1. Introduction
Acute hypoxic injury is a problem that scientists have been trying to solve. As altitude increases, the partial pressure of oxygen in the body gradually decreases, which may induce acute altitude sickness, high-altitude pulmonary edema, high-altitude cerebral edema, and even life-threatening conditions [
1,
2]. Recently, people have had an increasing need to enter extremely high-altitude areas (>7620 m), for example, for performing tasks, traveling, and climbing Mount Everest; thus, rapid adaptation to extreme hypoxia has attracted increasing attention. At an altitude of 7620 m (25,000 ft), the ambient oxygen partial pressure is 60 mmHg, and the alveolar oxygen partial pressure attains the critical value of 30 mmHg. Staying at this altitude for even a few minutes causes the alveolar oxygen partial pressure to drop below 30 mmHg and triggers loss of consciousness. Therefore, the area above the altitude of 7620 m has been called the “death zone” [
1,
3], and even after acclimation, the human body cannot stay at this altitude for a long time. Few studies have addressed the rapid entry of the body into an extremely hypoxic environment such as that found above 7620 m, and no effective regimen to achieve rapid adaptation to extreme hypoxia has been reported.
Withaferin A (WA), a chemical component extracted from the popular Indian herb
Withania somnifera, can block the activity of nuclear factor-κB (NF-κB) and inhibit angiogenesis and cell proliferation [
4]. WA has anti-inflammatory, antidiabetic, and antitumor effects, but the efficacy of WA in protection against hypoxia has not been reported. Promisingly, our study showed that WA pretreatment significantly increased the 24 h survival rate of Sprague-Dawley (SD) rats at 7620 m, suggesting that WA plays an important role in enhancing the rapid adaptability to extreme hypoxia. In this study, we confirmed that WA plays a protective role against extreme hypoxia by affecting mitochondrial quality control mechanisms.
2. Materials and Methods
2.1. Animals and Reagents
Adult male SD rats (10–12 weeks, >300 g) were used in this experiment. Rats were randomly divided into four groups, as follows: Con group, normal rats at the baseline altitude of 412 m; H group, rats exposed to a simulated altitude of 7620 m for 24 h; W group, rats given intraperitoneal injections of 2 mg/kg WA for 7 days at the baseline altitude of 412 m; WH group, rats given intraperitoneal injections of 2 mg/kg WA for 7 days and then exposed to 7620 m for 24 h. The rats in Con and H group were injected with vehicle before the corresponding treatment. WA (5119-48-2, IC-0201518) was purchased from InCellGene and was solubilized in 10% DMSO and 90% corn oil.
2.2. Simulation of Hypobaric Hypoxia
The hypobaric hypoxic environment was created in a hypobaric chamber system as previously described [
5]. After the height value was input into the control system, the hypobaric hypoxic environment was created in the cabin via the vacuum pump. The cabin was ventilated, and the speeds of ascent and descent were controlled at 10 m/s. The temperature and humidity of cabins were maintained at 25 ± 5 °C and 50 ± 5%. The survival of rats was recorded during 24 h hypoxia.
2.3. Hematoxylin-Eosin (HE) Staining and Reactive Oxygen Species (ROS)/Adenosine Triphosphate (ATP) Detection in Cardiac Tissue
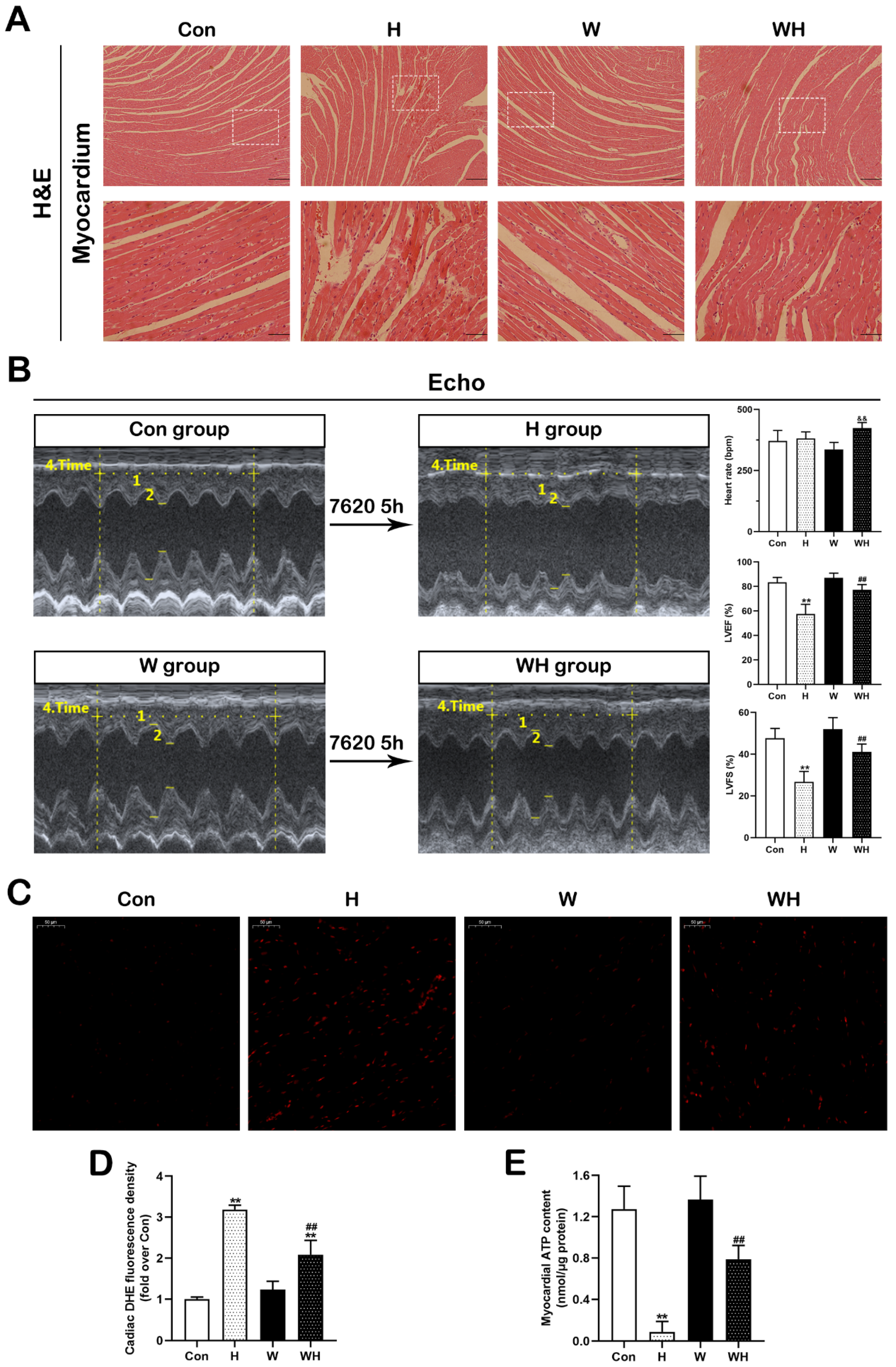
Rats were anesthetized with 1% pentobarbital sodium, and the cardiac tissue was removed, immediately frozen in liquid nitrogen, and stored at −80 °C for formalin-fixed section preparation, Western blot analysis, quantitative PCR (qPCR), and tissue ROS and ATP measurement. For HE staining, the cardiac tissue sections were stained with hematoxylin and eosin, and images were acquired by photomicroscopy.
