]
- David, D.C.; Hauptmann, S.; Scherping, I.; Schuessel, K.; Keil, U.; Rizzu, P.; Ravid, R.; Dröse, S.; Brandt, U.; Müller, W.E.; et al. Proteomic and Functional Analyses Reveal a Mitochondrial Dysfunction in P301L Tau Transgenic Mice *. J. Biol. Chem. 2005, 280, 23802–23814. [Google Scholar] [CrossRef] [PubMed]
- Rhein, V.; Song, X.; Wiesner, A.; Ittner, L.M.; Baysang, G.; Meier, F.; Ozmen, L.; Bluethmann, H.; Dröse, S.; Brandt, U.; et al. Amyloid-beta and tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer’s disease mice. Proc. Natl. Acad. Sci. USA 2009, 106, 20057–20062. [Google Scholar] [CrossRef]
- Esteras, N.; Rohrer, J.D.; Hardy, J.; Wray, S.; Abramov, A.Y. Mitochondrial hyperpolarization in iPSC-derived neurons from patients of FTDP-17 with 10 + 16 MAPT mutation leads to oxidative stress and neurodegeneration. Redox Biol. 2017, 12, 410–422. [Google Scholar] [CrossRef]
- Zhang, J.; Khvorostov, I.; Hong, J.S.; Oktay, Y.; Vergnes, L.; Nuebel, E.; Wahjudi, P.N.; Setoguchi, K.; Wang, G.; Do, A.; et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 2011, 30, 4860–4873. [Google Scholar] [CrossRef]
- Zhu, X.-H.; Lu, M.; Lee, B.-Y.; Ugurbil, K.; Chen, W. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc. Natl. Acad. Sci. USA 2015, 112, 2876–2881. [Google Scholar] [CrossRef] [PubMed]
- Lautrup, S.; Sinclair, D.A.; Mattson, M.P.; Fang, E.F. NAD+ in Brain Aging and Neurodegenerative Disorders. Cell Metab. 2019, 30, 630–655. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sauve, A.A. NAD(+) metabolism: Bioenergetics, signaling and manipulation for therapy. Biochim. Biophys. Acta 2016, 1864, 1787–1800. [Google Scholar] [CrossRef] [PubMed]
- Julien, C.; Tremblay, C.; Émond, V.; Lebbadi, M.; Salem, N., Jr.; Bennett, D.A.; Calon, F. Sirtuin 1 Reduction Parallels the Accumulation of Tau in Alzheimer Disease. J. Neuropathol. Exp. Neurol. 2009, 68, 48–58. [Google Scholar] [CrossRef]
- Min, S.W.; Cho, S.H.; Zhou, Y.; Schroeder, S.; Haroutunian, V.; Seeley, W.W.; Huang, E.J.; Shen, Y.; Masliah, E.; Mukherjee, C.; et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron 2010, 67, 953–966. [Google Scholar] [CrossRef]
- Rodgers, J.T.; Lerin, C.; Haas, W.; Gygi, S.P.; Spiegelman, B.M.; Puigserver, P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 2005, 434, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Kandimalla, R.; Manczak, M.; Fry, D.; Suneetha, Y.; Sesaki, H.; Reddy, P.H. Reduced dynamin-related protein 1 protects against phosphorylated Tau-induced mitochondrial dysfunction and synaptic damage in Alzheimer’s disease. Hum. Mol. Genet. 2016, 25, 4881–4897. [Google Scholar] [CrossRef]
