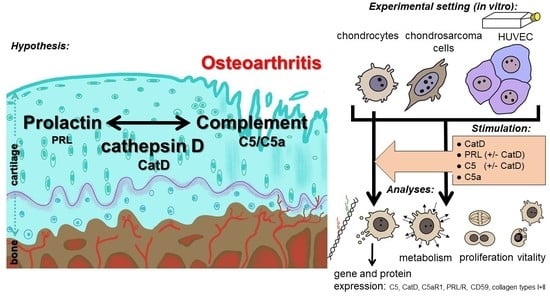
Complement Proteins C5/C5a, Cathepsin D and Prolactin in Chondrocytes: A Possible Crosstalk in the Pathogenesis of Osteoarthritis
Abstract
1. Introduction
1.1. Osteoarthritis
1.2. The Complement System
1.3. Prolactin
1.4. Cathepsin D
2. Methods and Materials
2.1. Isolation of the Chondrocytes and Cell-Culture
2.2. Stimulation of Chondrocytes
2.3. Viability Staining of the Chondrocytes
2.4. CyQUANT® NF Cell Proliferation Assay
2.5. The CellTiter-Blue® Cell Viability Assay
2.6. Gene Expression Analysis
2.6.1. RNA Isolation and cDNA Synthesis
2.6.2. qPCR
2.7. Immunofluorescence of Stimulated Primary Human Chondrocytes
2.8. Statistical Analysis
3. Results
3.1. Viability Staining
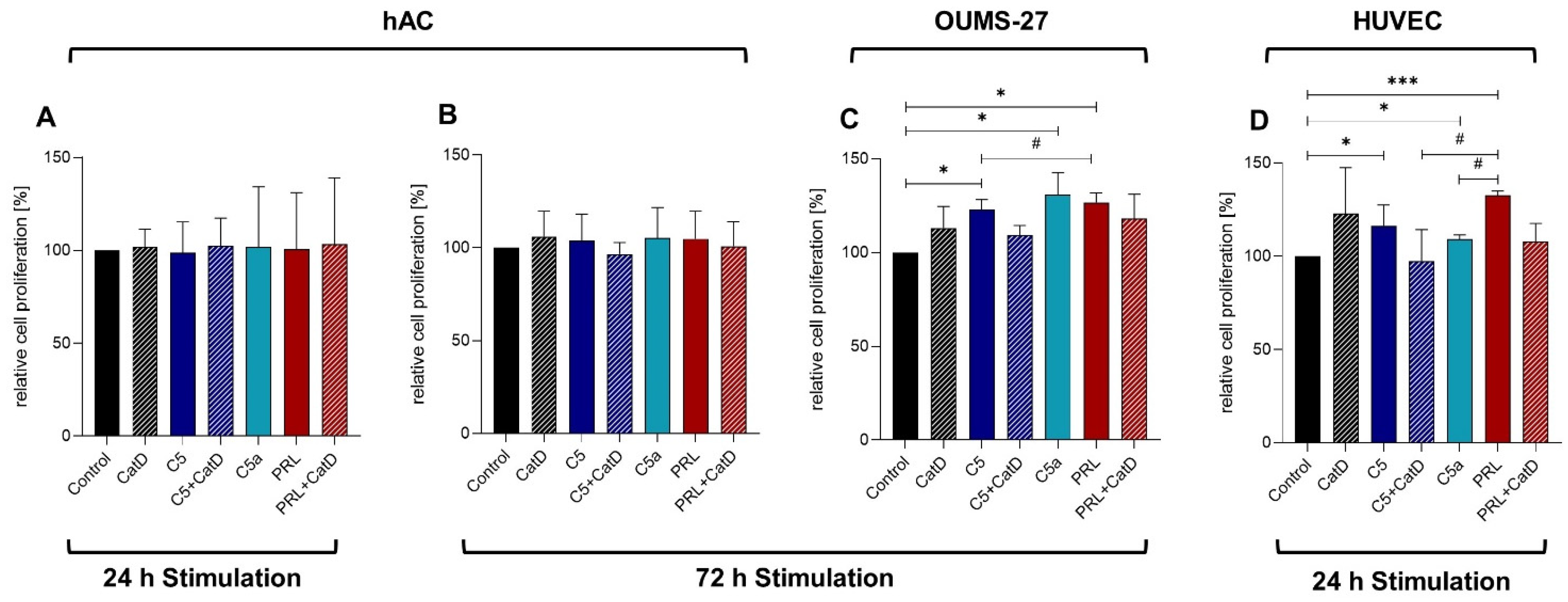
3.2. CyQUANT® NF Cell Proliferation Assay
3.3. CellTiter-Blue® Cell Viability Assay
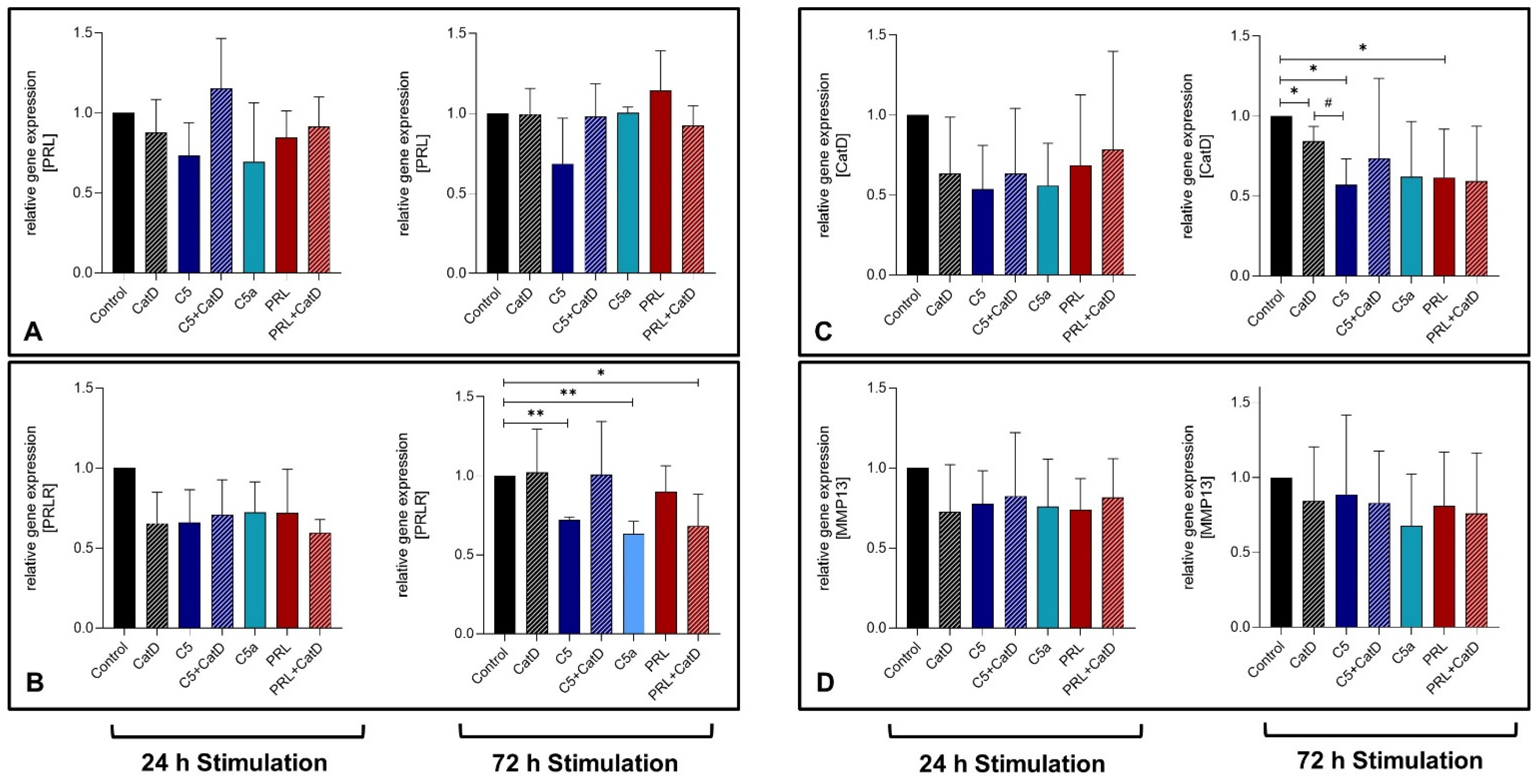
3.4. Gene Expression
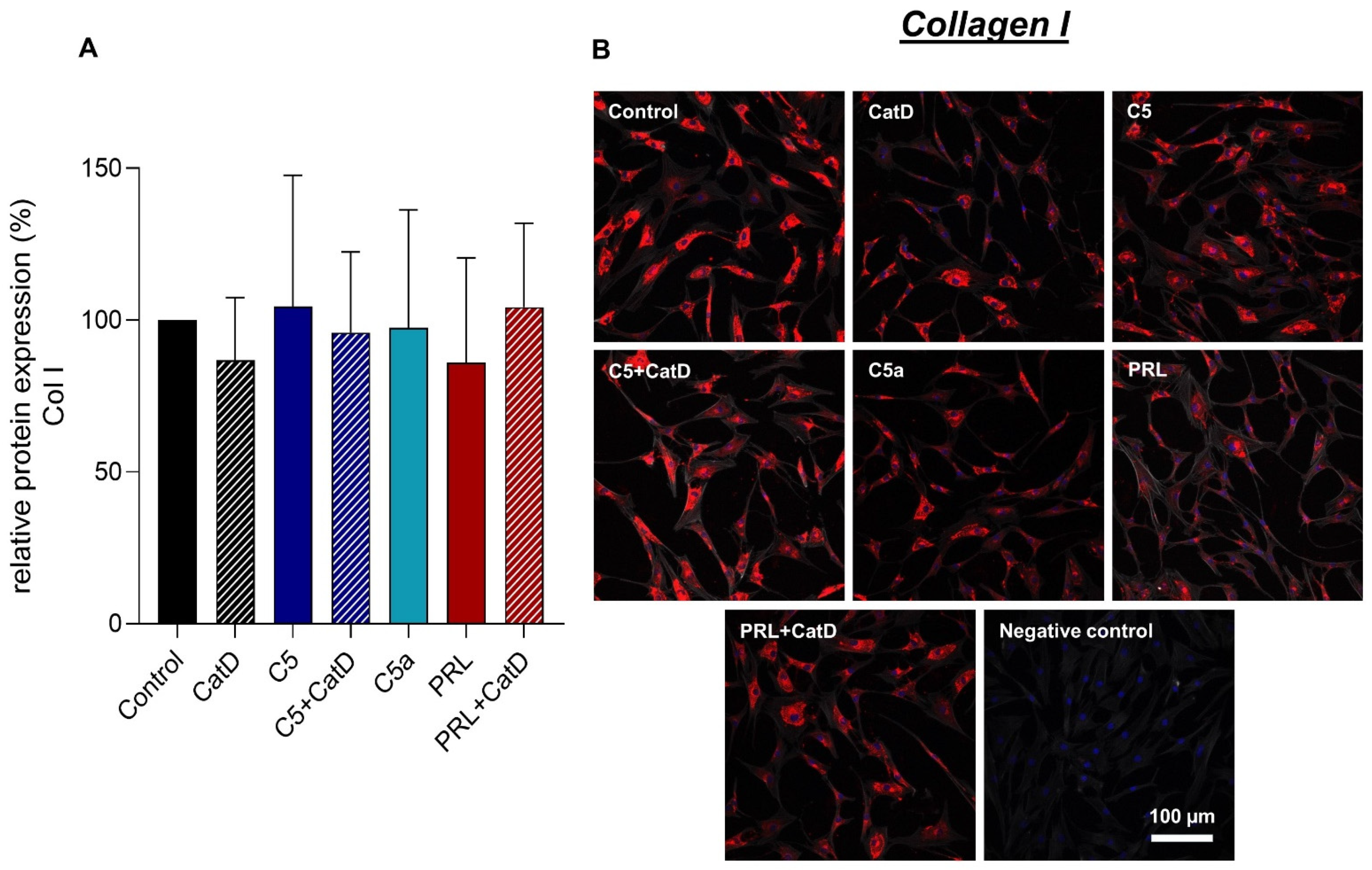
3.5. Protein Synthesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AP | Anaphylatoxin |
| C5aR1 | C5a receptor |
| CatD | Cathepsin D |
| Col I/II | Collagen type I/II |
| CRP | Complement regulatory protein |
| CS | Complement system |
| EDTA | Ethylenediaminetetraacetic acid |
| FCS | Fetal calf serum |
| hAC | Human articular chondrocytes |
| HUVEC | Human umbilical vein endothelial cells |
| MAC | Membrane Attack Complex |
| MMP | Matrix metalloproteinase |
| OA | Osteoarthritis |
| OUMS-27 | Okayama University Medical School-27 |
| PRL | Prolactin |
| PRLR | Prolactin receptor |
References
- Felson, D.T.; Zhang, Y. An update on the epidemiology of knee and hip osteoarthritis with a view to prevention. Arthritis Rheum. 1998, 41, 1343–1355. [Google Scholar] [CrossRef]
- Woolf, A.D.; Pfleger, B. Burden of major musculoskeletal conditions. Bull. World Health Organ. 2003, 81, 646–656. [Google Scholar] [PubMed]
