Determination of WWOX Function in Modulating Cellular Pathways Activated by AP-2α and AP-2γ Transcription Factors in Bladder Cancer
Abstract
1. Introduction
2. Materials and Methods
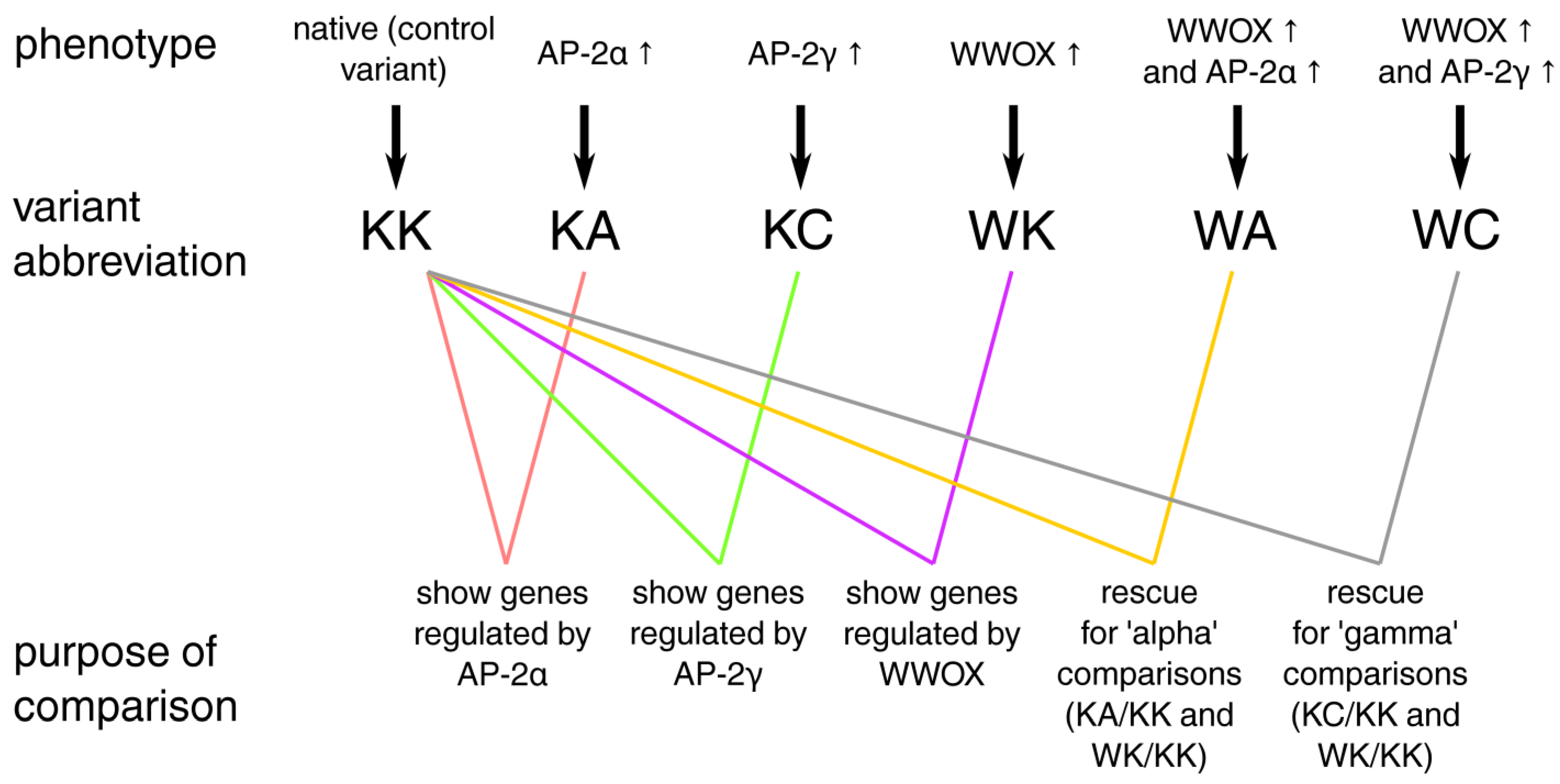
2.1. Cell Lines, Culture Conditions and Stable Transductions
2.2. Protein Extraction and Western Blot
2.3. Statistical Analysis
2.4. RNA Isolation, CAGE Library Preparation, Sequencing, Mapping and Gene Expression
2.5. Identification of Target Genes Regulated by AP-2 Transcription Factors
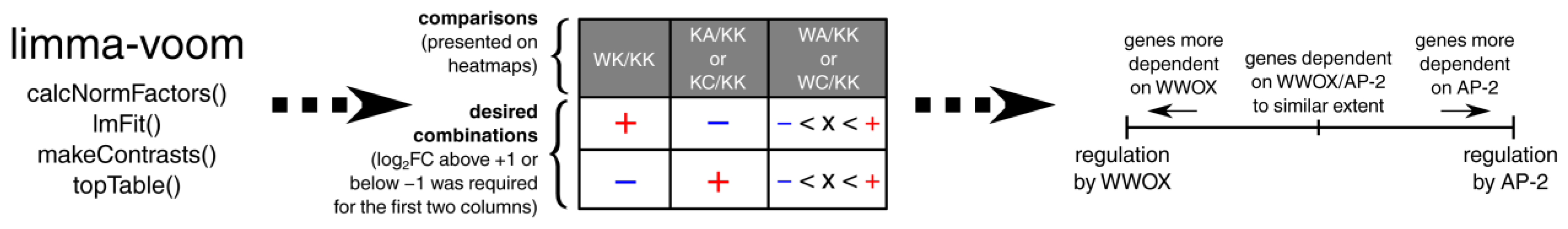
2.6. Differential Expression Analysis
2.7. Identification of Genes Inversely Regulated by WWOX and AP-2 and Their Functional Annotation
2.8. Pathway Visualization and Integration
2.9. Protein Association Networks Supported by Molecular Signatures
2.10. Principal Component Analysis
3. Results
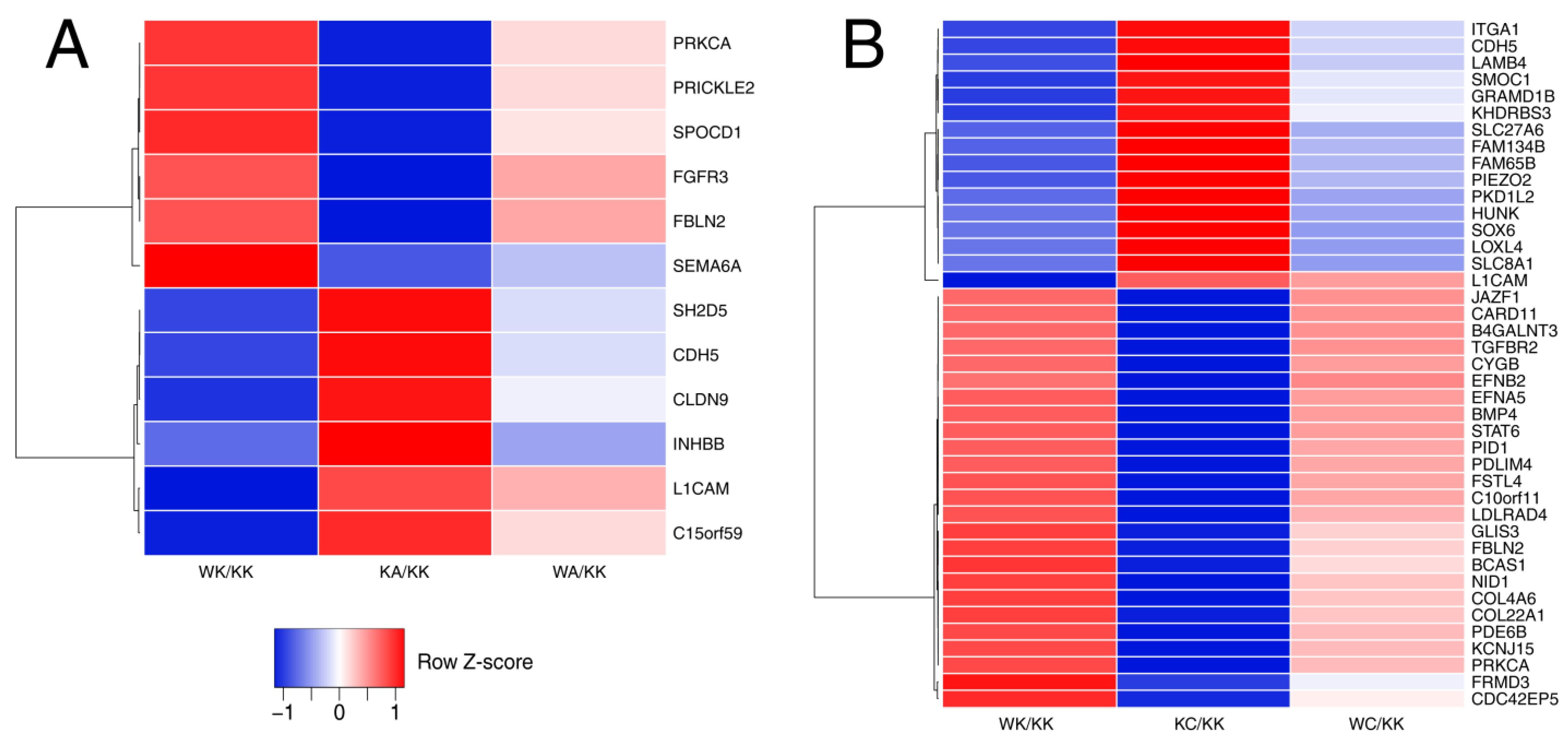
3.1. WWOX Can Overexpress or Downregulate the Genes Which Are Inversely Regulated by AP-2 Factor
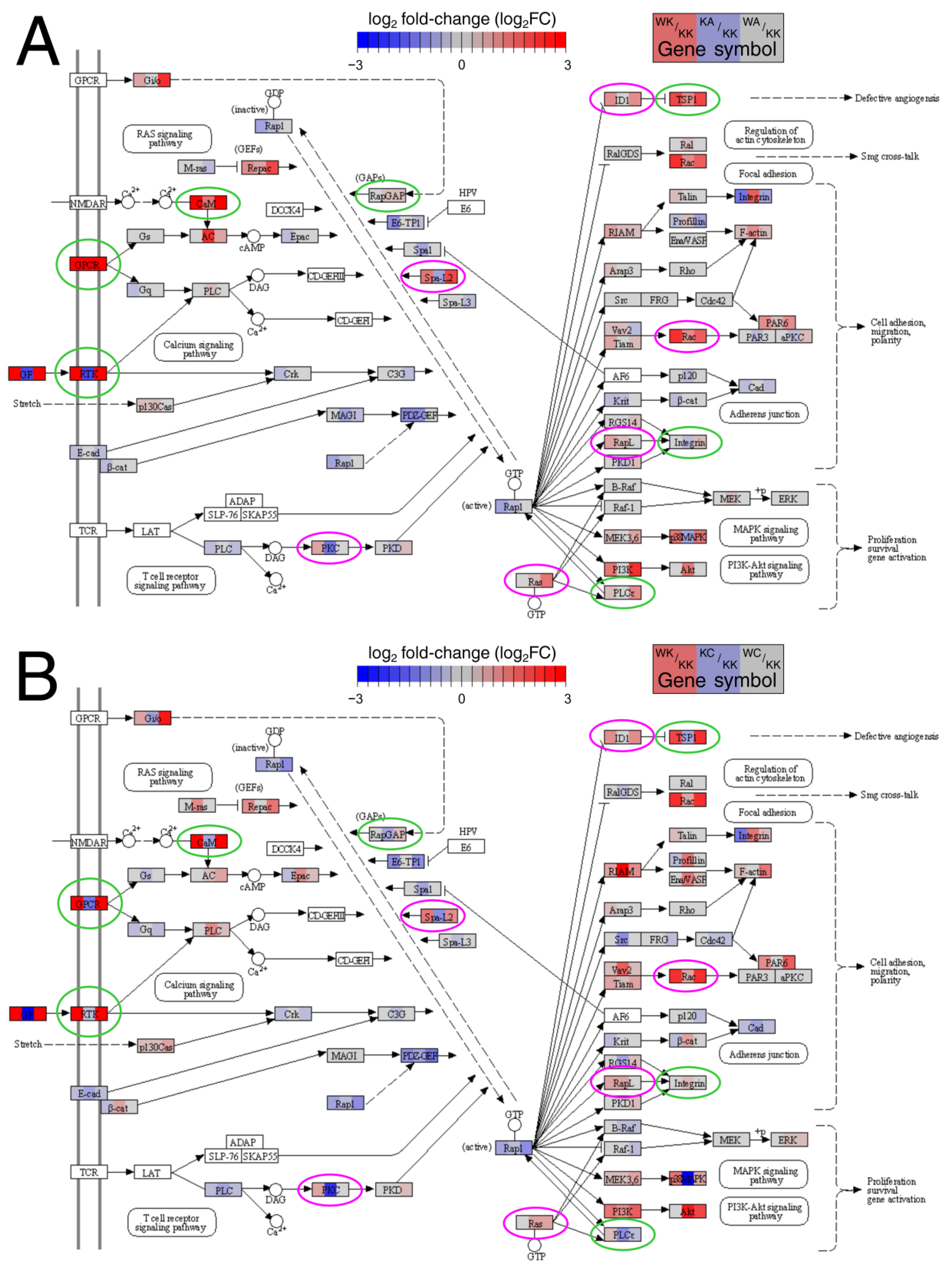
3.2. Many Important Pathways Were Denoted, with Rap1 and MAPK Repeating between Comparisons
3.3. Altered Signaling Was Found to Be Involved in the Regulation of Multiple Biological Processes
3.4. Observations Were Validated on the Protein Level
3.5. Definitive Network Was of High Interconnectivity and Phenotypes Were Explained by Dimensional Grouping
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Decamps, C.; Prive, F.; Bacher, R.; Jost, D.; Waguet, A.; Consortium, H.; Houseman, E.A.; Lurie, E.; Lutsik, P.; Milosavljevic, A.; et al. Guidelines for cell-type heterogeneity quantification based on a comparative analysis of reference-free DNA methylation deconvolution software. BMC Bioinform. 2020, 21, 16. [Google Scholar] [CrossRef] [PubMed]
- Hynds, R.E.; Vladimirou, E.; Janes, S.M. The secret lives of cancer cell lines. Dis. Model. Mech. 2018, 11, dmm037366. [Google Scholar] [CrossRef] [PubMed]
- Gabor, A.; Tognetti, M.; Driessen, A.; Tanevski, J.; Guo, B.; Cao, W.; Shen, H.; Yu, T.; Chung, V.; Single Cell Signaling in Breast Cancer DREAM Consortium Members; et al. Cell-to-cell and type-to-type heterogeneity of signaling networks: Insights from the crowd. Mol. Syst. Biol. 2021, 17, e10402. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Schmidt, B.U.S.; Kubitschke, H.; Morawetz, E.W.; Wolf, B.; Kas, J.A.; Losert, W. Detecting heterogeneity in and between breast cancer cell lines. Cancer Converg. 2020, 4, 1. [Google Scholar] [CrossRef]
- Qin, S.S.; Han, B.J.; Williams, A.; Jackson, K.M.; Jewell, R.; Chacon, A.C.; Lord, E.M.; Linehan, D.C.; Kim, M.; Reuben, A.; et al. Intertumoral Genetic Heterogeneity Generates Distinct Tumor Microenvironments in a Novel Murine Synchronous Melanoma Model. Cancers 2021, 13, 2293. [Google Scholar] [CrossRef]
- Kolat, D.; Kaluzinska, Z.; Pluciennik, E. Fragile Gene WWOX Guides TFAP2A/TFAP2C-Dependent Actions Against Tumor Progression in Grade II Bladder Cancer. Front. Oncol. 2021, 11, 621060. [Google Scholar] [CrossRef]
- Kolat, D.; Kaluzinska, Z.; Bednarek, A.K.; Pluciennik, E. WWOX Loses the Ability to Regulate Oncogenic AP-2gamma and Synergizes with Tumor Suppressor AP-2alpha in High-Grade Bladder Cancer. Cancers 2021, 13, 2957. [Google Scholar] [CrossRef]
- Aqeilan, R.I.; Palamarchuk, A.; Weigel, R.J.; Herrero, J.J.; Pekarsky, Y.; Croce, C.M. Physical and functional interactions between the Wwox tumor suppressor protein and the AP-2gamma transcription factor. Cancer Res. 2004, 64, 8256–8261. [Google Scholar] [CrossRef]
- Salah, Z.; Aqeilan, R.; Huebner, K. WWOX gene and gene product: Tumor suppression through specific protein interactions. Future Oncol. 2010, 6, 249–259. [Google Scholar] [CrossRef]
- Blainey, P.; Krzywinski, M.; Altman, N. Points of significance: Replication. Nat. Methods 2014, 11, 879–880. [Google Scholar] [CrossRef]
