Multiformin-Type Azaphilones Prevent SARS-CoV-2 Binding to ACE2 Receptor
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Human ACE2
2.3. Spectral Equipment
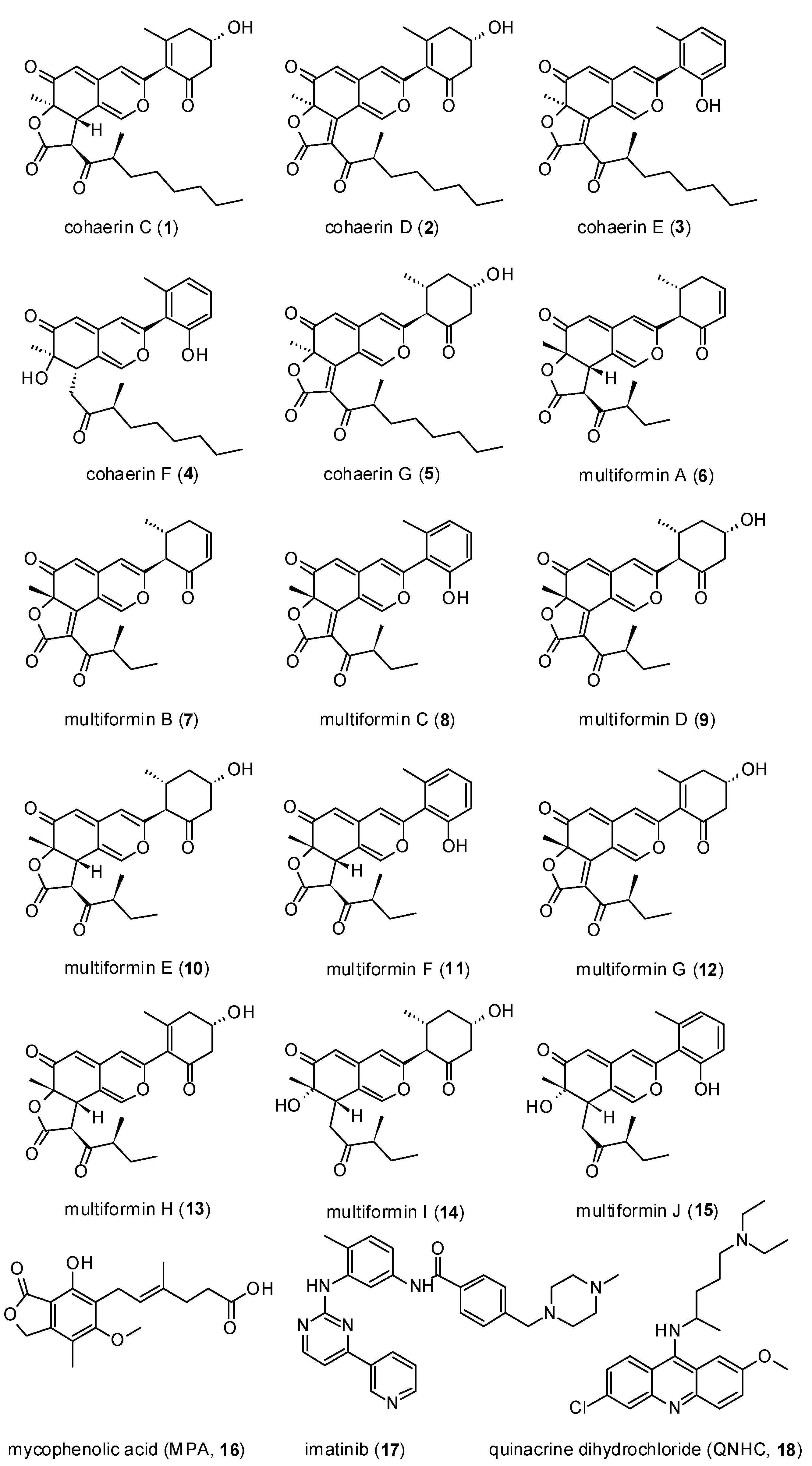
2.4. Isolation of Azaphilones
2.5. Spectral Data
2.6. Mosher’s Analysis of Multiformin E (10)
2.7. Expression and Purification of Human ACE2
2.8. Cy5 labeling of ACE2 Protein
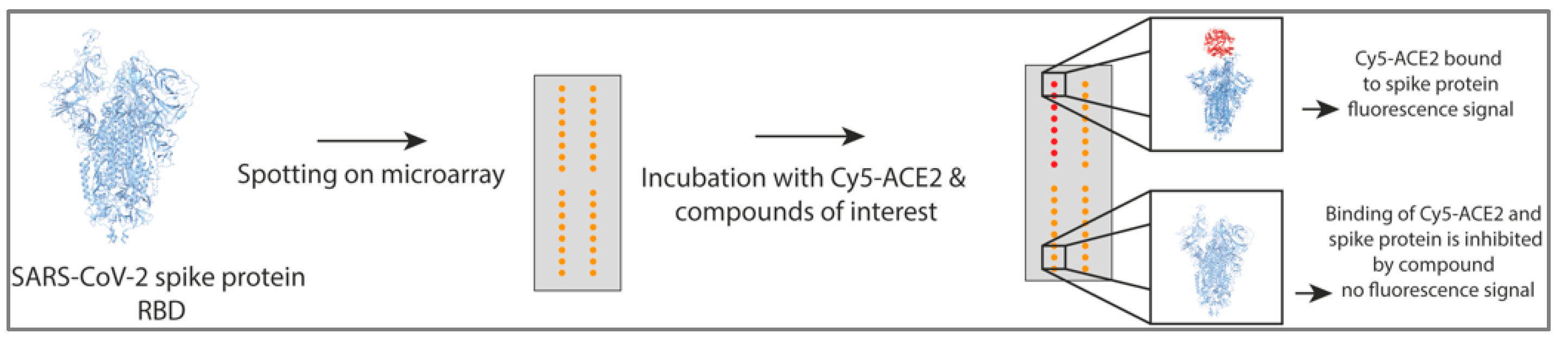
2.9. Protein Microarray Detecting ACE2 and Spike Protein Interaction
2.10. Analysis of the Antiviral Activity of Compounds on Spike-GFP Production
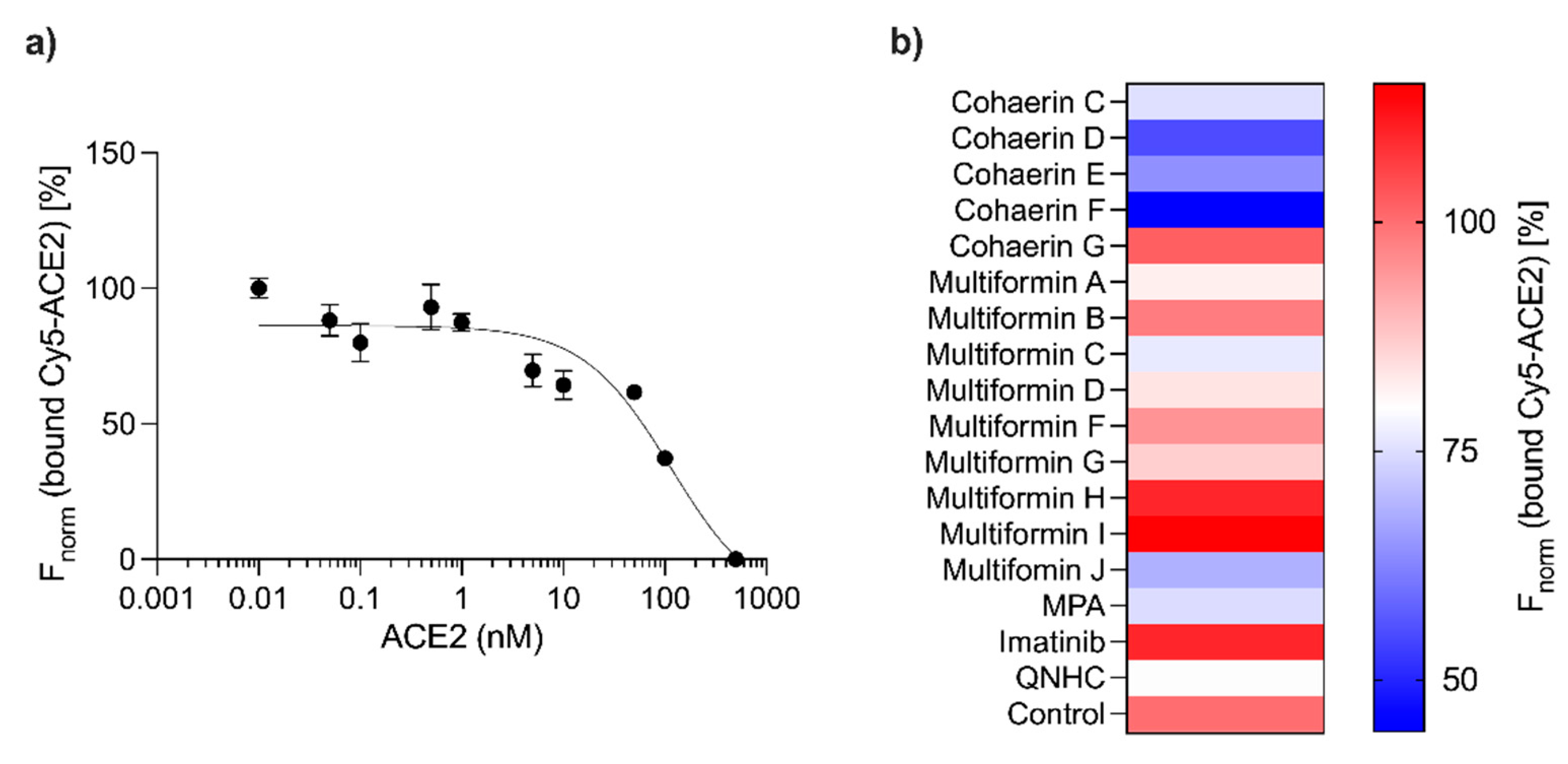
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e286. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Tian, E.K.; He, B.; Tian, L.; Han, R.; Wang, S.; Xiang, Q.; Zhang, S.; El Arnaout, T.; Cheng, W. Overview of lethal human coronaviruses. Signal Transduct. Target. Ther. 2020, 5, 89. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef]
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 2018, 100, 163–188. [Google Scholar] [CrossRef] [PubMed]
- Kirtipal, N.; Bharadwaj, S.; Kang, S.G. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect. Genet. Evol. 2020, 85, 104502. [Google Scholar] [CrossRef]
- Hamming, I.; Cooper, M.E.; Haagmans, B.L.; Hooper, N.M.; Korstanje, R.; Osterhaus, A.D.; Timens, W.; Turner, A.J.; Navis, G.; van Goor, H. The emerging role of ACE2 in physiology and disease. J. Pathol. 2007, 212, 1–11. [Google Scholar] [CrossRef]
- Crackower, M.A.; Sarao, R.; Oudit, G.Y.; Yagil, C.; Kozieradzki, I.; Scanga, S.E.; Oliveira-dos-Santos, A.J.; da Costa, J.; Zhang, L.; Pei, Y.; et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature 2002, 417, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Díez-Freire, C.; Vázquez, J.; Correa de Adjounian, M.F.; Ferrari, M.F.; Yuan, L.; Silver, X.; Torres, R.; Raizada, M.K. ACE2 gene transfer attenuates hypertension-linked pathophysiological changes in the SHR. Physiol. Genom. 