Plant Transglutaminases: New Insights in Biochemistry, Genetics, and Physiology
Abstract
1. Introduction
2. Biochemical Features of TGases
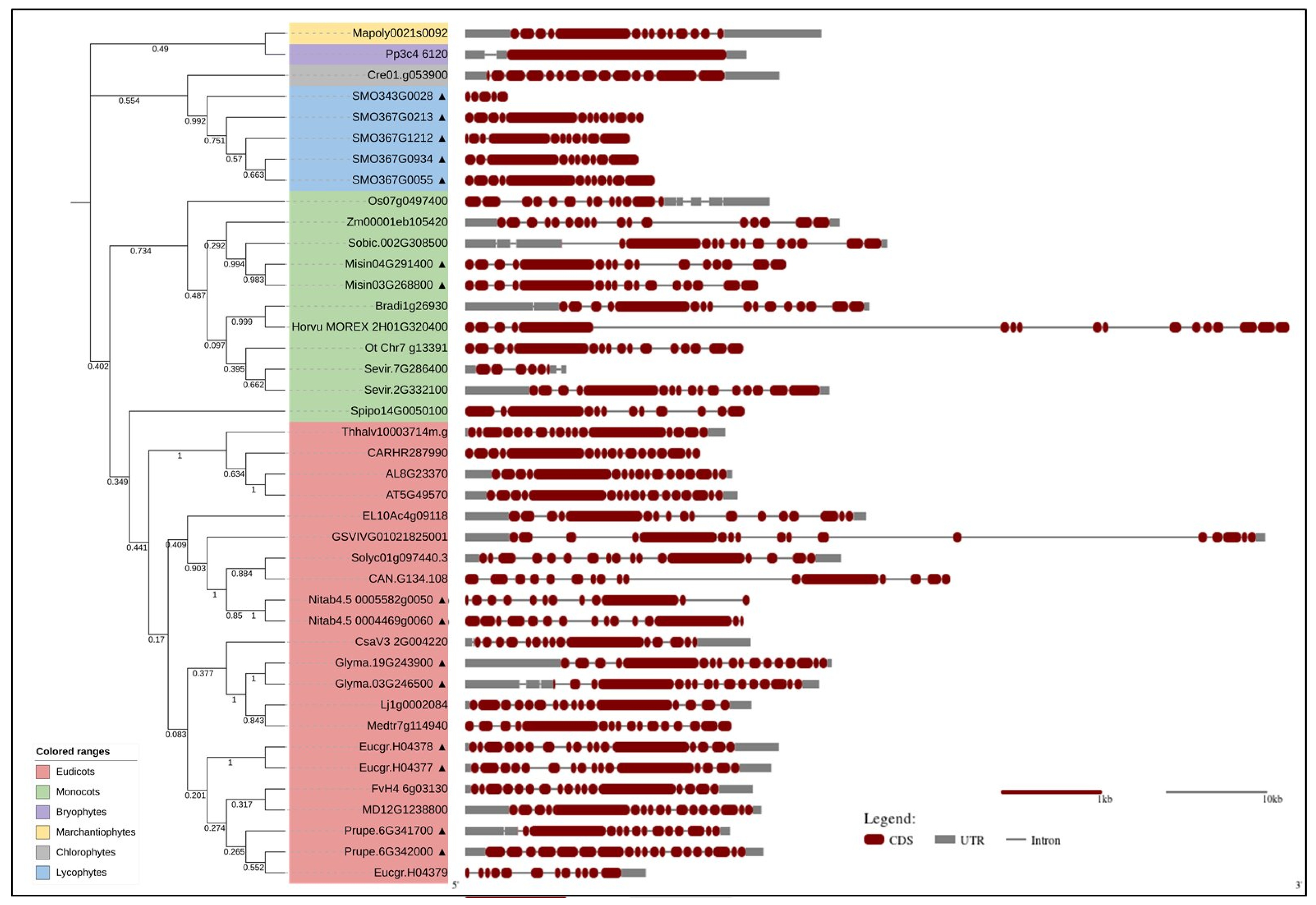
3. Bioinformatics Analyses
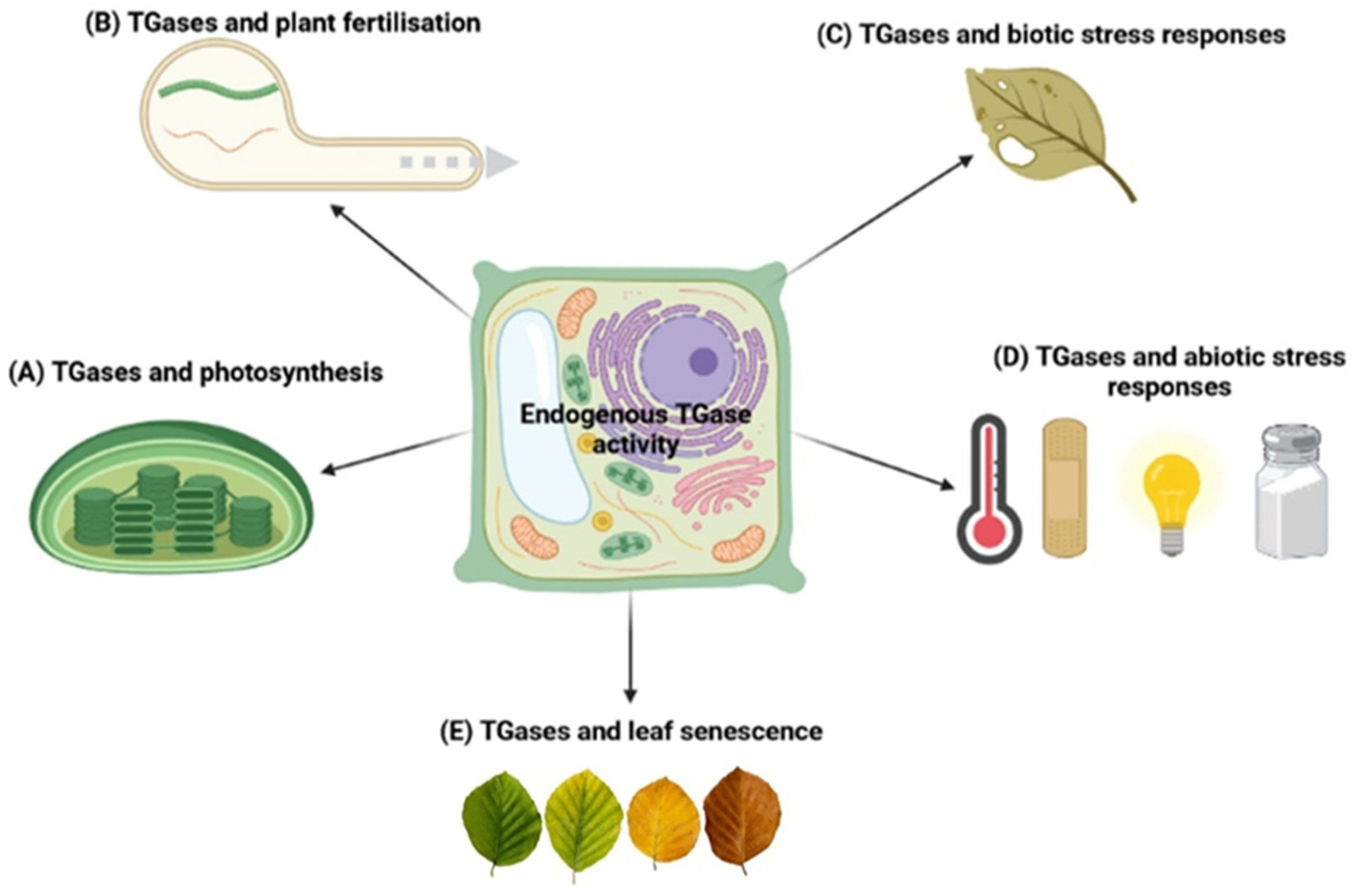
4. Physiological Role of TGases in Plants
4.1. TGases and Photosynthesis
4.2. TGases and Plant Fertilisation
4.3. TGases and Biotic Stress Responses
4.4. TGases and Abiotic Stress Responses
4.5. TGases and Leaf Senescence
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Clarke, D.; Mycek, M.; Neidle, A.; Waelsch, H. The incorporation of amines into protein. Arch. Biochem. Biophys. 1959, 79, 338–354. [Google Scholar] [CrossRef]
- Duarte, L.; Matte, C.R.; Bizarro, C.V.; Ayub, M.A.N.Z. Transglutaminases: Part I—Origins, sources, and biotechnological characteristics. World J. Microb. Biot. 2020, 36, 15. [Google Scholar] [CrossRef] [PubMed]
- Sobieszczuk-Nowicka, E.; Wieczorek, P.; Legocka, J. Kinetin affects the level of chloroplast polyamines and transglutaminase activity during senescence of barley leaves. Acta Biochim. Pol. 2009, 56, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Del Duca, S.; Serafini-Fracassini, D. Transglutaminases of higher, lower plants and fungi. Prog. Exp. Tumor Res. 2005, 38, 223–247. [Google Scholar]
- Serafini-Fracassini, D.; Del Duca, S. Transglutaminases: Widespread cross-linking enzymes in plants. Ann. Bot. Lond. 2008, 102, 145–152. [Google Scholar] [CrossRef]
- Icekson, I.; Apelbaum, A. Evidence for Transglutaminase Activity in Plant-Tissue. Plant Physiol. 1987, 84, 972–974. [Google Scholar] [CrossRef]
- Campos, A.; Carvajal-Vallejos, P.K.; Villalobos, E.; Franco, C.F.; Almeida, A.M.; Coelho, A.V.; Torne, J.M.; Santos, M. Characterisation of Zea mays L. plastidial transglutaminase: Interactions with thylakoid membrane proteins. Plant Biol. 2010, 12, 708–716. [Google Scholar] [CrossRef]
- Sobieszczuk-Nowicka, E.; Legocka, J. Plastid-associated polyamines: Their role in differentiation, structure, functioning, stress response and senescence. Plant Biol. 2014, 16, 297–305. [Google Scholar] [CrossRef]
- Del Duca, S.; Cai, G.; Di Sandro, A.; Serafini-Fracassini, D. Compatible and self-incompatible pollination in Pyrus communis displays different polyamine levels and transglutaminase activity. Amino Acids 2010, 38, 659–667. [Google Scholar] [CrossRef]
- Del Duca, S.; Faleri, C.; Iorio, R.A.; Cresti, M.; Serafini-Fracassini, D.; Cai, G. Distribution of Transglutaminase in Pear Pollen Tubes in Relation to Cytoskeleton and Membrane Dynamics. Plant Physiol. 2013, 161, 1706–1721. [Google Scholar] [CrossRef]
- Del Duca, S.; Serafini-Fracassini, D.; Cai, G. Senescence and programmed cell death in plants: Polyamine action mediated by transglutaminase. Front. Plant Sci. 2014, 5, 120. [Google Scholar] [CrossRef] [PubMed]
- Del Duca, S.; Aloisi, I.; Parrotta, L.; Cai, G. Cytoskeleton, Transglutaminase and Gametophytic Self-Incompatibility in the Malinae (Rosaceae). Int. J. Mol. Sci. 2019, 20, 209. [Google Scholar] [CrossRef] [PubMed]