- Henley, M.J.; Koehler, A.N. Advances in targeting ‘undruggable’ transcription factors with small molecules. Nat. Rev. Drug Discov. 2021, 20, 669–688. [Google Scholar] [CrossRef] [PubMed]
- Shiroma, Y.; Takahashi, R.U.; Yamamoto, Y.; Tahara, H. Targeting DNA binding proteins for cancer therapy. Cancer Sci. 2020, 111, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Islam, Z.; Ali, A.M.; Naik, A.; Eldaw, M.; Decock, J.; Kolatkar, P.R. Transcription Factors: The Fulcrum Between Cell Development and Carcinogenesis. Front. Oncol. 2021, 11, 681377. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.; Jambon, S.; Depauw, S.; David-Cordonnier, M.H. Targeting Transcription Factors for Cancer Treatment. Molecules 2018, 23, 1479. [Google Scholar] [CrossRef]
- Ohmiya, H.; Vitezic, M.; Frith, M.C.; Itoh, M.; Carninci, P.; Forrest, A.R.; Hayashizaki, Y.; Lassmann, T.; Consortium, F. RECLU: A pipeline to discover reproducible transcriptional start sites and their alternative regulation using capped analysis of gene expression (CAGE). BMC Genom. 2014, 15, 269. [Google Scholar] [CrossRef]
- Nones, K.; Patch, A.M. The Impact of Next Generation Sequencing in Cancer Research. Cancers 2020, 12, 2928. [Google Scholar] [CrossRef]
- Cui, W.; Xue, H.; Wei, L.; Jin, J.; Tian, X.; Wang, Q. High heterogeneity undermines generalization of differential expression results in RNA-Seq analysis. Hum. Genom. 2021, 15, 7. [Google Scholar] [CrossRef]
- Schurch, N.J.; Schofield, P.; Gierlinski, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.G.; Owen-Hughes, T.; et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef]
- Woodfield, G.W.; Chen, Y.; Bair, T.B.; Domann, F.E.; Weigel, R.J. Identification of primary gene targets of TFAP2C in hormone responsive breast carcinoma cells. Genes Chromosomes Cancer 2010, 49, 948–962. [Google Scholar] [CrossRef]
- Orso, F.; Jager, R.; Calogero, R.A.; Schorle, H.; Sismondi, P.; De Bortoli, M.; Taverna, D. AP-2alpha regulates migration of GN-11 neurons via a specific genetic programme involving the Axl receptor tyrosine kinase. BMC Biol. 2009, 7, 25. [Google Scholar] [CrossRef]
- Kaluzinska, Z.; Kolat, D.; Bednarek, A.K.; Pluciennik, E. PLEK2, RRM2, GCSH: A Novel WWOX-Dependent Biomarker Triad of Glioblastoma at the Crossroads of Cytoskeleton Reorganization and Metabolism Alterations. Cancers 2021, 13, 2955. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wang, X.M.; Yuan, H.Y.; Liu, Z.H.; Gao, S.; Peng, L. Exploring the mechanism of WWOX growth inhibitory effects on oral squamous cell carcinoma. Oncol. Lett. 2017, 13, 3198–3204. [Google Scholar] [CrossRef]
- Wang, X.; Bolotin, D.; Chu, D.H.; Polak, L.; Williams, T.; Fuchs, E. AP-2alpha: A regulator of EGF receptor signaling and proliferation in skin epidermis. J. Cell Biol. 2006, 172, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Kim, E.; Lee, S.; Kim, D.; Chun, J.; Park, K.H.; Youn, H.; Youn, B. TFAP2C-mediated upregulation of TGFBR1 promotes lung tumorigenesis and epithelial-mesenchymal transition. Exp. Mol. Med. 2016, 48, e273. [Google Scholar] [CrossRef] [PubMed]
- Celebi, A.; Orhan, C.; Seyhan, B.; Buyru, N. Silencing of Wwox Increases Nuclear Import of Dvl proteins in Head and Neck Cancer. J. Cancer 2020, 11, 4030–4036. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Chen, L.; Yang, L.; Ye, Z.; Huang, W.; Li, X.; Liu, Q.; Qiu, J.; Ding, X. KCTD1 mutants in scalpearnipple syndrome and AP2alpha P59A in Char syndrome reciprocally abrogate their interactions, but can regulate Wnt/betacatenin signaling. Mol. Med. Rep. 2020, 22, 3895–3903. [Google Scholar] [CrossRef]
- Do, H.; Kim, D.; Kang, J.; Son, B.; Seo, D.; Youn, H.; Youn, B.; Kim, W. TFAP2C increases cell proliferation by downregulating GADD45B and PMAIP1 in non-small cell lung cancer cells. Biol. Res. 2019, 52, 35. [Google Scholar] [CrossRef]
