Synergism of Feeding and Digestion Regulated by the Neuropeptide F System in Ostrinia furnacalis Larvae
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Drosophila Strains and Rearing
2.3. RNA Extraction and cDNA Synthesis
2.4. Synthesis of Double-Stranded RNA for Artificial Diet Treatment
2.5. Synthesis of Double-Stranded RNA for Injection Treatment
2.6. Quantitative Real-Time PCR
2.7. Western Blot
2.8. Determination of Midgut Enzyme Activities
2.9. Digestion and Feeding Assays
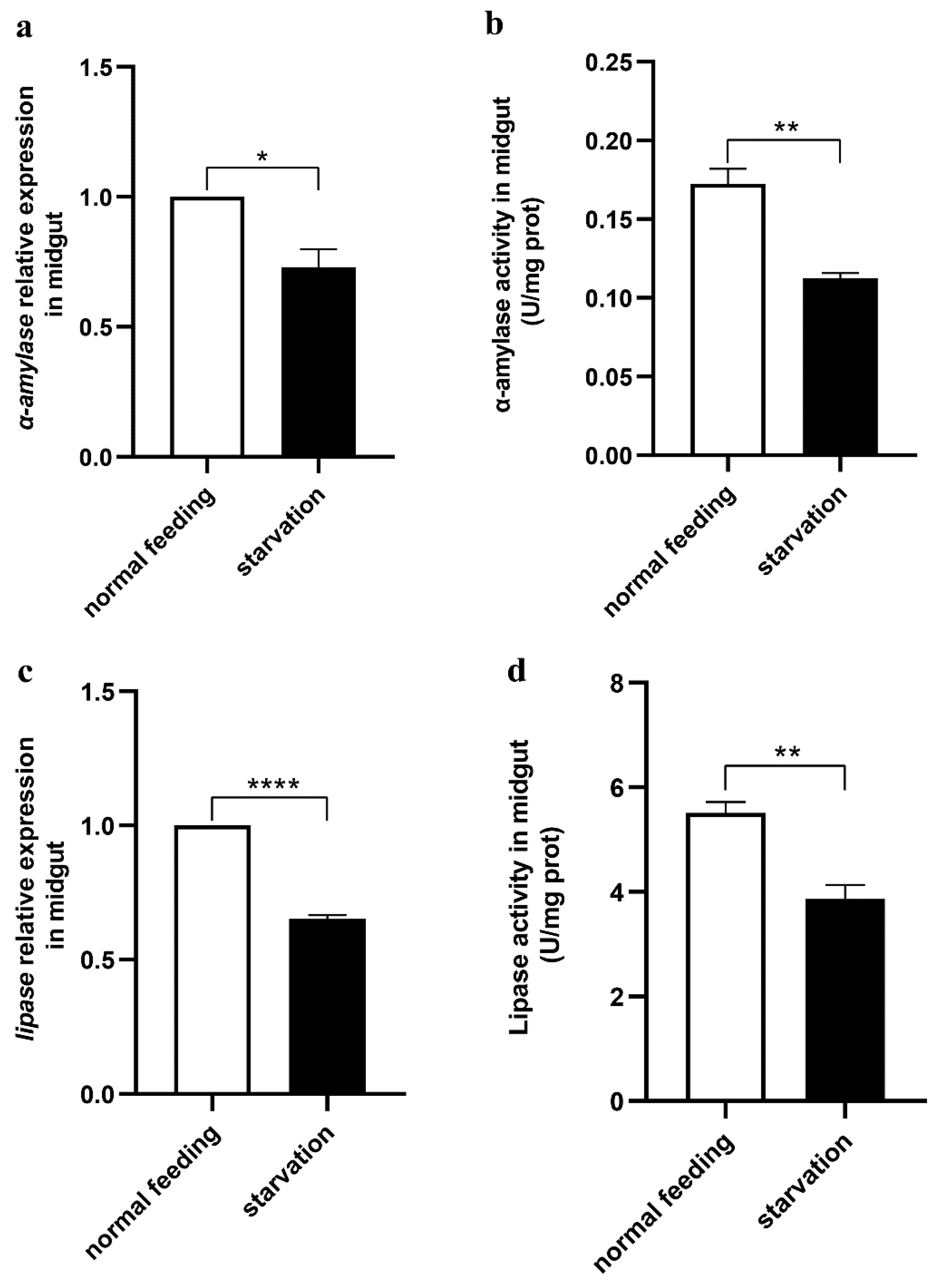
2.10. Starvation Treatment of Larvae
2.11. Rescue Experiments
2.12. Dual Luciferase Reporter Assay
3. Results
3.1. Construction and Expression of dsRNA Vectors
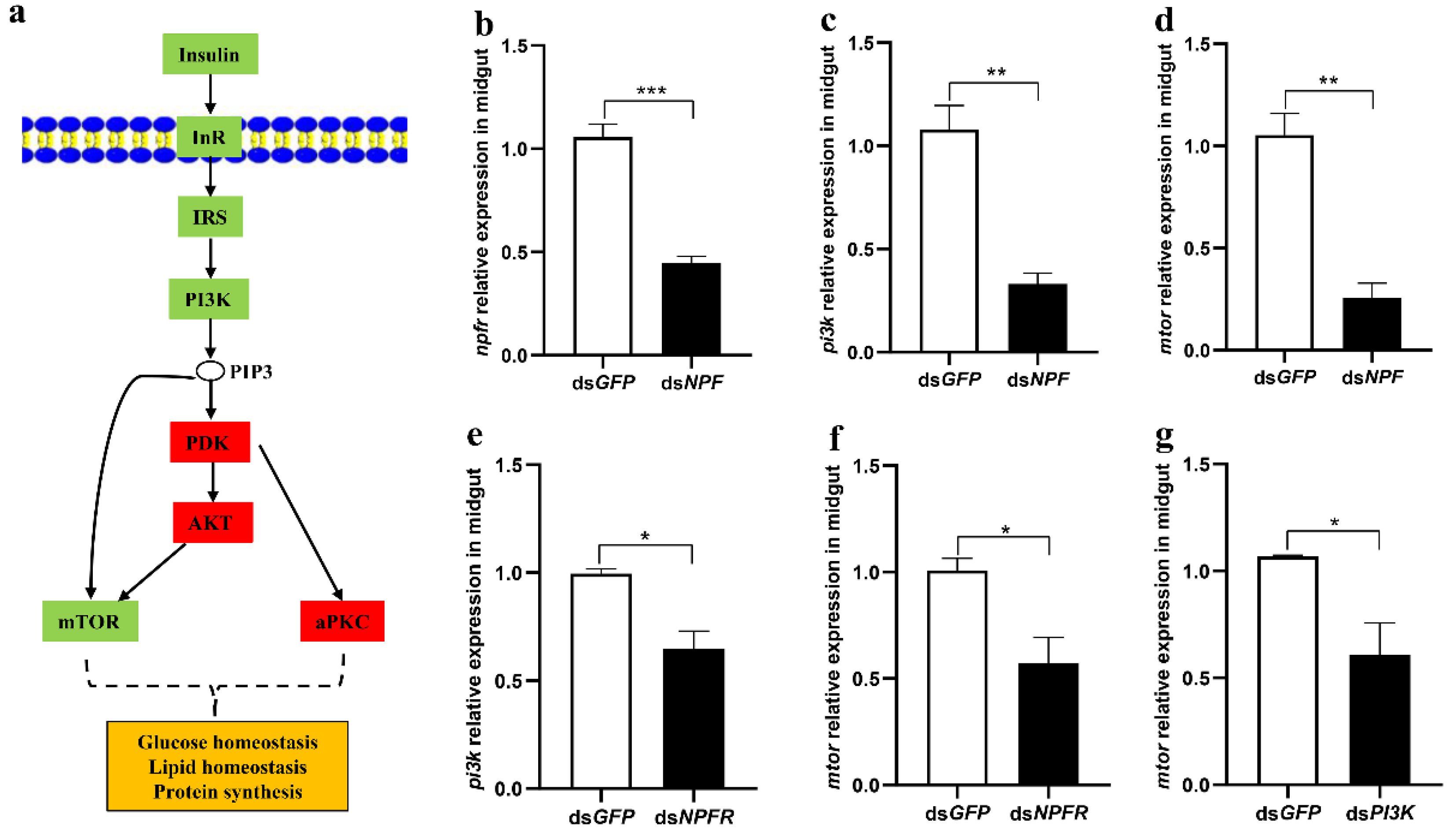
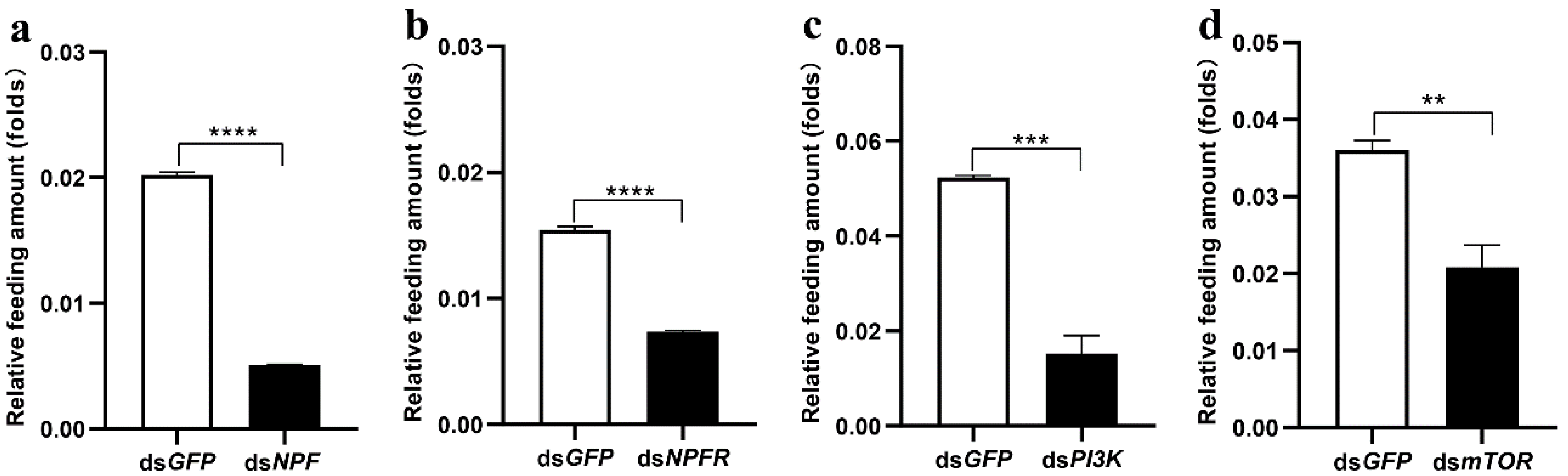
3.2. The Insulin Pathway in the Midgut Is Involved in NPF/NPFR-Mediated Feeding Regulation
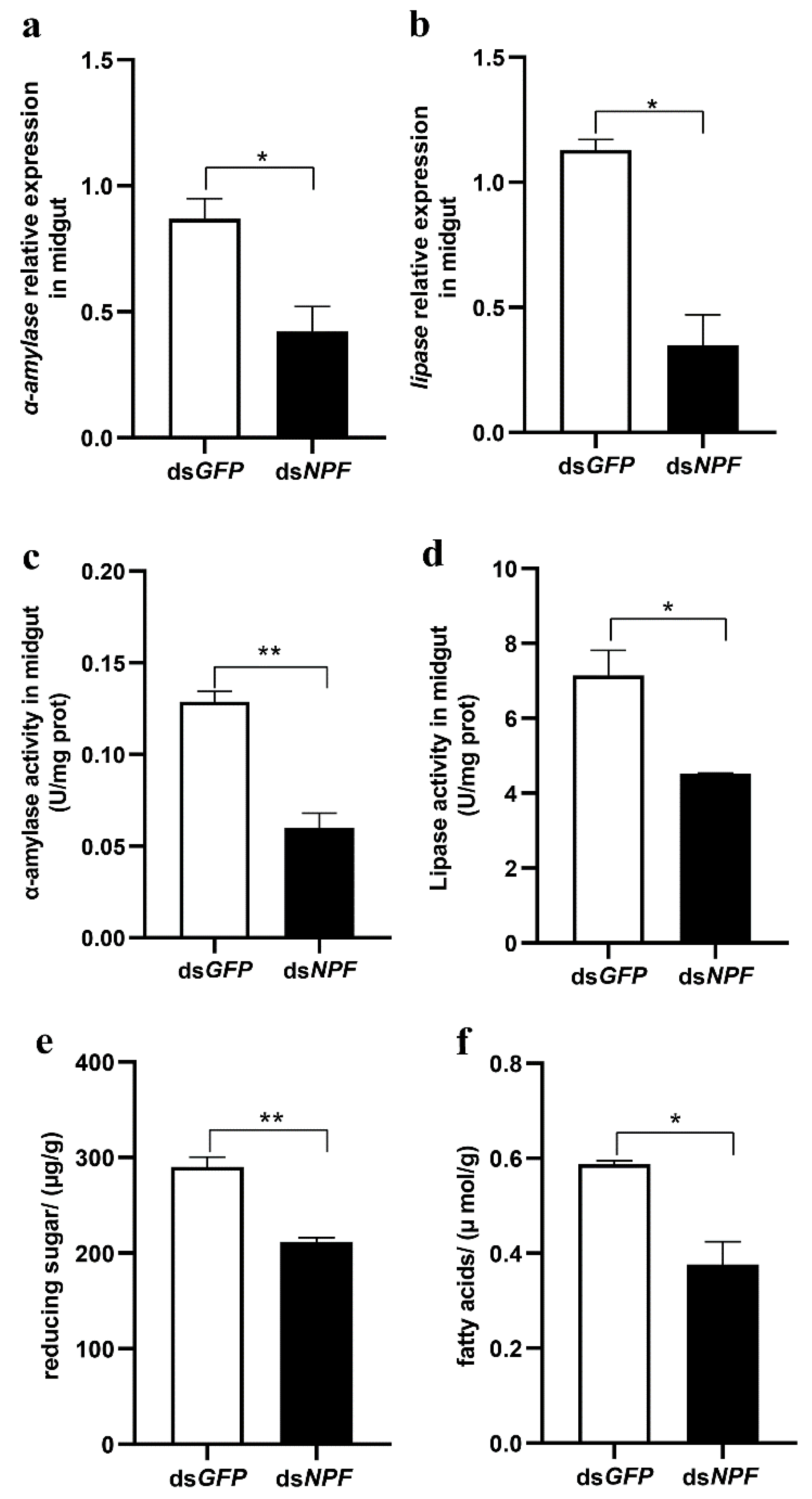
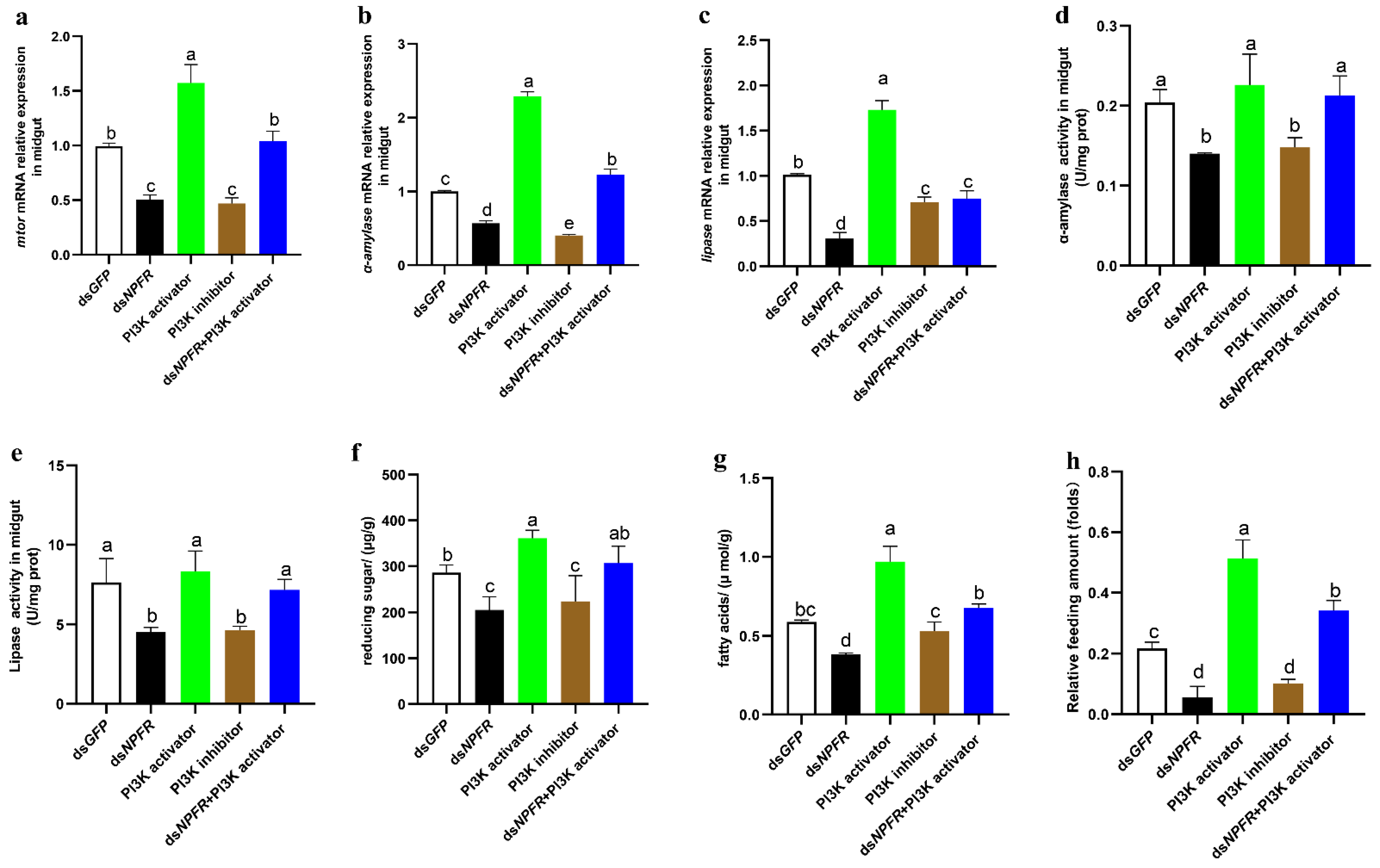
3.3. NPF/NPFR Regulates Digestive Enzymes via the Insulin Pathway
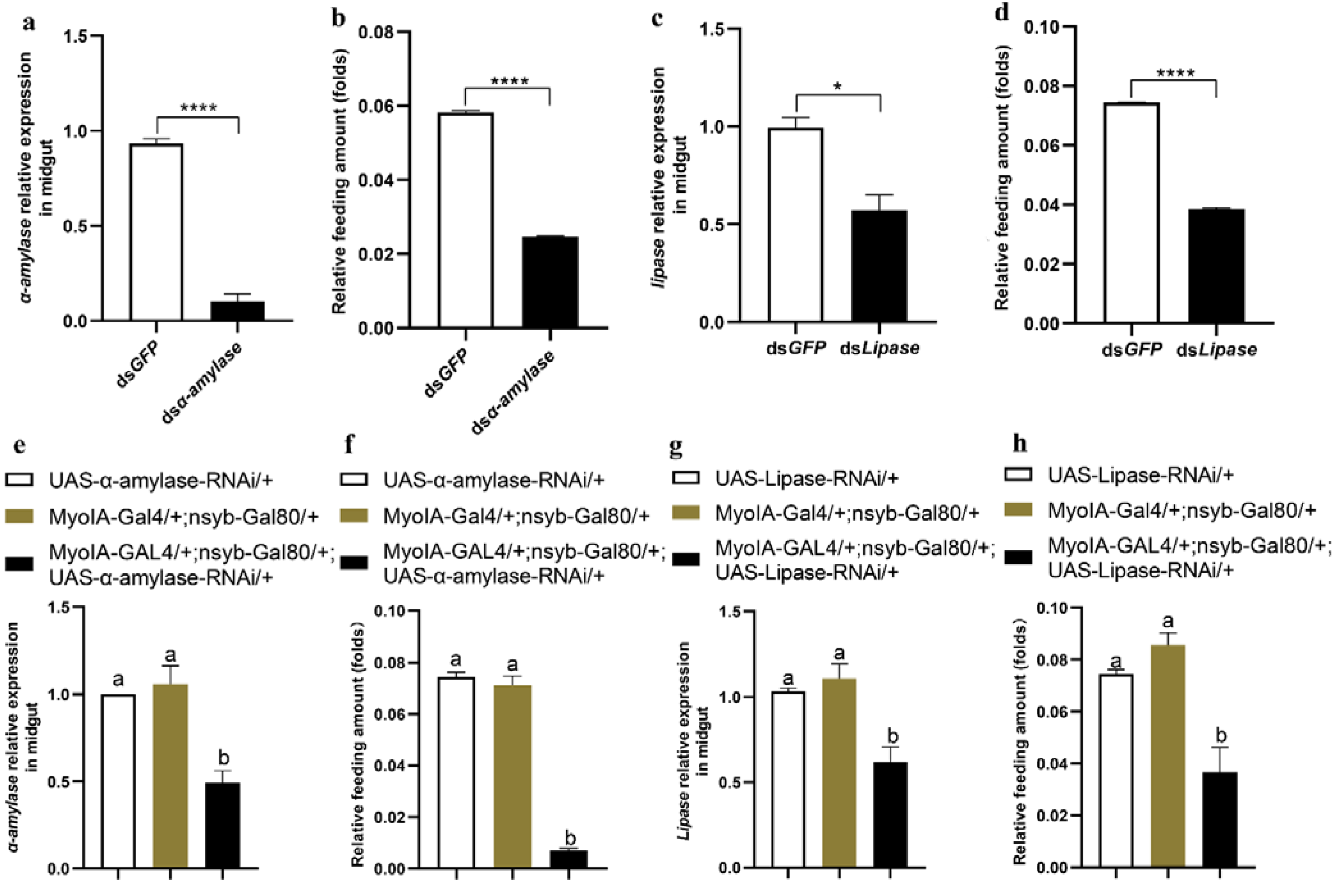
3.4. Midgut Digestive Enzymes Are Closely Related to Larval Feeding
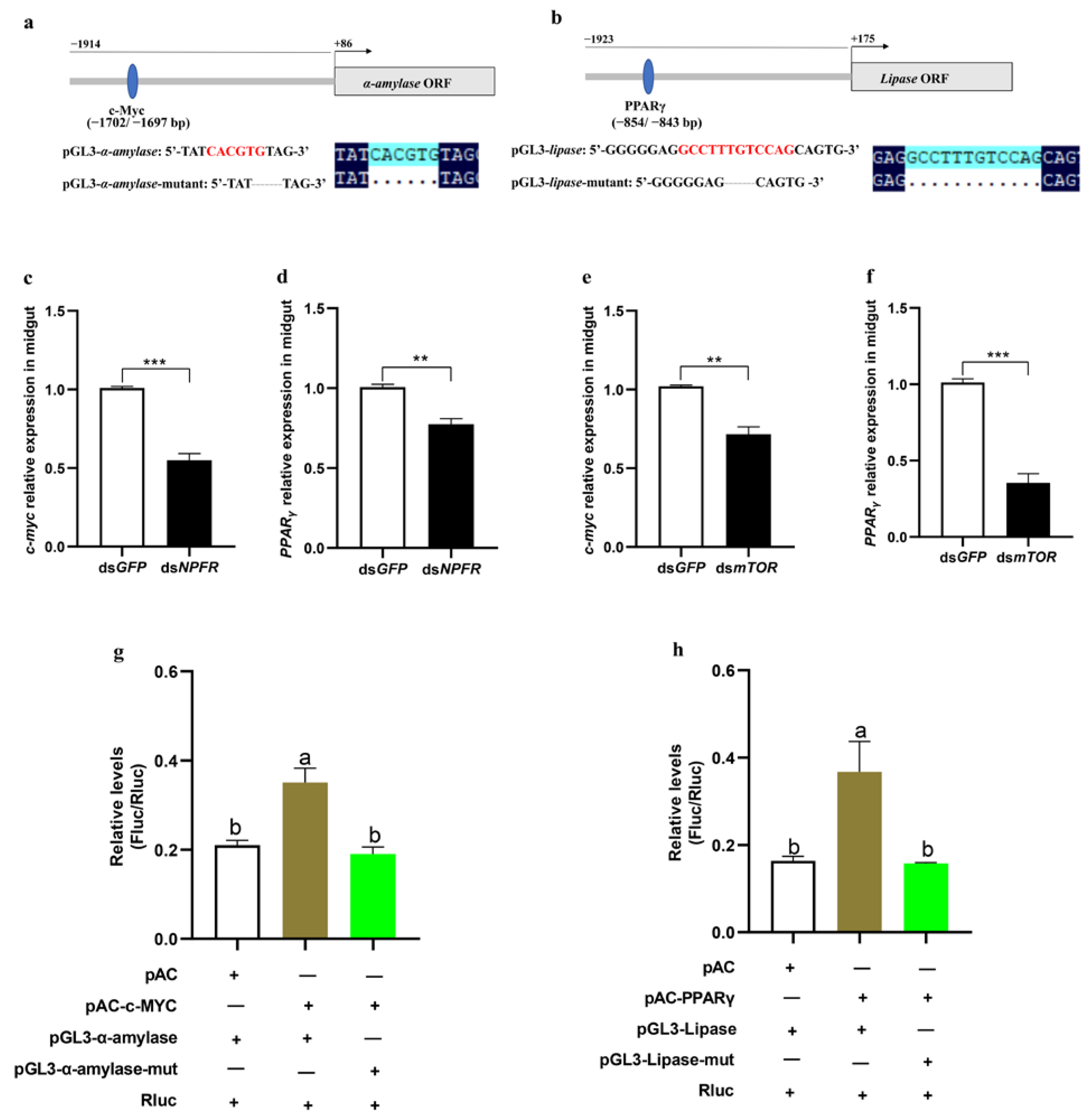
3.5. mTOR Regulates α-Amylase and Lipase Separately via the Transcription Factors c-Myc and PPARγ
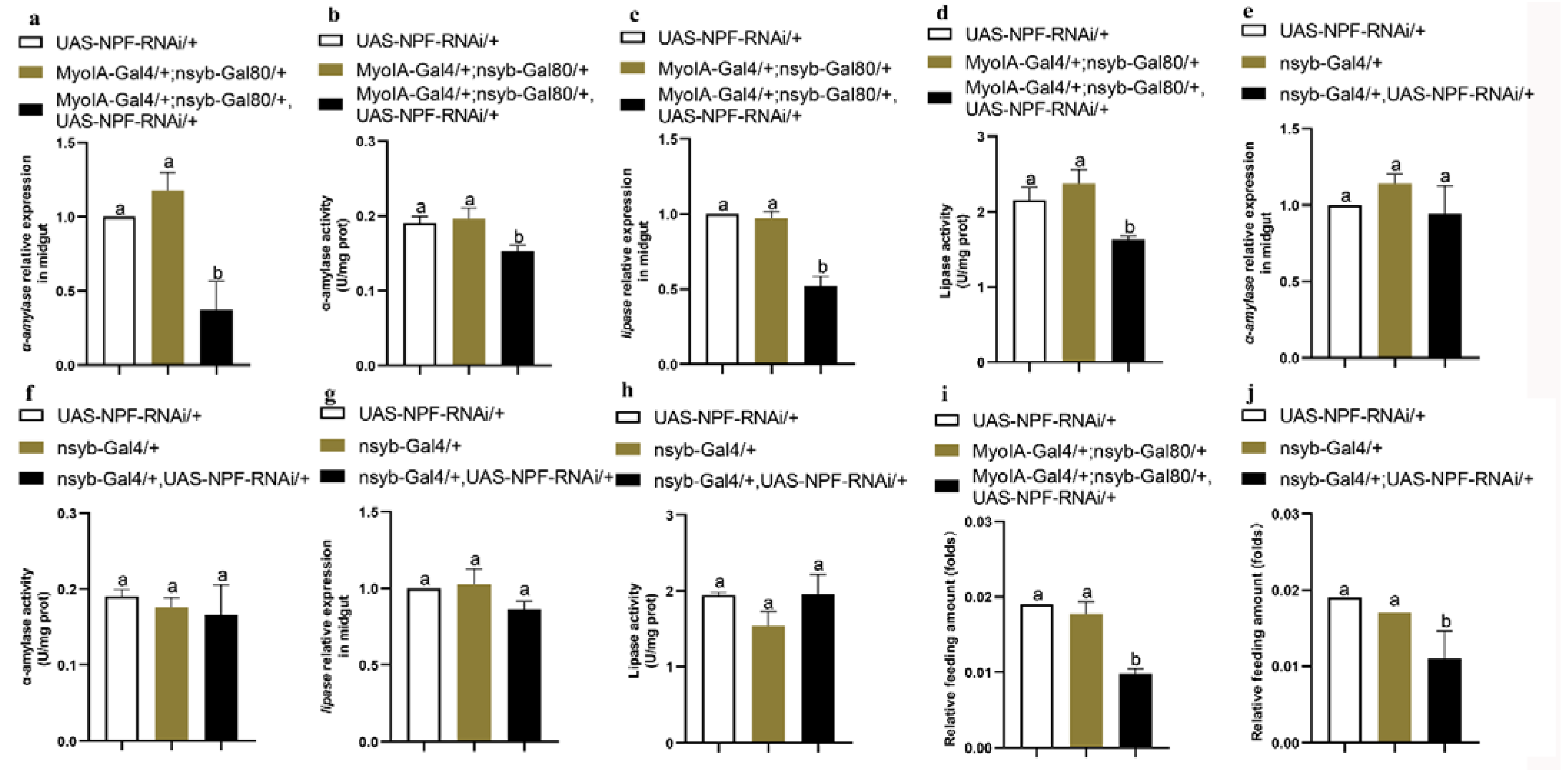
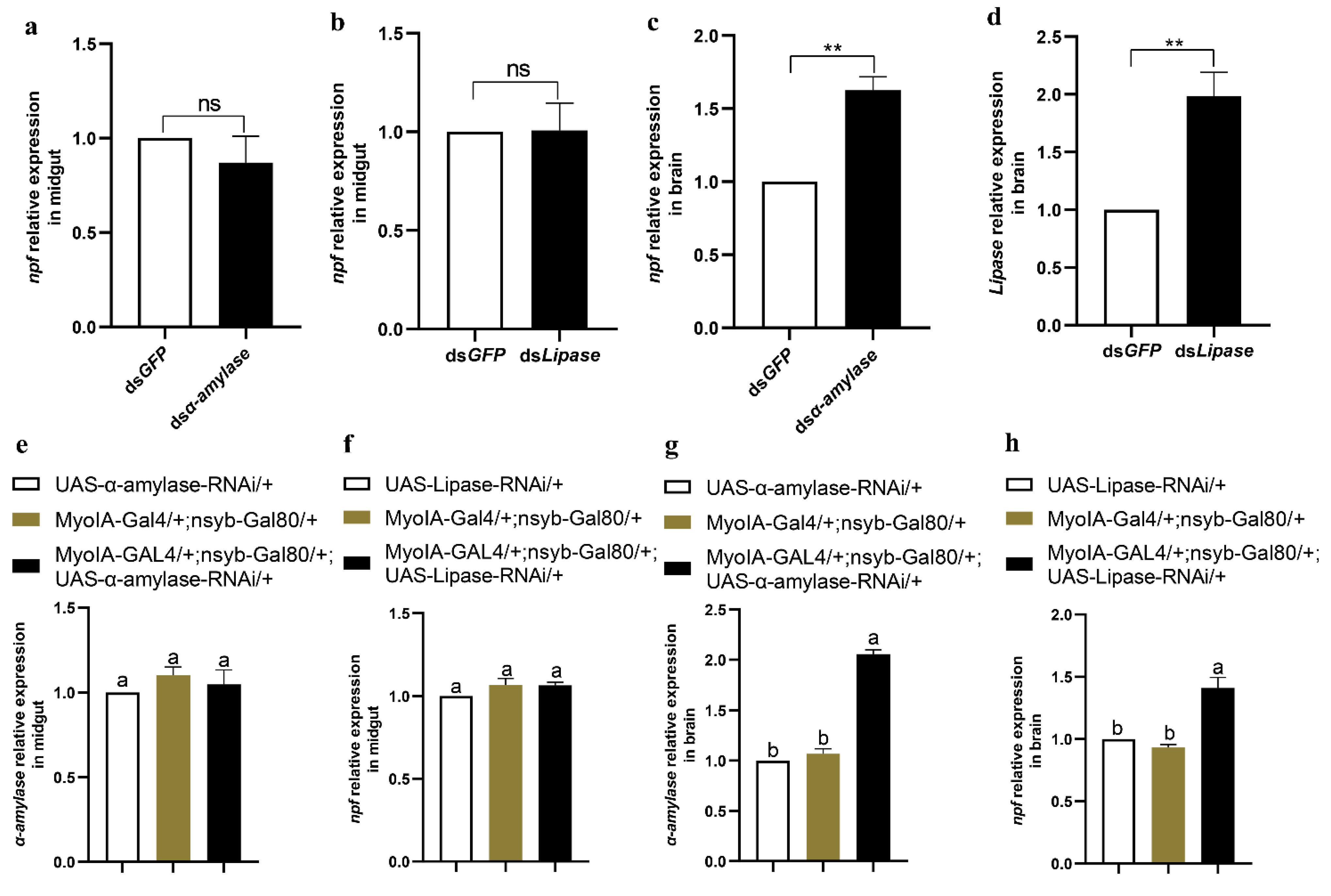
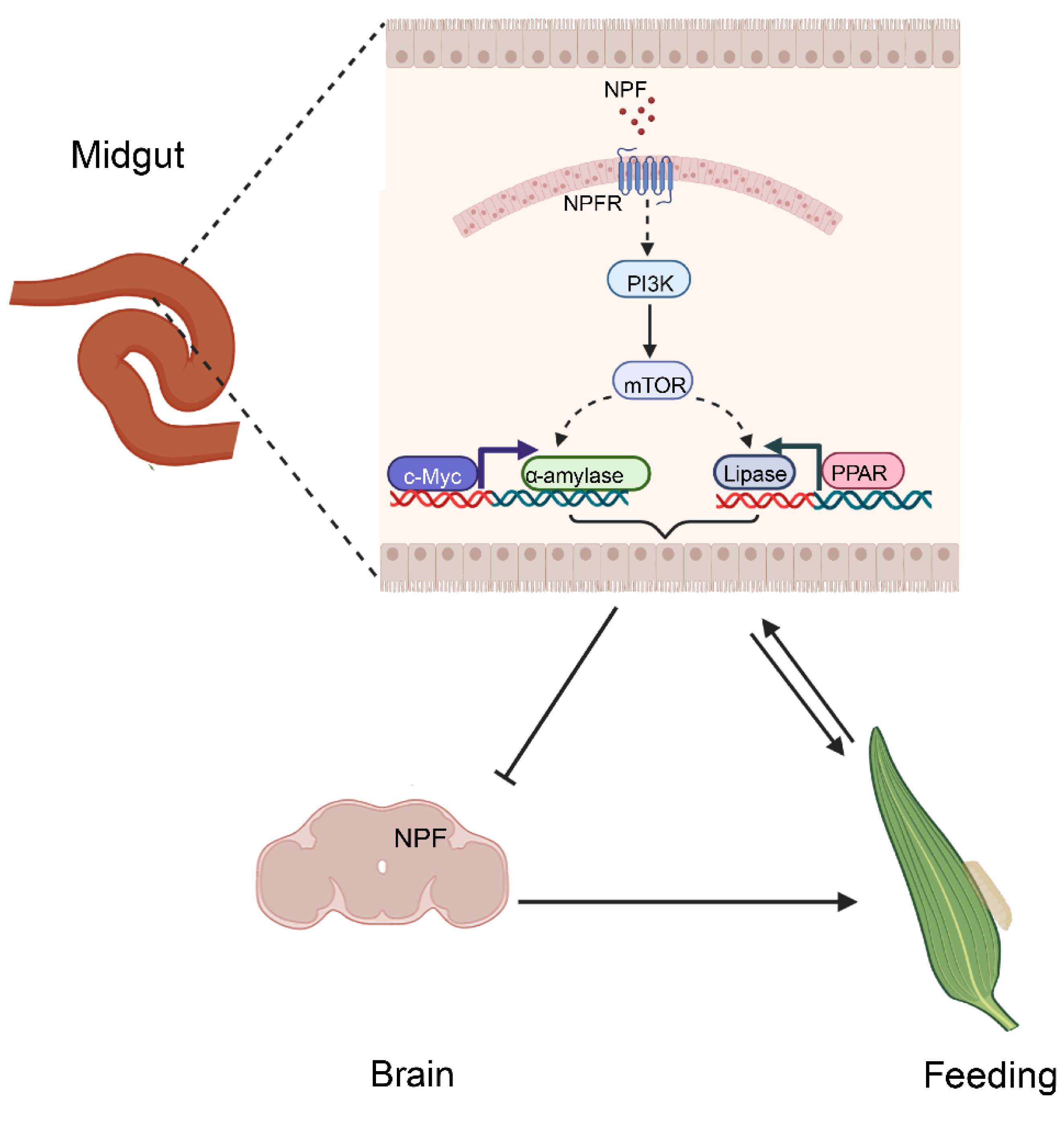
3.6. Feeding and Digestion Are Regulated via Feedback between Gut and Brain NPF
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Charnov, E.L. Optimal foraging, the marginal value theorem. Theor. Popul. Biol. 1976, 9, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zhao, Z.; Shen, P. Regulation of aversion to noxious food by Drosophila neuropeptide Y- and insulin-like systems. Nat. Nneurosci. 2005, 8, 1350–1355. [Google Scholar] [CrossRef] [PubMed]