- Mandrone, M.; Antognoni, F.; Aloisi, I.; Potente, G.; Poli, F.; Cai, G.; Faleri, C.; Parrotta, L.; Del Duca, S. Compatible and Incompatible Pollen-Styles Interaction in Pyrus communis L. Show Different Transglutaminase Features, Polyamine Pattern and Metabolomics Profiles. Front. Plant Sci. 2019, 10, 741. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, N.E.; Ortigosa, S.M.; Veramendi, J.; Pinto-Marijuan, M.; Fleck, I.; Carvajal, P.; Kotzabasis, K.; Santos, M.; Torne, J.M. Remodeling of tobacco thylakoids by over-expression of maize plastidial transglutaminase. Biochim. Biophys. Acta-Bioenerg. 2009, 1787, 1215–1222. [Google Scholar] [CrossRef]
- Lorand, L.; Iismaa, S.E. Transglutaminase diseases: From biochemistry to the bedside. Faseb J. 2019, 33, 3–12. [Google Scholar] [CrossRef]
- Brunner, F.; Rosahl, S.; Lee, J.; Rudd, J.J.; Geiler, C.; Kauppinen, S.; Rasmussen, G.; Scheel, D.; Nurnberger, T. Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. Embo J. 2002, 21, 6681–6688. [Google Scholar] [CrossRef]
- Serafini-Fracassini, D.; Del Duca, S.; D’Orazi, D. First evidence for polyamine conjugation mediated by an enzymic activity in plants. Plant Physiol. 1988, 87, 757–761. [Google Scholar] [CrossRef]
- Suzuki, T.; Park, H.; Hollingsworth, N.M.; Sternglanz, R.; Lennarz, W.J. PNG1, a yeast gene encoding a highly conserved peptide: N-glycanase. J. Cell Biol. 2000, 149, 1039–1051. [Google Scholar] [CrossRef]
- Della Mea, M.; Caparros-Ruiz, D.; Claparols, I.; Serafini-Fracassini, D.; Rigau, J. AtPng1p. The first plant transglutaminase. Plant Physiol. 2004, 135, 2046–2054. [Google Scholar] [CrossRef]
- Diepold, A.; Li, G.; Lennarz, W.J.; Nurnberger, T.; Brunner, F. The Arabidopsis AtPNG1 gene encodes a peptide: N-glycanase. Plant J. 2007, 52, 94–104. [Google Scholar] [CrossRef]
- Hou, K.; Wang, Y.; Tao, M.Q.; Jahan, M.S.; Shu, S.; Sun, J.; Guo, S.R. Characterization of the CsPNG1 gene from cucumber and its function in response to salinity stress. Plant Physiol. Bioch. 2020, 150, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Lilley, G.R.; Skill, J.; Griffin, M.; Bonner, P.L.R. Detection of Ca2+-dependent transglutaminase activity in root and leaf tissue of monocotyledonous and dicotyledonous plants. Plant Physiol. 1998, 117, 1115–1123. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Del Duca, S.; Tidu, V.; Bassi, R.; Serafini-Fracassini, D.; Esposito, C. Identification of transglutaminase activity and its substrates in isolated chloroplast of Helianthus tuberosus. Planta 1994, 193, 283–289. [Google Scholar] [CrossRef]
- Del Duca, S.; Dondini, L.; Della Mea, M.; Munoz de Rueda, P.; Serafini-Fracassini, D. Factors affecting transglutaminase activity catalysing polyamine conjugation to endogenous substrates in the entire chloroplast. Plant Physiol. Bioch. 2000, 38, 429–439. [Google Scholar] [CrossRef]
- Siepaio, M.P.; Meunier, J.C.F. Diamine Oxidase and Transglutaminase Activities in White Lupine Seedlings with Respect to Cross-Linking of Proteins. J. Agr. Food Chem. 1995, 43, 1151–1156. [Google Scholar] [CrossRef]
- Campos, N.; Castanon, S.; Urreta, I.; Santos, M.; Torne, J.M. Rice transglutaminase gene: Identification, protein expression, functionality, light dependence and specific cell location. Plant Sci. 2013, 205, 97–110. [Google Scholar] [CrossRef]
- Waffenschmidt, S.; Kusch, T.; Woessner, J.P. A transglutaminase immunologically related to tissue transglutaminase catalyzes cross-linking of cell wall proteins in Chlamydomonas reinhardtii. Plant Physiol. 1999, 121, 1003–1015. [Google Scholar] [CrossRef]
