The Puf-A Protein Is Required for Primordial Germ Cell Development
Abstract
1. Introduction
- I.
- the classical PUF proteins which control mRNA translation by binding to 3′UTR,
- II.
- the Nop9 orthologues for small subunit pre-rRNA processing, and
- III.
- the PUM3 orthologues for the biogenesis of the large ribosomal subunit [2].
2. Materials and Methods
2.1. Construction of Plasmids
- I.
- the promoter of phosphoglycerate kinase promoter (PPGK)
- II.
- PPGK for the expression of tetracycline-controlled transcriptional suppressor (tTS)
- III.
- IV.
- β-globin polyA
- V.
- the modified Tet-responsive Pol III hybrid promoter (PTREmod/U6) with tetO sequences (Figure 1A).
2.2. Transgenic Mice Carrying the Inducible Puf-A Knockdown Construct
2.3. Generation and Genotyping of Transgenic Mice
2.4. Determination of Zygosity by Real-Time Quantitative PCR (qPCR) of Genomic DNA
2.5. Puf-A shRNA Induction
2.6. Using Oct4-Driven EGFP Expression for Tracing PGCs
2.7. Analysis of PGC Distribution in Embryos with Time-Lapse Video Microscopy
2.8. Isolation of PGCs
2.9. Cytospin and Immunofluorescence Staining
2.10. BrdU Labeling
2.11. FACS Analysis of Cell Cycle
2.12. Analysis of Cell Death
2.13. Flow Cytometric Analysis of Cell Apoptosis
2.14. Sorting of PGCs and Quantitative RT-PCR
2.15. Statistical Analyses
3. Results
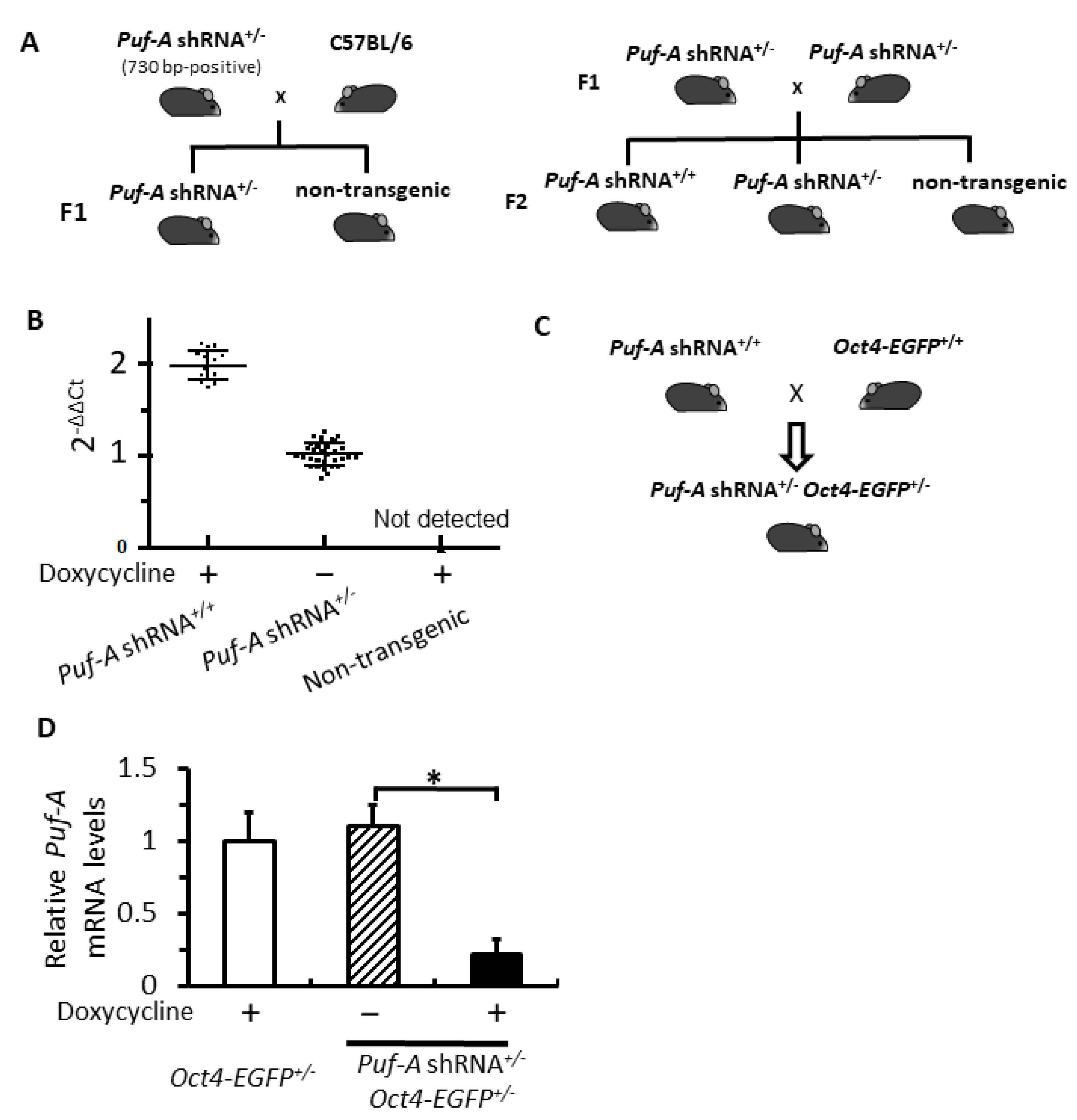
3.1. Generation of Transgenic Mice Carrying the Inducible Puf-A shRNA Construct