- Maule, A.G.; Shaw, C.; Halton, D.W.; Thim, L.; Johnston, C.F.; Fairweather, I.; Buchanan, K.D. Neuropeptide F: A novel parasitic flatworm regulatory peptide from Moniezia expansa (Cestoda: Cyclophyllidea). Parasitology 1991, 102, 309–316. [Google Scholar] [CrossRef]
- Van Wielendaele, P.; Wynant, N.; Dillen, S.; Zels, S.; Badisco, L.; Broeck, J.V. Neuropeptide F regulates male reproductive processes in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2013, 43, 252–259. [Google Scholar] [CrossRef]
- Xu, W.H. Advances in insect neuropeptides. Prog. Biochem. Biophy. 1997, 2, 116–120. [Google Scholar]
- Huang, Y.; Brown, M.R.; Lee, T.D.; Crim, J.W. RF-amide peptides isolated from the midgut of the corn earworm, Helicoverpa zea, resemble pancreatic polypeptide. Insect Biochem. Mol. Biol. 1998, 28, 345–356. [Google Scholar] [CrossRef]
- Brown, M.R.; Crim, J.W.; Arata, R.C.; Cai, H.N.; Chun, C.; Shen, P. Identification of a Drosophila brain-gut peptide related to the neuropeptide Y family. Peptides 1999, 20, 1035–1042. [Google Scholar] [CrossRef]
- De Loof, A.; Baggerman, G.; Breuer, M.; Claey, I.; Cerstiaens, A.; Clynen, E.; Janssen, T.; Schoofs, L.; Vanden Broeck, J. Gonadotropins in insects: An overview. Arch. Insect Biochem. Physiol. 2001, 47, 129–138. [Google Scholar] [CrossRef]
- Stanek, D.M.; Pohl, J.; Crim, W.; Brown, M.R. Neuropeptide F and its expression in the yellow fever mosquito, Aedes aegypti. Peptides 2002, 23, 1367–1378. [Google Scholar] [CrossRef]
- Garczynski, S.F.; Crim, J.W.; Brown, M.R. Characterization of neuropeptide F and its receptor from the African malaria mosquito, Anopheles gambiae. Peptides 2005, 26, 99–107. [Google Scholar] [CrossRef]
- Clynen, E.; Huybrechts, J.; Verleyen, P.; De Loof, A.; Schoofs, L. Annotation of novel neuropeptide precursors in the migratory locust based on transcript screening of a public EST database and mass spectrometry. BMC Genom. 2006, 7, 201. [Google Scholar] [CrossRef][Green Version]
