The Regulatory Functions and the Mechanisms of Long Non-Coding RNAs in Cervical Cancer
Abstract
:1. Introduction
2. Long Non-Coding RNAs and Potential Therapeutic Targeting in Cancer
3. LncRNAs-Regulated Pathways in Cervical Cancer
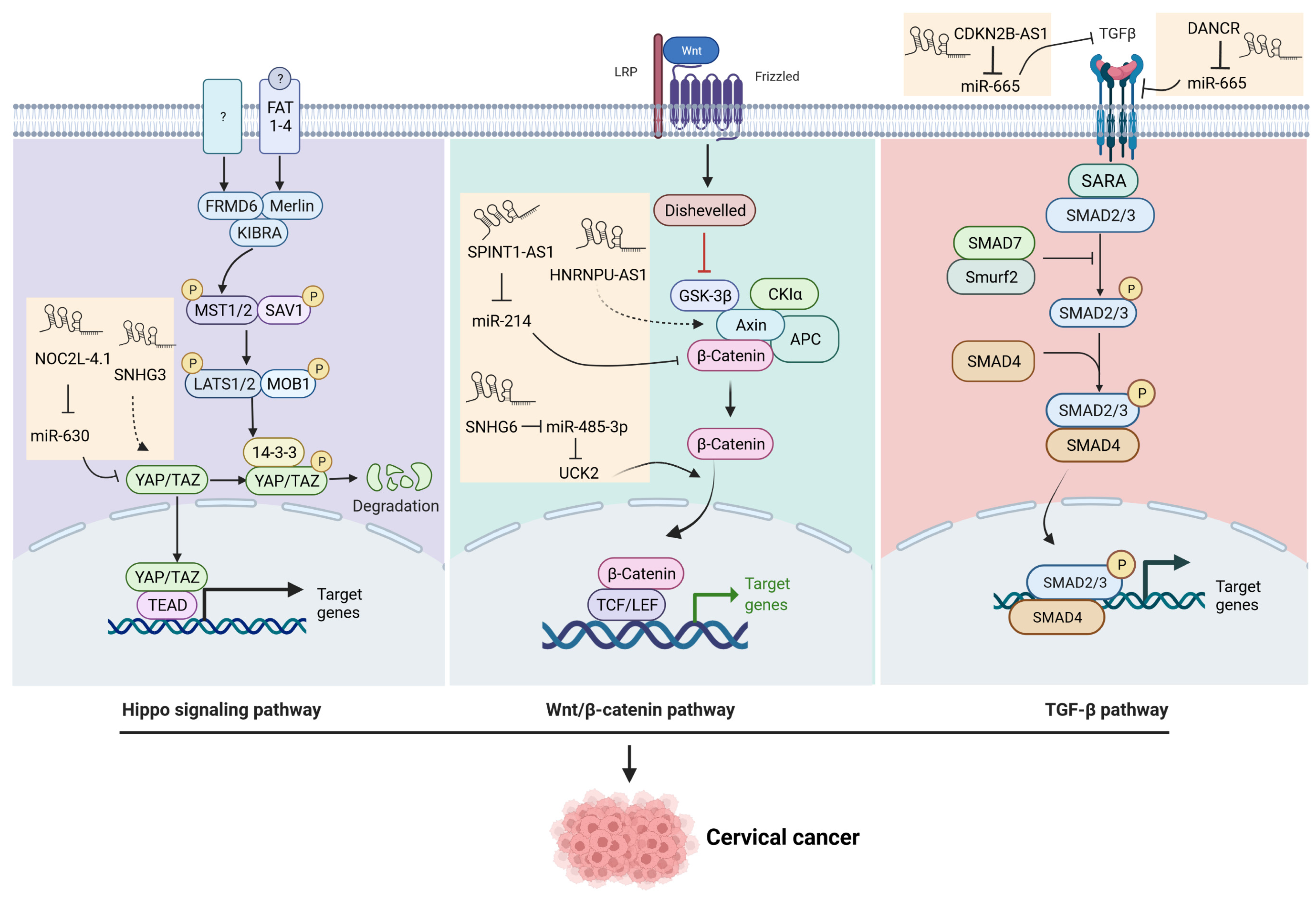
3.1. The Wnt Signaling Pathway
3.2. The Mitogen-Activated Protein Kinase (MAPK) Pathway
3.3. The TGF-β Signaling Pathway
3.4. The Hippo Signaling Pathway
3.5. DNA Damage Repair (DDR) and Genomic Integrity
3.6. The Phosphatidylinositol 3- kinase/Protein Kinase B (PI3K/AKT) Pathway
3.7. The Hypoxia Signaling Pathway
3.8. The p53 Pathway
3.9. Other lncRNA-Related Targets in Cervical Cancer
| LncRNA | Biological Samples | Interaction with RNA | Targets/Pathway | Biological Processes | Publication Time | Ref. |
|---|---|---|---|---|---|---|
| AC010198.2 | Cells | miR-34b-3p | STC2 | Drug resistance | Oct. 2021 | [103] |
| AFAP1-AS1 | Cells | miR-27b-3p | VEGF-C | stemness | July 2021 | [104] |
| AL592284.1 | Cells and tissues | miR-30a-5p | Vimentin/EMT | proliferation, metastasis | Nov. 2021 | [105] |
| ALOX12-AS1 | Tissues | miR-3171 | NA | proliferation | Jan. 2022 | [106] |
| ANXA2P2 | Cells and tissues | miR-361-3p | SOX9 | cisplatin-resistant | Jan. 2022 | [107] |
| CASC9-1 | Cells | miR-383-5p | MAPKAP1 | proliferation, migration, invasion, apoptosis | Nov. 2021 | [108] |
| CCAT2 | Cells and tissues | miR-493-5p | CREB1 | proliferation, EMT, tumor growth | Dec. 2021 | [109] |
| DANCR | Cells and tissues | miR-145-3p | ZEB1 | tumor growth, metastasis | July 2021 | [110] |
| DARS-AS1 | Cells and tissues | NA | ATP1B2, cGMP-PKG | proliferation, invasion, migration | June 2021 | [111] |
| DLEU2 | Cells | NA | ZFP36, p53, notch signaling, p53 | proliferation, cell cycle | May 2021 | [112] |
| DUXAP8 | Cells | miR-1297 | RCN2 | malignancy, tumor growth | Nov. 2021 | [113] |
| EGFR-AS1 | Cells | miR-2355-5p | ACTN4-mediated Wnt | proliferation, migration, invasion, apoptosis | Jan. 2022 | [38] |
| FBX19-AS1 | Cells | miR-193a-5p | COL1A1 | proliferation, migration, invasion, EMT, apoptosis, metastasis | Aug. 2021 | [114] |
| FEZF1-AS1 | Cells and tissues | miR-1254 | NA | proliferation, migration, invasion | July 2021 | [115] |
| FEZF1-AS1 | Cells | miR-367-3p | SLS12AS | proliferation, migration, invasion, apoptosis | Jan. 2022 | [115] |
| FGD5-AS1 | Cells | miR-129-5p | BST2 | macrophage M1 polarization | Oct. 2021 | [116] |
| FOXD2-AS1 | Cells and tissues | NA | METTL3, LSD1/p21 | proliferation, migration, tumor growth | July 2021 | [102] |
| FOXD3-AS1 | Cells and tissues | miR-128-3p | LIMK1 | proliferation, migration, invasion | May 2021 | [117] |
| HAND2-AS1 | Tissues | miR-21-5p | TIMP3/VEGFA | proliferation, migration, invasion, apoptosis, tumor growth | June 2021 | [118] |
| HNRNPU-AS1 | Cells and tissues | miR-205-5p | AXIN2, Wnt | proliferation, apoptosis, tumor growth | Sep. 2021 | [40] |
| HOTAIR | Cells and tissues | NA | Wnt | drug resistance | Oct. 2021 | [119] |
| HOTAIR | Cells | miR-29b | PTEN/PI3K | proliferation, drug resistance, EMT | Sep. 2021 | [120] |
| HOTAIR | Stem cells | miR-203 | ZEB1 | EMT | Sep. 2021 | [121] |
| HOTAIR | Cells | miR-214-3p | NA | proliferation, apoptosis | July 2021 | [122] |
| HOTAIR HOXA-AS2 | Cells and tissues | miR-509-3p | BTN3A1 | proliferation, migration, invasion, tumor growth | Sep. 2021 | [123] |
| HOXC13-AS | Cells, and tissues | NA | FTO, Wnt, FZD | proliferation, invasion, EMT | July 2021 | [124] |
| HOXC-AS3 | Cells and tissues | miR-105-5p | SOS1 | proliferation, migration, invasion, apoptosis | Oct. 2021 | [125] |
| HOXD-AS1 | Cells | NA | FRRS1 | proliferation, apoptosis, tumor growth | Jan. 2022 | [126] |
| ILF3-AS1 | Cells and tissues | miR-454-3p | PTEN | survival rate, migration, apoptosis, EMT | Aug. 2021 | [127] |
| KCNQ1OT1 | Cells | miR-1270 | LOXL2, PI3K/Akt | viability, apoptosis | Feb. 2022 | [67] |
| KCNQ1OT1 | Cells and tissues | miR-296-5p | HYOU1 | proliferation, migration, invasion, tumor growth | Dec. 2021 | [128] |
| LIN01006 | Cells and tissues | miR-28-5p | PAK2 | proliferation, migration, invasion, tumor growth | April 2021 | [129] |
| LINC00313 | Cells and tissues | miR-4677-3p | CDK6 | migration, EMT | Mar. 2021 | [130] |
| LINC00514 | Cells and tissues | miR-708-5p | HOXB3 | Proliferation, invasion | Jan. 2022 | [131] |
| LINC00662 | Cells | miR-103a | PDK4 | proliferation, apoptosis | June 2021 | [132] |
| LINC00665 | Cells | NA | CTNNB1, Wnt signaling | proliferation, migration, invasion, EMT | April 2021 | [41] |
| LINC00673 | Cells and serum | NA | cell cycle, p53 pathway | proliferation, cell cycle, tumor growth | May 2021 | [133] |
| LINC00707 | Cells and tissues | miR-382-5p | VEGFA | proliferation, tumor growth | June 2021 | [134] |
| LINC00885 | Cells and tissues | miR-3150b-3p | BAZ2A | Proliferation, apoptosis, tumor growth | Jan. 2022 | [101] |
| LINC00885 | Cells and tissues | NA | NA | proliferation, invasion, EMT | June 2021 | [135] |
| LINC00899 | Cells and tissues | miR-944 | ESR1 | proliferation, migration, invasion | June 2021 | [136] |
| LINC00997 | Cells | miR-574-3p | CUL2, MAPK | proliferation, migration, invasion, autophagy | July 2021 | [44] |
| LINC01133 | Cells and tissues | miR-30a-5p | FOXD1 | proliferation, metastasis, apoptosis | June 2021 | [137] |
| LNMAS | Tissues | NA | TWIST, STC1 | metastasis, EMT, immune evasion | Feb. 2022 | [138] |
| LOXL1-AS1 | Cells and tissues | miR-423-5p | ENC1, MEK/ERK pathway | proliferation, migration, invasion, angiogenesis, tumor growth | Jan. 2022 | [43] |
| MAG12-AS3 | Tissues | miR-15b | CCNE1 | proliferation, invasion | Jan. 2022 | [139] |
| MALAT1 | Cells and tissues | NA | NA | proliferation, invasion, migration | Aug. 2021 | [140] |
| MALAT1 | Cells | miR-485-5p | MAT2A | proliferation | Nov. 2021 | [141] |
| MALAT1 | Cells and tissues | miR-124-5p | NA | proliferation, tumor growth | Nov. 2021 | [142] |