3.2. Generation of the Inducible Puf-A shRNA and Oct4-EGFP Double Transgenic Mice
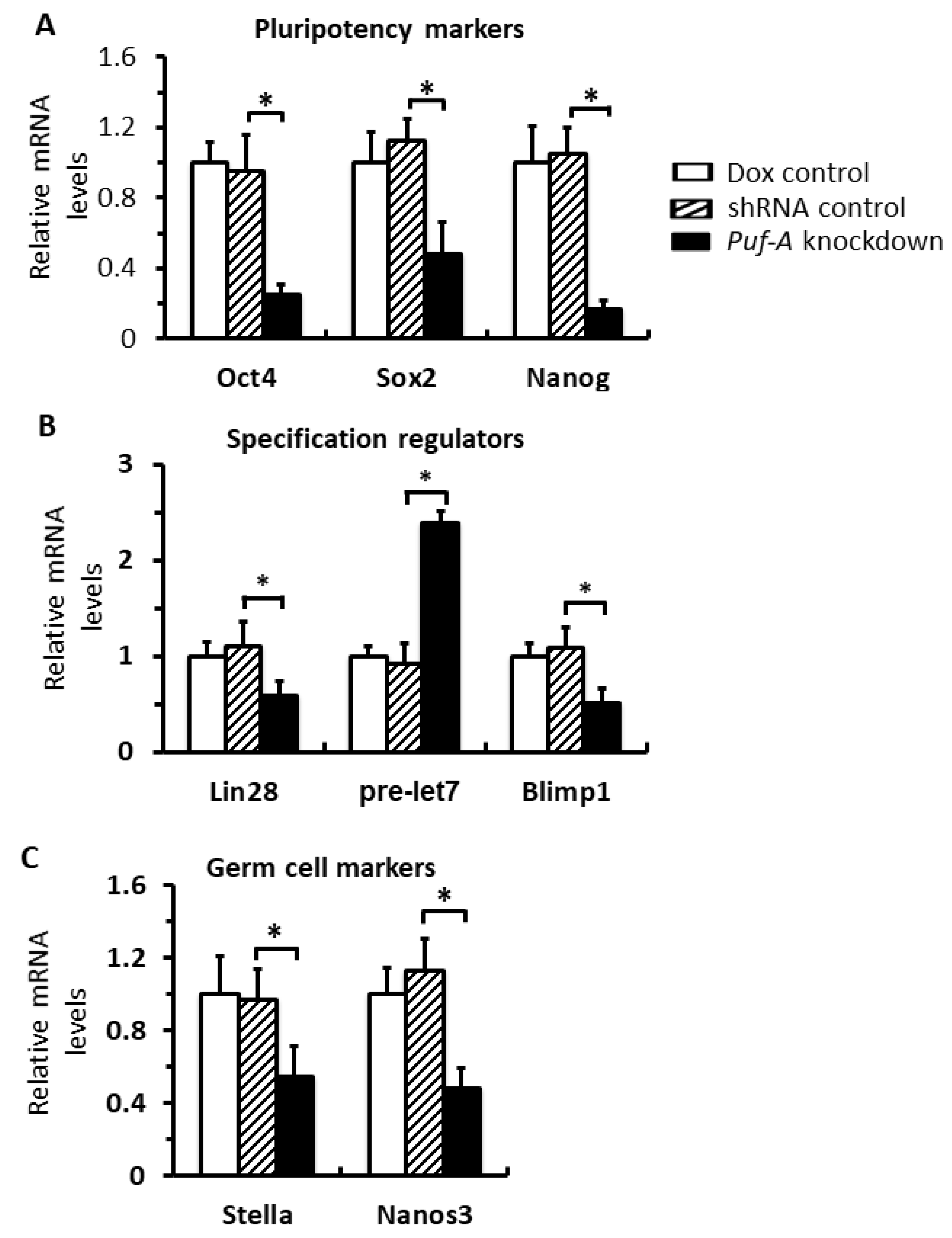
3.3. Puf-A Is Required for PGC Growth during Gonad Development
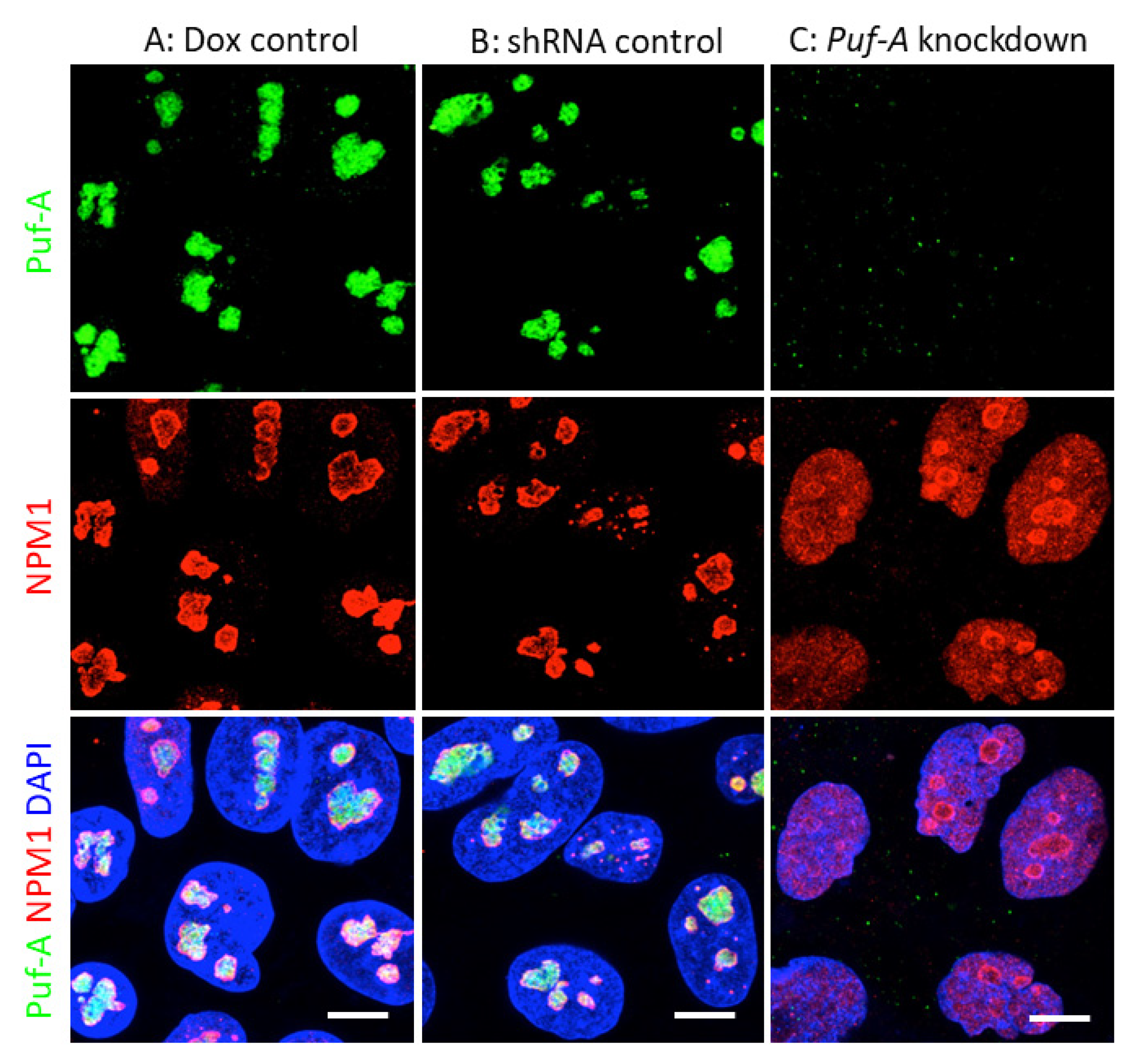
3.4. NPM1 Translocation to Nucleoplasm upon Puf-A Knockdown in PGCs
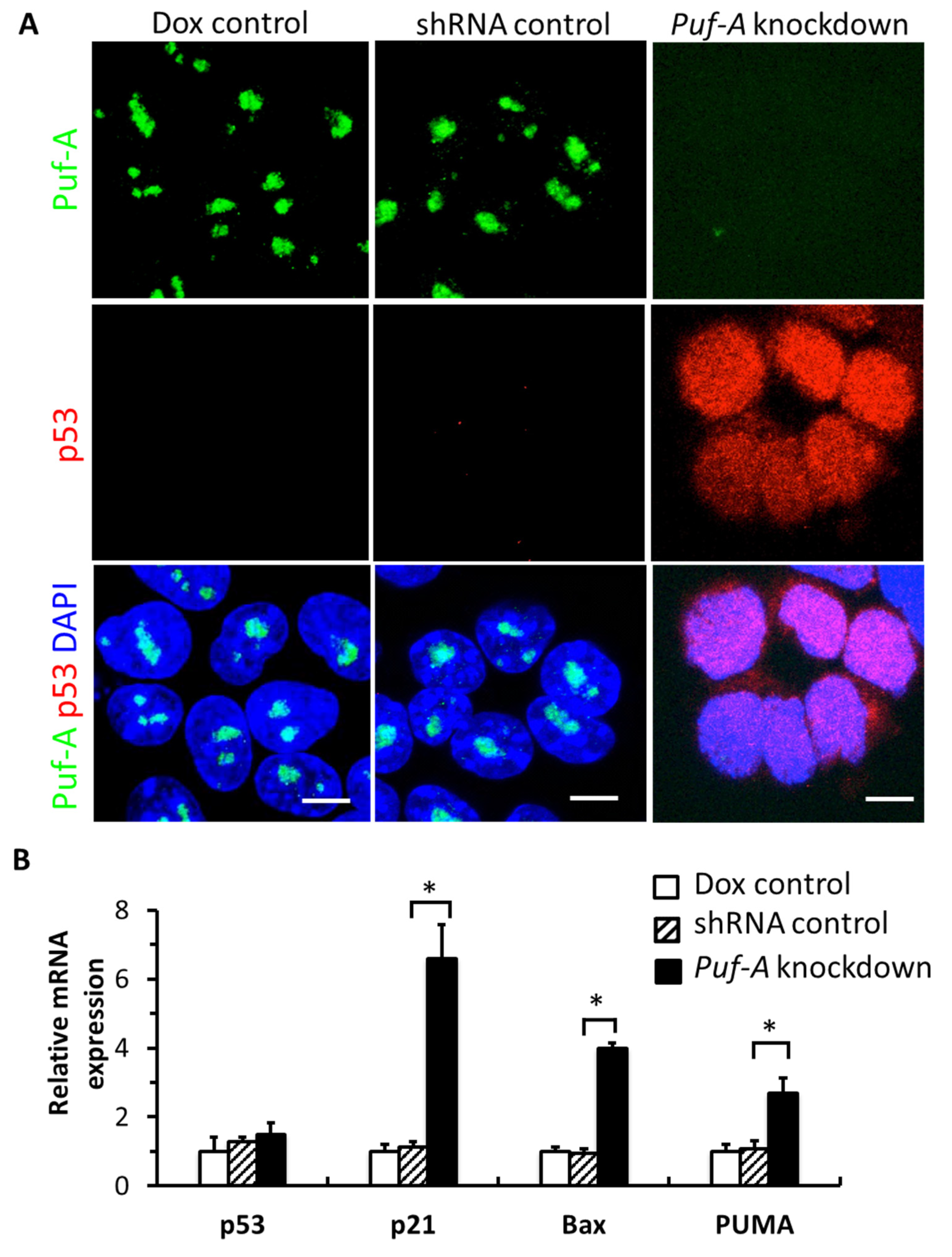
3.5. Knockdown of Puf-A Activates p53 Signaling in PGCs
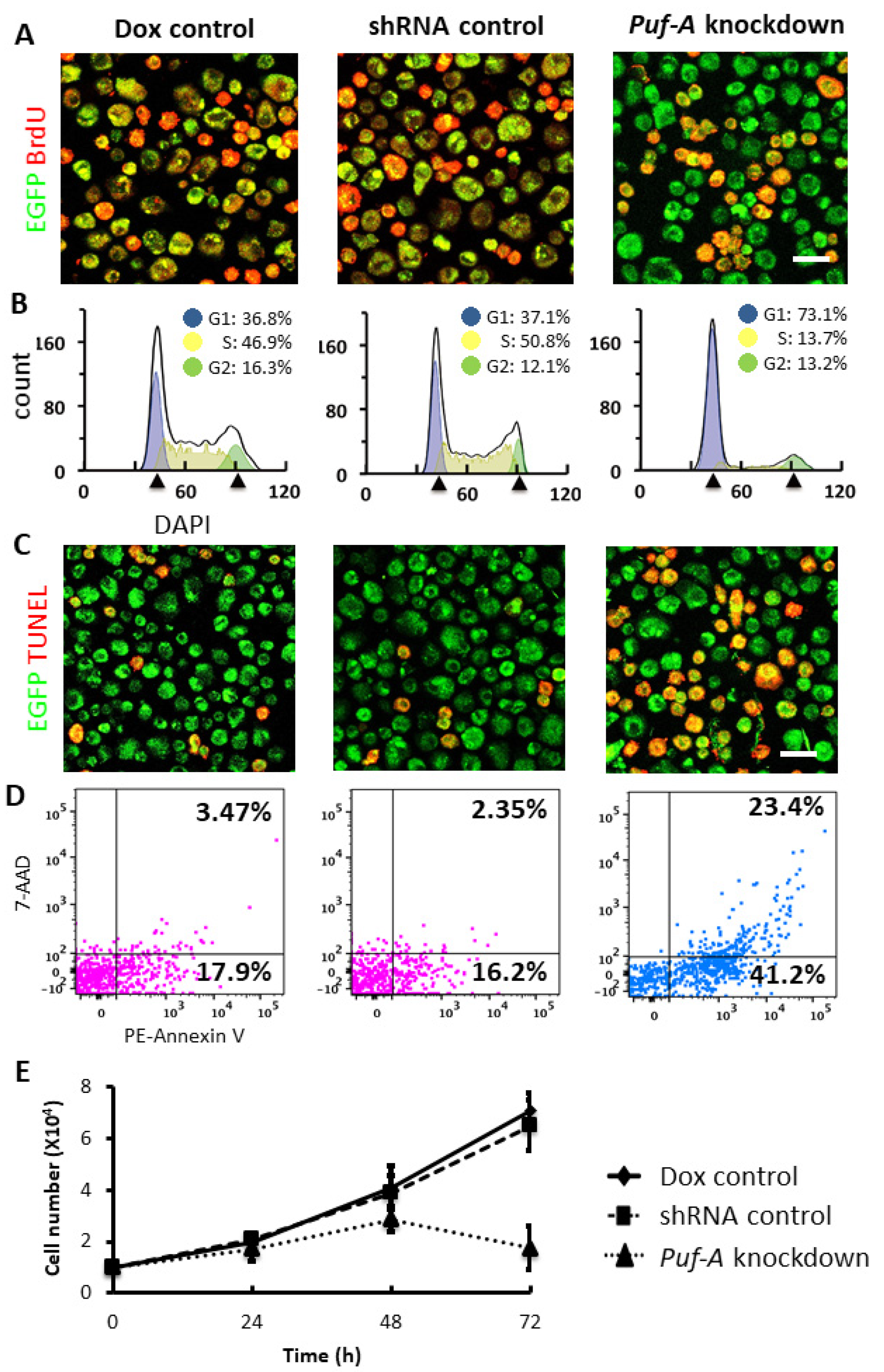
3.6. Puf-A Knockdown Impairs the Growth of PGCs
3.7. Puf-A Protects Germ Cell Fate during Early Gonad Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wickens, M.; Bernstein, D.S.; Kimble, J.; Parker, R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002, 18, 150–157. [Google Scholar] [CrossRef]
- Najdrová, V.; Stairs, C.W.; Vinopalová, M.; Voleman, L.; Doležal, P. The evolution of the Puf superfamily of proteins across the tree of eukaryotes. BMC Biol. 2020, 18, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Shiffman, D.; Mevarech, M.; Jensen, S.E.; Cohen, G.; Aharonowitz, Y. Cloning and comparative sequence analysis of the gene coding for isopenicillin N synthase in Streptomyces. Mol. Gen. Genet. 1988, 214, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Uyhazi, K.E.; Yang, Y.; Liu, N.; Qi, H.; Huang, X.A.; Mak, W.; Weatherbee, S.D.; de Prisco, N.; Gennarino, V.A.; Song, X.; et al. Pumilio proteins utilize distinct regulatory mechanisms to achieve complementary functions required for pluripotency and embryogenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 7851–7862. [Google Scholar] [CrossRef] [PubMed]