- Huang, S.S.; Su, W.P.; Lin, H.P.; Kuo, H.L.; Wei, H.L.; Chang, N.S. Role of WW Domain-containing Oxidoreductase WWOX in Driving T Cell Acute Lymphoblastic Leukemia Maturation. J. Biol. Chem. 2016, 291, 17319–17331. [Google Scholar] [CrossRef]
- Kolat, D.; Kaluzinska, Z.; Orzechowska, M.; Bednarek, A.K.; Pluciennik, E. Functional genomics of AP-2alpha and AP-2gamma in cancers: In silico study. BMC Med. Genom. 2020, 13, 174. [Google Scholar] [CrossRef]
- Kuckenberg, P.; Buhl, S.; Woynecki, T.; van Furden, B.; Tolkunova, E.; Seiffe, F.; Moser, M.; Tomilin, A.; Winterhager, E.; Schorle, H. The transcription factor TCFAP2C/AP-2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol. Cell Biol. 2010, 30, 3310–3320. [Google Scholar] [CrossRef]
- Han, J.B.; Huang, M.L.; Li, F.; Yang, R.; Chen, S.M.; Tao, Z.Z. MiR-214 Mediates Cell Proliferation and Apoptosis of Nasopharyngeal Carcinoma Through Targeting Both WWOX and PTEN. Cancer Biother. Radiopharm. 2020, 35, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.C.; Cho, E.; White, J.R.; Paemka, L.; Li, T.; Gu, V.W.; Thompson, D.T.; Koch, K.E.; Franke, C.; Gosse, M.; et al. AP-2alpha Regulates S-Phase and Is a Marker for Sensitivity to PI3K Inhibitor Buparlisib in Colon Cancer. Mol. Cancer Res. 2021, 19, 1156–1167. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Wang, H.; Liu, L.; Song, H.; Zhang, Y.; Wang, J.; Liu, L.; Xu, T.; Fan, R.; Xu, Y.; et al. CircIL4R activates the PI3K/AKT signaling pathway via the miR-761/TRIM29/PHLPP1 axis and promotes proliferation and metastasis in colorectal cancer. Mol. Cancer 2021, 20, 167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Q.; Li, L.; Song, H.L.; Paige, A.; Gabra, H. Effect of WWOX gene on the attachment and adhesion of ovarian cancer cells. Zhonghua Fu Chan Ke Za Zhi 2009, 44, 529–532. [Google Scholar]
- Orso, F.; Fassetta, M.; Penna, E.; Solero, A.; De Filippo, K.; Sismondi, P.; De Bortoli, M.; Taverna, D. The AP-2alpha transcription factor regulates tumor cell migration and apoptosis. Adv. Exp. Med. Biol. 2007, 604, 87–95. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, S.; Jiang, Q.; Deng, J.; Cheng, F.; Lin, Y.; Cheng, L.; Ye, Y.; Chen, X.; Yao, Y.; et al. TFAP2C facilitates somatic cell reprogramming by inhibiting c-Myc-dependent apoptosis and promoting mesenchymal-to-epithelial transition. Cell Death Dis. 2020, 11, 482. [Google Scholar] [CrossRef]
- Taouis, K.; Driouch, K.; Lidereau, R.; Lallemand, F. Molecular Functions of WWOX Potentially Involved in Cancer Development. Cells 2021, 10, 1051. [Google Scholar] [CrossRef]
- Xiong, Y.; Feng, Y.; Zhao, J.; Lei, J.; Qiao, T.; Zhou, Y.; Lu, Q.; Jiang, T.; Jia, L.; Han, Y. TFAP2A potentiates lung adenocarcinoma metastasis by a novel miR-16 family/TFAP2A/PSG9/TGF-beta signaling pathway. Cell Death Dis. 2021, 12, 352. [Google Scholar] [CrossRef]
- Cheng, Y.Y.; Chou, Y.T.; Lai, F.J.; Jan, M.S.; Chang, T.H.; Jou, I.M.; Chen, P.S.; Lo, J.Y.; Huang, S.S.; Chang, N.S.; et al. Wwox deficiency leads to neurodevelopmental and degenerative neuropathies and glycogen synthase kinase 3beta-mediated epileptic seizure activity in mice. Acta Neuropathol. Commun. 2020, 8, 6. [Google Scholar] [CrossRef]
- Orso, F.; Cora, D.; Ubezio, B.; Provero, P.; Caselle, M.; Taverna, D. Identification of functional TFAP2A and SP1 binding sites in new TFAP2A-modulated genes. BMC Genom. 2010, 11, 355. [Google Scholar] [CrossRef]
- Park, D.; Wershof, E.; Boeing, S.; Labernadie, A.; Jenkins, R.P.; George, S.; Trepat, X.; Bates, P.A.; Sahai, E. Extracellular matrix anisotropy is determined by TFAP2C-dependent regulation of cell collisions. Nat. Mater. 2020, 19, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Schrock, M.S.; Huebner, K. WWOX: A fragile tumor suppressor. Exp. Biol. Med. 2015, 240, 296–304. [Google Scholar] [CrossRef]
