Tph2 Gene Expression Defines Ethanol Drinking Behavior in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Behavioral Studies
2.2.1. Drugs
2.2.2. Two-Bottle Free-Choice Ethanol Consumption
2.2.3. Forced Swim Test (FST)
2.3. Molecular Analyses
2.3.1. Tph2 Gene Expression Analysis in “High Ethanol-Drinking” and “Low Ethanol-Drinking” Mice
2.3.2. Sample Preparation
2.3.3. Real-Time PCR
2.4. Statistical Analyses
3. Results
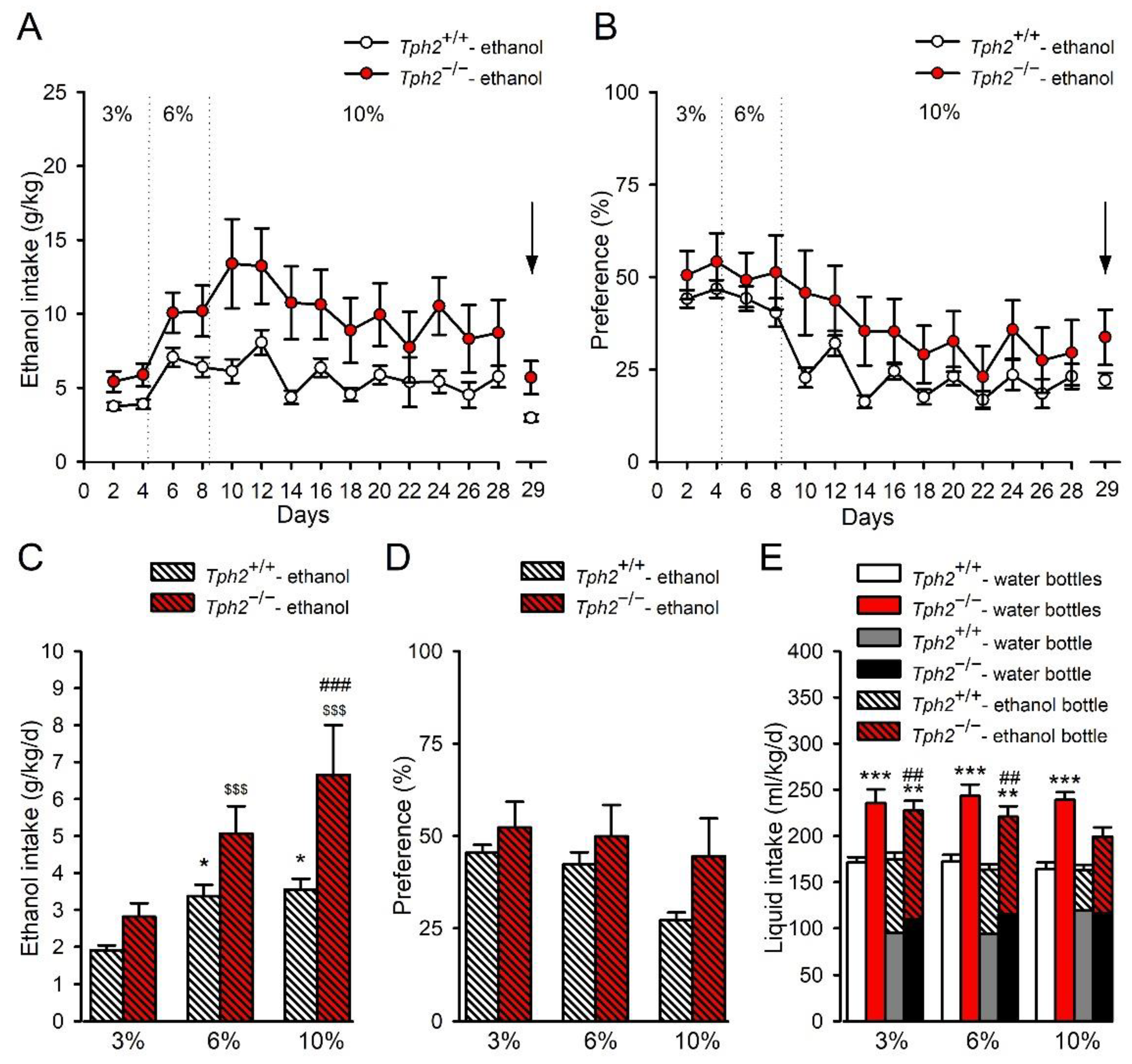
3.1. Tph2 Deficiency Leads to Increased Ethanol Consumption in Mice
3.2. Ethanol Exposure in Tph2−/− Mice Induces Antidepressant Effects
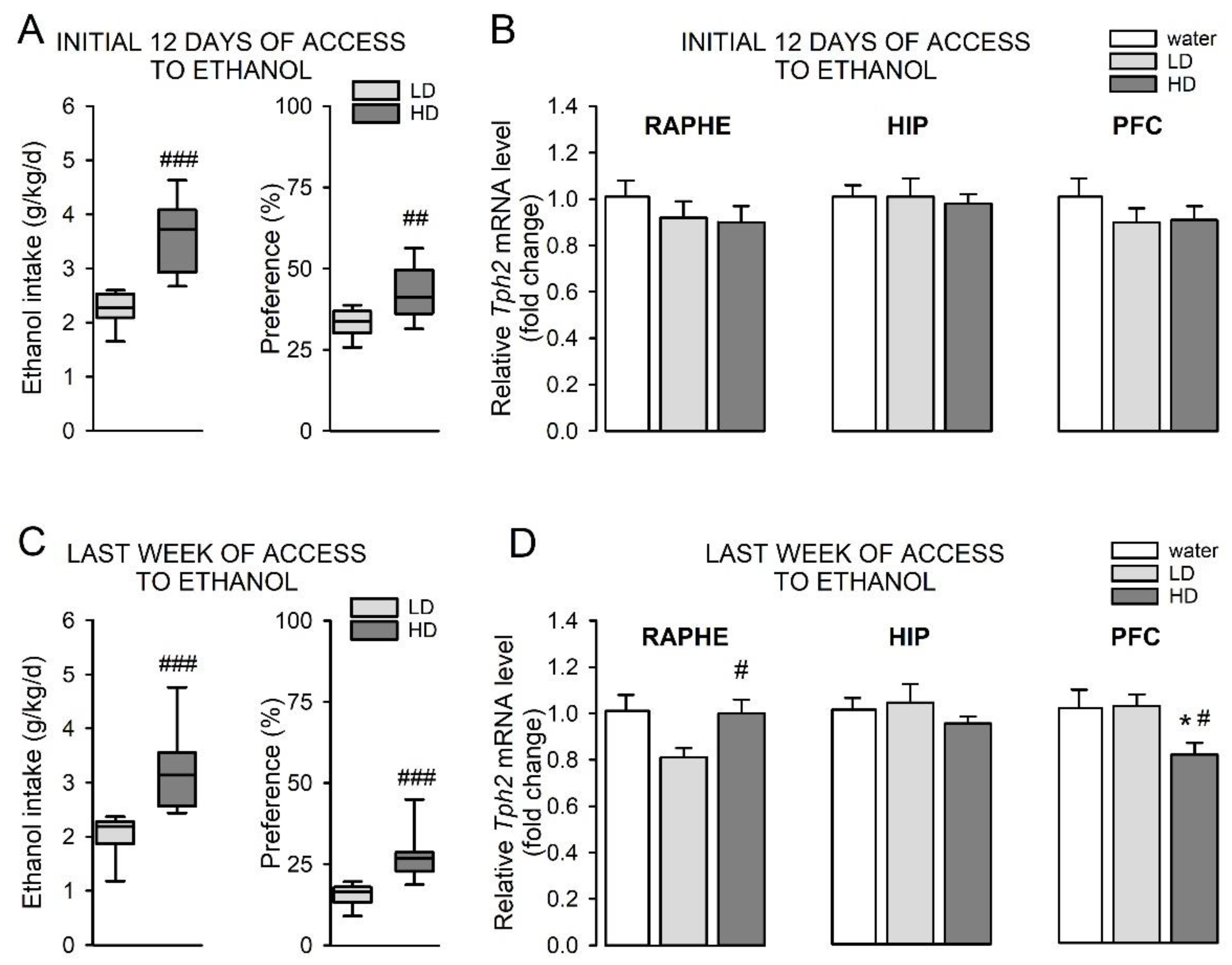
3.3. Wild-Type “High Ethanol-Drinking” Mice Display Alterations in Tph2 Expression Pattern Compared to “Low Ethanol-Drinking” Animals
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crowley, P. Long-term drug treatment of patients with alcohol dependence. Aust. Prescr. 2015, 38, 41. [Google Scholar] [CrossRef] [PubMed]
- Morley, K.; Perry, C.; Watt, J.; Hurzeler, T.; Leggio, L.; Lawrence, A.; Haber, P. New approved and emerging pharmacological approaches to alcohol use disorder: A review of clinical studies. Expert Opin. Pharmacother. 2021, 22, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Shield, K.D.; Parry, C.; Rehm, J. Chronic Diseases and Conditions Related to Alcohol Use. Alcohol Res. Curr. Rev. 2014, 35, 155. [Google Scholar]
- Polimanti, R.; Peterson, R.; Ong, J.; MacGregor, S.; Edwards, A.; Clarke, T.; Frank, J.; Gerring, Z.; Gillespie, N.; Lind, P.; et al. Evidence of causal effect of major depression on alcohol dependence: Findings from the psychiatric genomics consortium. Psychol. Med. 2019, 49, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- Boden, J.; Fergusson, D. Alcohol and depression. Addiction 2011, 106, 906–914. [Google Scholar] [CrossRef]
- Ruhé, H.G.; Mason, N.S.; Schene, A.H. Mood is indirectly related to serotonin, norepinephrine and dopamine levels in humans: A meta-analysis of monoamine depletion studies. Mol. Psychiatry 2007, 12, 331–359. [Google Scholar] [CrossRef] [PubMed]
- Marcinkiewcz, C. Serotonergic Systems in the Pathophysiology of Ethanol Dependence: Relevance to Clinical Alcoholism. ACS Chem. Neurosci. 2015, 6, 1026–1039. [Google Scholar] [CrossRef]