- Moore, F.L.; Jaruzelska, J.; Fox, M.S.; Urano, J.; Firpo, M.T.; Turek, P.J.; Dorfman, D.M.; Pera, R.A.R. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc. Natl. Acad. Sci. USA 2003, 100, 538–543. [Google Scholar] [CrossRef]
- Siemen, H.; Colas, D.; Heller, H.C.; Brüstle, O.; Pera, R.A.R. Pumilio-2 function in the mouse nervous system. PLoS ONE 2011, 6, e25932. [Google Scholar] [CrossRef]
- Lin, K.; Qiang, W.; Zhu, M.; Ding, Y.; Shi, Q.; Chen, X.; Zsiros, E.; Wang, K.; Yang, X.; Kurita, T.; et al. Mammalian Pum1 and Pum2 Control Body Size via Translational Regulation of the Cell Cycle Inhibitor Cdkn1b. Cell Rep. 2019, 26, 2434–2450.e6. [Google Scholar] [CrossRef]
- Chen, D.; Zheng, W.; Lin, A.; Uyhazi, K.; Zhao, H.; Lin, H. Pumilio 1 suppresses multiple activators of p53 to safeguard spermatogenesis. Curr. Biol. 2012, 22, 420–425. [Google Scholar] [CrossRef]
- Gennarino, V.A.; Singh, R.K.; White, J.J.; De Maio, A.; Han, K.; Kim, J.-Y.; Jafar-Nejad, P.; di Ronza, A.; Kang, H.; Sayegh, L.S.; et al. Pumilio1 haploinsufficiency leads to SCA1-like neurodegeneration by increasing wild-type Ataxin1 levels. Cell 2015, 160, 1087–1098. [Google Scholar] [CrossRef]
- Gennarino, V.A.; Palmer, E.E.; McDonell, L.M.; Wang, L.; Adamski, C.J.; Koire, A.; See, L.; Chen, C.A.; Schaaf, C.P.; Rosenfeld, J.A.; et al. A Mild PUM1 Mutation Is Associated with Adult-Onset Ataxia, whereas Haploinsufficiency Causes Developmental Delay and Seizures. Cell 2018, 172, 924–936.e11. [Google Scholar] [CrossRef]