| MEF2C-AS1 | Cells and tissues | miR-592 | RSPO1 | proliferation, migration, invasion | June 2021 | [143] |
| MEG3 | Cells and tissues | miR-7-5p | STC1 | ERs-mediated apoptosis | May 2021 | [144] |
| MiIR503HG | Cells and tissues | miR-191 | CEBPB | proliferation, metastasis, apoptosis | April 2021 | [145] |
| NEAT1 | Cells and tissues | miR-377 | FGFR1 | proliferation, migration, apoptosis | Jan. 2022 | [146] |
| NEAT1 | Cells and tissues | miR-34a | LDHA | drug-resistant, glycolysis rate | July 2021 | [147] |
| OIP5-AS1 | Cells and tissues | miR-124-5p | IDH2/HIF1a | proliferation, Hypoxia, Warburg effect | Aug. 2021 | [73] |
| OIP5-AS1 | Cells and tissues | MiR-147a | IGF1R, E-cadherin | migration, invasion, EMT | Mar. 2022 | [148] |
| OTUD6B-AS1 | Cells | miR-206 | cyclinD2 | drug-resistant | Oct. 2021 | [149] |
| RPL34-AS1 | Cells | NA | RPL34 | proliferation, migration, invasion, | May 2021 | [85] |
| SNHG15 | Cells and tissues | miR-4735-39 | HIF1a | tumor progression | Jan. 2022 | [72] |
| SNHG17 | Serum | miR-375-3p | NA | proliferation, migration, invasion | June 2021 | [150] |
| SNHG3 | Cells and tissues | NA | YAP1 | proliferation, migration, invasion, tumor growth | Jan. 2022 | [56] |
| SNHG5 | Cells and tissues | miR-132 | SOX4 | proliferation, migration, invasion | Mar. 2021 | [151] |
| SNHG6 | Cells and tissues | miR-485-3p | UCK2, Wnt | proliferation, migration, invasion, EMT | Nov. 2021 | [37] |
| SPINT1-AS1 | Cells and tissues | miR-214 | Wnt | proliferation, migration, invasion, tumor growth | July 2021 | [39] |
| TDRG1 | Cells and tissues | miR-214-5p | SEMA4C | invasion, tumor growth, hypoxia-induced glycolysis | April 2021 | [74] |
| UCA1 | Cells and tissues | miR-299-3p | NA | proliferation, invasion | Nov. 2021 | [152] |
| UNC5B-AS1 | Cells and tissues | miR-4455 | RSPO4 | proliferation, migration, invasion, apoptosis | Dec. 2021 | [153] |
| USP30-AS1 | Cells and tissues | miR-299-3p | PTP4A1 | proliferation, migration, invasion, tumor growth | July 2021 | [154] |
| WT1-AS | Cells and tissues | NA | SPL1/PIK3AP1 | proliferation, autophagy, apoptosis, tumor progression | July 2021 | [155] |
| WTA-AS | Cells and tissues | miR-205 | NA | cell cycle, apoptosis, migration, invasion, EMT, tumor growth | Dec. 2021 | [156] |
| ZFAS1 | Cells | MiR-190a-3p | KLF6 | proliferation, invasion, migration | Feb. 2022 | [100] |
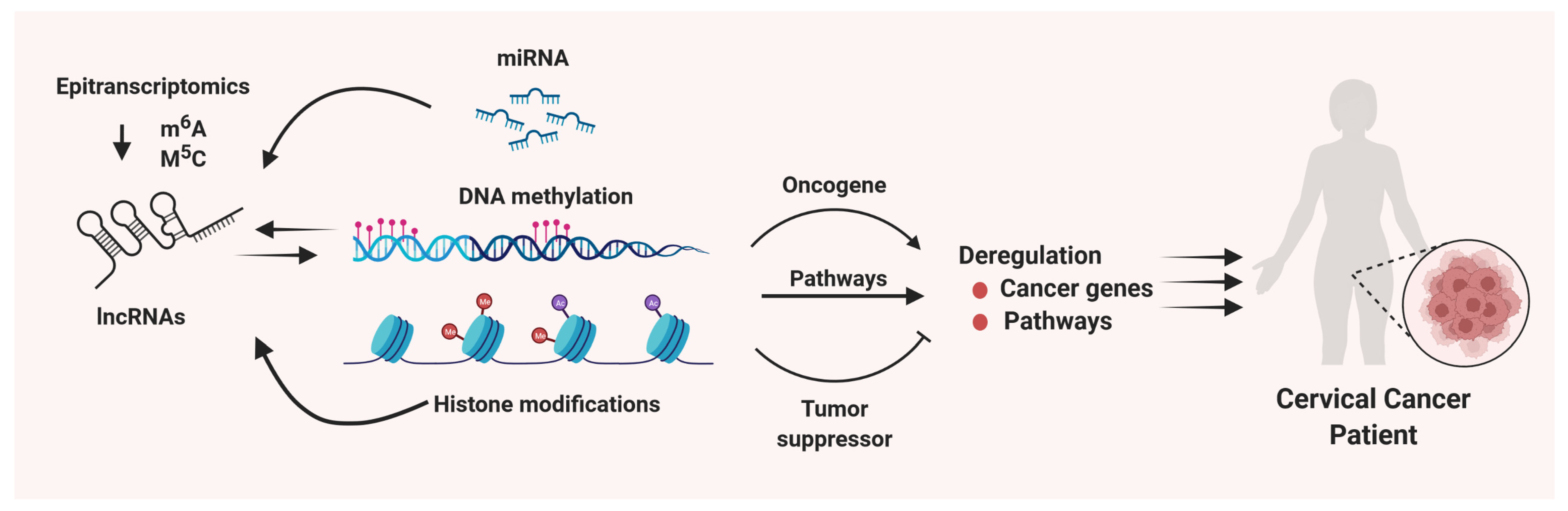
4. Epitranscriptomic Regulation of lncRNAs in Cervical Cancer
5. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ijff, M.; Crezee, J.; Oei, A.L.; Stalpers, L.J.; Westerveld, H. The role of hyperthermia in the treatment of locally advanced cervical cancer: A comprehensive review. Int. J. Gynecol. Cancer 2022, 32, 288–296. [Google Scholar] [CrossRef]
- Muthusami, S.; Sabanayagam, R.; Periyasamy, L.; Muruganantham, B.; Park, W.Y. A review on the role of epidermal growth factor signaling in the development, progression and treatment of cervical cancer. Int. J. Biol. Macromol. 2021, 194, 179–187. [Google Scholar] [CrossRef]
- Regauer, S.; Reich, O. The origin of Human Papillomavirus (HPV)—induced cervical squamous cancer. Curr. Opin. Virol. 2021, 51, 111–118. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Ghosh, S.; Jayaram, P.; Kabekkodu, S.P.; Satyamoorthy, K. Targeted drug delivery in cervical cancer: Current perspectives. Eur. J. Pharmacol. 2022, 917, 174751. [Google Scholar] [CrossRef]
- Snijders, P.J.F.; Steenbergen, R.; Heideman, D.A.M.; Meijer, C.J.L.M. HPV-mediated cervical carcinogenesis: Concepts and clinical implications. J. Pathol. 2006, 208, 152–164. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Zhang, J.; Mao, L. E6 hijacks KDM5C/lnc_000231/miR-497-5p/CCNE1 axis to promote cervical cancer progression. J. Cell Mol. Med. 2020, 24, 11422–11433. [Google Scholar] [CrossRef]
- White, E.A.; Kramer, R.E.; Tan, M.J.A.; Hayes, S.D.; Harper, J.W.; Howley, P.M. Comprehensive Analysis of Host Cellular Interactions with Human Papillomavirus E6 Proteins Identifies New E6 Binding Partners and Reflects Viral Diversity. J. Virol. 2012, 86, 13174–13186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Chen, L. Non-coding RNAs-EZH2 regulatory mechanisms in cervical cancer: The current state of knowledge. Biomed. Pharmacother. 2021, 146, 112123. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef] [Green Version]
- Feng, T.; Wu, Q.F. A review of non-coding RNA related to NF-kappaB signaling pathway in the pathogenesis of osteoarthritis. Int. Immunopharmacol. 2022, 106, 108607. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Al-Hendy, A. Non-coding RNAs: An important regulatory mechanism in pathogenesis of uterine fibroids. Fertil. Steril. 2018, 109, 802–803. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lu, Z.; Li, L.; Ma, M.; Long, F.; Wu, R.; Huang, L.; Chou, J.; Yang, K.; Zhang, Y.; et al. Identification and Validation of Ferroptosis-Related LncRNA Signatures as a Novel Prognostic Model for Colon Cancer. Front. Immunol. 2022, 12, 783362. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, M.; He, L.; Qi, H.; Shen, J.; Ying, K. Exosomal lncRNA SCIRT/miR-665 Transferring Promotes Lung Cancer Cell Metastasis through the Inhibition of HEYL. J. Oncol. 2021, 2021, 9813773. [Google Scholar] [CrossRef]
- Wu, B.; Xue, X.; Lin, S.; Tan, X.; Shen, G. LncRNA LINC00115 facilitates lung cancer progression through miR-607/ITGB1 pathway. Environ Toxicol. 2022, 37, 7–16. [Google Scholar] [CrossRef]
- Wu, Q.-N.; Luo, X.-J.; Liu, J.; Lu, Y.-X.; Wang, Y.; Qi, J.; Liu, Z.-X.; Huang, Q.-T.; Lu, J.-B.; Jin, Y.; et al. MYC-Activated LncRNA MNX1-AS1 Promotes the Progression of Colorectal Cancer by Stabilizing YB1. Cancer Res. 2021, 81, 2636–2650. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, J.; Yin, Y.; Meng, Q.; Lyu, Y. The role of EMT-related lncRNA in the process of triple-negative breast cancer metastasis. Biosci. Rep. 2021, 41, BSR20203121. [Google Scholar] [CrossRef]