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012, 64, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Sokolove, J.; Lepus, C.M. Role of inflammation in the pathogenesis of osteoarthritis: Latest findings and interpretations. Ther. Adv. Musculoskelet. Dis. 2013, 5, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Martel-Pelletier, J.; Alaaeddine, N.; Pelletier, J.P. Cytokines and their role in the pathophysiology of osteoarthritis. Front. Biosci. 1999, 4, D694–D703. [Google Scholar] [CrossRef]
- Nakamura, H.; Yoshino, S.; Kato, T.; Tsuruha, J.; Nishioka, K. T-cell mediated inflammatory pathway in osteoarthritis. Osteoarthr. Cartil. 1999, 7, 401–402. [Google Scholar] [CrossRef]
- Schulze-Tanzil, G.; Kohl, B.; El Sayed, K.; Arens, S.; Ertel, W.; Stolzel, K.; John, T. Anaphylatoxin receptors and complement regulatory proteins in human articular and non-articular chondrocytes: Interrelation with cytokines. Cell Tissue Res. 2012, 350, 465–475. [Google Scholar] [CrossRef]
- Tetlow, L.C.; Adlam, D.J.; Woolley, D.E. Matrix metalloproteinase and proinflammatory cytokine production by chondrocytes of human osteoarthritic cartilage: Associations with degenerative changes. Arthritis Rheum. 2001, 44, 585–594. [Google Scholar] [CrossRef]
- Silawal, S.; Triebel, J.; Bertsch, T.; Schulze-Tanzil, G. Osteoarthritis and the Complement Cascade. Clin. Med. Insights Arthritis Musculoskelet Disord. 2018, 11, 1179544117751430. [Google Scholar] [CrossRef]
- Chien, S.-Y.; Huang, C.-Y.; Tsai, C.-H.; Wang, S.-W.; Lin, Y.-M.; Tang, C.-H. Interleukin-1β induces fibroblast growth factor 2 expression and subsequently promotes endothelial progenitor cell angiogenesis in chondrocytes. Clin. Sci. 2016, 130, 667–681. [Google Scholar] [CrossRef]
- MacDonald, I.J.; Liu, S.-C.; Su, C.-M.; Wang, Y.-H.; Tsai, C.-H.; Tang, C.-H. Implications of Angiogenesis Involvement in Arthritis. Int. J. Mol. Sci. 2018, 19, 2012. [Google Scholar] [CrossRef] [PubMed]
- Dunkelberger, J.R.; Song, W.C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010, 20, 34–50. [Google Scholar] [CrossRef] [PubMed]