- Thomson, E.; Rappsilber, J.; Tollervey, D. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA 2007, 13, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; McCann, K.; Qiu, C.; Gonzalez, L.; Baserga, S.J.; Hall, T.M.T. Nop9 is a PUF-like protein that prevents premature cleavage to correctly process pre-18S rRNA. Nat. Commun. 2016, 7, 13085. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-T.; Ting, Y.-H.; Liang, K.-J.; Lo, K.-Y. The Roles of Puf6 and Loc1 in 60S Biogenesis Are Interdependent, and Both Are Required for Efficient Accommodation of Rpl43. J. Biol. Chem. 2016, 291, 19312–19323. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lee, I.; Moradi, E.; Hung, N.-J.; Johnson, A.W.; Marcotte, E.M. Rational Extension of the Ribosome Biogenesis Pathway Using Network-Guided Genetics. PLoS Biol. 2009, 7, e1000213. [Google Scholar] [CrossRef]
- Gu, W.; Deng, Y.; Zenklusen, D.; Singer, R.H. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes. Dev. 2004, 18, 1452–1465. [Google Scholar] [CrossRef]
- Kuo, M.-W.; Wang, S.-H.; Chang, J.-C.; Chang, C.-H.; Huang, L.-J.; Lin, H.-H.; Yu, A.L.-T.; Li, W.-H.; Yu, J. A novel puf-A gene predicted from evolutionary analysis is involved in the development of eyes and primordial germ-cells. PLoS ONE 2009, 4, e4980. [Google Scholar] [CrossRef]