- McPherson, L.A.; Woodfield, G.W.; Weigel, R.J. AP2 transcription factors regulate expression of CRABPII in hormone responsive breast carcinoma. J. Surg. Res. 2007, 138, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Lal, G.; Contreras, P.G.; Kulak, M.; Woodfield, G.; Bair, T.; Domann, F.E.; Weigel, R.J. Human Melanoma cells over-express extracellular matrix 1 (ECM1) which is regulated by TFAP2C. PLoS ONE 2013, 8, e73953. [Google Scholar] [CrossRef] [PubMed]
- Khawaled, S.; Suh, S.S.; Abdeen, S.K.; Monin, J.; Distefano, R.; Nigita, G.; Croce, C.M.; Aqeilan, R.I. WWOX Inhibits Metastasis of Triple-Negative Breast Cancer Cells via Modulation of miRNAs. Cancer Res. 2019, 79, 1784–1798. [Google Scholar] [CrossRef]
- Kolat, D.; Kaluzinska, Z.; Bednarek, A.K.; Pluciennik, E. The biological characteristics of transcription factors AP-2alpha and AP-2gamma and their importance in various types of cancers. Biosci. Rep. 2019, 39, BSR20181928. [Google Scholar] [CrossRef]
- Bazzazi, H.; Zhang, Y.; Jafarnejad, M.; Isenberg, J.S.; Annex, B.H.; Popel, A.S. Computer Simulation of TSP1 Inhibition of VEGF-Akt-eNOS: An Angiogenesis Triple Threat. Front. Physiol. 2018, 9, 644. [Google Scholar] [CrossRef]
- Brooks, M.D.; Bennett, R.D.; Weaver, A.L.; Sebo, T.J.; Eckert, S.E.; Strehler, E.E.; Carr, A.B. Human Calmodulin-Like Protein CALML3: A Novel Marker for Normal Oral Squamous Mucosa That Is Downregulated in Malignant Transformation. Int. J. Dent. 2013, 2013, 592843. [Google Scholar] [CrossRef]
- Yang, B.; Li, M.; Tang, W.; Liu, W.; Zhang, S.; Chen, L.; Xia, J. Dynamic network biomarker indicates pulmonary metastasis at the tipping point of hepatocellular carcinoma. Nat. Commun. 2018, 9, 678. [Google Scholar] [CrossRef]
- Bennett, R.D.; Pittelkow, M.R.; Strehler, E.E. Immunolocalization of the tumor-sensitive calmodulin-like protein CALML3 in normal human skin and hyperproliferative skin disorders. PLoS ONE 2013, 8, e62347. [Google Scholar] [CrossRef]
- Wu, Q.; Finley, S.D. Predictive model identifies strategies to enhance TSP1-mediated apoptosis signaling. Cell Commun. Signal. 2017, 15, 53. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Xing, S.G.; Jia, W.D.; Huang, M.; Bian, N.N. Low PLCE1 levels are correlated with poor prognosis in hepatocellular carcinoma. OncoTargets Ther. 2017, 10, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Usami, Y.; Wu, Y.; Gottlinger, H.G. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 2015, 526, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Bossolasco, M.; Veillette, F.; Bertrand, R.; Mes-Masson, A.M. Human TDE1, a TDE1/TMS family member, inhibits apoptosis in vitro and stimulates in vivo tumorigenesis. Oncogene 2006, 25, 4549–4558. [Google Scholar] [CrossRef][Green Version]
- Krueger, W.H.; Gonye, G.E.; Madison, D.L.; Murray, K.E.; Kumar, M.; Spoerel, N.; Pfeiffer, S.E. TPO1, a member of a novel protein family, is developmentally regulated in cultured oligodendrocytes. J. Neurochem. 1997, 69, 1343–1355. [Google Scholar] [CrossRef]
- Ho, J.; Du, Y.; Wong, O.G.; Siu, M.K.; Chan, K.K.; Cheung, A.N. Downregulation of the gli transcription factors regulator Kif7 facilitates cell survival and migration of choriocarcinoma cells. PLoS ONE 2014, 9, e108248. [Google Scholar] [CrossRef]
- Fang, W.T.; Fan, C.C.; Li, S.M.; Jang, T.H.; Lin, H.P.; Shih, N.Y.; Chen, C.H.; Wang, T.Y.; Huang, S.F.; Lee, A.Y.; et al. Downregulation of a putative tumor suppressor BMP4 by SOX2 promotes growth of lung squamous cell carcinoma. Int. J. Cancer 2014, 135, 809–819. [Google Scholar] [CrossRef]
- Choi, D.; Han, J.Y.; Shin, J.H.; Ryu, C.M.; Yu, H.Y.; Kim, A.; Lee, S.; Lim, J.; Shin, D.M.; Choo, M.S. Downregulation of WNT11 is associated with bladder tissue fibrosis in patients with interstitial cystitis/bladder pain syndrome without Hunner lesion. Sci. Rep. 2018, 8, 9782. [Google Scholar] [CrossRef]
- Chandler, C.; Liu, T.; Buckanovich, R.; Coffman, L.G. The double edge sword of fibrosis in cancer. Transl. Res. 2019, 209, 55–67. [Google Scholar] [CrossRef]