The myocardial ROS level was determined by DHE staining (S0063, Beyotime Biotechnology, China). The freshly frozen myocardial tissue sections were immediately stained with DHE reaction solution. The sections were detected at 535 nm excitation, and the fluorescence intensity was analyzed using NIH ImageJ software.
The myocardial ATP content was determined by ATP Assay Kit (S0026, Beyotime Biotechnology, China) according to the instructions. Briefly, the myocardium was split into homogenates, and the supernatant was removed after being centrifuged, and then the luminescence value of the sample was detected using the Tecan microplate reader after the ATP detection solution was added.
2.4. Echocardiography
Echocardiography was performed as previously described [
6]. In brief, rats were anesthetized with 3% isoflurane, and the prethoracic area was exposed by depilation. Then, the heart was scanned, and M-mode images were acquired. The following parameters were recorded: heart rate (HR), left ventricular internal dimension at end-systole (LVIDs), left ventricular internal dimension at end-diastole (LVIDd), left ventricular end-diastolic volume (LVEDV), and left ventricular end-systolic volume (LVESV). Left ventricular ejection fraction (LVEF) and left ventricular fractional shortening (LVFS) were calculated as LVEF = [(LVEDV − LVESV)/(LVEDV)] and LVFS = [(LVIDd − LVIDs)/(LVIDd)]. In addition, the E and A peak velocities were recorded, and E/A ratio was calculated to reflect the diastolic function.
2.5. Cell Culture and Treatments
The human cardiomyocyte cell line AC16 was used in this experiment. Cells in the control (Con) group were cultured in DMEM supplemented with 10% FBS in 5% CO2 at 37 °C. Cells in the W group were treated with 0.5 μmol/L WA for 48 h. For acute hypoxia simulation, cells in the Con and W group were exposed to 1% O2 for 24 h (“H” group and “WH” group, respectively) using a tri-gas incubator (Heal Force-HF100, Shanghai, China).
For PGC-1α transduction experiment, cells in culture were transfected with empty vector adenovirus (Ad-Con) or PGC-1α expression adenovirus (Ad-PGC1α, Hanbio, Shanghai, China). At 36 h after transfection, cells were treated with hypoxia as above.
2.6. Western Blot Analysis
As described previously [
7], protein was extracted from myocardial tissue or cells after different treatments, and the protein concentration was measured by the BCA method [
7]. The protein samples were mixed with loading buffer and heated for denaturation. After electrophoresis and membrane transfer in an XCell SureLock Mini-Cell apparatus (Invitrogen, Carlsbad, CA, USA), proteins were transferred to a PVDF membrane. After blocking with 5% BSA, the membrane was incubated with the primary antibody and horseradish peroxidase (HRP)-conjugated secondary antibody. Bands were detected using ECL reagents (Millipore, Burlington, MA, USA). The gray values of the target protein bands were measured with NIH ImageJ software. The antibodies used in this experiment are described in the
Supplemental Material.
2.7. qRT-PCR
For measurement of mRNA levels, total RNA from cardiac tissue or AC16 cells was extracted using a TRIzol kit (Invitrogen, CA, USA) according to the protocol. One microgram of total RNA was reverse transcribed to cDNA using a PrimeScript RT Reagent Kit (Takara, Tokyo, Japan). RT-PCR analysis was performed using a CFX96 (Bio-Rad, CA, USA) instrument and SYBR Premix Ex Taq (Takara, Tokyo, Japan) following the instructions. The housekeeping gene β-actin was used as the internal reference. The 2−ΔΔCt method was used to determine the relative mRNA levels of target genes.
To determine the mitochondrial DNA (mtDNA) content, the level of the mitochondria-encoded gene NADH dehydrogenase subunit 1 (ND-1) was normalized to that of the nuclear housekeeping gene β-actin to reflect the number of mitochondria.
The primers specific for peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), nuclear factor erythroid 2-related factor 2 (NRF2), NRF1, mitochondrial transcription factor A (TFAM), BCL2-interacting protein 3 (BNIP3), ND-1, and β-actin mRNA used in this experiment are listed in the
Supplemental Material.