- Roller, L.; Yamanaka, N.; Watanabe, K.; Daubnerova, I.; Zitnan, D.; Kataoka, H.; Tanaka, Y. The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Nuss, A.B.; Forschler, B.T.; Crim, J.W.; TeBrugge, V.; Pohl, J.; Brown, M.R. Molecular characterization of neuropeptide F from the eastern subterranean termite Reticulitermes flavipes (Kollar) (Isoptera: Rhinotermitidae). Peptides 2010, 31, 419–428. [Google Scholar] [CrossRef]
- Liu, X.G.; Zhang, Y.F.; Zhou, Z.J.; Zhao, Z.W. Cloning and sequence analysis of neuropeptide F from the oriental tobacco budworm Helicoverpa assulta (Guenée). Arch. Insect Biochem. Physiol. 2013, 84, 115–129. [Google Scholar] [CrossRef]
- Fadda, M.; Hasakiogullari, I.; Temmerman, L.; Beets, I.; Zels, S.; Schoofs, L. Regulation of feeding and metabolism by neuropeptide F and short neuropeptide F in invertebrates. Front. Endocrinol. 2019, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Cai, H.N. Drosophila neuropeptide F mediates integration of chemosensory stimulation and conditioning of the nervous system by food. J. Neurobiol. 2001, 47, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Yue, Z.; Liu, X.G.; Zhou, Z.J.; Hou, G.M.; Hua, P.; Zhao, Z.W. Development of a novel-type transgenic cotton plant for control of cotton bollworm. Plant Biotechnol. J. 2016, 14, 1747–1755. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qu, M.J.; Zhang, Y.; Li, J.W.; Liu, T.X. Expression of neuropeptide F gene and its regulation of feeding behavior in the pea aphid, Acyrthosiphon pisum. Front. Physiol 2018, 9, 87. [Google Scholar] [CrossRef]
- Tan, S.Q.; Li, A.M.; Wang, Y.; Shi, W.P. Role of the neuropeptide F 1 in regulating the appetite for food in Locusta migratoria. Pest Manag. Sci. 2019, 75, 1304–1309. [Google Scholar] [CrossRef]