- Villalobos, E.; Santos, M.; Talavera, D.; Rodriguez-Falcon, M.; Torne, J.M. Molecular cloning and characterization of a maize transglutaminase complementary DNA. Gene 2004, 336, 93–104. [Google Scholar] [CrossRef]
- El-Hofi, M.; Ismail, A.; Nour, M.; Ibrahim, O. Isolation, harac cation and haracterization of transglutaminase from rosemary (Rosmarinus officinalis L.) leaves. Acta Sci. Pol. Technol. Aliment. 2014, 13, 267–278. [Google Scholar] [CrossRef][Green Version]
- Shu, S.; Tang, Y.Y.; Zhou, X.P.; Jahan, M.S.; Sun, J.; Wang, Y.; Guo, S.R. Physiological mechanism of transglutaminase-mediated improvement in salt tolerance of cucumber seedlings. Plant Physiol. Bioch. 2020, 156, 333–344. [Google Scholar] [CrossRef]
- Kim, H.C.; Lewis, M.S.; Gorman, J.J.; Park, S.C.; Girard, J.E.; Folk, J.E.; Chung, S.I. Protransglutaminase-E from Guinea-Pig Skin—Isolation and Partial Characterization. J. Biol. Chem. 1990, 265, 21971–21978. [Google Scholar] [CrossRef]
- Pasternack, R.; Dorsch, S.; Otterbach, J.T.; Robenek, I.R.; Wolf, S.; Fuchsbauer, H.L. Bacterial pro-transglutaminase from Streptoverticillium mobaraense—Purification, characterisation and sequence of the zymogen. Eur. J. Biochem. 1998, 257, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Margosiak, S.A.; Dharma, A.; Brucecarver, M.R.; Gonzales, A.P.; Louie, D.; Kuehn, G.D. Identification of the Large Subunit of Ribulose 1,5-Bisphosphate Carboxylase Oxygenase as a Substrate for Transglutaminase in Medicago-sativa L (Alfalfa). Plant Physiol. 1990, 92, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Kuehn, G.D.; Sotelo, M.; Morales, T.; Brucecarver, M.R.; Guzman, E.; Margosiak, S.A. Purification and Properties of Transglutaminase from Medicago-sativa L (Alfalfa). Faseb J. 1991, 5, A1510. [Google Scholar]
- Pinto-Marijuan, M.; de Agazio, M.; Zacchini, M.; Santos, M.A.; Torne, J.M.; Fleck, I. Response of transglutaminase activity and bound putrescine to changes in light intensity under natural or controlled conditions in Quercus ilex leaves. Physiol. Plant. 2007, 131, 159–169. [Google Scholar] [CrossRef]
- Campos, N.; Torne, J.M.; Bleda, M.J.; Manich, A.; Urreta, I.; Montalban, I.A.; Castanon, S.; Moncalean, P.; Santos, M. Proteomic and transcriptomic analysis of rice tranglutaminase and chloroplast-related proteins. Plant Sci. 2014, 229, 142–153. [Google Scholar] [CrossRef]
- Zhong, M.; Wang, Y.; Hou, K.; Shu, S.; Sun, J.; Guo, S.R. TGase positively regulates photosynthesis via activation of Calvin cycle enzymes in tomato. Hortic. Res.-Engl. 2019, 6. [Google Scholar] [CrossRef]
- Li, H.B.; Zhang, L.W.; Cui, Y.H.; Luo, X.; Xue, C.H.; Wang, S.M.; Jiao, Y.H.; Zhang, S.; Liu, W.L.; Fan, R.B.; et al. Characterization of recombinant Zea mays transglutaminase expressed in Pichia pastoris and its impact on full and non-fat yoghurts. J. Sci. Food Agr. 2014, 94, 1225–1230. [Google Scholar] [CrossRef]
- Sobieszczuk-Nowicka, E.; Di Sandro, A.; Del Duca, S.; Serafini-Fracassini, D.; Legocka, J. Plastid-membrane-associated polyamines and thylakoid transglutaminases during etioplast-to-chloroplast transformation stimulated by kinetin. Physiol. Plant 2007, 130, 590–600. [Google Scholar] [CrossRef]
- Santos, M.; Alché Ramírez, J.d.D.; Rodríguez García, M.I.; Torné, J.M. Transglutaminase activity and localization during microspore induction in maize. Appl. Biol. Med. 2007, 1, 280–286. [Google Scholar]