- Qiu, C.; McCann, K.L.; Wine, R.N.; Baserga, S.J.; Hall, T.M.T. A divergent Pumilio repeat protein family for pre-rRNA processing and mRNA localization. Proc. Natl. Acad. Sci. USA 2014, 111, 18554–18559. [Google Scholar] [CrossRef]
- Cho, H.-C.; Huang, Y.; Hung, J.-T.; Hung, T.-H.; Cheng, K.-C.; Liu, Y.-H.; Kuo, M.-W.; Wang, S.-H.; Yu, A.L.; Yu, J. Puf-A promotes cancer progression by interacting with nucleophosmin in nucleolus. Oncogene 2022, 41, 1155–1165. [Google Scholar] [CrossRef]
- Ohinata, Y.; Ohta, H.; Shigeta, M.; Yamanaka, K.; Wakayama, T.; Saitou, M. A signaling principle for the specification of the germ cell lineage in mice. Cell 2009, 137, 571–584. [Google Scholar] [CrossRef]
- Saitou, M.; Payer, B.; O’Carroll, D.; Ohinata, Y.; Surani, M.A. Blimp1 and the emergence of the germ line during development in the mouse. Cell Cycle 2005, 4, 1736–1740. [Google Scholar] [CrossRef]
- Ohinata, Y.; Payer, B.; O’Carroll, D.; Ancelin, K.; Ono, Y.; Sano, M.; Barton, S.C.; Obukhanych, T.; Nussenzweig, M.C.; Tarakhovsky, A.; et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 2005, 436, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Bendel-Stenzel, M.; Anderson, R.; Heasman, J.; Wylie, C. The origin and migration of primordial germ cells in the mouse. Semin. Cell Dev. Biol. 1998, 9, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Shamblott, M.J.; Kerr, C.L.; Axelman, J.; Littlefield, J.W.; Clark, G.O.; Patterson, E.S.; Addis, R.C.; Kraszewski, J.N.; Kent, K.C.; Gearhart, J.D. Derivation and Differentiation of Human Embryonic Germ Cells. In Essentials of Stem Cell Biology, 3rd ed.; Lanza, R., Atala, A., Eds.; Academic Press: Boston, MA, USA, 2014; Volume 30, pp. 435–451. [Google Scholar] [CrossRef]