- Yang, C.; Chen, K. Long Non-Coding RNA in Esophageal Cancer: A Review of Research Progress. Pathol. Oncol. Res. 2022, 28, 1–13. [Google Scholar] [CrossRef]
- Di Fiore, R.; Suleiman, S.; Felix, A.; O’Toole, S.; O’Leary, J.; Ward, M.; Beirne, J.; Sabol, M.; Ozretić, P.; Yordanov, A.; et al. An Overview of the Role of Long Non-Coding RNAs in Human Choriocarcinoma. Int. J. Mol. Sci. 2021, 22, 6506. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.-W.; Wang, Y.; Chen, L.-L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niinuma, T.; Suzuki, H.; Nojima, M.; Nosho, K.; Yamamoto, H.; Takamaru, H.; Yamamoto, E.; Maruyama, R.; Nobuoka, T.; Miyazaki, Y.; et al. Upregulation of miR-196a and HOTAIR Drive Malignant Character in Gastrointestinal Stromal Tumors. Cancer Res. 2012, 72, 1126–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long Noncoding RNA HOTAIR Regulates Polycomb-Dependent Chromatin Modification and Is Associated with Poor Prognosis in Colorectal Cancers. Cancer Res. 2011, 71, 6320–6326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naz, F.; Tariq, I.; Ali, S.; Somaida, A.; Preis, E.; Bakowsky, U. The Role of Long Non-Coding RNAs (lncRNAs) in Female Oriented Cancers. Cancers 2021, 13, 6102. [Google Scholar] [CrossRef]
- Kallen, A.N.; Zhou, X.-B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.-S.; Zhang, H.; et al. The Imprinted H19 LncRNA Antagonizes Let-7 MicroRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef] [Green Version]
- Sheng, X.; Li, J.; Yang, L.; Chen, Z.; Zhao, Q.; Tan, L. Promoter hypermethylation influences the suppressive role of maternally expressed 3, a long non-coding RNA, in the development of epithelial ovarian cancer. Oncol. Rep. 2014, 32, 277–285. [Google Scholar] [CrossRef] [Green Version]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Fan, B.; Zhang, Q.; Wang, N.; Wang, G. LncRNAs, the Molecules Involved in Communications with Colorectal Cancer Stem Cells. Front. Oncol. 2022, 12, 811374. [Google Scholar] [CrossRef]
- McCabe, E.M.; Rasmussen, T.P. lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Semin. Cancer Biol. 2020, 75, 38–48. [Google Scholar] [CrossRef]
- Wang, X.-C.; Liu, Y.; Long, F.-W.; Liu, L.-R.; Fan, C.-W. Identification of a lncRNA prognostic signature-related to stem cell index and its significance in colorectal cancer. Futur. Oncol. 2021, 17, 3087–3100. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, T.; Xia, L.; Zhang, M. LncRNA WDFY3-AS2 promotes cisplatin resistance and the cancer stem cell in ovarian cancer by regulating hsa-miR-139-5p/SDC4 axis. Cancer Cell Int. 2021, 21, 284. [Google Scholar] [CrossRef] [PubMed]
- Eptaminitaki, G.C.; Wolff, N.; Stellas, D.; Sifakis, K.; Baritaki, S. Long Non-Coding RNAs (lncRNAs) in Response and Resistance to Cancer Immunosurveillance and Immunotherapy. Cells 2021, 10, 3313. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, S.; Guo, X.; Ward, C.; Volpe, G.; Qin, B.; Esteban, M.A.; Bao, X. Role of Long Non-coding RNAs in Reprogramming to Induced Pluripotency. Genom. Proteom. Bioinform. 2020, 18, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Min, L.; Qiu, X.; Wu, X.; Liu, C.; Ma, J.; Zhang, D.; Zhu, L. Biological Function of Long Non-coding RNA (LncRNA) Xist. Front. Cell Dev. Biol. 2021, 9, 1447. [Google Scholar] [CrossRef]
- Cáceres-Durán, M.Á.; Ribeiro-Dos-Santos, Â.; Vidal, A.F. Roles and Mechanisms of the Long Noncoding RNAs in Cervical Cancer. Int. J. Mol. Sci. 2020, 21, 9742. [Google Scholar] [CrossRef]
- Hamidi, A.A.; Khalili-Tanha, G.; Navaei, Z.N.; Moghbeli, M. Long non-coding RNAs as the critical regulators of epithelial mesenchymal transition in colorectal tumor cells: An overview. Cancer Cell Int. 2022, 22, 1–15. [Google Scholar] [CrossRef]
- Wei, J.; Gao, Y.; Li, Z.; Jia, H.; Han, B. LncRNA SNHG6 facilitates cell proliferation, migration, invasion and EMT by upregulating UCK2 and activating the Wnt/beta-catenin signaling in cervical cancer. Bioorg Chem. 2021, 120, 105488. [Google Scholar] [CrossRef]
- Li, J.; Wang, H. H3K27ac-activated EGFR-AS1 promotes cell growth in cervical cancer through ACTN4-mediated WNT pathway. Biol. Direct 2022, 17, 3. [Google Scholar] [CrossRef]
- Song, H.; Liu, Y.; Liang, H.; Jin, X.; Liu, L. SPINT1-AS1 Drives Cervical Cancer Progression via Repressing miR-214 Biogenesis. Front. Cell Dev. Biol. 2021, 9, 1943. [Google Scholar] [CrossRef]
- Niu, Z.; Wang, F.; Lv, S.; Lv, Y.; Liu, M.; Fu, L. HNRNPU-AS1 Regulates Cell Proliferation and Apoptosis via the MicroRNA 205-5p/AXIN2 Axis and Wnt/beta-Catenin Signaling Pathway in Cervical Cancer. Mol. Cell Biol. 2021, 41, e0011521. [Google Scholar] [CrossRef]
- Xia, L.; Chen, Y.X.; Lian, J.B. LINC00665 promotes HeLa cell proliferation, migration, invasion and epithelial-mesenchymal transition by activating the WNT-CTNNB1/betacatenin signaling pathway. Sheng Li Xue Bao 2021, 73, 233–243. [Google Scholar]
- Yong, J.; Groeger, S.; Meyle, J.; Ruf, S. MAPK and beta-Catenin signaling: Implication and interplay in orthodontic tooth movement. Front. Biosci. 2022, 27, 54. [Google Scholar] [CrossRef]
- Zhang, P.; Zhao, F.; Jia, K.; Liu, X. The LOXL1 antisense RNA 1 (LOXL1-AS1)/microRNA-423-5p (miR-423-5p)/ectodermal-neural cortex 1 (ENC1) axis promotes cervical cancer through the mitogen-activated protein kinase (MEK)/extracellular signal-regulated kinase (ERK) pathway. Bioengineered 2022, 13, 2567–2584. [Google Scholar] [CrossRef]
- Chu, D.; Liu, T.; Yao, Y.; Luan, N. LINC00997/MicroRNA 574-3p/CUL2 Promotes Cervical Cancer Development via Mitogen-Activated Protein Kinase Signaling. Mol. Cell Biol. 2021, 41, e0005921. [Google Scholar] [CrossRef]