2.8. Transmission Electron Microscopy (TEM)
For tissue TEM, fresh cardiac tissues were immediately fixed with 4% glutaraldehyde for 24 h and were then trimmed into 0.1 × 0.1 × 0.1 cm
3 blocks for further fixation with 1% osmium tetroxide in deionized water. For TEM, the adherent cells were digested, washed twice with PBS, and fixed with 4% glutaraldehyde for 24 h. Subsequent steps were performed as previously described [
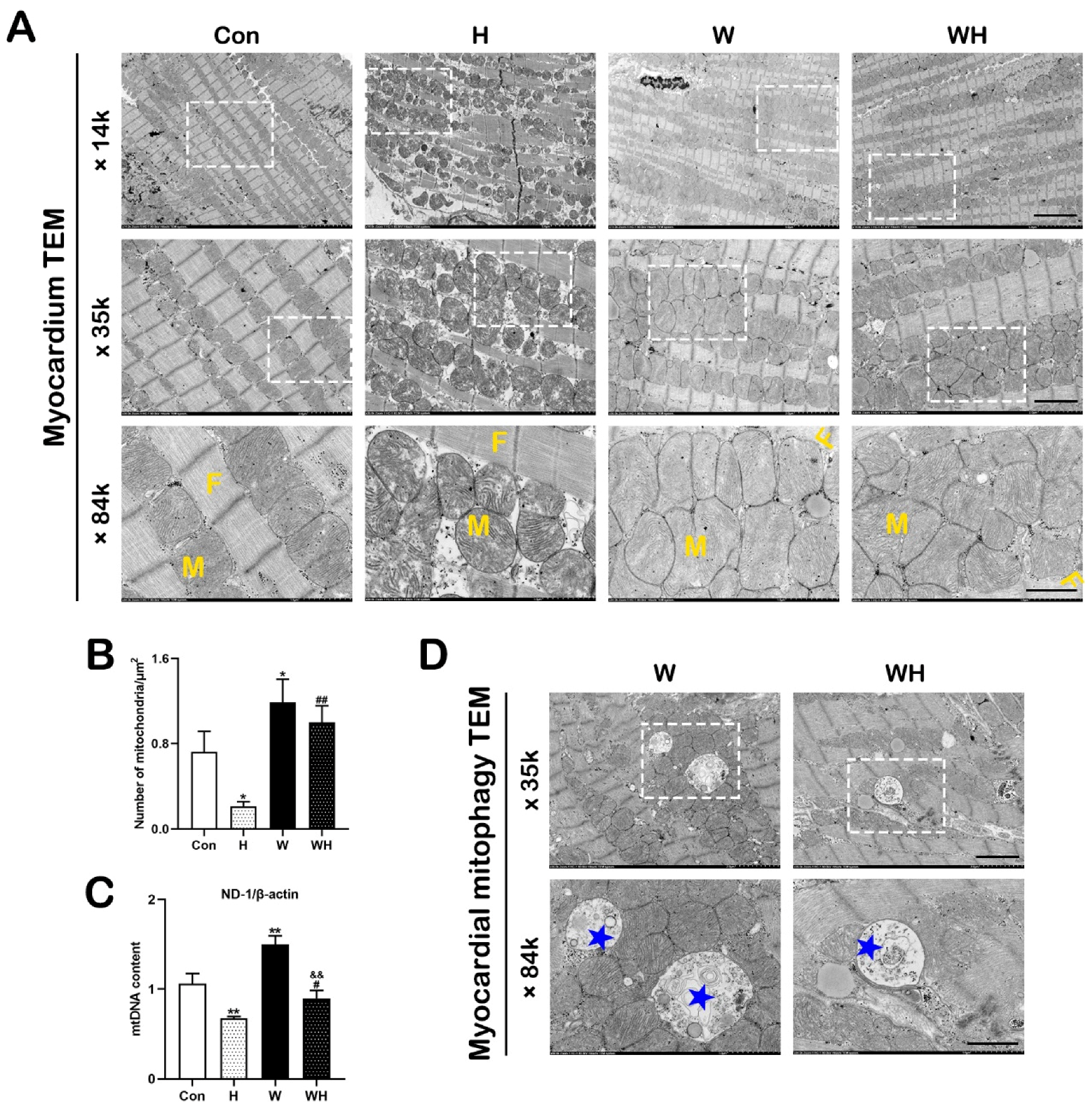
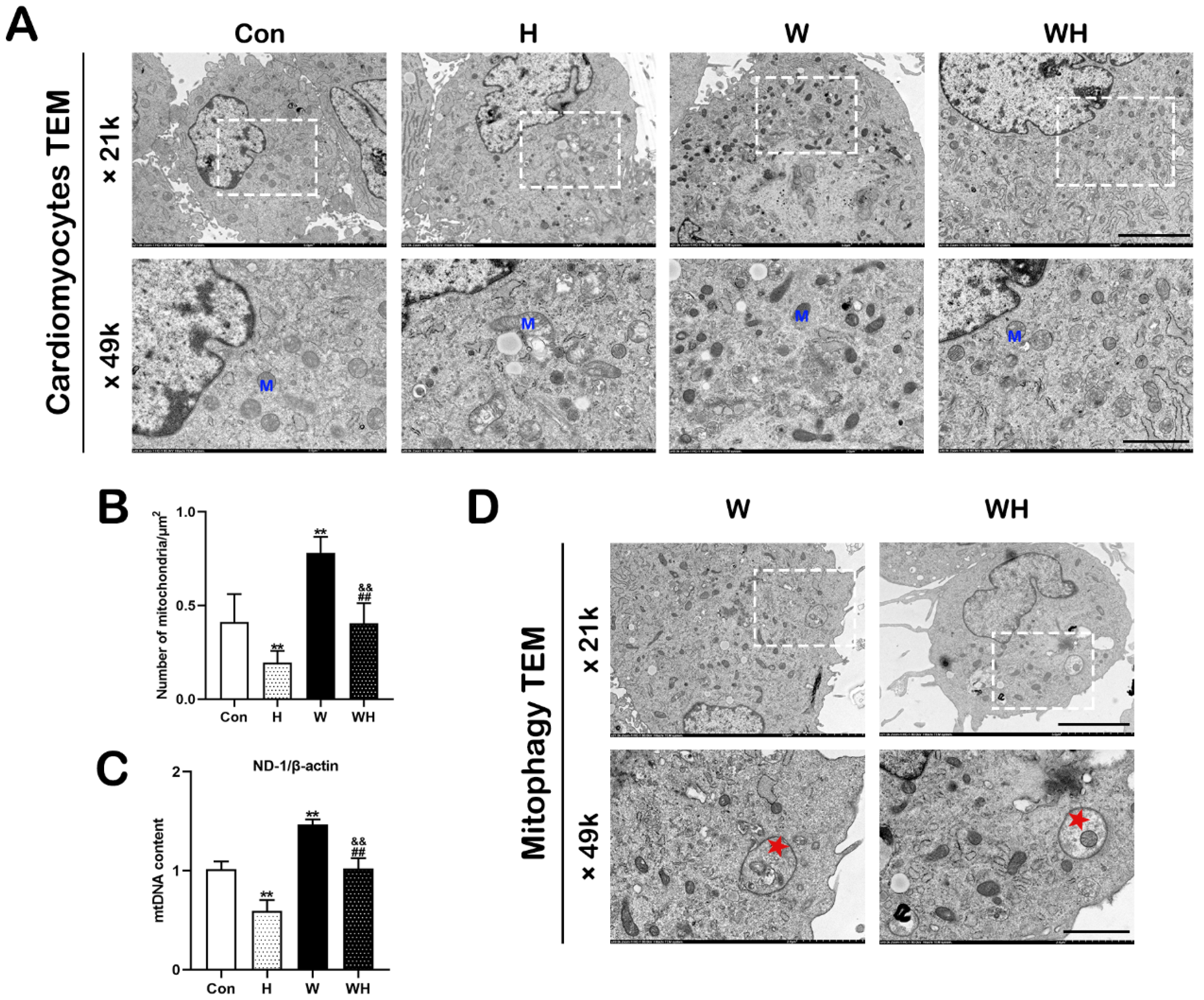
8]. Electron micrographs were acquired using a transmission electron microscope (HT7800 series, Hitachi, Tokyo, Japan) at 80 kV. The morphology of myocardial fibers, mitochondria, and autophagosomes was observed. At least five visual fields per sample were randomly selected to analyze the number of mitochondria.