- Wei, H.S.; Tan, S.Q.; Yan, S.; Li, Z.; Shen, J.; Liu, X.X. Nanocarrier-mediated transdermal dsRNA-NPF1 delivery system contributes to pest control via inhibiting feeding behavior in Grapholita molesta. J. Pest Sci. 2021, 95, 983–995. [Google Scholar] [CrossRef]
- Yue, Z.; Li, X.R.; Zhang, E.Y.; Liu, X.X.; Zhao, Z.W. A potential and novel type transgenic corn plant for control of the Corn Borer. Sci. Rep. 2017, 7, 44105. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.Y.; Wang, Y.; Peng, X.; Wang, Y.T.; Zhao, Z.W. Feeding effects of dsNPF interference in Ostrinia furnacalis. J. Integr. Agr. 2020, 19, 1475–1481. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, J.; Cui, H.Y.; Wang, C.Z.; Zhao, Z.W. Effects of NPF on larval taste responses and feeding behaviors in Ostrinia furnacalis. J. Insect Physiol. 2021, 133, 104276. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.F.; Shi, J.; Jiang, X.M.; Song, Y.; Du, J.; Zhao, Z.W. Neuropeptide F regulates feeding via the juvenile hormone pathway in Ostrinia furnacalis larvae. Pest Manag. Sci. Available online: https://onlinelibrary.wiley.com/doi/10.1002/ps.7289 (accessed on 17 November 2022).
- Gulia-Nuss, M.; Robertson, A.E.; Brown, M.R.; Strand, M.R. Insulin-like peptides and the target of rapamycin pathway coordinately regulate blood digestion and egg maturation in the mosquito Aedes aegypti. PLoS ONE 2011, 6, e20401. [Google Scholar] [CrossRef]
- Stafford, J.W.; Lynd, K.M.; Jung, A.Y.; Gordon, M.D. Integration of taste and calorie sensing in Drosophila. J. Neurosci. 2012, 32, 14767–14774. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.L.; Campos, A.R. Insulin signalling in mushroom body neurons regulates feeding behaviour in Drosophila larvae. J. Exp. Biol. 2012, 215, 2696–2702. [Google Scholar] [CrossRef]
- Van der Heide, L.P.; Ramakers, G.M.; Smidt, M.P. Insulin signaling in the central nervous system: Learning to survive. Prog. Neurobiol. 2006, 79, 205–221. [Google Scholar] [CrossRef]