- Tzavlaki, K.; Moustakas, A. TGF-beta Signaling. Biomolecules 2020, 10, 487. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wang, Q.; Quan, Z. Long non-coding RNA CASC9 enhances breast cancer progression by promoting metastasis through the meditation of miR-215/TWIST2 signaling associated with TGF-beta expression. Biochem. Biophys. Res. Commun. 2019, 515, 644–650. [Google Scholar] [CrossRef]
- Cao, L.; Jin, H.; Zheng, Y.; Mao, Y.; Fu, Z.; Li, X.; Dong, L. DANCR-mediated microRNA-665 regulates proliferation and metastasis of cervical cancer through the ERK/SMAD pathway. Cancer Sci. 2019, 110, 913–925. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, Y.; Lin, R.; Mao, B.; Wang, W.; Bai, Y.; He, W.; Liu, Q. Long Noncoding RNA loc285194 Expression in Human Papillomavirus-Positive and -Negative Cervical Squamous Cell Carcinoma, C33A, and SiHa Cells and Transforming Growth Factor-β1. Med. Sci. Monit. 2019, 25, 9012–9018. [Google Scholar] [CrossRef]
- Xu, J.; Liu, X.-Y.; Zhang, Q.; Liu, H.; Zhang, P.; Tian, Z.-B.; Zhang, C.-P.; Li, X.-Y. Crosstalk Among YAP, LncRNA, and Tumor-Associated Macrophages in Tumorigenesis Development. Front. Oncol. 2022, 11, 810893. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, Z.; Guo, X.; Dong, H.; Zhou, K.; Huang, Z.; Xiao, Z. lncRNA B4GALT1-AS1 promotes colon cancer cell stemness and migration by recruiting YAP to the nucleus and enhancing YAP transcriptional activity. J. Cell. Physiol. 2019, 234, 18524–18534. [Google Scholar] [CrossRef]
- Li, R.-H.; Tian, T.; Ge, Q.-W.; He, X.-Y.; Shi, C.-Y.; Li, J.-H.; Zhang, Z.; Liu, F.-Z.; Sang, L.-J.; Yang, Z.-Z.; et al. A phosphatidic acid-binding lncRNA SNHG9 facilitates LATS1 liquid–liquid phase separation to promote oncogenic YAP signaling. Cell Res. 2021, 31, 1088–1105. [Google Scholar] [CrossRef]
- Lou, C.; Zhao, J.; Gu, Y.; Li, Q.; Tang, S.; Wu, Y.; Tang, J.; Zhang, C.; Li, Z.; Zhang, Y. LINC01559 accelerates pancreatic cancer cell proliferation and migration through YAP-mediated pathway. J. Cell. Physiol. 2019, 235, 3928–3938. [Google Scholar] [CrossRef]
- Li, C.; Wang, S.; Xing, Z.; Lin, A.; Liang, K.; Song, J. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat. Cell Biol. 2017, 19, 106–119. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Bao, J.; Yao, J.; Li, J. lncRNA USP2-AS1 promotes colon cancer progression by modulating Hippo/YAP1 signaling. Am. J. Transl. Res. 2020, 12, 5670–5682. [Google Scholar]
- Wang, Q.; Ding, J.; Nan, G.; Lyu, Y.; Ni, G. LncRNA NOC2L-4.1 functions as a tumor oncogene in cervical cancer progression by regulating the miR-630/YAP1 pathway. J. Cell Biochem. 2019, 120, 16913–16920. [Google Scholar] [CrossRef]
- Zhu, H.; Zhu, C.; Feng, X.; Luo, Y. Long noncoding RNA SNHG3 promotes malignant phenotypes in cervical cancer cells via association with YAP1. Hum. Cell 2021, 35, 320–332. [Google Scholar] [CrossRef]
- Burgess, J.T.; Rose, M.; Boucher, D.; Plowman, J.; Molloy, C.; Fisher, M.; O’Leary, C.; Richard, D.J.; O’Byrne, K.J.; Bolderson, E. The Therapeutic Potential of DNA Damage Repair Pathways and Genomic Stability in Lung Cancer. Front. Oncol. 2020, 10, 1256. [Google Scholar] [CrossRef]
- Tehrani, S.S.; Karimian, A.; Parsian, H.; Majidinia, M.; Yousefi, B. Multiple Functions of Long Non-Coding RNAs in Oxidative Stress, DNA Damage Response and Cancer Progression. J. Cell Biochem. 2018, 119, 223–236. [Google Scholar] [CrossRef]
- Wang, X.; Liu, H.; Shi, L.; Yu, X.; Gu, Y.; Sun, X. LINP1 facilitates DNA damage repair through non-homologous end joining (NHEJ) pathway and subsequently decreases the sensitivity of cervical cancer cells to ionizing radiation. Cell Cycle 2018, 17, 439–447. [Google Scholar] [CrossRef]
- Wen, D.; Huang, Z.; Li, Z.; Tang, X.; Wen, X.; Liu, J.; Li, M. LINC02535 co-functions with PCBP2 to regulate DNA damage repair in cervical cancer by stabilizing RRM1 mRNA. J. Cell. Physiol. 2020, 235, 7592–7603. [Google Scholar] [CrossRef]
- Janku, F.; Yap, T.A.; Meric-Bernstam, F. Targeting the PI3K pathway in cancer: Are we making headway? Nat. Rev. Clin. Oncol. 2018, 15, 273–291. [Google Scholar] [CrossRef] [PubMed]
- Sanaei, M.; Khorasani, A.B.S.; Pourbagheri-Sigaroodi, A.; Shahrokh, S.; Zali, M.R.; Bashash, D. The PI3K/Akt/mTOR axis in colorectal cancer: Oncogenic alterations, non-coding RNAs, therapeutic opportunities, and the emerging role of nanoparticles. J. Cell. Physiol. 2021, 237, 1720–1752. [Google Scholar] [CrossRef]
- Presti, D.; Quaquarini, E. The PI3K/AKT/mTOR and CDK4/6 Pathways in Endocrine Resistant HR+/HER2- Metastatic Breast Cancer: Biological Mechanisms and New Treatments. Cancers 2019, 11, 1242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamcheu, J.C.; Roy, T.; Uddin, M.B.; Banang-Mbeumi, S.; Chamcheu, R.-C.N.; Walker, A.L.; Liu, Y.-Y.; Huang, S. Role and Therapeutic Targeting of the PI3K/Akt/mTOR Signaling Pathway in Skin Cancer: A Review of Current Status and Future Trends on Natural and Synthetic Agents Therapy. Cells 2019, 8, 803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, K.; Deng, C.; Du, P.; Liu, T.; Piao, J.; Piao, Y.; Yang, M.; Chen, L. G6PC indicated poor prognosis in cervical cancer and promoted cervical carcinogenesis in vitro and in vivo. Reprod. Biol. Endocrinol. 2022, 20, 50. [Google Scholar] [CrossRef]
- An, R.; Meng, S.; Qian, H. Identification of Key Pathways and Establishment of a Seven-Gene Prognostic Signature in Cervical Cancer. J. Oncol. 2022, 2022, 4748796. [Google Scholar] [CrossRef]
- Jiang, L.; Jin, H.; Gong, S.; Han, K.; Li, Z.; Zhang, W.; Tian, J. LncRNA KCNQ1OT1 -mediated cervical cancer progression by sponging miR -1270 as a ceRNA of LOXL2 through PI3k /Akt pathway. J. Obstet. Gynaecol. Res. 2022. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, L.; Ding, X.; Sui, X. LINC00861 inhibits the progression of cervical cancer cells by functioning as a ceRNA for miR-513b-5p and regulating the PTEN/AKT/mTOR signaling pathway. Mol. Med. Rep. 2020, 23, 1. [Google Scholar] [CrossRef]