- Vijayan, M.; Reddy, P.H. Reduced VDAC1, Maintained Mitochondrial Dynamics and Enhanced Mitochondrial Biogenesis in a Transgenic Tau Mouse Model of Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 8561. [Google Scholar] [CrossRef]
- Pradeepkiran, J.A.; Munikumar, M.; Reddy, A.P.; Reddy, P.H. Protective effects of a small molecule inhibitor ligand against hyperphosphorylated tau-induced mitochondrial and synaptic toxicities in Alzheimer disease. Hum. Mol. Genet. 2021, 31, 244–261. [Google Scholar] [CrossRef]
- Kshirsagar, S.; Sawant, N.; Morton, H.; Reddy, A.P.; Reddy, P.H. Mitophagy enhancers against phosphorylated Tau-induced mitochondrial and synaptic toxicities in Alzheimer disease. Pharmacol. Res. 2021, 174, 105973. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Tang, S.; Li, X. Sirtuins in Metabolic and Epigenetic Regulation of Stem Cells. Trends Endocrinol. Metab. 2019, 30, 177–188. [Google Scholar] [CrossRef]
- Li, S.; Yin, J.; Nielsen, M.; Beach, T.G.; Guo, L.; Shi, J. Sirtuin 3 Mediates Tau Deacetylation. J. Alzheimer’s Dis. 2019, 69, 355–362. [Google Scholar] [CrossRef]
- Farr, S.A.; Ripley, J.L.; Sultana, R.; Zhang, Z.; Niehoff, M.L.; Platt, T.L.; Murphy, M.P.; Morley, J.E.; Kumar, V.; Butterfield, D.A. Antisense oligonucleotide against GSK-3β in brain of SAMP8 mice improves learning and memory and decreases oxidative stress: Involvement of transcription factor Nrf2 and implications for Alzheimer disease. Free. Radic. Biol. Med. 2014, 67, 387–395. [Google Scholar] [CrossRef]
- Grimm, A. Impairments in Brain Bioenergetics in Aging and Tau Pathology: A Chicken and Egg Situation? Cells 2021, 10, 2531. [Google Scholar] [CrossRef] [PubMed]
- Dodson, M.; Anandhan, A.; Zhang, D.D.; Madhavan, L. An NRF2 Perspective on Stem Cells and Ageing. Front. Aging 2021, 2. [Google Scholar] [CrossRef]
- Dai, X.; Yan, X.; Wintergerst, K.A.; Cai, L.; Keller, B.B.; Tan, Y. Nrf2: Redox and Metabolic Regulator of Stem Cell State and Function. Trends Mol. Med. 2020, 26, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Merksamer, P.I.; Liu, Y.; He, W.; Hirschey, M.D.; Chen, D.; Verdin, E. The sirtuins, oxidative stress and aging: An emerging link. Aging 2013, 5, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef]
- Kuksal, N.; Chalker, J.; Mailloux, R.J. Progress in understanding the molecular oxygen paradox—Function of mitochondrial reactive oxygen species in cell signaling. Biol. Chem. 2017, 398, 1209–1227. [Google Scholar] [CrossRef]
- Grimm, A.; Mensah-Nyagan, A.G.; Eckert, A. Alzheimer, mitochondria and gender. Neurosci. Biobehav. Rev. 2016, 67, 89–101. [Google Scholar] [CrossRef]
- Qiu, X.; Brown, K.; Hirschey, M.D.; Verdin, E.; Chen, D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010, 12, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, N.R.; Gupta, M.; Kim, G.; Rajamohan, S.B.; Isbatan, A.; Gupta, M.P. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J. Clin. Invest. 2009, 119, 2758–2771. [Google Scholar] [CrossRef]