- Hui, J.; Wang, C.; Miao, Y.; Liu, R.; Xu, J. The pancancer landscape of Wnt family expression reveals potential biomarkers in urinary system tumors. Cancer Gene Ther. 2021, 28, 1035–1045. [Google Scholar] [CrossRef]
- Duan, X.; Iwanowycz, S.; Ngoi, S.; Hill, M.; Zhao, Q.; Liu, B. Molecular Chaperone GRP94/GP96 in Cancers: Oncogenesis and Therapeutic Target. Front. Oncol. 2021, 11, 629846. [Google Scholar] [CrossRef] [PubMed]
- Haase, M.; Fitze, G. HSP90AB1: Helping the good and the bad. Gene 2016, 575, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Chuang, J.J.; Dai, Y.C.; Lin, Y.L.; Chen, Y.Y.; Lin, W.H.; Chan, H.L.; Liu, Y.W. Downregulation of glutathione S-transferase M1 protein in N-butyl-N-(4-hydroxybutyl)nitrosamine-induced mouse bladder carcinogenesis. Toxicol. Appl. Pharmacol. 2014, 279, 322–330. [Google Scholar] [CrossRef]
- Schnekenburger, M.; Karius, T.; Diederich, M. Regulation of epigenetic traits of the glutathione S-transferase P1 gene: From detoxification toward cancer prevention and diagnosis. Front. Pharmacol. 2014, 5, 170. [Google Scholar] [CrossRef]
- Shan, J.; Shen, J.; Wu, M.; Zhou, H.; Feng, J.; Yao, C.; Yang, Z.; Ma, Q.; Luo, Y.; Wang, Y.; et al. Tcf7l1 Acts as a Suppressor for the Self-Renewal of Liver Cancer Stem Cells and Is Regulated by IGF/MEK/ERK Signaling Independent of beta-Catenin. Stem Cells 2019, 37, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Cidre-Aranaz, F.; Li, J.; Holting, T.L.B.; Orth, M.F.; Imle, R.; Kutschmann, S.; Ammirati, G.; Ceranski, K.; Carreno-Gonzalez, M.J.; Kasan, M.; et al. Integrative gene network and functional analyses identify a prognostically relevant key regulator of metastasis in Ewing sarcoma. Mol. Cancer 2022, 21, 1. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Fan, L.; Liao, X.; Cui, G.; Hu, H. CRTAC1 (Cartilage acidic protein 1) inhibits cell proliferation, migration, invasion and epithelial-mesenchymal transition (EMT) process in bladder cancer by downregulating Yin Yang 1 (YY1) to inactivate the TGF-beta pathway. Bioengineered 2021, 12, 9377–9389. [Google Scholar] [CrossRef]
- Li, Y.; Qiao, L.; Zang, Y.; Ni, W.; Xu, Z. Circular RNA FOXO3 Suppresses Bladder Cancer Progression and Metastasis by Regulating MiR-9-5p/TGFBR2. Cancer Manag. Res. 2020, 12, 5049–5056. [Google Scholar] [CrossRef]
- Wan, X.; Shi, L.; Ma, X.; Tang, H.; Wu, G. The Elevated ASK1 Expression Inhibits Proliferation and Invasion in Gastric Cancer HGC-27 Cells. Anat. Rec. 2018, 301, 1815–1819. [Google Scholar] [CrossRef]
- Han, T.; Gao, J.; Wang, L.; Qu, Y.; Sun, A.; Peng, K.; Zhu, J.; Liu, H.; Yang, W.; Shao, G.; et al. ASK1 inhibits proliferation and migration of lung cancer cells via inactivating TAZ. Am. J. Cancer Res. 2020, 10, 2785–2799. [Google Scholar]
- Hao, Y.; Li, G. Role of EFNA1 in tumorigenesis and prospects for cancer therapy. Biomed. Pharm. 2020, 130, 110567. [Google Scholar] [CrossRef] [PubMed]
- Depner, C.; Zum Buttel, H.; Bogurcu, N.; Cuesta, A.M.; Aburto, M.R.; Seidel, S.; Finkelmeier, F.; Foss, F.; Hofmann, J.; Kaulich, K.; et al. EphrinB2 repression through ZEB2 mediates tumour invasion and anti-angiogenic resistance. Nat. Commun. 2016, 7, 12329. [Google Scholar] [CrossRef] [PubMed]
- Kaidi, A.; Moorghen, M.; Williams, A.C.; Paraskeva, C. Is the downregulation of EphB2 receptor expression during colorectal tumorigenesis due to hypoxia? Gut 2007, 56, 1637–1638. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fox, B.P.; Kandpal, R.P. EphB6 receptor significantly alters invasiveness and other phenotypic characteristics of human breast carcinoma cells. Oncogene 2009, 28, 1706–1713. [Google Scholar] [CrossRef]
- Young, L.C.; Rodriguez-Viciana, P. MRAS: A Close but Understudied Member of the RAS Family. Cold Spring Harb. Perspect. Med. 2018, 8, a033621. [Google Scholar] [CrossRef]