- Krisinger, M.J.; Goebeler, V.; Lu, Z.; Meixner, S.C.; Myles, T.; Pryzdial, E.L.; Conway, E.M. Thrombin generates previously unidentified C5 products that support the terminal complement activation pathway. Blood 2012, 120, 1717–1725. [Google Scholar] [CrossRef] [PubMed]
- Leung, L.L.; Morser, J. Plasmin as a complement C5 convertase. eBioMedicine 2016, 5, 20–21. [Google Scholar] [CrossRef] [PubMed]
- Huber-Lang, M.; Denk, S.; Fulda, S.; Erler, E.; Kalbitz, M.; Weckbach, S.; Schneider, E.M.; Weiss, M.; Kanse, S.M.; Perl, M. Cathepsin D is released after severe tissue trauma in vivo and is capable of generating C5a in vitro. Mol. Immunol. 2012, 50, 60–65. [Google Scholar] [CrossRef]
- Wang, Q.; Rozelle, A.L.; Lepus, C.M.; Scanzello, C.R.; Song, J.J.; Larsen, D.M.; Crish, J.F.; Bebek, G.; Ritter, S.Y.; Lindstrom, T.M.; et al. Identification of a central role for complement in osteoarthritis. Nat. Med. 2011, 17, 1674–1679. [Google Scholar] [CrossRef]
- Walsh, D.A. Angiogenesis and arthritis. Rheumatology 1999, 38, 103–112. [Google Scholar] [CrossRef]
- Nozaki, M.; Raisler, B.J.; Sakurai, E.; Sarma, J.V.; Barnum, S.R.; Lambris, J.D.; Chen, Y.; Zhang, K.; Ambati, B.K.; Baffi, J.Z.; et al. Drusen complement components C3a and C5a promote choroidal neovascularization. Proc. Natl. Acad. Sci. USA 2006, 103, 2328–2333. [Google Scholar] [CrossRef]
- Kahr, W.H.A. Complement halts angiogenesis gone wild. Blood 2010, 116, 4393–4394. [Google Scholar] [CrossRef]
- Ben-Jonathan, N.; Mershon, J.L.; Allen, D.L.; Steinmetz, R.W. Extrapituitary prolactin: Distribution, regulation, functions, and clinical aspects. Endocr. Rev. 1996, 17, 639–669. [Google Scholar] [CrossRef]
- Freeman, M.E.; Kanyicska, B.; Lerant, A.; Nagy, G. Prolactin: Structure, function, and regulation of secretion. Physiol. Rev. 2000, 80, 1523–1631. [Google Scholar] [CrossRef] [PubMed]
- Ogueta, S.; Muñoz, J.; Obregon, E.; Delgado-Baeza, E.; García-Ruiz, J.P. Prolactin is a component of the human synovial liquid and modulates the growth and chondrogenic differentiation of bone marrow-derived mesenchymal stem cells. Mol. Cell. Endocrinol. 2002, 190, 51–63. [Google Scholar] [CrossRef]