- Esteras, N.; Kopach, O.; Maiolino, M.; Lariccia, V.; Amoroso, S.; Qamar, S.; Wray, S.; Rusakov, D.A.; Jaganjac, M.; Abramov, A.Y. Mitochondrial ROS control neuronal excitability and cell fate in frontotemporal dementia. Alzheimer’s Dement. 2022, 18, 318–338. [Google Scholar] [CrossRef]
- Chaudhari, P.; Ye, Z.; Jang, Y.Y. Roles of reactive oxygen species in the fate of stem cells. Antioxid. Redox Signal. 2014, 20, 1881–1890. [Google Scholar] [CrossRef] [PubMed]
- Ushio-Fukai, M. Reactive Oxygen Species (ROS) and Stem/Progenitor Cells. In Systems Biology of Free Radicals and Antioxidants; Laher, I., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 2471–2497. [Google Scholar] [CrossRef]
- Ding, Y.; Yang, H.; Wang, Y.; Chen, J.; Ji, Z.; Sun, H. Sirtuin 3 is required for osteogenic differentiation through maintenance of PGC-1ɑ-SOD2-mediated regulation of mitochondrial function. Int. J. Biol. Sci. 2017, 13, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Ahn, B.H.; Kim, H.S.; Song, S.; Lee, I.H.; Liu, J.; Vassilopoulos, A.; Deng, C.X.; Finkel, T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. USA 2008, 105, 14447–14452. [Google Scholar] [CrossRef]
- Finley, L.W.; Haas, W.; Desquiret-Dumas, V.; Wallace, D.C.; Procaccio, V.; Gygi, S.P.; Haigis, M.C. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PLoS ONE 2011, 6, e23295. [Google Scholar] [CrossRef] [PubMed]
- Scott, I.; Youle, R.J. Mitochondrial fission and fusion. Essays Biochem. 2010, 47, 85–98. [Google Scholar] [CrossRef]
- DuBoff, B.; Götz, J.; Feany, M.B. Tau Promotes Neurodegeneration via DRP1 Mislocalization In Vivo. Neuron 2012, 75, 618–632. [Google Scholar] [CrossRef] [PubMed]
- Cipolat, S.; de Brito, O.M.; Dal Zilio, B.; Scorrano, L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA 2004, 101, 15927–15932. [Google Scholar] [CrossRef] [PubMed]
- Olichon, A.; Baricault, L.; Gas, N.; Guillou, E.; Valette, A.; Belenguer, P.; Lenaers, G. Loss of OPA1 Perturbates the Mitochondrial Inner Membrane Structure and Integrity, Leading to Cytochrome c Release and Apoptosis *. J. Biol. Chem. 2003, 278, 7743–7746. [Google Scholar] [CrossRef]
- Frezza, C.; Cipolat, S.; Martins de Brito, O.; Micaroni, M.; Beznoussenko, G.V.; Rudka, T.; Bartoli, D.; Polishuck, R.S.; Danial, N.N.; De Strooper, B.; et al. OPA1 Controls Apoptotic Cristae Remodeling Independently from Mitochondrial Fusion. Cell 2006, 126, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Cogliati, S.; Frezza, C.; Soriano, M.E.; Varanita, T.; Quintana-Cabrera, R.; Corrado, M.; Cipolat, S.; Costa, V.; Casarin, A.; Gomes, L.C.; et