2006, 27, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Tikellis, C.; Pickering, R.; Tsorotes, D.; Du, X.J.; Kiriazis, H.; Nguyen-Huu, T.P.; Head, G.A.; Cooper, M.E.; Thomas, M.C. Interaction of diabetes and ACE2 in the pathogenesis of cardiovascular disease in experimental diabetes. Clin. Sci. 2012, 123, 519–529. [Google Scholar] [CrossRef]
- Velkoska, E.; Patel, S.K.; Burrell, L.M. Angiotensin converting enzyme 2 and diminazene: Role in cardiovascular and blood pressure regulation. Curr. Opin. Nephrol. Hypertens. 2016, 25, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Shults, N.V.; Harris, B.T.; Suzuki, Y.J. Angiotensin-converting enzyme 2 (ACE2) is upregulated in Alzheimer’s disease brain. bioRxiv 2020. [Google Scholar] [CrossRef]
- Dales, N.A.; Gould, A.E.; Brown, J.A.; Calderwood, E.F.; Guan, B.; Minor, C.A.; Gavin, J.M.; Hales, P.; Kaushik, V.K.; Stewart, M.; et al. Substrate-based design of the first class of angiotensin-converting enzyme-related carboxypeptidase (ACE2) inhibitors. J. Am. Chem. Soc. 2002, 124, 11852–11853. [Google Scholar] [CrossRef] [PubMed]
- Mores, A.; Matziari, M.; Beau, F.; Cuniasse, P.; Yiotakis, A.; Dive, V. Development of potent and selective phosphinic peptide inhibitors of angiotensin-converting enzyme 2. J. Med. Chem. 2008, 51, 2216–2226. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Damas, J.; Hughes, G.M.; Keough, K.C.; Painter, C.A.; Persky, N.S.; Corbo, M.; Hiller, M.; Koepfli, K.P.; Pfenning, A.R.; Zhao, H.; et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. USA 2020, 117, 22311–22322. [Google Scholar] [CrossRef]
- Jan, J.T.; Cheng, T.R.; Juang, Y.P.; Ma, H.H.; Wu, Y.T.; Yang, W.B.; Cheng, C.W.; Chen, X.; Chou, T.H.; Shie, J.J.; et al. Identification of existing pharmaceuticals and herbal medicines as inhibitors of SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA 2021, 118, e2021579118. [Google Scholar] [CrossRef]