- Zermeno, C.; Guzman-Morales, J.; Macotela, Y.; Nava, G.; Lopez-Barrera, F.; Kouri, J.B.; Lavalle, C.; de la Escalera, G.M.; Clapp, C. Prolactin inhibits the apoptosis of chondrocytes induced by serum starvation. J. Endocrinol. 2006, 189, R1–R8. [Google Scholar] [CrossRef][Green Version]
- Seriwatanachai, D.; Krishnamra, N.; Charoenphandhu, N. Chondroregulatory action of prolactin on proliferation and differentiation of mouse chondrogenic ATDC5 cells in 3-dimensional micromass cultures. Biochem. Biophys. Res. Commun. 2012, 420, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Piwnica, D.; Touraine, P.; Struman, I.; Tabruyn, S.; Bolbach, G.; Clapp, C.; Martial, J.A.; Kelly, P.A.; Goffin, V. Cathepsin D processes human prolactin into multiple 16K-like N-terminal fragments: Study of their antiangiogenic properties and physiological relevance. Mol. Endocrinol. 2004, 18, 2522–2542. [Google Scholar] [CrossRef]
- Macotela, Y.; Aguilar, M.B.; Guzman-Morales, J.; Rivera, J.C.; Zermeno, C.; Lopez-Barrera, F.; Nava, G.; Lavalle, C.; Martinez de la Escalera, G.; Clapp, C. Matrix metalloproteases from chondrocytes generate an antiangiogenic 16 kDa prolactin. J. Cell Sci. 2006, 119, 1790–1800. [Google Scholar] [CrossRef]
- Ge, G.; Fernández, C.A.; Moses, M.A.; Greenspan, D.S. Bone morphogenetic protein 1 processes prolactin to a 17-kDa antiangiogenic factor. Proc. Natl. Acad. Sci. USA 2007, 104, 10010–10015. [Google Scholar] [CrossRef]
- Ortiz, G.; Ledesma-Colunga, M.G.; Wu, Z.; García-Rodrigo, J.F.; Adan, N.; Martínez de la Escalera, G.; Clapp, C. Vasoinhibin reduces joint inflammation, bone loss, and the angiogenesis and vasopermeability of the pannus in murine antigen-induced arthritis. Lab. Investig. 2020, 100, 1068–1079. [Google Scholar] [CrossRef]
- Clapp, C.; Thebault, S.; Macotela, Y.; Moreno-Carranza, B.; Triebel, J.; Martinez de la Escalera, G. Regulation of blood vessels by prolactin and vasoinhibins. Adv. Exp. Med. Biol. 2015, 846, 83–95. [Google Scholar] [CrossRef]