- Signorini, M.; Beninati, S.; Bergamini, C.M. Identification of Transglutaminase Activity in the Leaves of Silver Beet (Beta vulgaris L.). J. Plant Physiol. 1991, 137, 547–552. [Google Scholar] [CrossRef]
- Piccini, C.; Parrotta, L.; Faleri, C.; Romi, M.; Del Duca, S.; Cai, G. Histomolecular responses in susceptible and resistant phenotypes of Capsicum annuum L. infected with Phytophthora capsici. Sci. Hortic. Amst. 2019, 244, 122–133. [Google Scholar] [CrossRef]
- Dondini, L.; Bonazzi, S.; Serafini-Fracassini, D. Recovery of growth capacity and of chloroplast transglutaminase activity induced by polyamines in a polyamine-deficient variant strain of Dunaliella salina. J. Plant Physiol. 2000, 157, 473–480. [Google Scholar] [CrossRef]
- Dondini, L.; Bonazzi, S.; Del Duca, S.; Bregoli, A.M.; Serafini-Fracassini, D. Acclimation of chloroplast transglutaminase to high NaCl concentration in a polyamine-deficient variant strain of Dunaliella salina and in its wild type. J. Plant Physiol. 2001, 158, 185–197. [Google Scholar] [CrossRef]
- Kang, H.; Cho, Y.D. Purification and properties of transglutaminase from soybean (Glycine max) leaves. Biochem. Biophys. Res. Commun. 1996, 223, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Lee, S.G.; Cho, Y.D. Identification of glycinin in vivo as a polyamine-conjugated protein via a gamma-glutamyl linkage. Biochem. J. 1998, 332, 467–473. [Google Scholar] [CrossRef]
- Dondini, L.; Del Duca, S.; Dall’Agata, L.; Bassi, R.; Gastaldelli, M.; Della Mea, M.; Di Sandro, A.; Claparols, I.; Serafini-Fracassini, D. Suborganellar localisation and effect of light on Helianthus tuberosus chloroplast transglutaminases and their substrates. Planta 2003, 217, 84–95. [Google Scholar] [CrossRef]
- Beninati, S.; Iorio, R.A.; Tasco, G.; Serafini-Fracassini, D.; Casadio, R.; Del Duca, S. Expression of different forms of transglutaminases by immature cells of Helianthus tuberosus sprout apices. Amino Acids 2013, 44, 271–283. [Google Scholar] [CrossRef]
- Di Sandro, A.; Del Duca, S.; Verderio, E.; Hargreaves, A.J.; Scarpellini, A.; Cai, G.; Cresti, M.; Faleri, C.; Iorio, R.A.; Hirose, S.; et al. An extracellular transglutaminase is required for apple pollen tube growth. Biochem. J. 2010, 429, 261–271. [Google Scholar] [CrossRef]
- Serafini-Fracassini, D.; Del Duca, S. Biochemistry and function of plant transglutaminases. Minerva Biotecnol. 2002, 14, 135–141. [Google Scholar]
- Della Mea, M.; De Filippis, F.; Genovesi, V.; Serafini-Fracassini, D.; Del Duca, S. The acropetal wave of developmental cell death of tobacco corolla is preceded by activation of transglutaminase in different cell compartments. Plant Physiol. 2007, 144, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Della Mea, M.; Faleri, C.; Fattorini, L.; Aloisi, I.; Serafini-Fracassini, D.; Del Duca, S. Spermine either delays or promotes cell death in Nicotiana tabacum L. corolla depending on the floral developmental stage and affects the distribution of transglutaminase. Plant Sci. 2015, 241, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Li, H.B.; Zhang, L.W.; Cui, Y.H.; Luo, X.; Xue, C.H.; Wang, S.M. Expression of soluble recombinant transglutaminase from Zea mays in Pichia pastoris. World J. Microb. Biot. 2013, 29, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Villalobos, E.; Torne, J.M.; Rigau, J.; Olles, I.; Claparols, I.; Santos, M. Immunogold localization of a transglutaminase related to grana development in different maize cell types. Protoplasma 2001, 216, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Della Mea, M.; Di Sandro, A.; Dondini, L.; Del Duca, S.; Vantini, F.; Bergamini, C.; Bassi, R.; Serafini-Fracassini, D. A Zea mays 39-kDa thylakoid transglutaminase catalyses the modification by polyamines of light-harvesting complex II in a light-dependent way. Planta 2004, 219, 754–764. [Google Scholar] [CrossRef]