- Buydens-Branchey, L.; Branchey, M.; Noumair, D.; Lieber, C. Age of alcoholism onset. II. Relationship to susceptibility to serotonin precursor availability. Arch. Gen. Psychiatry 1989, 46, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Patkar, A.A.; Gopalakrishnan, R.; Naik, P.C.; Murray, H.W.; Vergare, M.J.; Marsden, C.A. Changes in plasma noradrenaline and serotonin levels and craving during alcohol withdrawal. Alcohol Alcohol. 2003, 38, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Ballenger, J.C.; Goodwin, F.K.; Major, L.F.; Brown, G.L. Alcohol and Central Serotonin Metabolism in Man. Arch. Gen. Psychiatry 1979, 36, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, N.E.; Davenport, Y.; Cowdry, R.W.; Webster, M.H.; Goodwin, F.K. Monoamine metabolites in cerebrospinal fluid of depressive subgroups. Psychiatry Res. 1980, 2, 113–119. [Google Scholar] [CrossRef]
- Dougherty, D.M.; Marsh, D.M.; Mathias, C.W.; Dawes, M.A.; Bradley, D.M.; Morgan, C.J.; Badawy, A.A.B. The effects of alcohol on laboratory-measured impulsivity after L-Tryptophan depletion or loading. Psychopharmacology 2007, 193, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Petrakis, I.L.; Buonopane, A.; O’Malley, S.; Cermik, O.; Trevisan, L.; Boutros, N.N.; Limoncelli, D.; Krystal, J.H. The effect of tryptophan depletion on alcohol self-administration in non-treatment-seeking alcoholic individuals. Alcohol. Clin. Exp. Res. 2002, 26, 969–975. [Google Scholar] [CrossRef] [PubMed]
- Petrakis, I.; Trevisan, L.; Boutros, N.; Limoncelli, D.; Cooney, N.; Krysta, J. Effect of tryptophan depletion on alcohol cue-induced craving in abstinent alcoholic patients. Alcohol Clin. Exp. Res. 2001, 25, 1151–1155. [Google Scholar] [CrossRef]
- Kranzler, H.R.; Armeli, S.; Tennen, H.; Covault, J.; Feinn, R.; Arias, A.J.; Pettinati, H.; Oncken, C. A double-blind, randomized trial of sertraline for alcohol dependence: Moderation by age and 5-hydroxytryptamine transporter-linked promoter region genotype. J. Clin. Psychopharmacol. 2011, 31, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Cornelius, J.; Salloum, I.; Cornelius, M.; Perel, J.; Ehler, J.; Jarrett, P.; Levin, R.; Black, A.; Mann, J. Preliminary report: Double-blind, placebo-controlled study of fluoxetine in depressed alcoholics. Psychopharmacol. Bull. 1995, 31, 297–303. [Google Scholar] [PubMed]
- Agabio, R.; Trogu, E.; Pani, P.P. Antidepressants for the treatment of people with co-occurring depression and alcohol dependence. Cochrane Database Syst. Rev. 2018, 4. [Google Scholar] [CrossRef]