- Barrett, A.J. Cathepsin D: The lysosomal aspartic proteinase. Ciba Found. Symp. 1979, 75, 37–50. [Google Scholar]
- Lkhider, M.; Castino, R.; Bouguyon, E.; Isidoro, C.; Ollivier-Bousquet, M. Cathepsin D released by lactating rat mammary epithelial cells is involved in prolactin cleavage under physiological conditions. J. Cell Sci. 2004, 117, 5155–5164. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Handley, C.J.; Mok, M.T.; Ilic, M.Z.; Adcocks, C.; Buttle, D.J.; Robinson, H.C. Cathepsin D cleaves aggrecan at unique sites within the interglobular domain and chondroitin sulfate attachment regions that are also cleaved when cartilage is maintained at acid pH. Matrix Biol. 2001, 20, 543–553. [Google Scholar] [CrossRef]
- Troeberg, L.; Nagase, H. Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim. Et Biophys. Acta 2012, 1824, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Olszewska-Slonina, D.; Jung, S.; Matewski, D.; Olszewski, K.J.; Krzyzynska-Malinowska, E.; Braszkiewicz, A.; Kowaliszyn, B. Lysosomal enzymes in serum and synovial fluid in patients with osteoarthritis. Scand. J. Clin. Lab. Investig. 2015, 75, 145–151. [Google Scholar] [CrossRef]
- Sapolsky, A.I.; Altman, R.D.; Woessner, J.F.; Howell, D.S. The action of cathepsin D in human articular cartilage on proteoglycans. J. Clin. Investig. 1973, 52, 624–633. [Google Scholar] [CrossRef][Green Version]
- Rochefort, H.; Liaudet-Coopman, E. Cathepsin D in cancer metastasis: A protease and a ligand. APMIS 1999, 107, 86–95. [Google Scholar] [CrossRef]
- Fusek, M.; Vetvicka, V. Dual role of cathepsin D: Ligand and protease. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czechoslov. 2005, 149, 43–50. [Google Scholar] [CrossRef]
- Minarowska, A.; Minarowski, L.; Karwowska, A.; Gacko, M. Regulatory role of cathepsin D in apoptosis. Folia Histochem. Cytobiol. 2007, 45, 159–163. [Google Scholar]
- Pfaffl, M.W. Quantification strategies in real-time PCR. AZ Quant. PCR 2004, 1, 89–113. [Google Scholar]