2.9. Analysis of Apoptosis and Measurement of the Mitochondrial Membrane Potential (MMP)
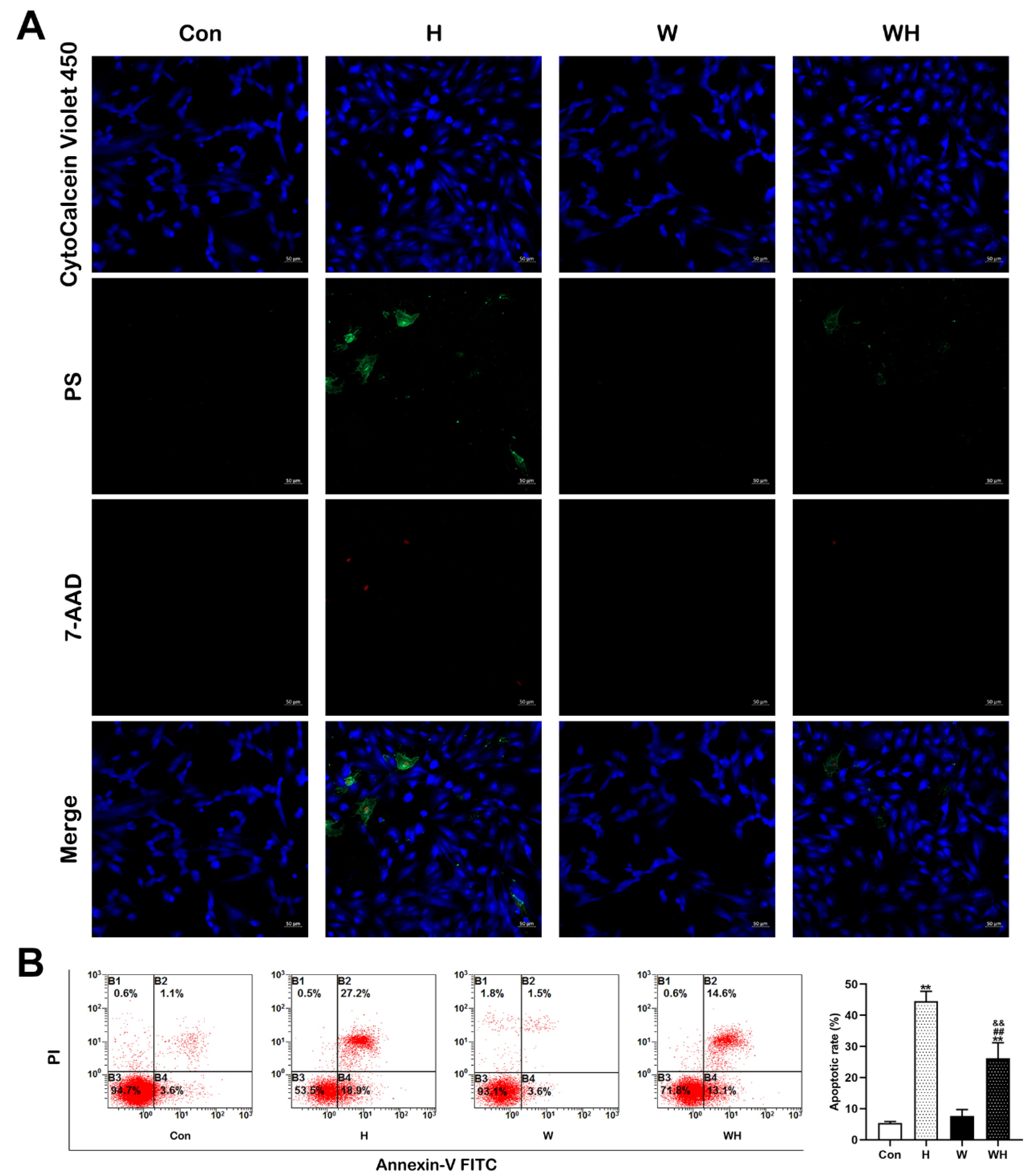
An apoptosis/necrosis detection kit (ab176749, Abcam, MA, USA) was used to simultaneously monitor apoptotic (green), necrotic (red), and healthy (blue) cells in strict accordance with the manufacturer’s instructions. The stained samples were analyzed by flow cytometry (Beckman Coulter, Brea, CA, USA) and fluorescence microscopy (LSM800, Carl Zeiss, Germany).
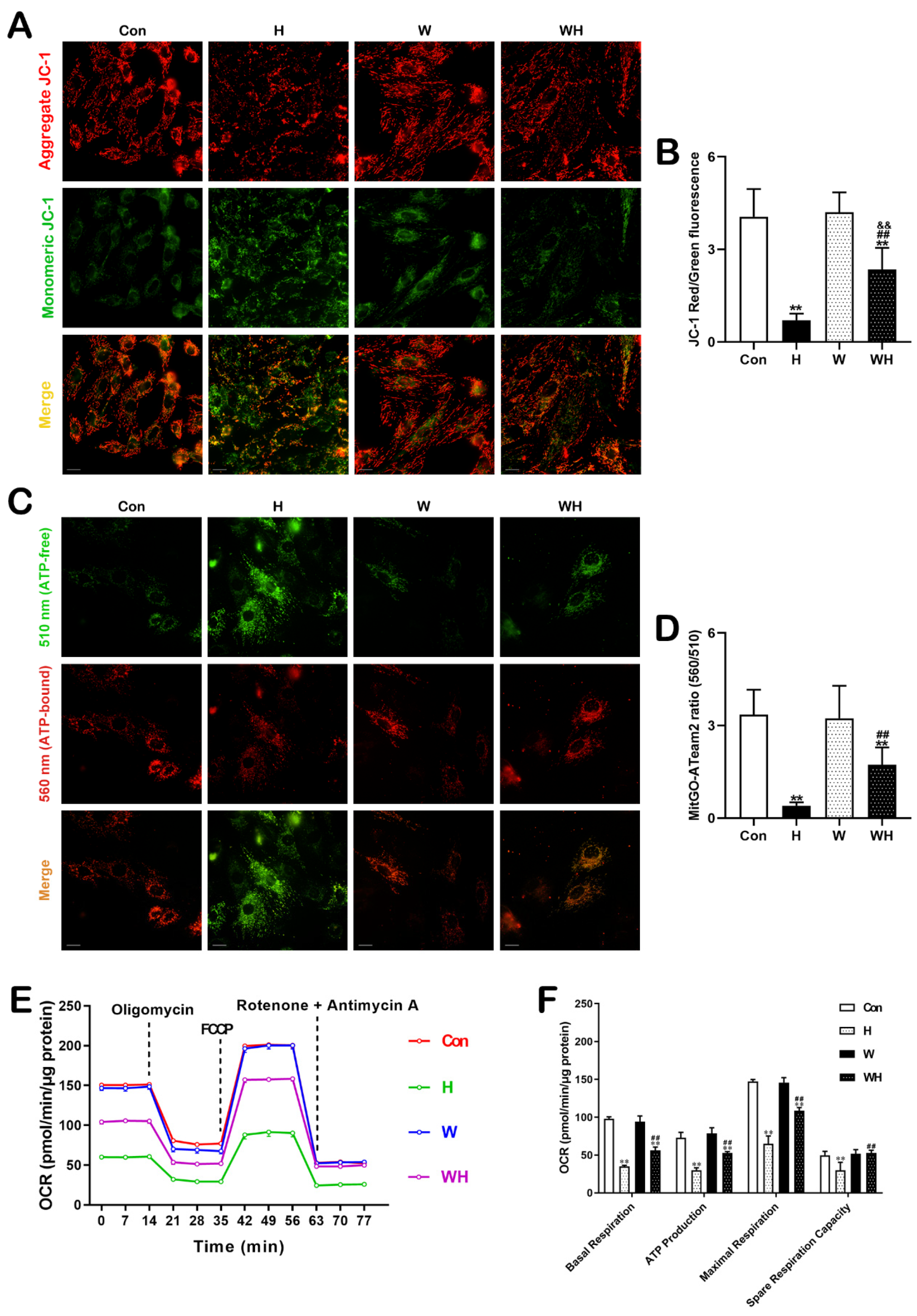
The MMP was measured using a JC-1 mitochondrial membrane potential assay kit (C2006, Beyotime Biotechnology, Shanghai, China). Cells were incubated with JC-1 solution at 37 °C for 20 min, and imaging was then performed using a high-content imaging system (Harmony, PerkinElmer, Rodgau, Germany). The ratio of red to green fluorescence intensity was used to determine the MMP.