al. Mitochondrial Cristae Shape Determines Respiratory Chain Supercomplexes Assembly and Respiratory Efficiency. Cell 2013, 155, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Kopeikina, K.J.; Carlson, G.A.; Pitstick, R.; Ludvigson, A.E.; Peters, A.; Luebke, J.I.; Koffie, R.M.; Frosch, M.P.; Hyman, B.T.; Spires-Jones, T.L. Tau Accumulation Causes Mitochondrial Distribution Deficits in Neurons in a Mouse Model of Tauopathy and in Human Alzheimer’s Disease Brain. Am. J. Pathol. 2011, 179, 2071–2082. [Google Scholar] [CrossRef] [PubMed]
- Vives-Bauza, C.; Zhou, C.; Huang, Y.; Cui, M.; de Vries, R.L.A.; Kim, J.; May, J.; Tocilescu, M.A.; Liu, W.; Ko, H.S.; et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA 2010, 107, 378–383. [Google Scholar] [CrossRef]
- Rambold, A.S.; Lippincott-Schwartz, J. Mechanisms of mitochondria and autophagy crosstalk. Cell Cycle 2011, 10, 4032–4038. [Google Scholar] [CrossRef] [PubMed]
- Cummins, N.; Tweedie, A.; Zuryn, S.; Bertran-Gonzalez, J.; Götz, J. Disease-associated tau impairs mitophagy by inhibiting Parkin translocation to mitochondria. EMBO J. 2019, 38, e99360. [Google Scholar] [CrossRef]
- Zhong, X.; Cui, P.; Cai, Y.; Wang, L.; He, X.; Long, P.; Lu, K.; Yan, R.; Zhang, Y.; Pan, X.; et al. Mitochondrial Dynamics Is Critical for the Full Pluripotency and Embryonic Developmental Potential of Pluripotent Stem Cells. Cell Metab. 2019, 29, 979–992. [Google Scholar] [CrossRef]
- Chen, H.; Detmer, S.A.; Ewald, A.J.; Griffin, E.E.; Fraser, S.E.; Chan, D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003, 160, 189–200. [Google Scholar] [CrossRef]
- Khacho, M.; Clark, A.; Svoboda, D.S.; Azzi, J.; MacLaurin, J.G.; Meghaizel, C.; Sesaki, H.; Lagace, D.C.; Germain, M.; Harper, M.-E.; et al. Mitochondrial Dynamics Impacts Stem Cell Identity and Fate Decisions by Regulating a Nuclear Transcriptional Program. Cell Stem Cell 2016, 19, 232–247. [Google Scholar] [CrossRef] [PubMed]
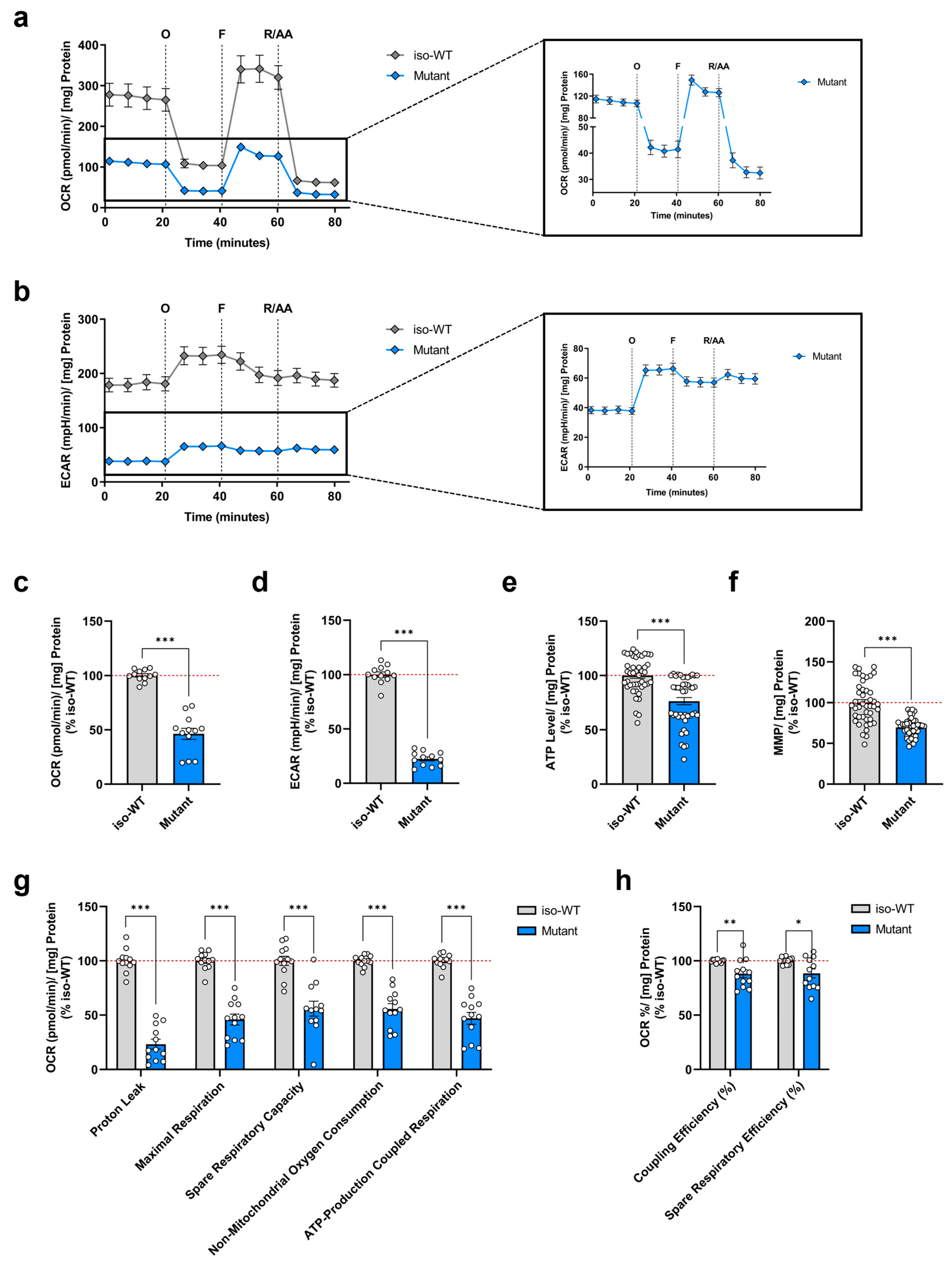
| Assay Parameter | Equation | Depicted in Figure |
|---|---|---|
| Non-Mitochondrial Oxygen Consumption | Minimum rate measurement after rotenone/antimycin A injection | 1g |
| Basal Respiration | (Last rate measurement before first injection)—(Non-Mitochondrial Oxygen Consumption) | |
| Maximal Respiration | (Maximum rate measurement after FCCP injection)—(Non-Mitochondrial Oxygen Consumption) | 1g |
| Proton Leak | (Minimum rate measurement after oligomycin injection)—(Non-Mitochondrial Oxygen Consumption) | 1g |
| ATP-Production Coupled Respiration | (Last rate measurement before oligomycin injection)—(Minimum rate measurement after oligomycin injection) | 1g |
| Spare Respiratory Capacity | (Maximal Respiration)—(Basal Respiration) | 1g |