- Carvajal-Vallejos, P.K.; Campos, A.; Fuentes-Prior, P.; Villalobos, E.; Almeida, A.M.; Barbera, E.; Torne, J.M.; Santos, M. Purification and in vitro refolding of maize chloroplast transglutaminase over-expressed in Escherichia coli. Biotechnol. Lett. 2007, 29, 1255–1262. [Google Scholar] [CrossRef]
- Chang, C.R.; Bowman, J.L.; Meyerowitz, E.M. Field Guide to Plant Model Systems. Cell 2016, 167, 325–339. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, U463–U465. [Google Scholar] [CrossRef]
- Prade, V.M.; Gundlach, H.; Twardziok, S.; Chapman, B.; Tan, C.; Langridge, P.; Schulman, A.H.; Stein, N.; Waugh, R.; Zhang, G. The pseudogenes of barley. Plant J. 2018, 93, 502–514. [Google Scholar] [CrossRef]
- Bureau, T.E.; Wessler, S.R. Mobile Inverted-Repeat Elements of the Tourist Family Are Associated with the Genes of Many Cereal Grasses. Proc. Natl. Acad. Sci. USA 1994, 91, 1411–1415. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Wicker, T.; Jenkins, J.; Plott, C.; Lux, T.; Koh, C.S.; Ens, J.; Gundlach, H.; Boston, L.B.; Tulpova, Z.; et al. Long-read sequence assembly: A technical evaluation in barley. Plant Cell 2021, 33, 1888–1906. [Google Scholar] [CrossRef] [PubMed]
- Ortigosa, S.M.; Díaz-Vivancos, P.; Clemente-Moreno, M.J.; Pintó-Marijuan, M.; Fleck, I.; Veramendi, J.; Santos, M.; Hernandez, J.A.; Torné, J.M. Oxidative stress induced in tobacco leaves by chloroplast over-expression of maize plastidial transglutaminase. Planta 2010, 232, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Campos, N.; Villalobos, E.; Fontanet, P.; Torné, J.M.; Santos, M. A peptide of 17 aminoacids from the N-terminal region of maize plastidial transglutaminase is essential for chloroplast targeting. Am. J. Mol. Biol. 2012, 2, 245–257. [Google Scholar] [CrossRef]
- Ioannidis, N.E.; Lopera, O.; Santos, M.; Torne, J.M.; Kotzabasis, K. Role of Plastid Transglutaminase in LHCII Polyamination and Thylakoid Electron and Proton Flow. PLoS ONE 2012, 7, e419791. [Google Scholar] [CrossRef]
- Serafini-Fracassini, D.; Della Mea, M.; Tasco, G.; Casadio, R.; Del Duca, S. Plant and animal transglutaminases: Do similar functions imply similar structures? Amino Acids 2009, 36, 643–657. [Google Scholar] [CrossRef]
- Ioannidis, N.E.; Malliarakis, D.; Torne, J.M.; Santos, M.; Kotzabasis, K. The Over-expression of the Plastidial Transglutaminase from Maize in Arabidopsis Increases the Activation Threshold of Photoprotection. Front. Plant Sci. 2016, 7, 635. [Google Scholar] [CrossRef]
- Shu, S.; Yuan, R.N.; Shen, J.L.; Chen, J.; Wang, L.J.; Wu, J.Q.; Sun, J.; Wang, Y.; Guo, S.R. The positive regulation of putrescine on light-harvesting complex II and excitation energy dissipation in salt-stressed cucumber seedlings. Envion. Exp. Bot. 2019, 162, 283–294. [Google Scholar] [CrossRef]
- Aloisi, I.; Cai, G.; Serafini-Fracassini, D.; Del Duca, S. Polyamines in Pollen: From Microsporogenesis to Fertilization. Front. Plant Sci. 2016, 7, 155. [Google Scholar] [CrossRef]