- Jensen, E.C. Quantitative Analysis of Histological Staining and Fluorescence Using ImageJ. Anat. Rec. 2013, 296, 378–381. [Google Scholar] [CrossRef]
- Van Coppenolle, F.; Skryma, R.; Ouadid-Ahidouch, H.; Slomianny, C.; Roudbaraki, M.; Delcourt, P.; Dewailly, E.; Humez, S.; Crépin, A.; Gourdou, I.; et al. Prolactin stimulates cell proliferation through a long form of prolactin receptor and K+ channel activation. Biochem. J. 2004, 377, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Yu-Lee, L.Y. Prolactin modulation of immune and inflammatory responses. Recent Prog. Horm. Res. 2002, 57, 435–455. [Google Scholar] [CrossRef] [PubMed]
- Ben--Jonathan, N.; Liby, K.; McFarland, M.; Zinger, M. Prolactin as an autocrine/paracrine growth factor in human cancer. Trends Endocrinol. Metab. TEM 2002, 13, 245–250. [Google Scholar] [CrossRef]
- Alkharusi, A.; Lesma, E.; Ancona, S.; Chiaramonte, E.; Nyström, T.; Gorio, A.; Norstedt, G. Role of Prolactin Receptors in Lymphangioleiomyomatosis. PLoS ONE 2016, 11, e0146653. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, S.; Walsh, D.A. Angiogenesis in osteoarthritis. Curr. Opin. Rheumatol. 2008, 20, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Reuwer, A.Q.; Nowak-Sliwinska, P.; Mans, L.A.; van der Loos, C.M.; von der Thüsen, J.H.; Twickler, M.T.B.; Spek, C.A.; Goffin, V.; Griffioen, A.W.; Borensztajn, K.S. Functional consequences of prolactin signalling in endothelial cells: A potential link with angiogenesis in pathophysiology? J. Cell. Mol. Med. 2012, 16, 2035–2048. [Google Scholar] [CrossRef]
- Kurihara, R.; Yamaoka, K.; Sawamukai, N.; Shimajiri, S.; Oshita, K.; Yukawa, S.; Tokunaga, M.; Iwata, S.; Saito, K.; Chiba, K.; et al. C5a promotes migration, proliferation, and vessel formation in endothelial cells. Inflamm. Res. 2010, 59, 659–666. [Google Scholar] [CrossRef]
- Huber-Lang, M.; Ekdahl, K.N.; Wiegner, R.; Fromell, K.; Nilsson, B. Auxiliary activation of the complement system and its importance for the pathophysiology of clinical conditions. Semin. Immunopathol. 2018, 40, 87–102. [Google Scholar] [CrossRef]