| Spare Respiratory Efficiency | (Maximal Respiration)/(Basal Respiration) × 100% | 1h |
| Coupling Efficiency | (ATP-Production Coupled Respiration)/(Basal Respiration) × 100% | 1h |
| Target Gene | Forward Primer | Reverse Primer |
|---|---|---|
| CAT | 5′-AGATGCAGCACTGGAAGGAG-3′ | 5′-GCATGCACAACTCTCTCAGG-3′ |
| DRP1 | 5′-TCACGAGACAAGTCTTCTAAAG-3′ | 5′-CCTCCAGATGCAACCTTG-3′ |
| ERRα | 5′-TAGTTACCTTGGGCACTGGG-3′ | 5′-GTTTGTGGACAGGCTGTGAG-3′ |
| FIS1 | 5′-GCTCAAGGAATACGAGAAGG-3′ | 5′-AGTCCATCTTTCTTCATGGC-3′ |
| GAPDH | 5′-CATGGTTTACATGTTCCAATATGA-3′ | 5′-GGATCTCGCTCCTGGAAG-3′ |
| GPX1 | 5′-GTGCTCGGCTTCCCGTGCAAC-3′ | 5′-CTCGAAGAGCATGAAGTTGGGC-3′ |
| MFN1 | 5′-CAGTCACCAAGTAAAACAACAAA-3′ | 5′-GGGTAATCTAGCAATTTCTTCTTC-3′ |
| MFN2 | 5′-CAGGATTCAGAAAGCCCAG-3′ | 5′-GATGCACTCCTCAAATCTCC-3′ |
| NRF2 | 5′-CCAGCACATCCAGTCAGAA-3′ | 5′-AGCCGAAGAAACCTCATTGT-3′ |
| OPA1 | 5′-CCAGGTGTGATTAATACTGTGA-3′ | 5′-CCATCTTGAATACACAGTATGATG-3′ |
| PGC1-α | 5′-TCCTCTTCAAGATCCTGCTATTAC-3′ | 5′-TCTCAGACTCTCGCTTCTCATA-3′ |
| SIRT1 | 5′--3′GTAGGCGGCTTGATGGTAAT-3 | 5′-GGGTTCTTCTAAACTTGGACTCT-3′ |
| SIRT3 | 5’-CATGAGCTGCAGTGACTGGT-3’ | 5’-GAGCTTGCCGTTCAACTAGG-3′ |
| SOD1 | 5′-GCTGGTTTGCGTCGTAGTCT-3′ | 5′-ATGCAGGCCTTCAGTCAGTC-3′ |
| SOD2 | 5′-AAGCACCACGCGGCCTACG-3′ | 5′-CCATTGAACTTCAGTGCAGGCTG-3′ |
| TFAM | 5′-CAAGTTGTCCAAAGAAACCTGTAA-3′ | 5′-GCCACTCCGCCCTATAAG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szabo, L.; Grimm, A.; García-León, J.A.; Verfaillie, C.M.; Eckert, A. Genetically Engineered Triple MAPT-Mutant Human-Induced Pluripotent Stem Cells (N279K, P301L, and E10+16 Mutations) Exhibit Impairments in Mitochondrial Bioenergetics and Dynamics. Cells 2023, 12, 1385. https://doi.org/10.3390/cells12101385
Szabo L, Grimm A, García-León JA, Verfaillie CM, Eckert A. Genetically Engineered Triple MAPT-Mutant Human-Induced Pluripotent Stem Cells (N279K, P301L, and E10+16 Mutations) Exhibit Impairments in Mitochondrial Bioenergetics and Dynamics. Cells. 2023; 12(10):1385. https://doi.org/10.3390/cells12101385
Chicago/Turabian StyleSzabo, Leonora, Amandine Grimm, Juan Antonio García-León, Catherine M. Verfaillie, and Anne Eckert. 2023. "Genetically Engineered Triple MAPT-Mutant Human-Induced Pluripotent Stem Cells (N279K, P301L, and E10+16 Mutations) Exhibit Impairments in Mitochondrial Bioenergetics and Dynamics" Cells 12, no. 10: 1385. https://doi.org/10.3390/cells12101385
APA StyleSzabo, L., Grimm, A., García-León, J. A., Verfaillie, C. M., & Eckert, A. (2023). Genetically Engineered Triple MAPT-Mutant Human-Induced Pluripotent Stem Cells (N279K, P301L, and E10+16 Mutations) Exhibit Impairments in Mitochondrial Bioenergetics and Dynamics. Cells, 12(10), 1385. https://doi.org/10.3390/cells12101385