- Freundlieb, S.; Schirra-Müller, C.; Bujard, H. A tetracycline controlled activation/repression system with increased potential for gene transfer into mammalian cells. J. Gene. Med. 1999, 1, 4–12. [Google Scholar] [CrossRef]
- Witzgall, R.; O’Leary, E.; Leaf, A.; Onaldi, D.; Bonventre, J.V. The Krüppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc. Natl. Acad. Sci. USA 1994, 91, 4514–4518. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Krog, G.R.; Clausen, F.B.; Dziegiel, M.H. Quantitation of RHD by real-time polymerase chain reaction for determination of RHD zygosity and RHD mosaicism/chimerism: An evaluation of four quantitative methods. Transfusion 2007, 47, 715–722. [Google Scholar] [CrossRef]
- Yoshimizu, T.; Sugiyama, N.; De Felice, M.; Yeom, Y.I.; Ohbo, K.; Masuko, K.; Obinata, M.; Abe, K.; Scholer, H.R.; Matsui, Y. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 1999, 41, 675–684. [Google Scholar] [CrossRef]
- Kurki, S.; Peltonen, K.D.; Latonen, L.; Kiviharju, T.M.; Ojala, P.M.; Meek, D.; Laiho, M. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 2004, 5, 465–475. [Google Scholar] [CrossRef]
- Nakano, K.; Vousden, K.H. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 2001, 7, 683–694. [Google Scholar] [CrossRef]
- Toshiyuki, M.; Reed, J.C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 1995, 80, 293–299. [Google Scholar] [CrossRef]