- Wittmann, A.C.; Benrabaa, S.A.M.; López-Cerón, D.A.; Chang, E.S.; Mykles, D.L. Effects of temperature on survival, moulting, and expression of neuropeptide and mTOR signalling genes in juvenile Dungeness crab (Metacarcinus magister). J. Exp. Biol. 2018, 221, jeb187492. [Google Scholar] [CrossRef]
- Ferreira, L.M.; Romão, T.P.; De-Melo-Neto, O.P.; Silva-Filha, M.H. The orthologue to the cpm1/cqm1 receptor in aedes aegypti is expressed as a midgut GPI-anchored α-glucosidase, which does not bind to the insecticidal binary toxin. Insect Biochem. Mol. Boil. 2010, 40, 604–610. [Google Scholar] [CrossRef]
- Kaur, R.; Kaur, N.; Gupta, A.K. Structural features, substrate specificity, kinetic properties of insect α-amylase and specificity of plant α-amylase inhibitors. Pesticide Biochem. Phys. 2014, 116, 83–93. [Google Scholar] [CrossRef]
- Terra, W.R.; Ferreira, C. Insect digestive enzymes: Properties, compartmentalization and function. Comp. Biochem. Physiol. B Comp. Biochem. 1994, 109, 1–62. [Google Scholar] [CrossRef]
- Terra, W.R.; Ferreira, C.; Baker, J.E. Compartmentalization of digestion. In Biology of the Insect Midgut; Springer: Dordrecht, The Netherlands, 1996. [Google Scholar]
- Terra, W.R.; Ferreira, C.; Jordo, B.P.; Dillon, R.J. Digestive enzymes. In Biology of the Insect Midgut; Springer: Dordrecht, The Netherlands, 1996. [Google Scholar]
- Baker, J.E.; Woo, S.M. Purification and partial characterization and postembryonic levels of amylases from Sitophilus oryzae and Sitophilus granarius. Arch. Insect Biochem. Physiol. 2010, 2, 415–428. [Google Scholar] [CrossRef]