- Glondu, M.; Coopman, P.; Laurent-Matha, V.; Garcia, M.; Rochefort, H.; Liaudet-Coopman, E. A mutated cathepsin-D devoid of its catalytic activity stimulates the growth of cancer cells. Oncogene 2001, 20, 6920–6929. [Google Scholar] [CrossRef]
- Laurent-Matha, V.; Maruani-Herrmann, S.; Prebois, C.; Beaujouin, M.; Glondu, M.; Noel, A.; Alvarez-Gonzalez, M.L.; Blacher, S.; Coopman, P.; Baghdiguian, S.; et al. Catalytically inactive human cathepsin D triggers fibroblast invasive growth. J. Cell Biol. 2005, 168, 489–499. [Google Scholar] [CrossRef]
- Pranjol, M.Z.I.; Gutowski, N.J.; Hannemann, M.; Whatmore, J.L. Cathepsin D non-proteolytically induces proliferation and migration in human omental microvascular endothelial cells via activation of the ERK1/2 and PI3K/AKT pathways. Biochim. Biophys. Et Acta (BBA) Mol. Cell Res. 2018, 1865, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Silawal, S.; Kohl, B.; Shi, J.; Schulze-Tanzil, G. Complement Regulation in Human Tenocytes under the Influence of Anaphylatoxin C5a. Int. J. Mol. Sci. 2021, 22, 3105. [Google Scholar] [CrossRef] [PubMed]
- Shlopov, B.V.; Lie, W.R.; Mainardi, C.L.; Cole, A.A.; Chubinskaya, S.; Hasty, K.A. Osteoarthritic lesions: Involvement of three different collagenases. Arthritis Rheum. 1997, 40, 2065–2074. [Google Scholar] [CrossRef] [PubMed]

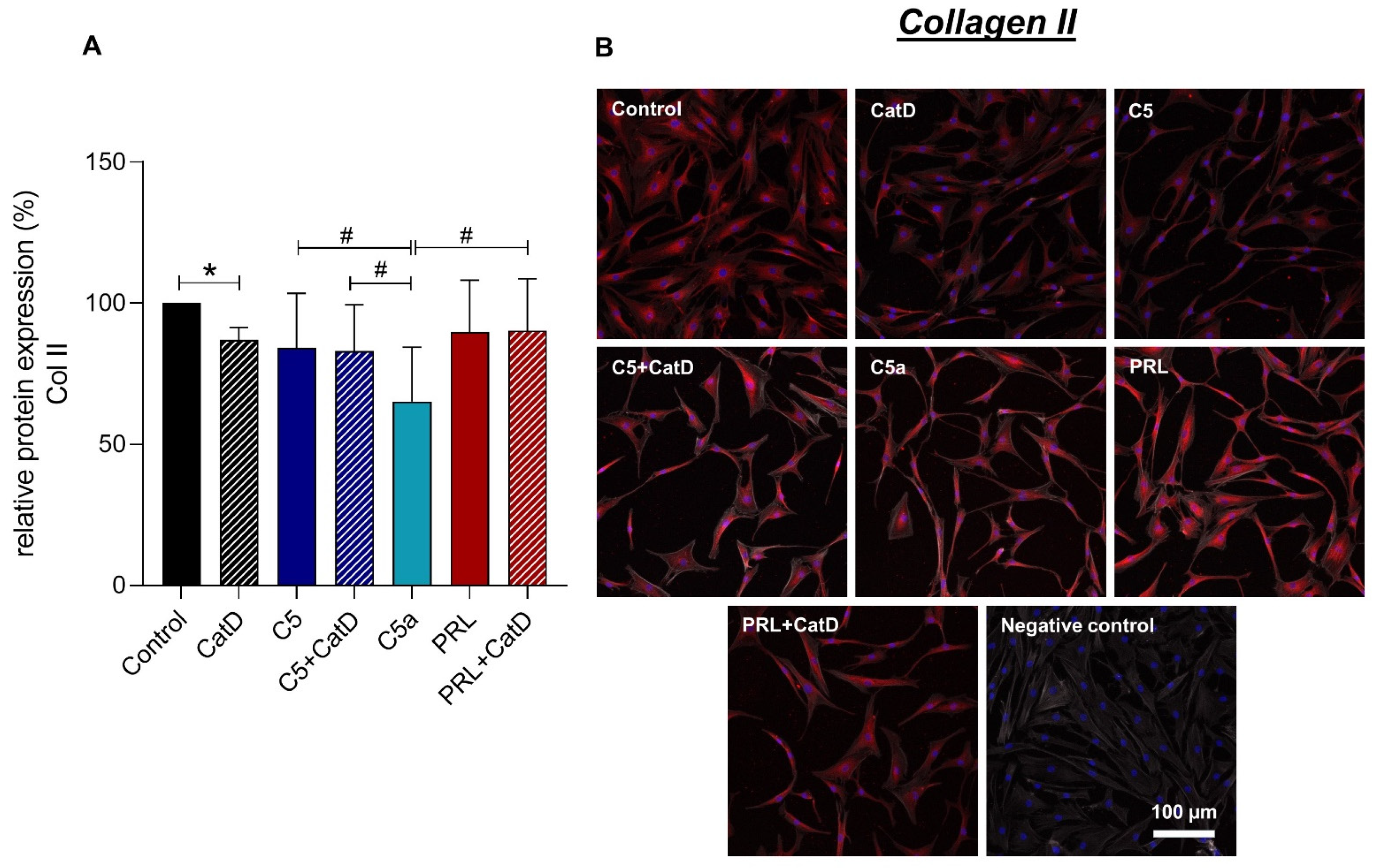
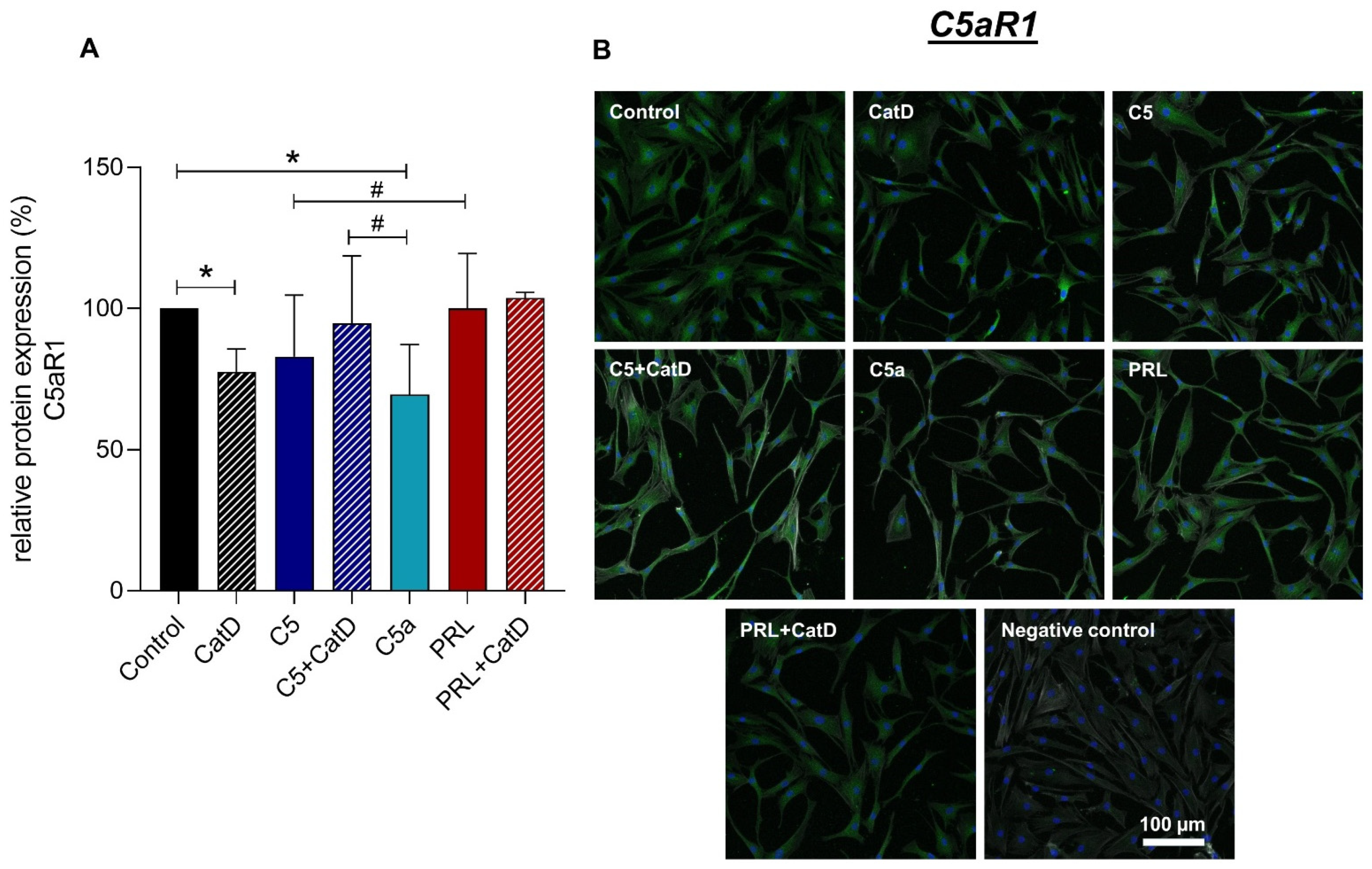
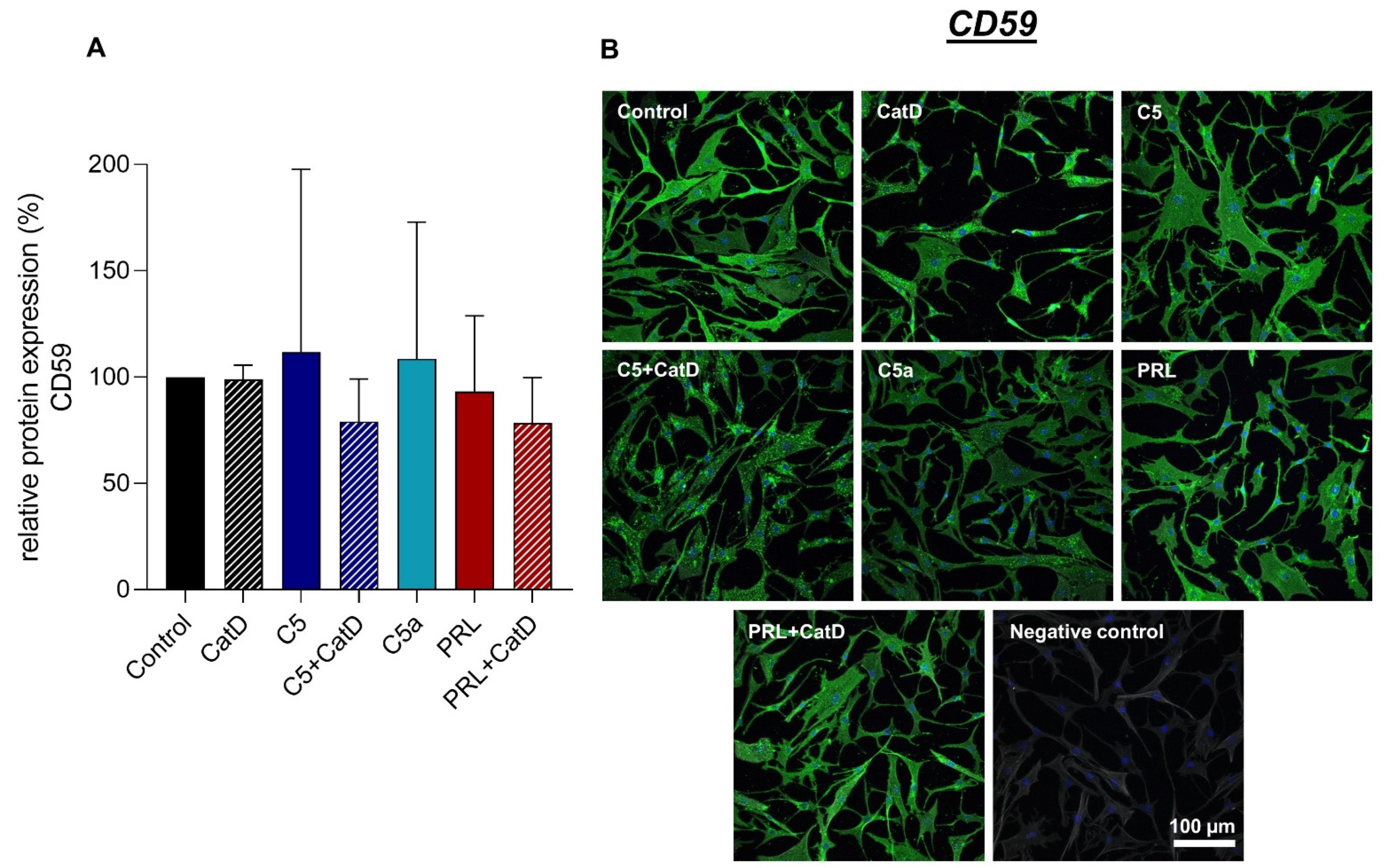
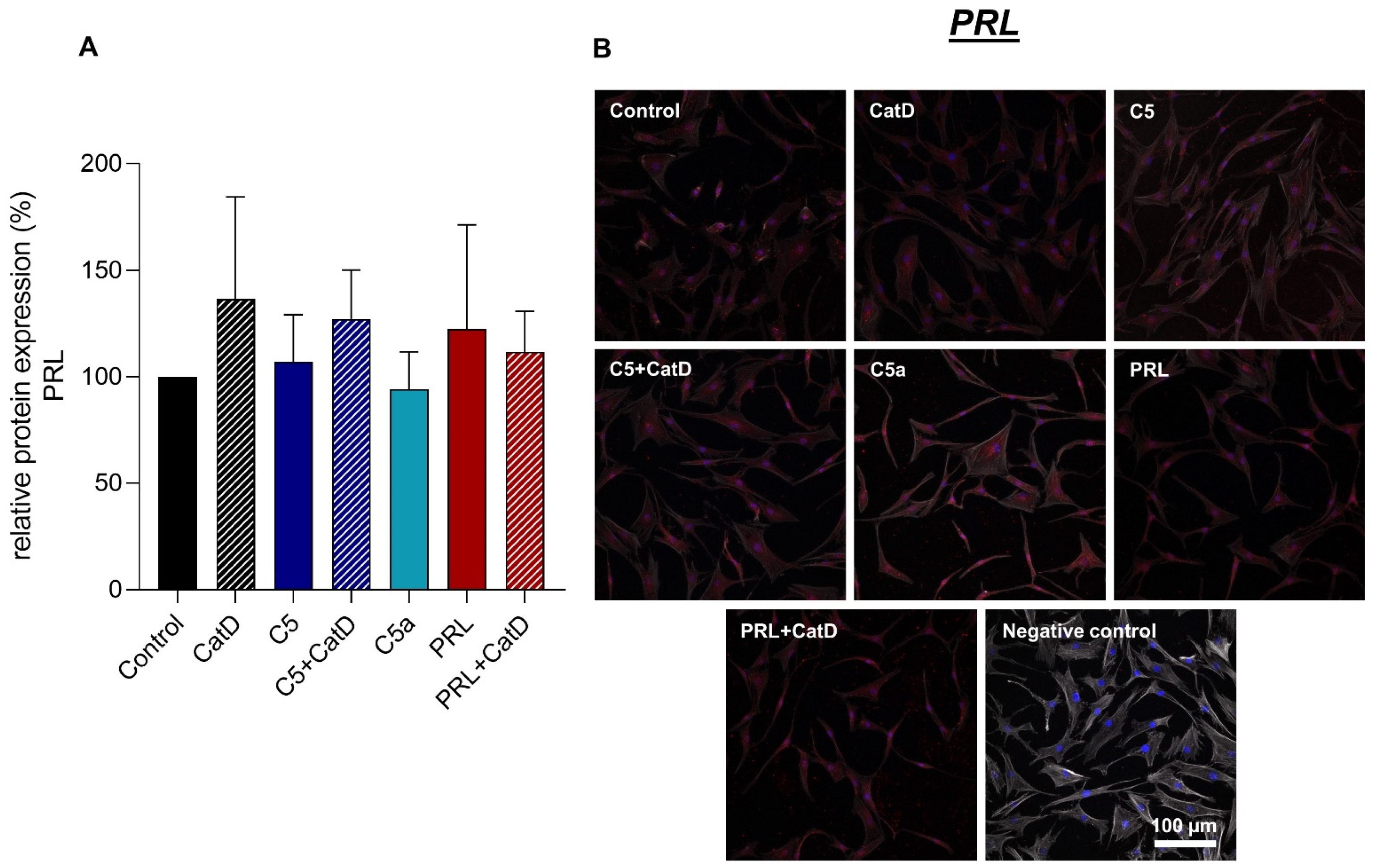
| Recombinant Proteins | Assay-ID | Company | Amplicon Length |
|---|---|---|---|
| HPRT | Hs99999909_m1 | Applied Biosystems, Foster City, CA, USA | 100 |
| C5aR1 | Hs00704891_s1 | 68 | |
| C5 | Hs01004342_m1 | 66 | |
| CD59 | Hs00174141_m1 | 70 | |
| PRL | Hs00168730_m1 | 106 | |
| PRLR | Hs01061477_m1 | 107 | |
| CatD | Hs00157205_m1 | 103 | |
| MMP-13 | Hs00233992_m1 | 91 |
| Primary Antibodies | Company | Type | Catalogue Number |