- Shi, W.-J.; Liu, H.; Ge, Y.-F.; Wu, D.; Tan, Y.-J.; Shen, Y.-C.; Wang, H.; Xu, H. LINC00673 exerts oncogenic function in cervical cancer by negatively regulating miR-126-5p expression and activates PTEN/PI3K/AKT signaling pathway. Cytokine 2020, 136, 155286. [Google Scholar] [CrossRef]
- Vito, A.; El-Sayes, N.; Mossman, K. Hypoxia-Driven Immune Escape in the Tumor Microenvironment. Cells 2020, 9, 992. [Google Scholar] [CrossRef]
- Shen, Y.; Liu, Y.; Sun, T.; Yang, W. LincRNA-p21 knockdown enhances radiosensitivity of hypoxic tumor cells by reducing autophagy through HIF-1/Akt/mTOR/P70S6K pathway. Exp. Cell Res. 2017, 358, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yang, M.; Lv, H.; Zhou, M.; Mao, X.; Qin, X.; Xu, Y.; Li, L.; Xing, H. lncRNA SNHG15 Induced by SOX12 Promotes the Tumorigenic Properties and Chemoresistance in Cervical Cancer via the miR-4735-3p/HIF1a Pathway. Oxidative Med. Cell. Longev. 2022, 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, Y.; Maerkeya, K.; Reyanguly, D.; Han, L. LncRNA OIP5-AS1 Regulates the Warburg Effect Through miR-124-5p/IDH2/HIF-1alpha Pathway in Cervical Cancer. Front Cell Dev. Biol. 2021, 9, 655018. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, C.; Tian, Y. Long non-coding RNA TDRG1 promotes hypoxia-induced glycolysis by targeting the miR-214-5p/SEMA4C axis in cervical cancer cells. Histochem. J. 2021, 52, 245–256. [Google Scholar] [CrossRef]
- Ta, W.; Zhang, Y.; Zhang, S.; Sun, P. LncRNA ANCR downregulates hypoxia-inducible factor 1α and inhibits the growth of HPV-negative cervical squamous cell carcinoma under hypoxic conditions. Mol. Med. Rep. 2019, 21, 413–419. [Google Scholar] [CrossRef]
- Ozaki, T.; Nakagawara, A. Role of p53 in Cell Death and Human Cancers. Cancers 2011, 3, 994–1013. [Google Scholar] [CrossRef]
- Harris, S.L.; Levine, A.J. The p53 pathway: Positive and negative feedback loops. Oncogene 2005, 24, 2899–2908. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Latif, M.; Riad, A.; Soliman, R.A.; Elkhouly, A.M.; Nafae, H.; Gad, M.Z. MALAT-1/p53/miR-155/miR-146a ceRNA circuit tuned by methoxylated quercitin glycoside alters immunogenic and oncogenic profiles of breast cancer. Mol. Cell Biochem. 2022, 477, 1281–1293. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Sohrabi, B.; Hussen, B.M.; Mehravaran, E.; Jamali, E.; Arsang-Jang, S.; Fathi, M.; Taheri, M.; Samsami, M. Down-regulation of MEG3, PANDA and CASC2 as p53-related lncRNAs in breast cancer. Breast Dis. 2022, 41, 137–143. [Google Scholar] [CrossRef]
- Xu, M.; Zhao, X.; Zhao, S.; Yang, Z.; Yuan, W.; Han, H. Landscape analysis of lncRNAs shows that DDX11-AS1 promotes cell-cycle progression in liver cancer through the PARP1/p53 axis. Cancer Lett. 2021, 520, 282–294. [Google Scholar] [CrossRef]
- Pal, S.; Garg, M.; Pandey, A.K. Deciphering the Mounting Complexity of the p53 Regulatory Network in Correlation to Long Non-Coding RNAs (lncRNAs) in Ovarian Cancer. Cells 2020, 9, 527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Li, Y.; Sheng, J.; Wu, F.; Li, K.; Huang, R.; Wang, X.; Jiao, T.; Guan, X.; Lu, Y.; et al. P53-R273H mutation enhances colorectal cancer stemness through regulating specific lncRNAs. J. Exp. Clin. Cancer Res. 2019, 38, 379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.L.; Subramanian, M.; Jones, M.F.; Chaudhary, R.; Singh, D.K.; Zong, X.; Gryder, B.; Sindri, S.; Mo, M.; Schetter, A.; et al. Long Noncoding RNA PURPL Suppresses Basal p53 Levels and Promotes Tumorigenicity in Colorectal Cancer. Cell Rep. 2017, 20, 2408–2423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhary, R.; Gryder, B.; Woods, W.S.; Subramanian, M.; Jones, M.F.; Li, X.L.; Jenkins, L.M.; Shabalina, S.A.; Mo, M.; Dasso, M.; et al. Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3. eLife 2017, 6, e23244. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Ren, C.; Jiang, D.; Yang, L.; Chen, Y.; Li, F. RPL34-AS1-induced RPL34 inhibits cervical cancer cell tumorigenesis via the MDM2-P53 pathway. Cancer Sci. 2021, 112, 1811–1821. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Munger, K. Expression of the Long Noncoding RNA DINO in Human Papillomavirus-Positive Cervical Cancer Cells Reactivates the Dormant TP53 Tumor Suppressor through ATM/CHK2 Signaling. mBio 2020, 11, e01190-20. [Google Scholar] [CrossRef]
- Cui, L.; Nai, M.; Zhang, K.; Li, L.; Li, R. lncRNA WT1-AS inhibits the aggressiveness of cervical cancer cell via regulating p53 expression via sponging miR-330-5p. Cancer Manag. Res. 2019, ume 11, 651–667. [Google Scholar] [CrossRef] [Green Version]
- Meza-Sosa, K.F.; Miao, R.; Navarro, F.; Zhang, Z.; Zhang, Y.; Hu, J.J.; Hartford, C.C.R.; Li, X.L.; Pedraza-Alva, G.; Pérez-Martínez, L.; et al. SPARCLE, a p53-induced lncRNA, controls apoptosis after genotoxic stress by promoting PARP-1 cleavage. Mol. Cell 2022, 82, 785–802.e10. [Google Scholar] [CrossRef]
- Sun, Y.F.; Wang, Y.; Li, X.D.; Wang, H. SNHG15, a p53-regulated lncRNA, suppresses cisplatin-induced apoptosis and ROS accumulation through the miR-335-3p/ZNF32 axis. Am. J. Cancer Res. 2022, 12, 816–828. [Google Scholar]
- Ou, X.; Zhou, X.; Li, J.; Ye, J.; Liu, H.; Fang, D.; Cai, Q.; Cai, S.; He, Y.; Xu, J. p53-Induced LINC00893 Regulates RBFOX2 Stability to Suppress Gastric Cancer Progression. Front. Cell Dev. Biol. 2022, 9, 3948. [Google Scholar] [CrossRef]
- Wang, C.; Yang, Y.; Wu, X.; Li, J.; Liu, K.; Fang, D.; Li, B.; Shan, G.; Mei, X.; Wang, F.; et al. Reciprocal modulation of long noncoding RNA EMS and p53 regulates tumorigenesis. Proc. Natl. Acad. Sci. USA 2022, 119, e2111409119. [Google Scholar] [CrossRef] [PubMed]