- Gorelick, D.A. Serotonin uptake blockers and the treatment of alcoholism. Recent Dev. Alcohol. 1989, 7, 267–281. [Google Scholar]
- Babor, T.; Hofmann, M.; DelBoca, F.; Hesselbrock, V.; Meyer, R.; Dolinsky, Z.; Rounsaville, B. Types of alcoholics, I. Evidence for an empirically derived typology based on indicators of vulnerability and severity. Arch. Gen. Psychiatry 1992, 49, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Pettinati, H.; Volpicelli, J.; Kranzler, H.; Luck, G.; Rukstalis, M.; Cnaan, A. Sertraline treatment for alcohol dependence: Interactive effects of medication and alcoholic subtype. Alcohol. Clin. Exp. Res. 2000, 24, 1041–1049. [Google Scholar] [CrossRef] [PubMed]
- Dundon, W.; Lynch, K.; Pettinati, H.; Lipkin, C. Treatment outcomes in type A and B alcohol dependence 6 months after serotonergic pharmacotherapy. Alcohol. Clin. Exp. Res. 2004, 28, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Kranzler, H.; Burleson, J.; Brown, J.; Babor, T. Fluoxetine treatment seems to reduce the beneficial effects of cognitive-behavioral therapy in type B alcoholics. Alcohol. Clin. Exp. Res. 1996, 20, 1534–1541. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.; McBride, W.; Lumeng, L.; Li, T. Regional brain levels of monoamines in alcohol-preferring and -nonpreferring lines of rats. Pharmacol. Biochem. Behav. 1982, 16, 145–149. [Google Scholar] [CrossRef]
- Ahtee, L.; Eriksson, K. 5-Hydroxytryptamine and 5-hydroxyindolylacetic acid content in brain of rat strains selected for their alcohol intake. Physiol. Behav. 1972, 8, 123–126. [Google Scholar] [CrossRef]
- Aulakh, C.; Tolliver, T.; Wozniak, K.; Hill, J.; Murphy, D. Functional and biochemical evidence for altered serotonergic function in the fawn-hooded rat strain. Pharmacol. Biochem. Behav. 1994, 49, 615–620. [Google Scholar] [CrossRef]
- Casu, M.; Pisu, C.; Lobina, C.; Pani, L. Immunocytochemical study of the forebrain serotonergic innervation in Sardinian alcohol-preferring rats. Psychopharmacology 2004, 172, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Weiss, F. Ethanol exposure differentially alters central monoamine neurotransmission in alcohol-preferring versus -nonpreferring rats. J. Pharm. Exp. Ther. 1999, 288, 1223–1228. [Google Scholar]
- Jessa, M.; Krzaścik, P.; Kostowski, W. Neonatal treatment with 5,7-dihydroxytryptamine induces decrease in alcohol drinking in adult animals. Pol. J. Pharmacol. 2001, 53, 109–116. [Google Scholar] [PubMed]