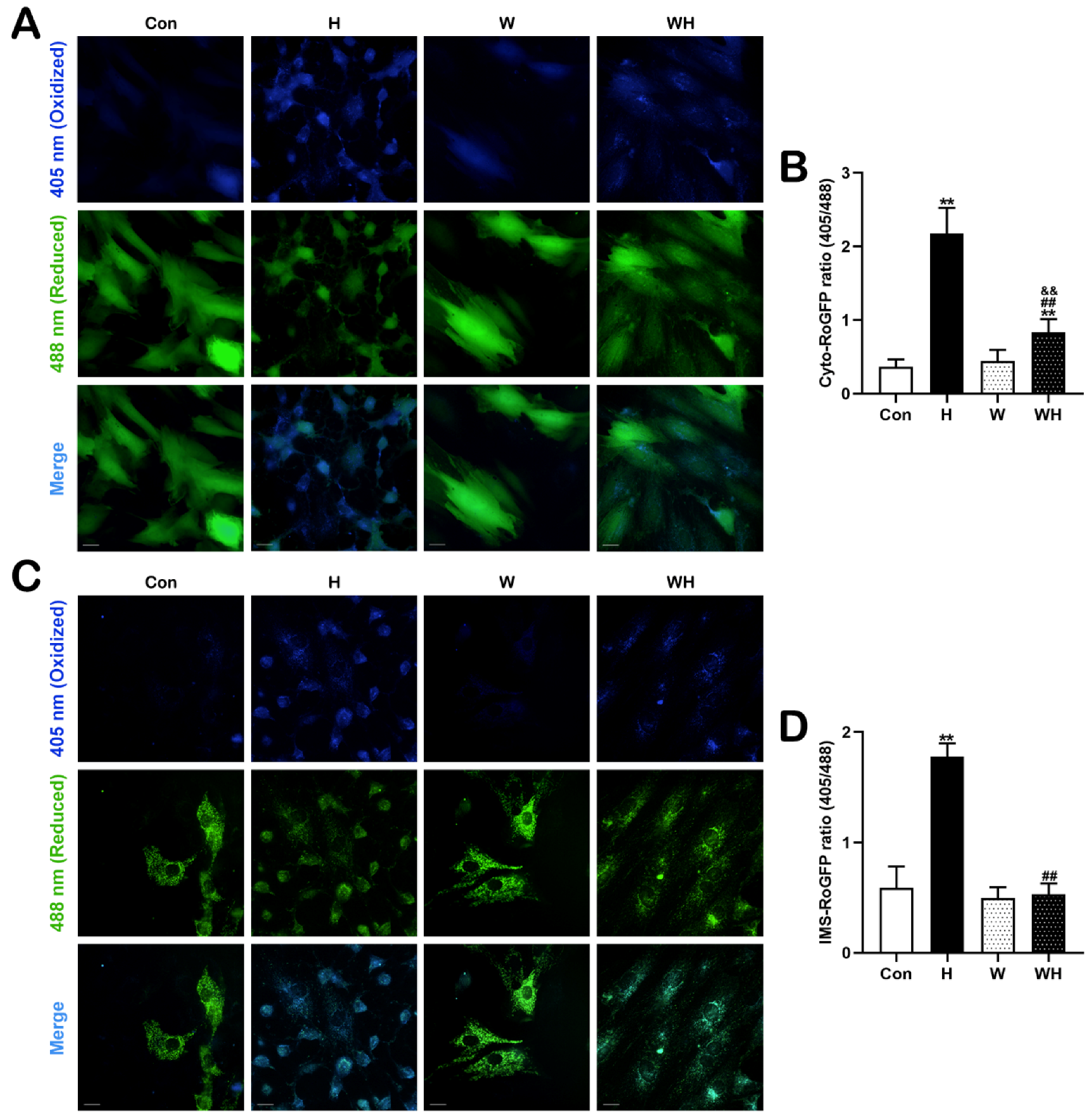
2.10. Measurement of ROS with RoGFP Sensors in Subcellular Compartments
Cyto-RoGFP (Addgene, #49435) and IMS-RoGFP (Addgene, #49436) were employed to determine ROS levels in the cytoplasm and mitochondrial intermembrane space (IMS). The principle of RoGFP sensors was previously described [
9,
10]. In RoGFP-transfected cells, blue fluorescence excited at 405 nm reflects the oxidation state, while green fluorescence excited at 488 nm reflects the reduction state. The ratio of the two fluorescence intensities indicates the intracellular ROS level. The fluorescent images were acquired at 36 h post-transfection as the oxidative stress status before hypoxia. Then, the cells were treated with 1% O
2 for 24 h followed by images capture to indicate the oxidative stress status after hypoxia. The fluorescent images were taken using high-content imaging and analysis software (Perkin Elmer, Woodbridge, ON, USA).