- Rong, H.; Chen, B.; Ma, K.; Wei, X.; Peng, J.; Zhu, J. Downregulation of lncRNA LINC-PINT Participates in the Recurrence of Esophageal Squamous Cell Carcinoma Possibly by Interacting miRNA-21. Cancer Biotherapy Radiopharm. 2021, 36, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Yang, X.; Li, J.; Yang, W.; Ma, H.; Zhang, Z. p53-targeted lincRNA-p21 acts as a tumor suppressor by inhibiting JAK2/STAT3 signaling pathways in head and neck squamous cell carcinoma. Mol. Cancer 2019, 18, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, E.-B.; Yin, D.-D.; Sun, M.; Kong, R.; Liu, X.-H.; You, L.-H.; Han, L.; Xia, R.; Wang, K.-M.; Yang, J.-S.; et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014, 5, e1243. [Google Scholar] [CrossRef]
- Mello, S.S.; Sinow, C.; Raj, N.; Mazur, P.K.; Bieging-Rolett, K.; Broz, D.K.; Imam, J.F.C.; Vogel, H.; Wood, L.D.; Sage, J.; et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017, 31, 1095–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitt, A.; Garcia, J.T.; Hung, T.; Flynn, R.; Shen, Y.; Qu, K.; Payumo, A.Y.; Peres-Da-Silva, A.; Broz, D.K.; Baum, R.; et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 2016, 48, 1370–1376. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Chen, X.; Hua, Z.; Shi, Z.; Zhou, Q.; Zhengzheng, S.; Wang, Y. Long non-coding RNA expression profile in cervical cancer tissues. Oncol. Lett. 2017, 14, 1379–1386. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, Y.; Chen, M.; Cui, J. Identification of Novel Long Non-coding and Circular RNAs in Human Papillomavirus-Mediated Cervical Cancer. Front. Microbiol. 2017, 8, 1720. [Google Scholar] [CrossRef] [Green Version]
- Olgun, G.; Sahin, O.; Tastan, O. Discovering lncRNA mediated sponge interactions in breast cancer molecular subtypes. BMC Genom. 2018, 19, 650. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Hou, W.; Zhang, C.; Ji, P.; Hu, R.; Zhang, Q.; Wang, Y.; Li, P.; Zhang, H.; Chen, Y.; et al. Long non-coding RNA ZFAS1 regulates cell proliferation and invasion in cervical cancer via the miR-190a-3p/KLF6 axis. Bioengineered 2022, 13, 3840–3851. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, J.; Zhou, L.; Yin, C. LINC00885 promotes cervical cancer progression through sponging miR-3150b-3p and upregulating BAZ2A. Biol. Direct 2022, 17, 4. [Google Scholar] [CrossRef] [PubMed]
- Ji, F.; Lu, Y.; Chen, S.; Lin, X.; Yu, Y.; Zhu, Y.; Luo, X. m6A methyltransferase METTL3-mediated lncRNA FOXD2-AS1 promotes the tumorigenesis of cervical cancer. Mol. Ther.-Oncolytics 2021, 22, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Jin, S.; Zou, B.; Liang, Y.; Xie, J.; Wu, S. Analyzing the whole-transcriptome profiles of ncRNAs and predicting the competing endogenous RNA networks in cervical cancer cell lines with cisplatin resistance. Cancer Cell Int. 2021, 21, 532. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Duan, L.J.; Lu, B.N.; Pang, Y.Z.; Pang, Z.R. LncRNA AFAP1-AS1/miR-27b-3p/VEGF-C axis modulates stemness characteristics in cervical cancer cells. Chin. Med. J. 2021, 134, 2091–2101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, H.-L.; Liu, J.-B.; Zhang, Y.; Li, Y.-H. LncRNA AL592284.1 facilitates proliferation and metastasis of cervical cancer cells via miR-30a-5p/Vimentin/EMT axis. Biochem. Biophys. Res. Commun. 2021, 577, 95–102. [Google Scholar] [CrossRef]
- Yang, W.; Wang, X.; Song, S.; Chu, Y.; Sun, D.; Yu, X.; Zou, Y. Long noncoding RNA ALOX12-AS1 inhibits cervical cancer cells proliferation via targeting miR-3171. Anti-Cancer Drugs, 2021; Publish Ahead. [Google Scholar] [CrossRef]
- He, S.; Feng, Y.; Zou, W.; Wang, J.; Li, G.; Xiong, W.; Xie, Y.; Ma, J.; Liu, X. The Role of the SOX9/lncRNA ANXA2P2/miR-361-3p/SOX9 Regulatory Loop in Cervical Cancer Cell Growth and Resistance to Cisplatin. Front Oncol. 2021, 11, 784525. [Google Scholar] [CrossRef]
- Gao, S.; Lv, Q.; Xu, F.; Li, H.; Guo, X. LncRNA CASC9-1 Facilitates Cell Malignant Behaviors in Cervical Squamous Cell Carcinoma by Targeting miR-383-5p to Up-regulate MAPKAP1. Arch. Med. Res. 2021, 53, 138–146. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Y.; Cai, H.; Jiang, H.; Li, W.; Shi, Y. Long coding RNA CCAT2 enhances the proliferation and epithelial-mesenchymal transition of cervical carcinoma cells via the microRNA-493-5p/CREB1 axis. Bioengineered 2021, 12, 6264–6274. [Google Scholar] [CrossRef]
- Hu, C.; Han, Y.; Zhu, G.; Li, G.; Wu, X. Krüppel-like factor 5-induced overexpression of long non-coding RNA DANCR promotes the progression of cervical cancer via repressing microRNA-145-3p to target ZEB1. Cell Cycle 2021, 20, 1441–1454. [Google Scholar] [CrossRef]
- Kong, X.; Wang, J.S.; Yang, H. Upregulation of lncRNA DARS-AS1 accelerates tumor malignancy in cervical cancer by activating cGMP-PKG pathway. J. Biochem Mol. Toxicol. 2021, 35, 1–11. [Google Scholar] [CrossRef]
- He, M.; Wang, Y.; Cai, J.; Xie, Y.; Tao, C.; Jiang, Y.; Li, H.; Song, F. LncRNA DLEU2 promotes cervical cancer cell proliferation by regulating cell cycle and NOTCH pathway. Exp. Cell Res. 2021, 402, 112551. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Liu, Y.; Qi, T.; Qian, W.; Hu, D.; Feng, W. Long non-coding RNA DUXAP8 elevates RCN2 expression and facilitates cell malignant behaviors and angiogenesis in cervical cancer via sponging miR-1297. Diagn. Pathol. 2021, 16, 105. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Shi, H.; Shi, X.; Jiang, X. LncRNA FBXL19-AS1 promotes proliferation and metastasis of cervical cancer through upregulating COL1A1 as a sponge of miR-193a-5p. J. Biol. Res. 2021, 28, 20. [Google Scholar] [CrossRef]
- Yang, X.; Qu, Y.; Zhang, J. Up-Regulated LncRNA FEZF1-AS1 Promotes the Progression of Cervical Carcinoma Cells via MiR-367-3p/SLC12A5 Signal Axis. Arch. Med. Res. 2021, 53, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Du, X.; Xiao, L.; Zeng, Q.; Liu, Q. Activation of FGD5-AS1 Promotes Progression of Cervical Cancer through Regulating BST2 to Inhibit Macrophage M1 Polarization. J. Immunol. Res. 2021, 2021, 5857214. [Google Scholar] [CrossRef]