- Gentile, A.; Antognoni, F.; Iorio, R.A.; Distefano, G.; Las Casas, G.; La Malfa, S.; Serafini-Fracassini, D.; Del Duca, S. Polyamines and transglutaminase activity are involved in compatible and self-incompatible pollination of Citrus grandis. Amino Acids 2012, 42, 1025–1035. [Google Scholar] [CrossRef]
- Del Duca, S.; Serafini-Fracassini, D.; Bonner, P.; Cresti, M.; Cai, G. Effects of post-translational modifications catalysed by pollen transglutaminase on the functional properties of microtubules and actin filaments. Biochem. J. 2009, 418, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Aloisi, I.; Cai, G.; Tumiatti, V.; Minarini, A.; Del Duca, S. Natural polyamines and synthetic analogs modify the growth and the morphology of Pyrus communis pollen tubes affecting ROS levels and causing cell death. Plant Sci. 2015, 239, 92–105. [Google Scholar] [CrossRef]
- Aloisi, I.; Cai, G.; Faleri, C.; Navazio, L.; Serafini-Fracassini, D.; Del Duca, S. Spermine Regulates Pollen Tube Growth by Modulating Ca2+-Dependent Actin Organization and Cell Wall Structure. Front. Plant Sci. 2017, 8, 1701. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.F.; Guo, J.Z.; Li, H.; Yang, Z.B. Signaling in Pollen Tube Growth: Crosstalk, Feedback, and Missing Links. Mol. Plant. 2013, 6, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Del Duca, S.; Serafini-Fracassini, D.; Cai, G. An unconventional road for the secretion of transglutaminase in pollen tubes? Plant Signal. Behav. 2013, 8, e24446. [Google Scholar] [CrossRef][Green Version]
- Greenberg, J.T.; Yao, N. The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol. 2004, 6, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.; Kato, N.; Lawton, M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature 2001, 411, 848–853. [Google Scholar] [CrossRef]
- Del Duca, S.; Betti, L.; Trebbi, G.; Serafini-Fracassini, D.; Torrigiani, P. Transglutaminase activity changes during the hypersensitive reaction, a typical defense response of tobacco NN plants to TMV. Physiol. Plant. 2007, 131, 241–250. [Google Scholar] [CrossRef]
- Halim, V.A.; Hunger, A.; Macioszek, V.; Landgraf, P.; Nürnberger, T.; Scheel, D.; Rosahl, S. The oligopeptide elicitor Pep-13 induces salicylic acid-dependent and-independent defense reactions in potato. Physiol. Mol. Plant Pathol. 2004, 64, 311–318. [Google Scholar] [CrossRef]
- Parker, J.E.; Schulte, W.; Hahlbrock, K.; Scheel, D. An extracellular glycoprotein from Phytophthora megasperma f. sp. glycinea elicits phytoalexin synthesis in cultured parsley cells and protoplasts. Mol. Plant-Microbe Interact. 1991, 4, 19–27. [Google Scholar] [CrossRef]
- Martins, I.M.; Matos, M.; Costa, R.; Silva, F.; Pascoal, A.; Estevinho, L.M.; Choupina, A.B. Transglutaminases: Recent achievements and new sources. Appl. Microbiol. Biot. 2014, 98, 6957–6964. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Akram, W.; Bashir, Z.; Shahzadi, I.; Wang, R.; Abbas, H.M.K.; Hu, D.; Ahmed, S.; Xu, X.M.; Li, G.H.; et al. Functional and Structural Analysis of a Novel Acyltransferase from Pathogenic Phytophthora melonis. ACS Omega 2021, 6, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- Jahan, M.S.; Shi, Z.R.; Zhong, M.; Zhang, Y.M.; Zhou, R.R.; El-Mogy, M.M.; Sun, J.; Shu, S.; Guo, S.R.; Wang, Y. Comparative transcriptome analysis reveals gene network regulation of TGase-induced thermotolerance in tomato. Not. Bot. Horti Agrobo. 2021, 49, 12208. [Google Scholar] [CrossRef]