2.11. Measurement of Mitochondrial ATP with mitGO-ATeam2
A FRET-based fluorescent ATP probe, mitGO-ATeam2 (a gift from Hiromi Imamura, Kyoto University), was used to monitor the mitochondrial ATP level in living cells. The probe was previously described [
10,
11]. Cardiomyocytes were transfected with the pcDNA-mitGo-ATeam2 plasmid using Lipo3000 (Invitrogen, CA, USA). The mitGO-ATeam2 sensor was excited at 488 nm, and emission was measured at 510 nm and 560 nm. The ratio of fluorescence emission intensity at 560 nm to that at 510 nm indicates the ATP level in cells. The fluorescent images were acquired similarly as described in
Section 2.10.
2.12. Measurement of the Mitochondrial Oxygen Consumption Rate (OCR)
An XF24 extracellular flux analyzer (Agilent Seahorse Bioscience, Santa Clara, CA, USA) was employed to measure the OCR, as previously described [
12]. Analysis was carried out according to the manufacturer’s instructions. Values were normalized to the intracellular protein concentration and are presented as pmol/min/μg protein. The OCR parameters, including basal respiration, ATP-linked respiration, maximal respiration rate, and spare respiratory capacity, were calculated and analyzed.
2.13. Statistical Analysis
All experiments were independently repeated at least 3 times, and results are presented as the means ± SEMs values. For comparisons among multiple groups, two-way ANOVA followed by Tukey’s multiple comparison test was used. p < 0.05 was considered to indicate a significant difference. Statistical analysis and graphs were achieved using GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA).
4. Discussion
Most studies on hypoxia adaptation have focused on rapid adaptation or acclimatization at mid–high altitudes (3000–5000 m). Few studies have addressed rapid adaptation to extreme hypoxia at altitudes above 7620 m. Our study, for the first time, showed that WA, which exerts anti-inflammatory and anticancer effects, can effectively promote rapid adaptation to extreme hypoxia. WA significantly increased the ability of SD rats to adapt to acute extreme hypoxia and increased the survival rate of SD rats after 24 h in an extremely hypoxic environment (at an altitude of 7620 m). Mechanistically, WA may play a protective role against extreme hypoxia by regulating cellular mitochondrial quality control mechanisms. With SD rats as the study’s subjects, we found that WA pretreatment significantly reduced myocardial tissue injury caused by extreme hypoxia, ensured the ATP supply to the heart, and improved cardiac function in the extremely hypoxic environment. By a combination of animal and cellular experiments, we confirmed that WA pretreatment impacted mitochondrial quality control mechanisms, which significantly enhanced mitochondrial biogenesis and BNIP3-mediated mitophagy in the myocardium and cardiomyocytes. The enhanced mitophagy could clear dysfunctional mitochondria damaged by hypoxia to prevent mitochondria-induced cell death, and the enhanced mitochondrial biogenesis could supply cells with an appropriate number of functioning mitochondria under extreme hypoxia. These two pathways, mitophagy and mitochondrial biogenesis, were complementary and maintained mitochondrial respiration to ensure the cellular ATP supply under extreme hypoxia. In summary, our study demonstrated for the first time that WA plays an important role in improving the body’s ability to rapidly adapt to extremely hypoxic environments. Regulation of myocardial mitochondrial quality control mechanisms—that is, upregulation of mitochondrial biogenesis and BNIP3-mediated mitophagy—may be an important mechanism by which WA exerts its anti-hypoxic effects.
Altitude is generally divided into three zones: 1500–3500 m is considered high altitude; 3500–5500 is considered very high altitude; and above 5500 m is considered extremely high altitude [
3]. Numerous studies have reported that acute hypoxic responses occur when an untrained body rapidly enters an altitude of more than 2500 m [
13]. Adequate acclimatization can significantly reduce the occurrence of acute hypoxic responses. It is generally recognized that adaptation is achieved by staying 6–7 days at an altitude of approximately 2000–3000 m or ascending less than 500 m per day [
14]. At an altitude of 7620 m, the ambient oxygen partial pressure is 60 mmHg, and the alveolar oxygen partial pressure falls to 30 mmHg, which can cause loss of consciousness. The human body, even after acclimation, cannot stay at this altitude for a long time. We used SD rats as the model and found that most unacclimated SD rats survived for only approximately 6 h at the simulated altitude of 7620 m, and the 24 h survival rate was only 9.5%. Achieving rapid adaptation to hypoxia in extreme altitude environments (>7620 m) is a challenging problem.
WA is a chemical component extracted from the Indian herb
Withania somnifera [
4]. WA has anti-inflammatory, antidiabetic, anticancer, and antiangiogenic effects, which have attracted widespread interest in this substance worldwide. WA exerts its anticancer effect by inhibiting the ubiquitin–proteasome pathway, inhibiting cell cycle progression, regulating oxidative stress, inducing cancer cell senescence, and inhibiting NF-κB [
4]. In vivo studies in mice have shown that WA has inhibitory effects on prostate cancer, breast cancer, pancreatic cancer, thyroid cancer, and ovarian cancer [
4,
15,
16,
17]. Other studies have shown that WA is a leptin sensitizer, exerting a strong antidiabetic effect in mice [
18]. More recently, reports have indicated that WA can also be used as a potential drug for the treatment of coronavirus disease 2019 (COVID-19) [
19].
As WA is a pleiotropic drug, we were encouraged to find that it significantly improved the hypoxia tolerance of SD rats. After intraperitoneal injection of WA for 7 days at a dosage of 2 mg/kg/d, we found that the survival of SD rats at the simulated altitude of 7620 m was significantly improved and that the 24 h survival rate increased to 85.7%. In addition, myocardial hypoxic injury was significantly reduced, and cardiac function after hypoxia was effectively improved in WA-pretreated rats. Moreover, our in vitro experiments confirmed that acute hypoxic injury was significantly reduced in cardiomyocytes pretreated with WA, as evidenced by the significant reductions in the apoptosis and necrosis rates. Previous research also confirmed that the therapeutic dose of WA has minimal toxic effects on normal tissues and cells [
20,
21], explaining why WA has a cytotoxic effect on cancer cells but does not damage—or even protects—normal cells. However, the reasons for these effects are still not clear.
The most fundamental cause of hypoxia-induced injury is the O
2 deficiency-induced lack of ATP supply to cells, especially those in cardiac and brain tissues, which have a great demand for energy. Extreme O
2 deficiency can easily cause the death of cardiac and brain tissues. Mitochondria are the energy factories of cells. Electrons carried by substrates are transferred to O
2 in the mitochondrial electron transport chain (ETC), which is coupled to the production of ATP via a process called mitochondrial respiration or oxidative phosphorylation [
22]. At 7620 m, although the oxygen partial pressure of human alveoli is reduced to 30 mmHg, the oxygen partial pressure of cellular mitochondria is still above 1 mmHg, and cells can still produce ATP through oxidative phosphorylation.