- Yang, X.; Du, H.; Bian, W.; Li, Q.; Sun, H. FOXD3AS1/miR1283p/LIMK1 axis regulates cervical cancer progression. Oncol. Rep. 2021, 45, 62. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zou, T.; Liang, W.; Zhang, Z.; Qie, M. Long non-coding RNA HAND2-AS1 delays cervical cancer progression via its regulation on the microRNA-21-5p/TIMP3/VEGFA axis. Cancer Gene Ther. 2021, 28, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Trujano-Camacho, S.; Cantu-de Leon, D.; Delgado-Waldo, I.; Coronel-Hernandez, J.; Millan-Catalan, O.; Hernandez-Sotelo, D.; Lopez-Camarillo, C.; Perez-Plasencia, C.; Campos-Parra, A.D. Inhibition of Wnt-beta-Catenin Signaling by ICRT14 Drug Depends of Post-Transcriptional Regulation by HOTAIR in Human Cervical Cancer HeLa Cells. Front. Oncol. 2021, 11, 729228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wu, Q.; Liu, Y.; Wang, X.; Ma, C.; Zhu, W. LncRNA HOTAIR Promotes Chemoresistance by Facilitating Epithelial to Mesenchymal Transition through miR-29b/PTEN/PI3K Signaling in Cervical Cancer. Cells Tissues Organs 2021, 211, 16–29. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, J.; Wu, Q.; Liu, Y.; Ma, C. HOTAIR Contributes to Stemness Acquisition of Cervical Cancer through Regulating miR-203 Interaction with ZEB1 on Epithelial-Mesenchymal Transition. J. Oncol. 2021, 2021, 4190764. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Y.; Lin, M.; Wu, D.; Zhao, M. LncRNA HOTAIR promotes proliferation and inhibits apoptosis by sponging miR-214-3p in HPV16 positive cervical cancer cells. Cancer Cell Int. 2021, 21, 400. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; He, P. Long noncoding RNA HOXA-AS2 accelerates cervical cancer by the miR-509-3p/BTN3A1 axis. J. Pharm. Pharmacol. 2021, 73, 1387–1396. [Google Scholar] [CrossRef]
- Wang, T.; Li, W.; Ye, B.; Zhang, S.; Lei, X.; Zhang, D. FTO-stabilized lncRNA HOXC13-AS epigenetically upregulated FZD6 and activated Wnt/beta-catenin signaling to drive cervical cancer proliferation, invasion, and EMT. J. BUON. 2021, 26, 1279–1291. [Google Scholar] [PubMed]
- Zhao, R.; Song, J.; Jin, Y.; Liu, Y. Long noncoding RNA HOXC-AS3 enhances the progression of cervical cancer via activating ErbB signaling pathway. Histochem. J. 2021, 52, 991–1006. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, L.; Liu, Q.; He, F.; Zhu, H. LncRNA HOXD-AS1 affects proliferation and apoptosis of cervical cancer cells by promoting FRRS1 expression via transcription factor ELF1. Cell Cycle 2022, 21, 416–426. [Google Scholar] [CrossRef]
- Zhu, L.; Chen, R.; Jiang, C.; Xie, Q.; Zhao, W.; Gao, X. Mechanism underlying long noncoding RNA ILF3AS1mediated inhibition of cervical cancer cell proliferation, invasion and migration, and promotion of apoptosis. Mol. Med. Rep. 2021, 24, 554. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Y. Long non-coding RNA KCNQ1OT1 facilitates the progression of cervical cancer and tumor growth through modulating miR-296-5p/HYOU1 axis. Bioengineered 2021, 12, 8753–8767. [Google Scholar] [CrossRef]
- Tian, L.; Han, F.; Yang, J.; Ming, X.; Chen, L. Long non-coding RNA LINC01006 exhibits oncogenic properties in cervical cancer by functioning as a molecular sponge for microRNA-28-5p and increasing PAK2 expression. Int. J. Mol. Med. 2021, 47, 1. [Google Scholar] [CrossRef]
- Zhai, Y.; Liu, Y.; Wang, Z.; Wang, W.; Zhou, J.; Lu, J. Long Non-Coding RNA LINC00313 Accelerates Cervical Carcinoma Progression by miR-4677-3p/CDK6 Axis. OncoTargets Ther. 2021, ume 14, 2213–2226. [Google Scholar] [CrossRef]
- Liu, X.; Shen, X.; Zhang, J. Long non-coding RNA LINC00514 promotes the proliferation and invasion through the miR -708-5p/ HOXB3 axis in cervical squamous cell carcinoma. Environ. Toxicol. 2021, 37, 161–170. [Google Scholar] [CrossRef]
- Liu, Y.; Qiu, S.; Zheng, X.; Qiu, Y.; Yao, S.; Ge, Y. LINC00662 modulates cervical cancer cell proliferation, invasion, and apoptosis via sponging miR-103a-3p and upregulating PDK4. Mol. Carcinog. 2021, 60, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-K.; Ni, R.-X.; Wang, W.-J.; Wang, D.; Zhao, M.; Lei, C.-Z.; Sun, X.-J.; Huang, C.-Z.; Bai, P.; Che, Y.-Q.; et al. Overexpression of LINC00673 Promotes the Proliferation of Cervical Cancer Cells. Front. Oncol. 2021, 11, 1834. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Li, J.; Fan, F.; Zhou, P. LINC00707 Regulates miR-382-5p/VEGFA Pathway to Enhance Cervical Cancer Progression. J. Immunol. Res. 2021, 2021, 5524632. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tu, H.; Zhang, L.; Xiong, J.; Li, L. FOXP3-induced LINC00885 promotes the proliferation and invasion of cervical cancer cells. Mol. Med. Rep. 2021, 23, 458. [Google Scholar] [CrossRef]
- Chen, Y.; Gu, Y.; Gu, Y.; Wu, J. Long Noncoding RNA LINC00899/miR-944/ESR1 Axis Regulates Cervical Cancer Cell Proliferation, Migration, and Invasion. J. Interferon Cytokine Res. 2021, 41, 220–233. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, Y.; Sun, X. LINC01133 promotes the progression of cervical cancer via regulating miR-30a-5p/FOXD1. Asia-Pacific J. Clin. Oncol. 2020, 17, 253–263. [Google Scholar] [CrossRef]
- Liao, Y.; Huang, J.; Liu, P.; Zhang, C.; Liu, J.; Xia, M.; Shang, C.; Ooi, S.; Chen, Y.; Qin, S.; et al. Downregulation of LNMAS orchestrates partial EMT and immune escape from macrophage phagocytosis to promote lymph node metastasis of cervical cancer. Oncogene 2022, 41, 1931–1943. [Google Scholar] [CrossRef]
- Chai, Y.; Wang, L.; Qu, Y.; Hu, Z. LncRNA MAGI2-As3 Suppresses the Proliferation and Invasion of Cervical Cancer by Sponging MiR-15b. J. Heal. Eng. 2022, 2022, 9707206. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, W.; Huang, W.; Hua, Z.; Li, S. LncRNA MALAT1 was regulated by HPV16 E7 independently of pRB in cervical cancer cells. J. Cancer 2021, 12, 6344–6355. [Google Scholar] [CrossRef]
- Tie, W.; Ge, F. MALAT1 Inhibits Proliferation of HPV16-Positive Cervical Cancer by Sponging miR-485-5p to Promote Expression of MAT2A. DNA Cell Biol. 2021, 40, 1407–1417. [Google Scholar] [CrossRef]