- Ducker, C.; Chow, L.K.Y.; Saxton, J.; Handwerger, J.; McGregor, A.; Strahl, T.; Layfield, R.; Shaw, P.E. De-ubiquitination of ELK-1 by USP17 potentiates mitogenic gene expression and cell proliferation. Nucleic Acids Res. 2019, 47, 4495–4508. [Google Scholar] [CrossRef]
- Dos Reis, P.P.; Bharadwaj, R.R.; Machado, J.; Macmillan, C.; Pintilie, M.; Sukhai, M.A.; Perez-Ordonez, B.; Gullane, P.; Irish, J.; Kamel-Reid, S. Claudin 1 overexpression increases invasion and is associated with aggressive histological features in oral squamous cell carcinoma. Cancer 2008, 113, 3169–3180. [Google Scholar] [CrossRef]
- Suh, Y.; Yoon, C.H.; Kim, R.K.; Lim, E.J.; Oh, Y.S.; Hwang, S.G.; An, S.; Yoon, G.; Gye, M.C.; Yi, J.M.; et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene 2013, 32, 4873–4882. [Google Scholar] [CrossRef]
- Katoh, M.; Katoh, M. CLDN23 gene, frequently down-regulated in intestinal-type gastric cancer, is a novel member of CLAUDIN gene family. Int J. Mol. Med. 2003, 11, 683–689. [Google Scholar] [CrossRef]
- Shu, C.; Han, S.; Hu, C.; Chen, C.; Qu, B.; He, J.; Dong, S.; Xu, P. Integrin beta1 regulates proliferation, apoptosis, and migration of trophoblasts through activation of phosphoinositide 3 kinase/protein kinase B signaling. J. Obstet. Gynaecol. Res. 2021, 47, 2406–2416. [Google Scholar] [CrossRef]
- Ma, Q.; Song, J.; Ma, H.; Gao, K.; Yang, Y.; He, N. Synergistic anticancer effect of Grb2 and ITGA1 on cancer cells highly expressing Grb2 through suppressing ERK phosphorylation. Int. J. Clin. Exp. Pathol. 2019, 12, 182–189. [Google Scholar] [PubMed]
- Wang, Q.; Cao, T.; Guo, K.; Zhou, Y.; Liu, H.; Pan, Y.; Hou, Q.; Nie, Y.; Fan, D.; Lu, Y.; et al. Regulation of Integrin Subunit Alpha 2 by miR-135b-5p Modulates Chemoresistance in Gastric Cancer. Front. Oncol. 2020, 10, 308. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Kong, Y.; Zhang, L.; He, T.; Zhou, X.; Yan, Y.; Zhang, L.; Zhou, D.; Lu, S.; Zhou, J.; et al. High Expression of ITGA3 Promotes Proliferation and Cell Cycle Progression and Indicates Poor Prognosis in Intrahepatic Cholangiocarcinoma. Biomed. Res. Int. 2018, 2018, 2352139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.H.; Yang, Z.L.; Zhou, E.X.; Miao, X.Y.; Zou, Q.; Li, J.H.; Liang, L.F.; Zeng, G.X.; Chen, S.L. Overexpression of Thy1 and ITGA6 is associated with invasion, metastasis and poor prognosis in human gallbladder carcinoma. Oncol. Lett. 2016, 12, 5136–5144. [Google Scholar] [CrossRef][Green Version]
- Qu, J.; Zheng, B.; Ohuchida, K.; Feng, H.; Chong, S.J.F.; Zhang, X.; Liang, R.; Liu, Z.; Shirahane, K.; Mizumoto, K.; et al. PIK3CB is involved in metastasis through the regulation of cell adhesion to collagen I in pancreatic cancer. J. Adv. Res. 2021, 33, 127–140. [Google Scholar] [CrossRef]
- Wang, G.; Yang, X.; Li, C.; Cao, X.; Luo, X.; Hu, J. PIK3R3 induces epithelial-to-mesenchymal transition and promotes metastasis in colorectal cancer. Mol. Cancer Ther. 2014, 13, 1837–1847. [Google Scholar] [CrossRef]
- Lin, H.P.; Lin, C.Y.; Huo, C.; Jan, Y.J.; Tseng, J.C.; Jiang, S.S.; Kuo, Y.Y.; Chen, S.C.; Wang, C.T.; Chan, T.M.; et al. AKT3 promotes prostate cancer proliferation cells through regulation of Akt, B-Raf, and TSC1/TSC2. Oncotarget 2015, 6, 27097–27112. [Google Scholar] [CrossRef]
- Zboray, K.; Mohrherr, J.; Stiedl, P.; Pranz, K.; Wandruszka, L.; Grabner, B.; Eferl, R.; Moriggl, R.; Stoiber, D.; Sakamoto, K.; et al. AKT3 drives adenoid cystic carcinoma development in salivary glands. Cancer Med. 2018, 7, 445–453. [Google Scholar] [CrossRef]
- Xiong, Y.; Ju, L.; Yuan, L.; Chen, L.; Wang, G.; Xu, H.; Peng, T.; Luo, Y.; Xiao, Y.; Wang, X. KNSTRN promotes tumorigenesis and gemcitabine resistance by activating AKT in bladder cancer. Oncogene 2021, 40, 1595–1608. [Google Scholar] [CrossRef]
- Pu, Q.; Lu, L.; Dong, K.; Geng, W.W.; Lv, Y.R.; Gao, H.D. The Novel Transcription Factor CREB3L4 Contributes to the Progression of Human Breast Carcinoma. J. Mammary Gland. Biol. Neoplasia 2020, 25, 37–50. [Google Scholar] [CrossRef]