- Mendiola-Olaya, E.; Valencia-Jimenez, A.; Valdes-Rodriguez, S.; Delano-Frier, J.; Blanco-Labra, A. Digestive amylase from the larger grain borer, Prostephanus truncates Horn. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2000, 126, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Neto, O.B.; Batista, J.A.N.; Rigden, D.J.; Franco, O.L.; Falcão, R.; Fragoso, R.R.; Mello, L.V.; dos Santos, R.C.; Grossi-de-Sá, M.F. Molecular cloning of alpha-amylases from cotton boll weevil, anthonomus grandis and structural relations to plant inhibitors: An approach to insect resistance. J. Protein Chem. 2003, 22, 77–87. [Google Scholar] [CrossRef]
- Terra, W.R.; Espinoza-Fuentes, F.P.; Ferreira, C. Midgut amylase, lysozyme, aminopeptidase, and trehalase from larvae and adults of Musca domestica. Arch. Insect Biochemi. Physiol. 1988, 9, 283–297. [Google Scholar] [CrossRef]
- Zeng, F.; Cohen, A.C. Comparison of α-amylase and protease activities of a zoophytophagus and two phytophagous Heteroptera. Com. Biochem. Physiol. A 2000, 126, 101–106. [Google Scholar] [CrossRef]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzym. Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Kapoor, M.; Gupta, M.N. Lipase promiscuity and its biochemical applications. Process Biochem. 2012, 47, 555–569. [Google Scholar] [CrossRef]
- Jaeger, K.E.; Reetz, M.T. Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 1998, 16, 396–403. [Google Scholar] [CrossRef]
- Shambaugh, G.F. Protease stimulation by foods in adult Aedes aegypti linn. Ohio J. Sci. 1954, 54, 151–160. [Google Scholar]
- Engelmann, F. Control of intestinal proteolytic enzymes in a cockroach. Naturwissenschaften 1966, 53, 113–114. [Google Scholar] [CrossRef] [PubMed]
- Weidlich, S.; Hoffmann, K.H.; Woodring, J. Secretion of lipases in the digestive tract of the cricket Gryllus bimaculatus. Arch. Insect Biochem. Physiol. 2015, 90, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.Z.; Zhou, H.; Wei, Y.L.; Yan, S.; Shen, J. A novel plasmid—Escherichia coli system produces large batch dsRNAs for insect gene silencing. Pest Manag. Sci. 2020, 76, 2505–2512. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Edgecomb, R.S.; Harth, C.E.; Schneiderman, A.M. Regulation of feeding behavior in adult Drosophila melanogaster varies with feeding regime and nutritional state. J. Exp. Biol. 1994, 197, 215–235. [Google Scholar] [CrossRef]