- Fehr, A.R.; Perlman, S. Coronaviruses: An Overview of Their Replication and Pathogenesis; Springer: New York, NY, USA, 2015. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Leist, S.R.; Schäfer, A.; Won, J.; Brown, A.J.; Montgomery, S.A.; Hogg, A.; Babusis, D.; Clarke, M.O.; et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020, 11, 222. [Google Scholar] [CrossRef]
- Tanner, J.A.; Watt, R.M.; Chai, Y.B.; Lu, L.Y.; Lin, M.C.; Peiris, J.M.; Poon, L.L.; Kung, H.F.; Huang, J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003, 278, 39578–39582. [Google Scholar] [CrossRef]
- Yadav, R.; Imran, M.; Dhamija, P.; Suchal, K.; Handu, S. Virtual screening and dynamics of potential inhibitors targeting RNA binding domain of nucleocapsid phosphoprotein from SARS-CoV-2. J. Biomol. Struct. Dyn. 2021, 39, 4433–4448. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020, 178, 104787. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Duan, X.; Yang, L.; Nilsson-Payant, B.E.; Wang, P.; Duan, F.; Tang, X.; Yaron, T.M.; Zhang, T.; Uhl, S.; et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 2021, 589, 270–275. [Google Scholar] [CrossRef]
- Bakhshandeh, B.; Sorboni, S.G.; Javanmard, A.R.; Mottaghi, S.S.; Mehrabi, M.R.; Sorouri, F.; Abbasi, A.; Jahanafrooz, Z. Variants in ACE2; potential influences on virus infection and COVID-19 severity. Infect. Genet. Evol. 2021, 90, 104773. [Google Scholar] [CrossRef]
- Gan, H.H.; Twaddle, A.; Marchand, B.; Gunsalus, K.C. Structural Modeling of the SARS-CoV-2 Spike/Human ACE2 Complex Interface can Identify High-Affinity Variants Associated with Increased Transmissibility. J. Mol. Biol. 2021, 433, 167051. [Google Scholar] [CrossRef]
- Wendt, L.; Sir, E.B.; Kuhnert, E.; Heitkämper, S.; Lambert, C.; Hladki, A.I.; Romero, A.I.; Luangsa-ard, J.J.; Srikitikulchai, P.; Peršoh, D.; et al. Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycol. Prog. 2018, 17, 115–154. [Google Scholar] [CrossRef]
- Surup, F.; Mohr, K.I.; Jansen, R.; Stadler, M. Cohaerins G-K, azaphilone pigments from Annulohypoxylon cohaerens and absolute stereochemistry of cohaerins C-K. Phytochemistry 2013, 95, 252–258. [Google Scholar] [CrossRef]