- Saitou, M.; Yamaji, M. Primordial germ cells in mice. Cold Spring Harb. Perspect. Biol. 2012, 4, 1–19. [Google Scholar] [CrossRef] [PubMed]
- West, J.A.; Viswanathan, S.R.; Yabuuchi, A.; Cunniff, K.; Takeuchi, A.; Park, I.-H.; Sero, J.E.; Zhu, H.; Perez-Atayde, A.; Frazier, A.L.; et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 2009, 460, 909–913. [Google Scholar] [CrossRef] [PubMed]
- Matzuk, M.M. LIN28 lets BLIMP1 take the right course. Dev. Cell 2009, 17, 160–161. [Google Scholar] [CrossRef][Green Version]
- Kehler, J.; Tolkunova, E.; Koschorz, B.; Pesce, M.; Gentile, L.; Boiani, M.; Lomelí, H.; Nagy, A.; McLaughlin, K.J.; Schöler, H.; et al. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004, 5, 1078–1083. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Kurimoto, K.; Yabuta, Y.; Sasaki, H.; Nakatsuji, N.; Saitou, M.; Tada, T. Conditional knockdown of Nanog induces apoptotic cell death in mouse migrating primordial germ cells. Development 2009, 136, 4011–4020. [Google Scholar] [CrossRef]
- Campolo, F.; Gori, M.; Favaro, R.; Nicolis, S.; Pellegrini, M.; Botti, F.; Rossi, P.; Jannini, E.A.; Dolci, S. Essential role of Sox2 for the establishment and maintenance of the germ cell line. Stem Cells 2013, 31, 1408–1421. [Google Scholar] [CrossRef]
- Kanamori, M.; Oikawa, K.; Tanemura, K.; Hara, K. Mammalian germ cell migration during development, growth, and homeostasis. Reprod. Med. Biol. 2019, 18, 247–255. [Google Scholar] [CrossRef]