- Myers, R.; Melchior, C. Alcohol drinking in the rat after destruction of serotonergic and catecholaminergic neurons in the brain. Res. Commun. Chem. Pathol. Pharmacol. 1975, 10, 363–378. [Google Scholar] [PubMed]
- Vasudeva, R.K.; Hobby, A.R.; Kirby, L.G. Ethanol consumption in the Sprague-Dawley rat increases sensitivity of the dorsal raphe nucleus to 5,7-dihydroxytryptamine. Behav. Brain Res. 2015, 295, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Adell, A.; Myers, R. Selective destruction of midbrain raphe nuclei by 5,7-DHT: Is brain 5-HT involved in alcohol drinking in Sprague-Dawley rats? Brain Res. 1995, 693, 70–79. [Google Scholar] [CrossRef]
- Wang, J.; Shum, A.; Lin, T.; Wang, Y. Central serotonergic lesions increase voluntary alcohol consumption in Sprague Dawley rats: Moderation by long-term ethanol administration. Alcohol. Clin. Exp. Res. 1996, 20, 1252–1259. [Google Scholar] [CrossRef] [PubMed]
- Baumgarten, H.; Lachenmayer, L. 5,7-dihydroxytryptamine: Improvement in chemical lesioning of indoleamine neurons in the mammalian brain. Z. Zellforsch. Mikrosk. Anat. 1972, 135, 399–414. [Google Scholar] [CrossRef] [PubMed]
- Neal, K.B.; Parry, L.J.; Bornstein, J.C. Strain-specific genetics, anatomy and function of enteric neural serotonergic pathways in inbred mice. J. Physiol. 2009, 587, 567–586. [Google Scholar] [CrossRef] [PubMed]
- Walther, D.J.; Peter, J.U.; Bashammakh, S.; Hörtnagl, H.; Voits, M.; Fink, H.; Bader, M. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science 2003, 299, 76. [Google Scholar] [CrossRef] [PubMed]
- Gutknecht, L.; Kriegebaum, C.; Waider, J.; Schmitt, A.; Lesch, K. Spatio-temporal expression of tryptophan hydroxylase isoforms in murine and human brain: Convergent data from Tph2 knockout mice. Eur. Neuropsychopharmacol. 2009, 19, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Bach, H.; Arango, V.; Kassir, S.A.; Tsaava, T.; Dwork, A.J.; Mann, J.J.; Underwood, M.D. Alcoholics have more tryptophan hydroxylase 2 mRNA and protein in the dorsal and median raphe nuclei. Alcohol. Clin. Exp. Res. 2014, 38, 1894–1901. [Google Scholar] [CrossRef]