- Zhong, M.; Wang, Y.; Zhang, Y.M.; Shu, S.; Sun, J.; Guo, S.R. Overexpression of Transglutaminase from Cucumber in Tobacco Increases Salt Tolerance through Regulation of Photosynthesis. Int. J. Mol. Sci. 2019, 20, 894. [Google Scholar] [CrossRef]
- Tang, Y.-Y.; Yuan, Y.-H.; Shu, S.; Guo, S.-R. Regulatory mechanism of NaCl stress on photosynthesis and antioxidant capacity mediated by transglutaminase in cucumber (Cucumis sativus L.) seedlings. Sci. Hortic. Amst. 2018, 235, 294–306. [Google Scholar] [CrossRef]
- García-Jiménez, P.; Just, P.M.; Delgado, A.M.; Robaina, R.R. Transglutaminase activity decrease during acclimation to hyposaline conditions in marine seaweed Grateloupia doryphora (Rhodophyta, Halymeniaceae). J. Plant Physiol. 2007, 164, 367–370. [Google Scholar] [CrossRef][Green Version]
- Serafini-Fracassini, D.; Del Duca, S.; Torrigiani, P. Polyamine conjugation during the cell cycle of Helianthus tuberosus: Non enzymatic and transglutaminase-like binding activity. Plant Physiol. Biochem. 1989, 27, 659–668. [Google Scholar]
- Serafini-Fracassini, D.; Della Mea, M.; Parrotta, L.; Faleri, C.; Cai, G.; Del Duca, S.; Aloisi, I. AtPng1 knockout mutant of Arabidopsis thaliana shows a juvenile phenotype, morpho-functional changes, altered stress response and cell wall modifications. Plant Physiol. Bioch. 2021, 167, 11–21. [Google Scholar] [CrossRef]
- Carter, R.; Woolfenden, H.; Baillie, A.; Amsbury, S.; Carroll, S.; Healicon, E.; Sovatzoglou, S.; Braybrook, S.; Gray, J.E.; Hobbs, J. Stomatal opening involves polar, not radial, stiffening of guard cells. Curr. Biol. 2017, 27, 2974–2983.e2972. [Google Scholar] [CrossRef]
- Telci, D.; Griffin, M. Tissue transglutaminase (TG2)—A wound response enzyme. Front. Biosci. Landmrk 2006, 11, 867–882. [Google Scholar] [CrossRef]
- Qin, Y.; Guo, X.W.; Li, L.; Wang, H.W.; Kim, W. The Antioxidant Property of Chitosan Green Tea Polyphenols Complex Induces Transglutaminase Activation in Wound Healing. J. Med. Food 2013, 16, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Paluch-Lubawa, E.; Stolarska, E.; Sobieszczuk-Nowicka, E. Dark-Induced Barley Leaf Senescence–A Crop System for Studying Senescence and Autophagy Mechanisms. Front. Plant Sci. 2021, 12, 425. [Google Scholar] [CrossRef]
- Sarwat, M.; Naqvi, A.R.; Ahmad, P.; Ashraf, M.; Akram, N.A. Phytohormones and microRNAs as sensors and regulators of leaf senescence: Assigning macro roles to small molecules. Biotechnol. Adv. 2013, 31, 1153–1171. [Google Scholar] [CrossRef] [PubMed]
- Sobieszczuk-Nowicka, E.; Zmienko, A.; Samelak-Czajka, A.; Łuczak, M.; Pietrowska-Borek, M.; Iorio, R.; Del Duca, S.; Figlerowicz, M.; Legocka, J. Dark-induced senescence of barley leaves involves activation of plastid transglutaminases. Amino Acids 2015, 47, 825–838. [Google Scholar] [CrossRef] [PubMed]
- van der Graaff, E.; Schwacke, R.; Schneider, A.; Desimone, M.; Flügge, U.-I.; Kunze, R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006, 141, 776–792. [Google Scholar] [CrossRef]
- Iqbal, N.; Nazar, R.; Khan, M.I.R.; Masood, A.; Khan, N.A. Role of gibberellins in regulation of source–sink relations under optimal and limiting environmental conditions. Curr. Sci. 2011, 100, 998–1007. [Google Scholar]