In the human body, the heart is the organ with the greatest need for energy. Almost all ATP production in the adult mammalian myocardium is via mitochondrial oxidative phosphorylation; thus, mitochondria are particularly important in myocardial tissues. Diverse molecular pathways, including mitochondrial fission and fusion, mitophagy, and mitochondrial biogenesis, are involved in the regulation of mitochondrial quality; these pathways are collectively referred to as mitochondrial quality control mechanisms [
23,
24]. The mitochondrial fission process in mammalian cells is regulated mainly by DRP1, which is localized in the cytoplasm. Under the action of mitochondrial fission-inducing factors, DRP1 binds to the outer mitochondrial membrane protein FIS1 to induce mitochondrial fission [
25]. Mitochondrial fusion involves the fusion of the inner membrane and the outer membrane. MFN1 and MFN2 mediate mitochondrial outer membrane fusion, and OPA1 is involved mainly in mitochondrial inner membrane fusion [
25]. Mitophagy can remove damaged, aged, and dysfunctional mitochondria from cells. Mitophagy is activated mainly via three receptor-mediated pathways: the PINK1/Parkin-mediated, FUNDC1-mediated, and BNIP3-mediated mitophagy pathways [
26]. Studies have demonstrated that mitophagy induced under hypoxia depends mainly on the activation of BNIP3 [
26,
27]. PGC-1α is a transcriptional cofactor that plays a core regulatory role in mitochondrial biogenesis [
28]. PGC-1α promotes the expression of TFAM by activating the downstream transcription factors NRF1 and NRF2, which can promote the transcription of mitochondrial enzyme-related genes and interact with TFAM to drive mtDNA transcription and replication [
28].
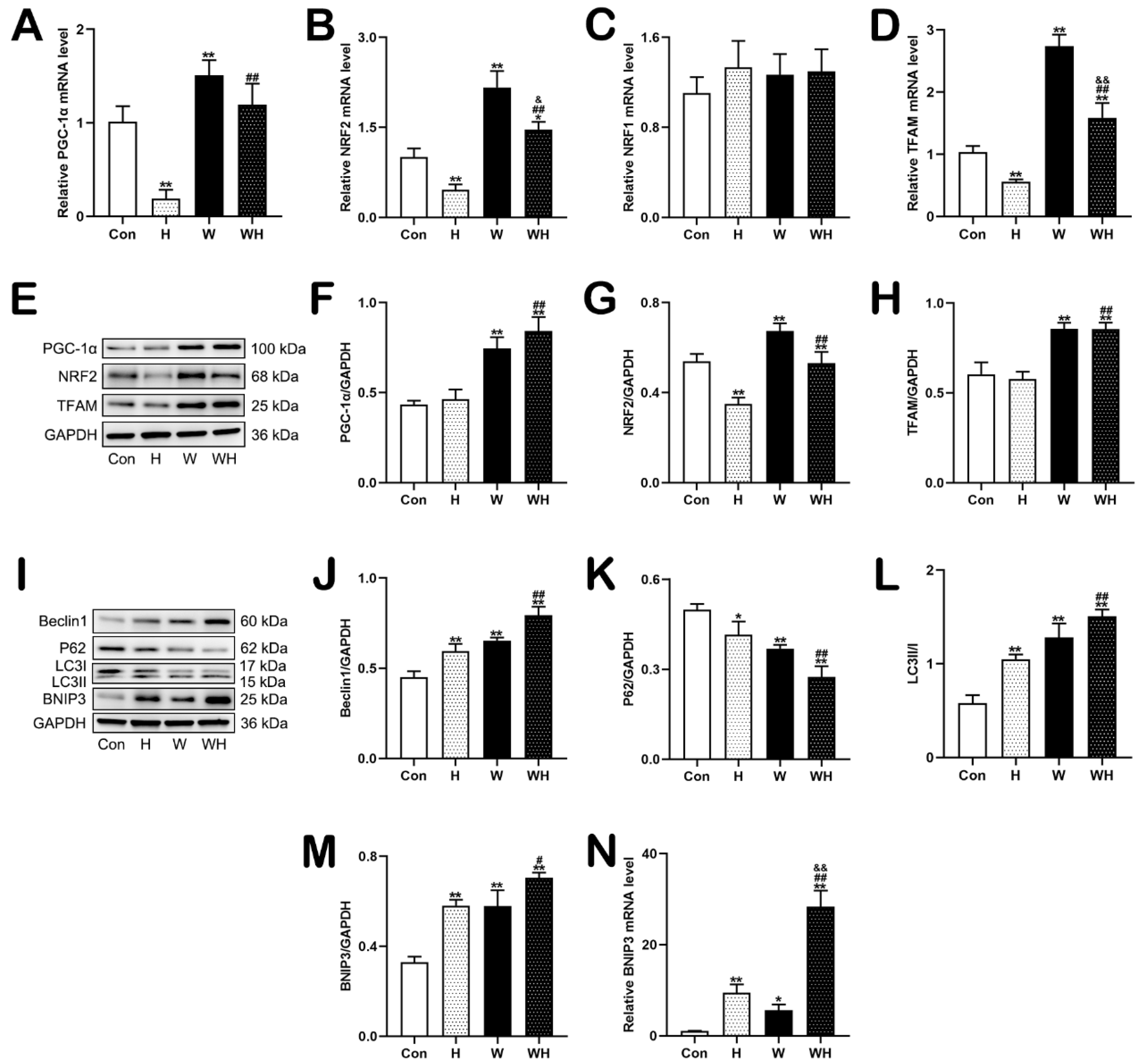
This study showed that the transcriptional levels of the mitochondrial biogenesis-related genes PGC-1α, NRF2, and TFAM, as well as the levels of the encoded proteins, were significantly increased in myocardial tissues of rats pretreated with WA. The mRNA and protein levels of the mitophagy-related gene BNIP3 were also significantly increased, whereas the expression of proteins associated with mitochondrial fission and fusion did not change, suggesting that WA has an impact on mitochondrial quality control mechanisms—mainly regulating the processes of mitochondrial biogenesis and mitophagy—but has no significant effect on mitochondrial fission or fusion. Although the protein expression levels of PGC-1α, NRF2, and TFAM in myocardial tissues of the rats in the WH group did not change compared with those in the W group, the corresponding mRNA levels were significantly reduced. The results of our in vitro experiments confirmed this finding. WA pretreatment significantly enhanced mitochondrial biogenesis and BNIP3-mediated mitophagy in cardiomyocytes in the W group, and the mRNA levels of PGC-1α, NRF2, and TFAM were significantly decreased in cardiomyocytes in the WH group (after acute hypoxia) compared with those in the W group (before hypoxia). These results suggest that acute hypoxia may inhibit the process of mitochondrial biogenesis. Our previous study has verified that the BNIP3-mediated mitophagy plays an important role in clearing damaged mitochondria, further reduces the production of mitochondrial-derived ROS, and prevents further damage caused by increased ROS [
29,
30]. In this study, we confirmed that mitochondrial damage was significantly reduced in myocardial tissues and cells after WA pretreatment and that the production of ROS in tissues and cells was significantly reduced. Both in vivo and in vitro, qPCR analysis of mtDNA and TEM imaging showed that the H group had significantly fewer mitochondria than the Con group, while the WH group had significantly fewer mitochondria than the W group; moreover, the few remaining mitochondria in the H group were dysfunctional. Both of these results suggest a significant decrease in the number of mitochondria in myocardial tissue and cells after hypoxia. On the one hand, the decrease in the number of mitochondria after hypoxia may be due to hypoxia-induced inhibition of mitochondrial biogenesis. On the other hand, hypoxia-induced rupture and dissolution of mitochondria is also an important cause of the reduction in mitochondria. The protection role of PGC-1α in acute hypoxia was confirmed in this study. Because WA pretreatment upregulated PGC-1α-mediated mitochondrial biogenesis, even though the number of mitochondria decreased after hypoxia, the number of mitochondria in myocardial tissues and cells in the WH group was still similar to that in the Con group. Most importantly, these new mitochondria exhibited normal function. The results of mitochondrial function assays suggested that the reduction in the MMP in cardiomyocytes under acute hypoxia was alleviated after WA pretreatment and that the level of mitochondrial respiration was maintained to ensure the production of ATP under acute hypoxia.