- Quang, D.N.; Hashimoto, T.; Nomura, Y.; Wollweber, H.; Hellwig, V.; Fournier, J.; Stadler, M.; Asakawa, Y. Cohaerins A and B, azaphilones from the fungus Hypoxylon cohaerens, and comparison of HPLC-based metabolite profiles in Hypoxylon sect. Annulata Phytochem. 2005, 66, 797–809. [Google Scholar] [CrossRef]
- Hoye, T.R.; Jeffrey, C.S.; Shao, F. Mosher ester analysis for the determination of absolute configuration of stereogenic (chiral) carbinol carbons. Nat. Protoc. 2007, 2, 2451–2458. [Google Scholar] [CrossRef] [PubMed]
- Quang, D.N.; Hashimoto, T.; Stadler, M.; Radulović, N.; Asakawa, Y. Antimicrobial azaphilones from the fungus Hypoxylon multiforme. Planta Med. 2005, 71, 1058–1062. [Google Scholar] [CrossRef] [PubMed]
- Pyser, J.B.; Baker Dockrey, S.A.; Benítez, A.R.; Joyce, L.A.; Wiscons, R.A.; Smith, J.L.; Narayan, A.R.H. Stereodivergent, chemoenzymatic synthesis of azaphilone natural products. J. Am. Chem. Soc. 2019, 141, 18551–18559. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Feng, X.; Xiao, Z.; Liu, L.; Li, H.; Ma, L.; Lu, Y.; Ju, J.; She, Z.; Lin, Y. Azaphilones and p-terphenyls from the mangrove endophytic fungus Penicillium chermesinum (ZH4-E2) isolated from the South China Sea. J. Nat. Prod. 2011, 74, 997–1002. [Google Scholar] [CrossRef]

| POS | 10 | 11 | 12 | 13 | 14 | 15 a |
|---|---|---|---|---|---|---|
| 1 | 7.36, d (0.9) | 7.46, s | 8.75, d (1.3) | 7.38, d (1.3) | 6.61, s | 7.43, br s |
| 4 | 6.26, s | 6.47, s | 6.41, s | 6.24, s | 6.21, d (2.8) | 6.27, m |
| 5 | 5.3, d (1.3) | 5.42, s | 5.35, d (0.86) | 5.35, d (1.3) | 5.29, m | 5.44, s |
| 8 | 3.91, d (12.1) | 3.98, d (12.1) | 3.94, d (12.1) | 3.32, m | 3.47, m | |
| 9 | 1.51, s | 1.56, s | 1.68, s | 1.53, s | 1.21, s | 1.36, s |
| 10 | 3.32, d (12.1) | 3.27, m | ||||
| 12 | 2.63, ddd (13.4, 5.0, 1.9) | 6.85, d (8.2) | 2.73, dt (15.9, 1.8) | 2.69, dd (15.9, 4.0) | 2.62, m | 6.75, d (7.9) |