- Chen, M.; Du, Q.; Zhang, H.Y.; Wang, X.; Liang, Z. High-throughput screening using siRNA (RNAi) libraries. Expert Rev. Mol. Diagn. 2007, 7, 281–291. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.Q.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Kuttenkeuler, D.; Boutros, M. Genome-wide RNAi as a route to gene function in Drosophila. Brief. Funct. Genomics. 2004, 3, 168–176. [Google Scholar] [CrossRef]
- Price, D.R.; Gatehouse, J.A. RNAi-mediated crop protection against insects. Trends. Biotechnol. 2008, 26, 393–400. [Google Scholar] [CrossRef]
- Claeys, I.; Simonet, G.; Poels, J.; Loy, T.V.; Vercammen, L.; De Loof, A.; Broeck, J.V. Insulin-related peptides and their conserved signal transduction pathway. Peptides 2002, 23, 807–816. [Google Scholar] [CrossRef]
- Teleman, A.A. Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem. J. 2010, 425, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zhu, Y.T.; Shi, D.H. Research progress in cell metabolism of glucose and lipid regulated by transcription factors via mTOR. Basic. Clin. Med. 2014, 34, 1574–1577. [Google Scholar]
- Fletcher, S.; Prochownik, E.V. Small-molecule inhibitors of the Myc oncoprotein. BBA-Gene Regul. Mech. 2015, 1849, 525–543. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Jin, K.K.; Mortensen, R.M.; Mutyaba, P.L.; Krebsbach, P.H. Osteoblast-Targeted suppression of PPAR gamma increases osteogenesis through activation of mTOR signaling. Stem Cells. 2013, 31, 2183–2192. [Google Scholar] [CrossRef]
- Muraleedharan, D.; Prabhu, V.K.K. Role of the median neurosecretory cells in secretion of protease and invertase in the red cotton bug, dysdercus cingulatus. J. Insect Physiol. 1979, 25, 237–240. [Google Scholar] [CrossRef]