| Col1 | SouthernBiotech, Birmingham, AL, USA | Goat-anti-human | 1310-01 |
| Col2 | Origene Technologies, Rockville, MD, USA | Rabbit-anti-human | R1039X |
| C5aR1 | GeneTex, Eching, Germany | Mouse-anti-human | GTX74845 |
| CD59 | Bio-Rad, Feldkirchen, Germany | Mouse-anti-human | MCA1054GA |
| PRL | Abbexa, Cambridge, UK | Rabbit-anti-human | abx100284 |
| Secondary Antibodies | Company | Type | |
| Cy3 | Dianova GmbH, Hamburg, Germany | Donkey-anti-goat | 705-165-147 |
| Alexa Fluor 555 | Life Technologies Corp., Carlsbad, CA, USA | Donkey-anti-rabbit | A-31572 |
| Alexa Fluor 488 | Invitrogen, Waltham, MA, USA | Donkey-anti-mouse | A21202 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silawal, S.; Wagner, M.; Roth, D.; Bertsch, T.; Schwarz, S.; Willauschus, M.; Gesslein, M.; Triebel, J.; Schulze-Tanzil, G. Complement Proteins C5/C5a, Cathepsin D and Prolactin in Chondrocytes: A Possible Crosstalk in the Pathogenesis of Osteoarthritis. Cells 2022, 11, 1134. https://doi.org/10.3390/cells11071134
Silawal S, Wagner M, Roth D, Bertsch T, Schwarz S, Willauschus M, Gesslein M, Triebel J, Schulze-Tanzil G. Complement Proteins C5/C5a, Cathepsin D and Prolactin in Chondrocytes: A Possible Crosstalk in the Pathogenesis of Osteoarthritis. Cells. 2022; 11(7):1134. https://doi.org/10.3390/cells11071134
Chicago/Turabian StyleSilawal, Sandeep, Miriam Wagner, Dominik Roth, Thomas Bertsch, Silke Schwarz, Maximilian Willauschus, Markus Gesslein, Jakob Triebel, and Gundula Schulze-Tanzil. 2022. "Complement Proteins C5/C5a, Cathepsin D and Prolactin in Chondrocytes: A Possible Crosstalk in the Pathogenesis of Osteoarthritis" Cells 11, no. 7: 1134. https://doi.org/10.3390/cells11071134
APA StyleSilawal, S., Wagner, M., Roth, D., Bertsch, T., Schwarz, S., Willauschus, M., Gesslein, M., Triebel, J., & Schulze-Tanzil, G. (2022). Complement Proteins C5/C5a, Cathepsin D and Prolactin in Chondrocytes: A Possible Crosstalk in the Pathogenesis of Osteoarthritis. Cells, 11(7), 1134. https://doi.org/10.3390/cells11071134