| 2.48, dd (13.4,11.6) | 2.541 ddd (15.9, 7.7, 0.9) | 2.50, ddd (15.9, 8.0, 0.9) | 2.46, m | |||
| 13 | 3.92, m | 7.19, m | 4.34, ddt (7.8, 6.1, 4.0) | 4.30, ddt (8.0, 5.4, 4.0) | 3.91, m | 7.20, t (7.9) |
| 14 | 1.60, m | 6.79, s | 2.63, dd (18.5, 6.5) | 2.86, m | 2.23 dtd (13.0, 3.8, 2.2) | 6.82, d (7.9) |
| 2.23, m | 2.91, m | 2.59, m | 1.59, m | |||
| 15 | 2.11, m | 2.08, m | ||||
| 16 | 1.04, d (6.5) | 2.22, m | 2.11, m | 2.02, br s | 1.02, d (6.5) | 2.22, s |
| 18 | 4.41, d (12.1) | 4.47, d (12.1) | 4.43, d (12.1) | 2.85, m | 3.02, m | |
| 2.49, m | 2.51, m | |||||
| 20 | 2.95, dqd (7.3, 7.2, 6.0) | 2.96, m | 3.50, sxt (6.9) | 2.95, m | 2.39, m | 2.33 m |
| 21 | 1.74, ddq (14.0, 7.3, 7.2) | 1.75, m | 1.82, m | 1.74, m | 1.59, m | 1.72, m |
| 1.38, dqd (14.0, 7.3, 6.0) | 1.38, m | 1.35, dqd (13.5, 7.3, 6.9) | 1.37, m | 1.29, m | 1.36, m | |
| 22 | 0.87, t (7.3) | 0.87, t (7.5) | 0.94, t (7.3) | 0.87, t (7.5) | 0.82, t (7.3) | 0.87, d (7.5) |
| 23 | 1.06, d (7.3) | 1.06, d (7.3) | 1.00, d (6.9) | 1.05 d (7.3) | 0.95, d (7.0) | 0.96, d (6.9) |
| POS | 10 | 11 | 12 | 13 | 14 | 15 a |
|---|---|---|---|---|---|---|
| 1 | 147.6, CH | 148.1, CH | 154.3, CH | 147.9, CH | 145.4, CH | 147.0, CH |
| 3 | 159.2, C | 156.7, C | 154.7, C | 155.3, C | 158.7, C | 154.7, C |
| 4 | 110.9, CH | 112.2, CH | 113.7, CH | 112.3, CH | 110.8, CH | 111.6, CH |
| 5 | 106.7, CH | 106.9, CH | 106.3, CH | 107.1, CH | 104.0, CH | 104.4, CH |
| 6 | 191.5, C | 191.5, C | 190.5, C | 191.5, C | 198.6, C | 198.8, C |
| 7 | 83.5, C | 83.7, C | 88.4, C | 83.5, C | 73.6, C | 73.2, C |
| 8 | 44.8, CH | 44.5, CH | 167.0, C | 44.8, CH | 42.0, CH | 40.8, CH |
| 9 | 23.2, CH3 | 23.3, CH3 | 26.2, CH3 | 23.3, CH3 | 27.0, CH3 | 26.9, CH3 |
| 10 | 61.7, CH | 120.8, C | 130.6, C | 131.1, C | 61.7, CH | 119.6, C |
| 11 | 204.2, C | 156.5, C | 194.6, C | 194.7, C | 204.2, C | 153.8, C |
| 12 | 51.4, CH2 | 114.3, CH | 46.9, CH2 | 46.9, CH2 | 51.4, CH2 | 113.8, CH |