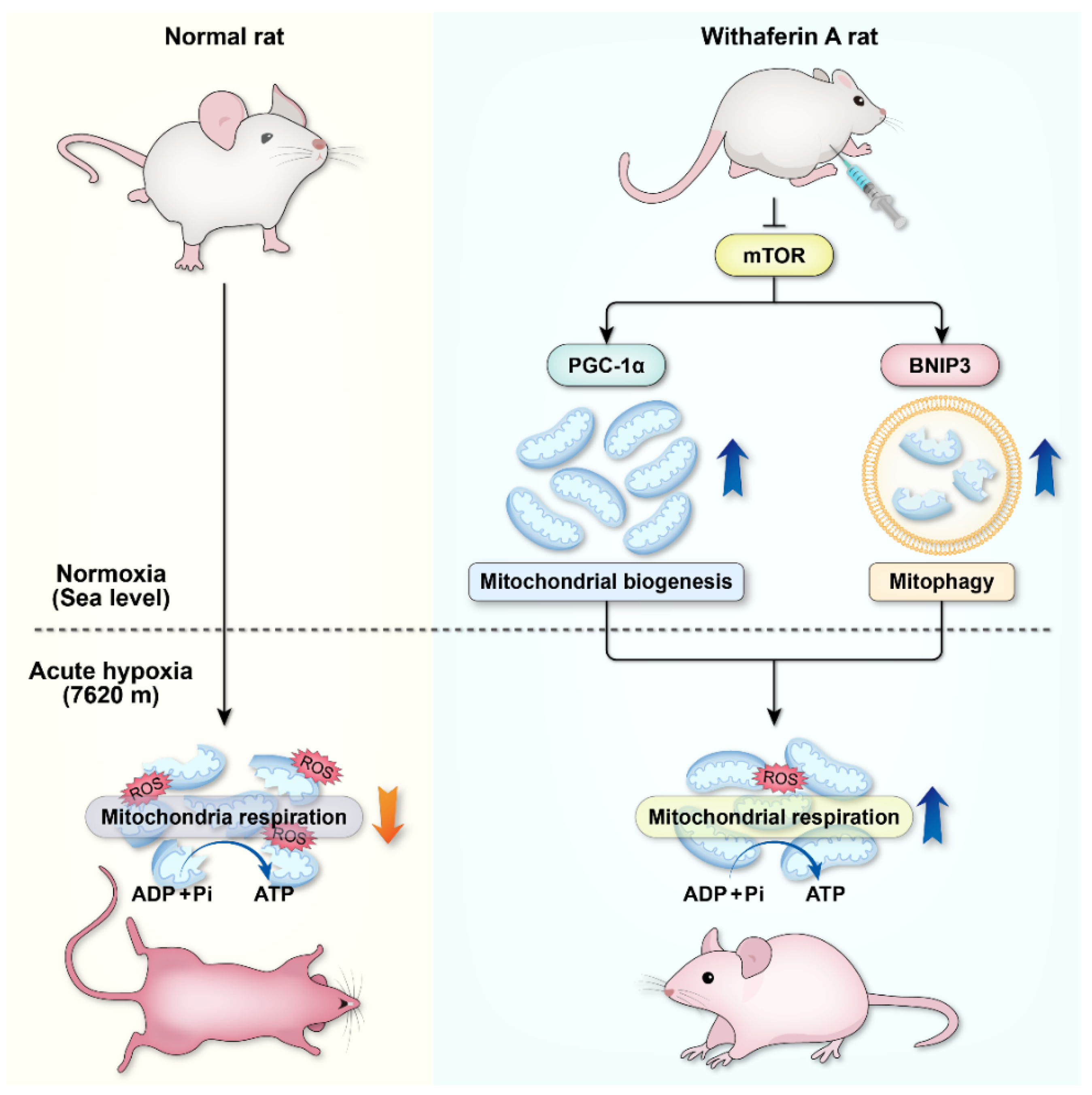
In summary, this study has demonstrated that acute hypoxia can cause severe mitochondrial damage and decrease the number of mitochondria, making it difficult to maintain effective mitochondrial respiration. WA can improve the body’s extreme hypoxia tolerance. Our previous findings highlight the protective role of mTOR in acute hypoxic adaptation, and targeted regulation of mTOR could be a new strategy to improve acute hypoxic tolerance in the body [
30]. As the upstream regulator of PGC1α and BNIP3, mTOR should be an important target for WA to play its role. By inhibiting the mTOR, on the one hand, WA activates BNIP3-mediated mitophagy for rapid clearance of mitochondria damaged by extreme hypoxia, thereby preventing the increase in ROS and the activation of apoptosis caused by dysfunctional mitochondria. On the other hand, WA activates mitochondrial biogenesis, and the cleared mitochondria are continuously replaced by newly formed mitochondria, which exhibit normal mitochondrial respiratory function. Mitophagy and mitochondrial biogenesis coordinate with each other to ensure that the energy supply of the body is maintained in an extremely hypoxic environment (
Figure 9). Therefore, WA can be used as an effective drug to improve the body’s adaptability to extreme hypoxia. Mitophagy and mitochondrial biogenesis play an important role in the rapid adaptation to extreme hypoxia. Targeted regulation of mitophagy or mitochondrial biogenesis could be a novel strategy to improve tolerance to extreme hypoxia.