- Lu, E.; Hu, X.; Pan, C.; Chen, J.; Xu, Y.; Zhu, X. Up-regulation of peroxiredoxin-1 promotes cell proliferation and metastasis and inhibits apoptosis in cervical cancer. J. Cancer 2020, 11, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Luo, P.; Wang, L.; Yang, S.; Qin, S.; Wei, Z.; Liu, J. CTNNB1 Knockdown Inhibits Cell Proliferation and Aldosterone Secretion Through Inhibiting Wnt/beta-Catenin Signaling in H295R Cells. Technol. Cancer Res. Treat. 2020, 19, 1533033820979685. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Liu, D.; Qiu, Z.; Huang, Y.; Chen, B.; Wang, L.; Xu, H.; Huang, N.; Liu, L.; Li, W. GSK3beta overexpression indicates poor prognosis and its inhibition reduces cell proliferation and survival of non-small cell lung cancer cells. PLoS ONE 2014, 9, e91231. [Google Scholar] [CrossRef]
- Maier, H.J.; Wirth, T.; Beug, H. Epithelial-mesenchymal transition in pancreatic carcinoma. Cancers 2010, 2, 2058–2083. [Google Scholar] [CrossRef]
- Roca, H.; Hernandez, J.; Weidner, S.; McEachin, R.C.; Fuller, D.; Sud, S.; Schumann, T.; Wilkinson, J.E.; Zaslavsky, A.; Li, H.; et al. Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS ONE 2013, 8, e76773. [Google Scholar] [CrossRef] [PubMed]
- Abdeen, S.K.; Aqeilan, R.I. Decoding the link between WWOX and p53 in aggressive breast cancer. Cell Cycle 2019, 18, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Bogachek, M.V.; De Andrade, J.P.; Weigel, R.J. Regulation of epithelial-mesenchymal transition through SUMOylation of transcription factors. Cancer Res. 2015, 75, 11–15. [Google Scholar] [CrossRef]
- Kantarci, H.; Edlund, R.K.; Groves, A.K.; Riley, B.B. Tfap2a promotes specification and maturation of neurons in the inner ear through modulation of Bmp, Fgf and notch signaling. PLoS Genet. 2015, 11, e1005037. [Google Scholar] [CrossRef]
- Latos, P.A.; Goncalves, A.; Oxley, D.; Mohammed, H.; Turro, E.; Hemberger, M. Fgf and Esrrb integrate epigenetic and transcriptional networks that regulate self-renewal of trophoblast stem cells. Nat. Commun. 2015, 6, 7776. [Google Scholar] [CrossRef]
- Li, L.; Lian, B.; Li, C.; Li, W.; Li, J.; Zhang, Y.; He, X.; Li, Y.; Xie, L. Integrative analysis of transcriptional regulatory network and copy number variation in intrahepatic cholangiocarcinoma. PLoS ONE 2014, 9, e98653. [Google Scholar] [CrossRef]
- Wang, H.Y.; Juo, L.I.; Lin, Y.T.; Hsiao, M.; Lin, J.T.; Tsai, C.H.; Tzeng, Y.H.; Chuang, Y.C.; Chang, N.S.; Yang, C.N.; et al. WW domain-containing oxidoreductase promotes neuronal differentiation via negative regulation of glycogen synthase kinase 3beta. Cell Death Differ. 2012, 19, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Ho, P.C.; Lee, I.T.; Chen, Y.A.; Chu, C.H.; Teng, C.C.; Wu, S.N.; Sze, C.I.; Chiang, M.F.; Chang, N.S. WWOX Phosphorylation, Signaling, and Role in Neurodegeneration. Front. Neurosci. 2018, 12, 563. [Google Scholar] [CrossRef] [PubMed]
- Antonicelli, F.; Lorin, J.; Kurdykowski, S.; Gangloff, S.C.; Le Naour, R.; Sallenave, J.M.; Hornebeck, W.; Grange, F.; Bernard, P. CXCL10 reduces melanoma proliferation and invasiveness in vitro and in vivo. Br. J. Dermatol. 2011, 164, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Backx, E.; Wauters, E.; Baldan, J.; Van Bulck, M.; Michiels, E.; Heremans, Y.; De Paep, D.L.; Kurokawa, M.; Goyama, S.; Bouwens, L.; et al. MECOM permits pancreatic acinar cell dedifferentiation avoiding cell death under stress conditions. Cell Death Differ. 2021, 28, 2601–2615. [Google Scholar] [CrossRef]
- Korur, S.; Huber, R.M.; Sivasankaran, B.; Petrich, M.; Morin, P., Jr.; Hemmings, B.A.; Merlo, A.; Lino, M.M. GSK3beta regulates differentiation and growth arrest in glioblastoma. PLoS ONE 2009, 4, e7443. [Google Scholar] [CrossRef]
- Okabe, H.; Satoh, S.; Furukawa, Y.; Kato, T.; Hasegawa, S.; Nakajima, Y.; Yamaoka, Y.; Nakamura, Y. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 2003, 63, 3043–3048. [Google Scholar]