| 13 | 68.2, CH | 131.8, CH | 65.6, CH | 65.6, CH | 68.2, CH | 131.3, CH |
| 14 | 43.4, CH2 | 122.1, CH | 41.8, CH2 | 41.7, CH2 | 43.4, CH2 | 122.9, CH |
| 15 | 31.6, CH | 139.4, C | 162.5, C | 161.7, C | 31.5, C | 138.8, C |
| 16 | 20.5, CH3 | 19.7, CH3 | 22.9, CH3 | 23.2, CH3 | 20.5, CH3 | 19.9, CH3 |
| 17 | 170.3, C | 170.4, C | 169.0, C | 170.3, C | - | - |
| 18 | 55.7, CH | 55.8, CH | 123.9, C | 55.7, CH | 39.9, CH2 | 39.0, CH2 |
| 19 | 207.2, C | 207.2, C | 201.1, C | 207.2, C | 213.1, C | 213.5, C |
| 20 | 48.5, CH | 48.6, CH | 45.5, CH | 44.5, CH | 49.1, CH | 48.7, CH |
| 21 | 25.2, CH2 | 25.2, CH2 | 25.9, CH2 | 25.2, CH2 | 26.1, CH2 | 25.7, CH2 |
| 22 | 11.9, CH3 | 11.9, CH3 | 12.0, CH3 | 11.9, CH3 | 11.9, CH3 | 11.7, CH3 |
| 23 | 16.0, CH3 | 15.9, CH3 | 16.2, CH3 | 15.9, CH3 | 15.9, CH3 | 15.7, CH3 |
| 4A | 144.3, C | 144.8, C | 144.5, C | 144.4, C | 147.0, C | 144.9, C |
| 8A | 115.5, C | 115.5, C | 111.9, C | 115.5, C | 121.2, C | 120.6, C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jansen-Olliges, L.; Chatterjee, S.; Jia, L.; Stahl, F.; Bär, C.; Stadler, M.; Surup, F.; Zeilinger, C. Multiformin-Type Azaphilones Prevent SARS-CoV-2 Binding to ACE2 Receptor. Cells 2023, 12, 83. https://doi.org/10.3390/cells12010083
Jansen-Olliges L, Chatterjee S, Jia L, Stahl F, Bär C, Stadler M, Surup F, Zeilinger C. Multiformin-Type Azaphilones Prevent SARS-CoV-2 Binding to ACE2 Receptor. Cells. 2023; 12(1):83. https://doi.org/10.3390/cells12010083
Chicago/Turabian StyleJansen-Olliges, Linda, Shambhabi Chatterjee, Lili Jia, Frank Stahl, Christian Bär, Marc Stadler, Frank Surup, and Carsten Zeilinger. 2023. "Multiformin-Type Azaphilones Prevent SARS-CoV-2 Binding to ACE2 Receptor" Cells 12, no. 1: 83. https://doi.org/10.3390/cells12010083
APA StyleJansen-Olliges, L., Chatterjee, S., Jia, L., Stahl, F., Bär, C., Stadler, M., Surup, F., & Zeilinger, C. (2023). Multiformin-Type Azaphilones Prevent SARS-CoV-2 Binding to ACE2 Receptor. Cells, 12(1), 83. https://doi.org/10.3390/cells12010083