- Spit, J.; Badisco, L.; Verlinden, H.; Van Wielendaele, P.; Zels, S.; Dillen, S.; Vanden Broeck, J. Peptidergic control of food intake and digestion in insects. Can. J. Zoo. 2012, 90, 489–506. [Google Scholar] [CrossRef]
- Canavoso, L.E.; Jouni, Z.E.; Karnas, K.J.; Pennington, J.E.; Wells, M.A. Fat metabolism in insects. Annu. Rev. Nutr. 2001, 21, 23–46. [Google Scholar] [CrossRef]
- Franco, O.L.; Rigden, D.J.; Melo, F.R.; Grossi, M.F. Plant α-amylase inhibitors and their interaction with insect α-amylase structure, function and potential for crop protection. Eur. J. Biochem. 2002, 269, 397–412. [Google Scholar] [CrossRef]
- Chung, B.Y.; Hutter, S.A.; Miller, K.M.; Guduguntla, L.S.; Kondo, S.; Pletcher, S.D. Drosophila Neuropeptide F signaling independently regulates feeding and sleep-wake behavior. Cell Rep. 2017, 19, 2441–2450. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Song, Y.; Jiang, X.; He, L.; Wei, L.; Zhao, Z. Synergism of Feeding and Digestion Regulated by the Neuropeptide F System in Ostrinia furnacalis Larvae. Cells 2023, 12, 194. https://doi.org/10.3390/cells12010194
Zhao J, Song Y, Jiang X, He L, Wei L, Zhao Z. Synergism of Feeding and Digestion Regulated by the Neuropeptide F System in Ostrinia furnacalis Larvae. Cells. 2023; 12(1):194. https://doi.org/10.3390/cells12010194
Chicago/Turabian StyleZhao, Jiajia, Yu Song, Xuemin Jiang, Lei He, Liya Wei, and Zhangwu Zhao. 2023. "Synergism of Feeding and Digestion Regulated by the Neuropeptide F System in Ostrinia furnacalis Larvae" Cells 12, no. 1: 194. https://doi.org/10.3390/cells12010194
APA StyleZhao, J., Song, Y., Jiang, X., He, L., Wei, L., & Zhao, Z. (2023). Synergism of Feeding and Digestion Regulated by the Neuropeptide F System in Ostrinia furnacalis Larvae. Cells, 12(1), 194. https://doi.org/10.3390/cells12010194