- Strome, S.; Updike, D. Specifying and protecting germ cell fate. Nat. Reviews. Mol. Cell Biol. 2015, 16, 406–416. [Google Scholar] [CrossRef]
- Nicholls, P.K.; Schorle, H.; Naqvi, S.; Hu, Y.-C.; Fan, Y.; Carmell, M.A.; Dobrinski, I.; Watson, A.L.; Carlson, D.F.; Fahrenkrug, S.C.; et al. Mammalian germ cells are determined after PGC colonization of the nascent gonad. Proc. Natl. Acad. Sci. USA 2019, 116, 25677–25687. [Google Scholar] [CrossRef]
- Yang, K.; Wang, M.; Zhao, Y.; Sun, X.; Yang, Y.; Li, X.; Zhou, A.; Chu, H.; Zhou, H.; Xu, J.; et al. A redox mechanism underlying nucleolar stress sensing by nucleophosmin. Nat. Commun. 2016, 7, 13599. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ko, C.-F.; Chang, Y.-C.; Cho, H.-C.; Yu, J. The Puf-A Protein Is Required for Primordial Germ Cell Development. Cells 2022, 11, 1476. https://doi.org/10.3390/cells11091476
Ko C-F, Chang Y-C, Cho H-C, Yu J. The Puf-A Protein Is Required for Primordial Germ Cell Development. Cells. 2022; 11(9):1476. https://doi.org/10.3390/cells11091476
Chicago/Turabian StyleKo, Chi-Fong, Yi-Chieh Chang, Huan-Chieh Cho, and John Yu. 2022. "The Puf-A Protein Is Required for Primordial Germ Cell Development" Cells 11, no. 9: 1476. https://doi.org/10.3390/cells11091476
APA StyleKo, C.-F., Chang, Y.-C., Cho, H.-C., & Yu, J. (2022). The Puf-A Protein Is Required for Primordial Germ Cell Development. Cells, 11(9), 1476. https://doi.org/10.3390/cells11091476