- Cejudo, F.J.; Ferrández, J.; Cano, B.; Puerto-Galán, L.; Guinea, M. The function of the NADPH thioredoxin reductase C-2-Cys peroxiredoxin system in plastid redox regulation and signalling. FEBS Lett. 2012, 586, 2974–2980. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]

| Plant Species | Source | Molecular Weight (kDa) | Optimum pH Assay | Localisation | References |
|---|---|---|---|---|---|
| Arabidopsis thaliana | Recombinant enzyme | 86 | 7.5–8.5 | Microsomal fraction | [19] |
| Entire plant | n.d. | 8.4 | Entire cell | [20] | |
| Beta vulgaris | Leaf | n.d. | 7.8 | Entire cell | [41] |
| Capsicum annuum | Entire plant | 70, 60, 56 64, 34 | n.d. | Root | [42] |
| Chlamydomonas reinhardtii | Cell | 72 | 7.4 | Cell wall | [27] |
| Cucumber sativus | Cotyledon | 77, 58, 50, 30 | 8.5 | Thylakoid | [39] |
| Dunaliella salina | Cell and chloroplast | 70, 50, 25 | 8.5 | Chloroplast | [43,44] |
| Glycine max | Leaf and seedling | 80 | 7.6 | Entire cell | [45,46] |
| Helianthus tuberosus | Leaf | 58, 22, 19 | 8.0–9.5 | Chloroplast | [24] |
| Chloroplast | 58, 24, 150 | n.d. | Thylakoid and stroma | [47] | |
| Chloroplast | 58 | 8.5 | Thylakoid and stroma | [23] | |
| Etiolated apex | 85, 75, 58 | 8.5 | Immature cell | [48] | |
| Hordeum vulgare | Leaf | 33, 58, 75 | 8.5 | Thylakoid | [3] |
| Malus domestica | Pollen | 70, 75 | 6.5 | Chloroplast, microsomal, and cell wall | [49] |
| Nicotiana tabacum | Flower corolla | 38, 58 | 7.5–8.5 | Microsome, plastid, and cell wall | [50,51] |
| Flower corolla | n.d. | 6.5 | Epidermis and cell wall | [52] | |
| Oryza sativa | Entire plant | 40 | 6.5 | Thylakoid | [26] |
| Entire plant | 72, 39 | n.d. | Chloroplast | [36] | |
| Rosmarinus officinalis | Entire plant | n.d. | 7.0 | n.d. | [29] |
| Zea mays | Pollen | 58–50; 160–180 | n.d. | Pollen | [40] |
| Chloroplast | 58, 61–67, 77, 150 | n.d. | Thylakoid and grana | [28] | |
| Chloroplast | 58 | n.d. | Thylakoid | [7] | |
| Recombinant enzyme | 58 | 8.0 | Chloroplast | [14] | |
| Recombinant enzyme | 47 | 8.0 | n.d. | [53] | |
| Meristematic callus | 58, 34 | n.d. | Chloroplast and adult leaf | [54] | |
| Chloroplast | 39 | 8.5 | Thylakoid | [19,55] | |
| Recombinant enzyme | 55 | 8.5 | Chloroplast | [56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parrotta, L.; Tanwar, U.K.; Aloisi, I.; Sobieszczuk-Nowicka, E.; Arasimowicz-Jelonek, M.; Del Duca, S. Plant Transglutaminases: New Insights in Biochemistry, Genetics, and Physiology. Cells 2022, 11, 1529. https://doi.org/10.3390/cells11091529
Parrotta L, Tanwar UK, Aloisi I, Sobieszczuk-Nowicka E, Arasimowicz-Jelonek M, Del Duca S. Plant Transglutaminases: New Insights in Biochemistry, Genetics, and Physiology. Cells. 2022; 11(9):1529. https://doi.org/10.3390/cells11091529
Chicago/Turabian StyleParrotta, Luigi, Umesh Kumar Tanwar, Iris Aloisi, Ewa Sobieszczuk-Nowicka, Magdalena Arasimowicz-Jelonek, and Stefano Del Duca. 2022. "Plant Transglutaminases: New Insights in Biochemistry, Genetics, and Physiology" Cells 11, no. 9: 1529. https://doi.org/10.3390/cells11091529
APA StyleParrotta, L., Tanwar, U. K., Aloisi, I., Sobieszczuk-Nowicka, E., Arasimowicz-Jelonek, M., & Del Duca, S. (2022). Plant Transglutaminases: New Insights in Biochemistry, Genetics, and Physiology. Cells, 11(9), 1529. https://doi.org/10.3390/cells11091529