- Xu, D.; McKee, C.M.; Cao, Y.; Ding, Y.; Kessler, B.M.; Muschel, R.J. Matrix metalloproteinase-9 regulates tumor cell invasion through cleavage of protease nexin-1. Cancer Res. 2010, 70, 6988–6998. [Google Scholar] [CrossRef]
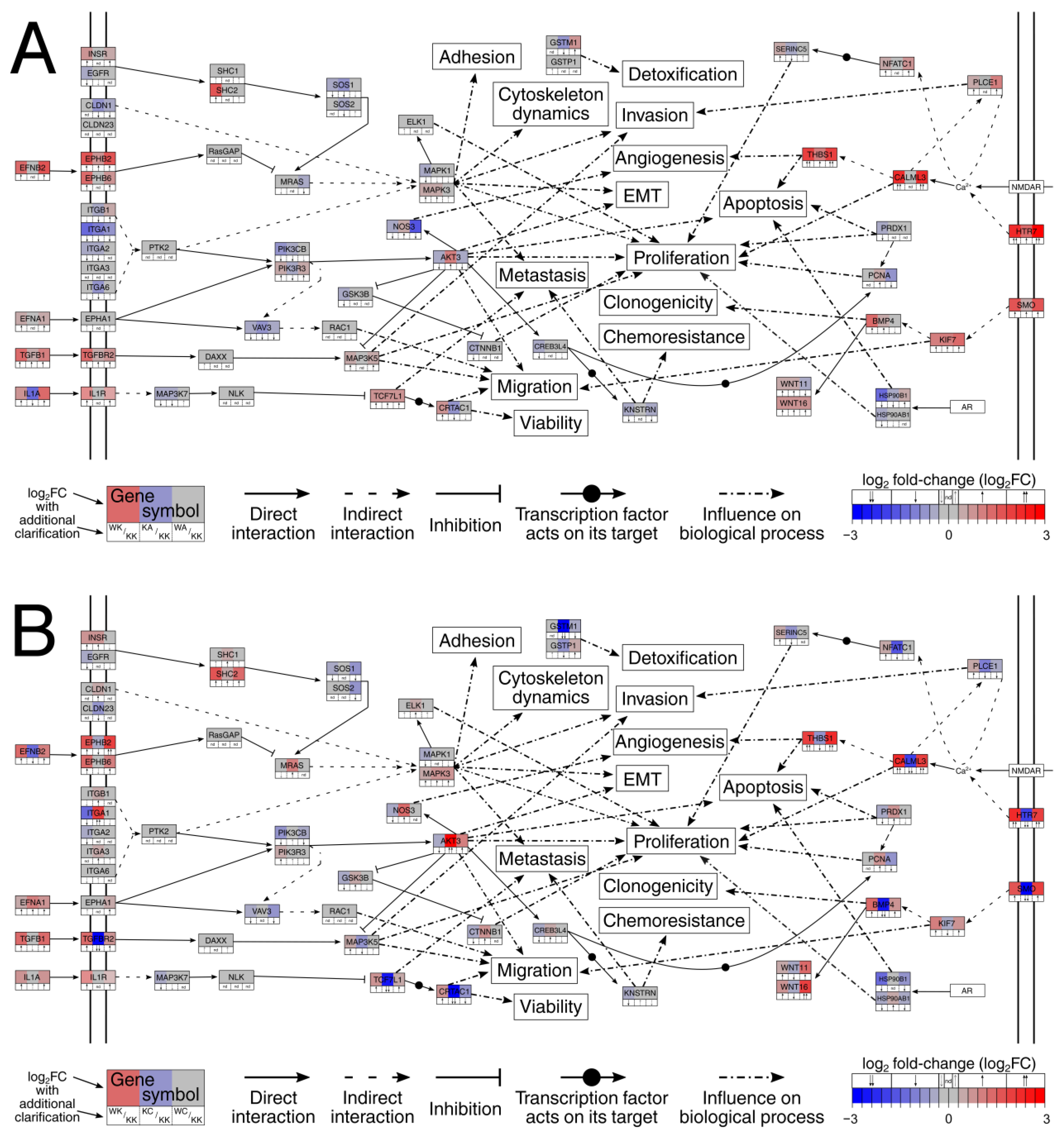
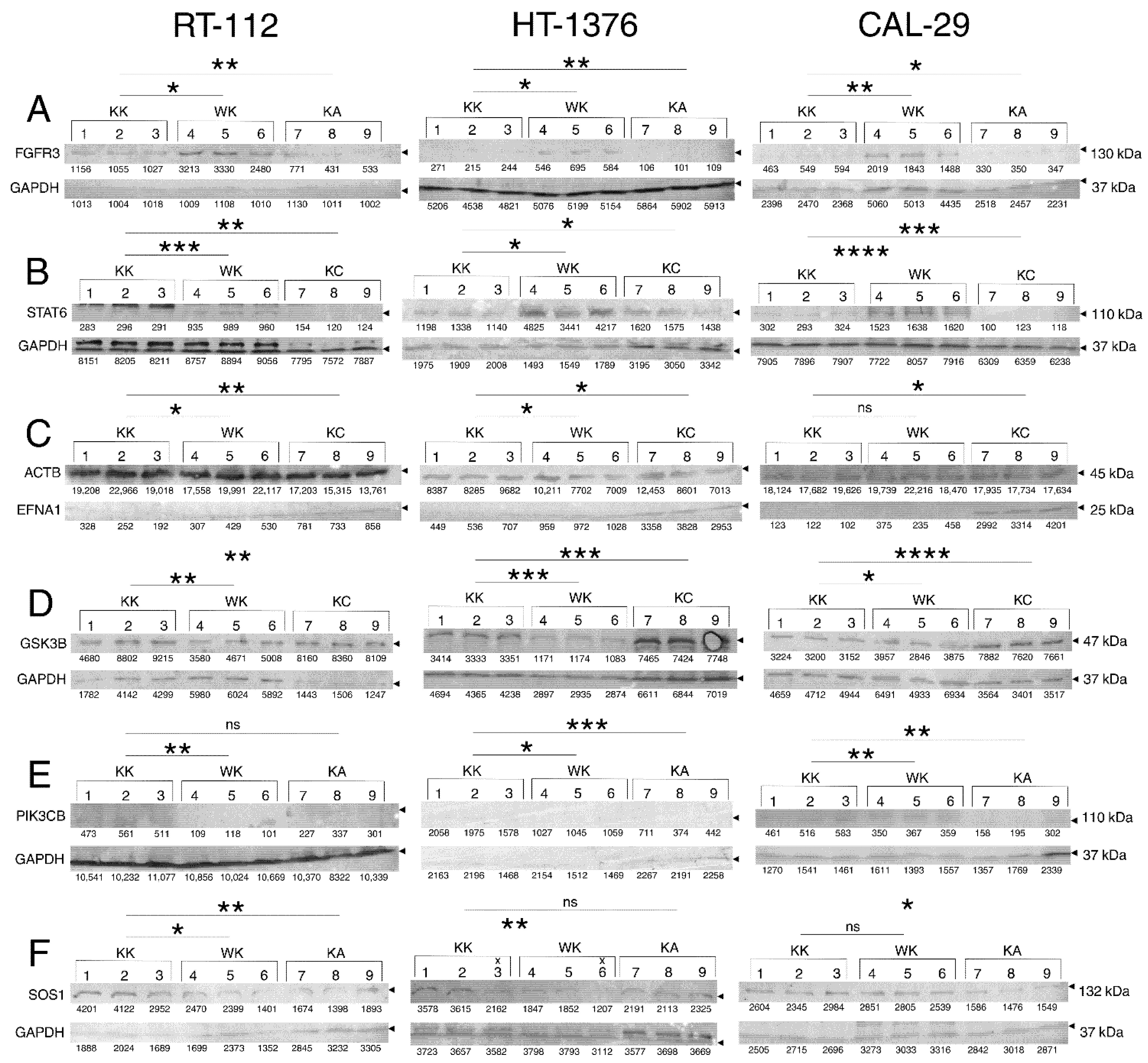
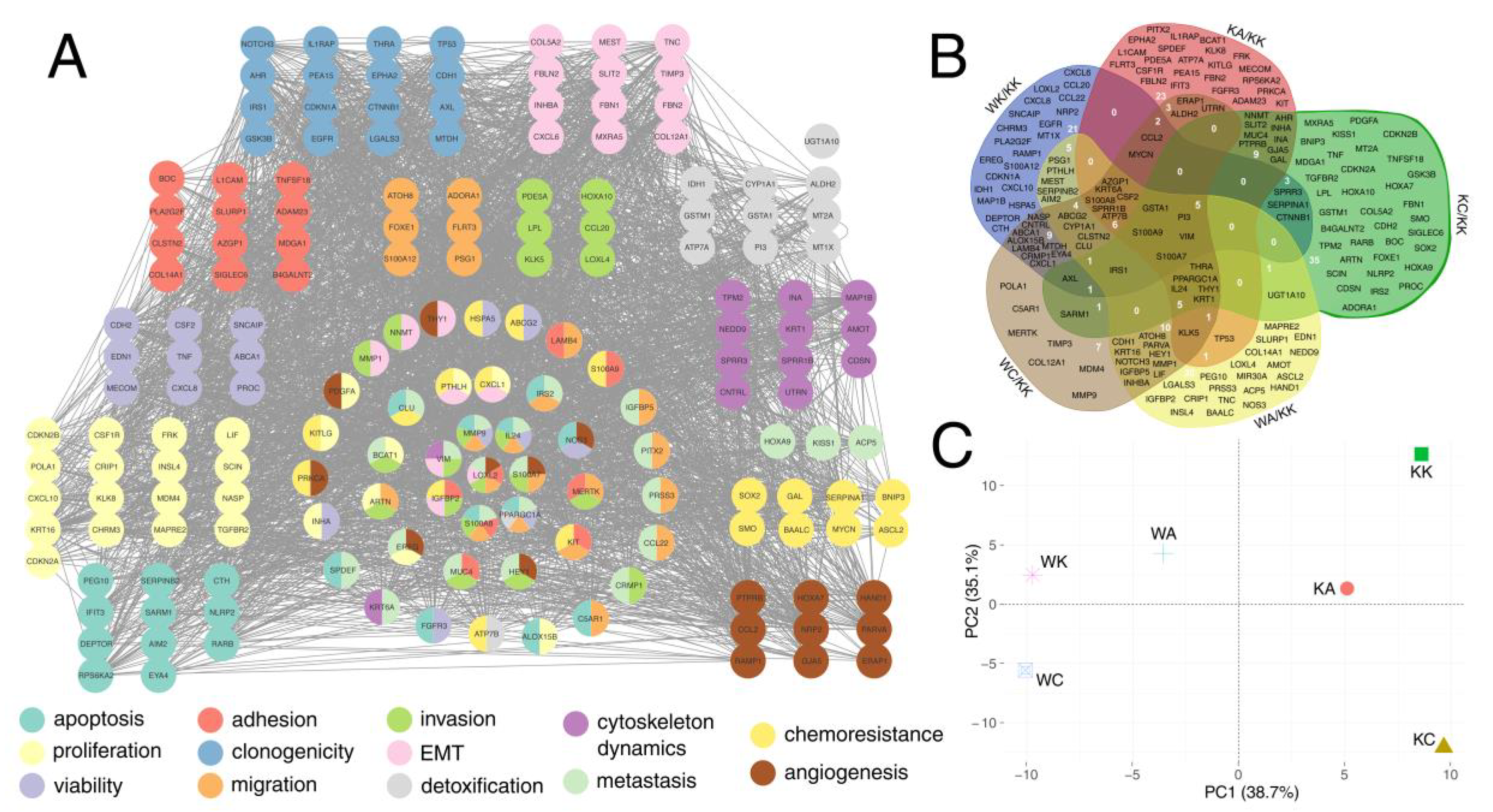
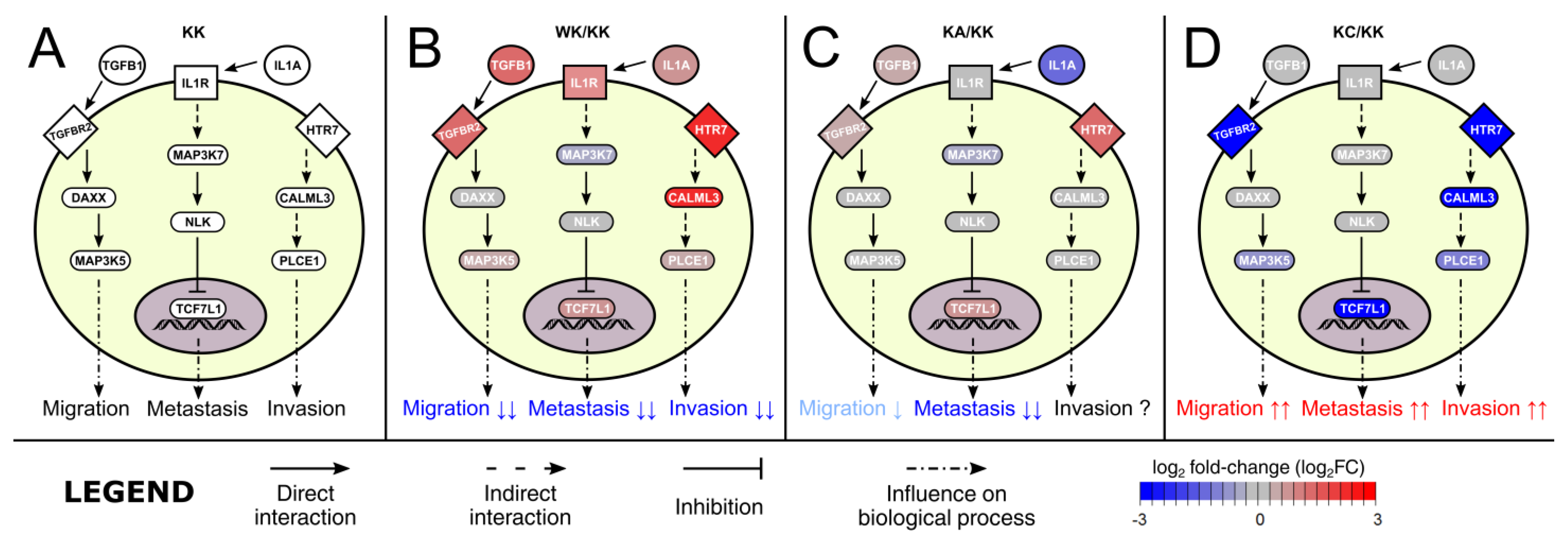
| Biological Process | Gene Set Accession Number or Direct Link |
|---|---|
| MSigDB | |
| Apoptosis | GO:0006915 |
| Proliferation | GO:0008283 |
| Adhesion | GO:0022610 |
| Migration | GO:0016477 |
| Epithelial-to-Mesenchymal Transition | M5930 |
| Cytoskeleton dynamics | GO:0005856 |
| GeneRIF | |
| Viability | https://maayanlab.cloud/Harmonizome/gene_set/ viability/GeneRIF + Biological + Term + Annotations (accessed on 26 January 2022). |
| Clonogenicity | https://maayanlab.cloud/Harmonizome/gene_set/ clonogenicity/GeneRIF + Biological + Term + Annotations (accessed on 26 January 2022). |
| Invasion | https://maayanlab.cloud/Harmonizome/gene_set/ invasion/GeneRIF + Biological + Term + Annotations (accessed on 26 January 2022). |
| Detoxification | https://maayanlab.cloud/Harmonizome/gene_set/ detoxification/GeneRIF + Biological + Term + Annotations (accessed on 26 January 2022). |
| Metastasis | https://maayanlab.cloud/Harmonizome/gene_set/ metastasis/GeneRIF + Biological + Term + Annotations (accessed on 26 January 2022). |
| Chemoresistance | https://maayanlab.cloud/Harmonizome/gene_set/ chemoresistance/GeneRIF + Biological + Term + Annotations (accessed on 26 January 2022). |
| Angiogenesis | https://maayanlab.cloud/Harmonizome/gene_set/ angiogenesis/GO + Biological + Process + Annotations (accessed on 26 January 2022). |
| Comp. | Pathview Web | DAVID | dbEMT |
|---|---|---|---|
| Alpha | MAPK signaling pathway (hsa04010); Cell adhesion molecules (hsa04514); Leukocyte transendothelial migration (hsa04670) | Wnt signaling pathway (hsa04310); Rap1 signaling pathway (hsa04015); Signaling pathways regulating pluripotency of stem cells (hsa04550) Cell adhesion molecules (hsa04514); Leukocyte transendothelial migration (hsa04670); Axon guidance (hsa04360); | Pathways in cancer (hsa05200); MicroRNAs in cancer (hsa05206) |
| Gamma | PI3K-Akt signaling pathway (hsa04151); MAPK signaling pathway (hsa04010); Focal adhesion (hsa04510) | PI3K-Akt signaling pathway (hsa04151); Rap1 signaling pathway (hsa04015); Ras signaling pathway (hsa04014); Focal adhesion (hsa04510); ECM-receptor interaction (hsa04512); Protein digestion and absorption (hsa04974); Amoebiasis (hsa05146); Axon guidance (hsa04360); Pathways in cancer (hsa05200) | MicroRNAs in cancer (hsa05206) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kołat, D.; Kałuzińska, Ż.; Bednarek, A.K.; Płuciennik, E. Determination of WWOX Function in Modulating Cellular Pathways Activated by AP-2α and AP-2γ Transcription Factors in Bladder Cancer. Cells 2022, 11, 1382. https://doi.org/10.3390/cells11091382
Kołat D, Kałuzińska Ż, Bednarek AK, Płuciennik E. Determination of WWOX Function in Modulating Cellular Pathways Activated by AP-2α and AP-2γ Transcription Factors in Bladder Cancer. Cells. 2022; 11(9):1382. https://doi.org/10.3390/cells11091382
Chicago/Turabian StyleKołat, Damian, Żaneta Kałuzińska, Andrzej K. Bednarek, and Elżbieta Płuciennik. 2022. "Determination of WWOX Function in Modulating Cellular Pathways Activated by AP-2α and AP-2γ Transcription Factors in Bladder Cancer" Cells 11, no. 9: 1382. https://doi.org/10.3390/cells11091382
APA StyleKołat, D., Kałuzińska, Ż., Bednarek, A. K., & Płuciennik, E. (2022). Determination of WWOX Function in Modulating Cellular Pathways Activated by AP-2α and AP-2γ Transcription Factors in Bladder Cancer. Cells, 11(9), 1382. https://doi.org/10.3390/cells11091382