- Liang, T.; Wang, Y.; Jiao, Y.; Cong, S.; Jiang, X.; Dong, L.; Zhang, G.; Xiao, D. LncRNA MALAT1 Accelerates Cervical Carcinoma Proliferation by Suppressing miR-124 Expression in Cervical Tumor Cells. J. Oncol. 2021, 2021, 8836078. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, C.; Gong, M.; Jiang, C. A Novel Identified Long Non-coding RNA, lncRNA MEF2C-AS1, Inhibits Cervical Cancer via Regulation of miR-592/RSPO1. Front. Mol. Biosci. 2021, 8, 687113. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Cao, Y.-M.; Liu, J.-H.; Ding, J.; Xie, X.-Y.; Cao, P.-G. MEG3 Induces Cervical Carcinoma Cells’ Apoptosis Through Endoplasmic Reticulum Stress by miR-7-5p/STC1 Axis. Cancer Biotherapy Radiopharm. 2021, 36, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.L.; Zhang, Y.X.; Liu, N.; Liu, H.; Yuan, Y.C. LncRNA MIR503HG regulated cell viability, metastasis and apoptosis of cervical cancer via miR-191/CEBPB axis. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 3200–3210. [Google Scholar]
- Geng, F.; Jia, W.C.; Li, T.; Li, N.; Wei, W. Knockdown of lncRNA NEAT1 suppresses proliferation and migration, and induces apoptosis of cervical cancer cells by regulating the miR377/FGFR1 axis. Mol. Med. Rep. 2022, 25, 10. [Google Scholar]
- Shao, X.; Zheng, X.; Ma, D.; Liu, Y.; Liu, G. Inhibition of lncRNA-NEAT1 sensitizes 5-Fu resistant cervical cancer cells through de-repressing the microRNA-34a/LDHA axis. Biosci. Rep. 2021, 41, BSR20200533. [Google Scholar] [CrossRef]
- Zhang, L.; Cai, Y.; Tian, C.; Li, Y.; Ma, K.; Gao, X. LncRNA Opa interacting protein 5-antisense RNA 1 (OIP5-AS1) promotes the migration, invasion and epithelial-mesenchymal transition (EMT) through targeting miR-147a/insulin-like growth factor 1 receptor (IGF1R) pathway in cervical cancer tissues and cell model. J. Obstet. Gynaecol. Res. 2022. [Google Scholar] [CrossRef]
- Hou, H.; Yu, R.; Zhao, H.; Yang, H.; Hu, Y.; Hu, Y.; Guo, J. LncRNA OTUD6B-AS1 Induces Cisplatin Resistance in Cervical Cancer Cells Through Up-Regulating Cyclin D2 via miR-206. Front. Oncol. 2021, 11, 777220. [Google Scholar] [CrossRef]
- Cao, S.; Li, H.; Li, L. LncRNA SNHG17 Contributes to the Progression of Cervical Cancer by Targeting microRNA-375-3p. Cancer Manag. Res. 2021, ume 13, 4969–4978. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, X.; Li, Y.; Teng, X.; Zou, L.; Yu, B. LncRNA SNHG5 promotes cervical cancer progression by regulating the miR-132/SOX4 pathway. Autoimmunity 2021, 54, 88–96. [Google Scholar] [CrossRef]
- An, M.; Xing, X.; Chen, T. Long non-coding RNA UCA1 enhances cervical cancer cell proliferation and invasion by regulating microRNA-299-3p expression. Oncol. Lett. 2021, 22, 772. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Zhang, Y.; Wang, M.; Hu, J.; Fang, Y. Inhibition of the long non-coding RNA UNC5B-AS1/miR-4455/RSPO4 axis reduces cervical cancer growth in vitro and in vivo. J. Gene Med. 2021, 23, e3382. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chi, Y.; Chen, H.; Zhao, L. Long non-coding RNA USP30-AS1 aggravates the malignant progression of cervical cancer by sequestering microRNA-299-3p and thereby overexpressing PTP4A1. Oncol. Lett. 2021, 22, 505. [Google Scholar] [CrossRef]
- Tong, W.; Zhang, H. Overexpression of long non-coding RNA WT1-AS or silencing of PIK3AP1 are inhibitory to cervical cancer progression. Cell Cycle 2021, 20, 2583–2596. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Zhang, J.; He, Z.; Wu, S. Long Noncoding RNA WT1-AS Inhibits the Progression of Cervical Cancer by Sponging miR-205. Cancer Biotherapy Radiopharm. 2021, 36, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.; Shi, H.; He, C. Epitranscriptomic influences on development and disease. Genome Biol. 2017, 18, 197. [Google Scholar] [CrossRef] [Green Version]
- Rauch, S.; He, C.; Dickinson, B.C. Targeted m6A Reader Proteins to Study Epitranscriptomic Regulation of Single RNAs. J. Am. Chem. Soc. 2018, 140, 11974–11981. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; He, C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2012, 29, 108–115. [Google Scholar] [CrossRef] [Green Version]
- Motorin, Y.; Helm, M. RNA nucleotide methylation: 2021 update. Wiley Interdiscip. Rev. RNA 2021, 13, e1691. [Google Scholar] [CrossRef]
- Yin, L.; Zhu, X.; Novák, P.; Zhou, L.; Gao, L.; Yang, M.; Zhao, G.; Yin, K. The epitranscriptome of long noncoding RNAs in metabolic diseases. Clin. Chim. Acta 2021, 515, 80–89. [Google Scholar] [CrossRef]
- Zhang, H.; Kong, W.; Zhao, X.; Han, C.; Liu, T.; Li, J.; Song, D. N6-Methyladenosine-Related lncRNAs as potential biomarkers for predicting prognoses and immune responses in patients with cervical cancer. BMC Genom. Data 2022, 23, 1925–1937. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, D.; Wu, D.; Zhang, D.; Sun, M. Long Noncoding RNA KCNMB2-AS1 Stabilized by N6-Methyladenosine Modification Promotes Cervical Cancer Growth Through Acting as a Competing Endogenous RNA. Cell Transplant. 2020, 29, 0963689720964382. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, J.; Wang, Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am. J. Transl. Res. 2019, 11, 4909–4921. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Q.; Al-Hendy, A. The Regulatory Functions and the Mechanisms of Long Non-Coding RNAs in Cervical Cancer. Cells 2022, 11, 1149. https://doi.org/10.3390/cells11071149
Yang Q, Al-Hendy A. The Regulatory Functions and the Mechanisms of Long Non-Coding RNAs in Cervical Cancer. Cells. 2022; 11(7):1149. https://doi.org/10.3390/cells11071149
Chicago/Turabian StyleYang, Qiwei, and Ayman Al-Hendy. 2022. "The Regulatory Functions and the Mechanisms of Long Non-Coding RNAs in Cervical Cancer" Cells 11, no. 7: 1149. https://doi.org/10.3390/cells11071149
APA StyleYang, Q., & Al-Hendy, A. (2022). The Regulatory Functions and the Mechanisms of Long Non-Coding RNAs in Cervical Cancer. Cells, 11(7), 1149. https://doi.org/10.3390/cells11071149