- Underwood, M.; Mann, J.; Arango, V. Morphometry of dorsal raphe nucleus serotonergic neurons in alcoholism. Alcohol. Clin. Exp. Res. 2007, 31, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Bonkale, W.; Turecki, G.; Austin, M. Increased tryptophan hydroxylase immunoreactivity in the dorsal raphe nucleus of alcohol-dependent, depressed suicide subjects is restricted to the dorsal subnucleus. Synapse 2006, 60, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gainetdinov, R.; Beaulieu, J.; Sotnikova, T.; Burch, L.; Williams, R.; Schwartz, D.; Krishnan, K.; Caron, M. Loss-of-function mutation in tryptophan hydroxylase-2 identified in unipolar major depression. Neuron 2005, 45, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Matthes, S.; Mosienko, V.; Bashammakh, S.; Alenina, N.; Bader, M. Tryptophan Hydroxylase as Novel Target for the Treatment of Depressive Disorders. Pharmacology 2010, 85, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Lynskey, M.; Todorov, A.; Schrage, A.; Littlefield, A.; Grant, J.; Zhu, Q.; Nelson, E.; Madden, P.; Bucholz, K.; et al. A candidate gene association study of alcohol consumption in young women. Alcohol. Clin. Exp. Res. 2011, 35, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Plemenitaš, A.; Kores Plesničar, B.; Kastelic, M.; Porcelli, S.; Serretti, A.; Dolžan, V. Genetic variability in tryptophan hydroxylase 2 gene in alcohol dependence and alcohol-related psychopathological symptoms. Neurosci. Lett. 2015, 604, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Zill, P.; Preuss, U.; Koller, G.; Bondy, B.; Soyka, M. SNP- and haplotype analysis of the tryptophan hydroxylase 2 gene in alcohol-dependent patients and alcohol-related suicide. Neuropsychopharmacology 2007, 32, 1687–1694. [Google Scholar] [CrossRef] [PubMed]
- Gacek, P.; Conner, T.; Tennen, H.; Kranzler, H.; Covault, J. Tryptophan hydroxylase 2 gene and alcohol use among college students. Addict. Biol. 2008, 13, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, J.; Siesser, W.; Sachs, B.; Peterson, S.; Cools, M.; Setola, V.; Folgering, J.; Flik, G.; Caron, M. Deficient serotonin neurotransmission and depression-like serotonin biomarker alterations in tryptophan hydroxylase 2 (Tph2) loss-of-function mice. Mol. Psychiatry 2012, 17, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Sachs, B.D.; Rodriguiz, R.M.; Siesser, W.B.; Kenan, A.; Royer, E.L.; Jacobsen, J.P.R.; Wetsel, W.C.; Caron, M.G. The effects of brain serotonin deficiency on behavioural disinhibition and anxiety-like behaviour following mild early life stress. Int. J. Neuropsychopharmacol. 2013, 16, 2081–2094. [Google Scholar] [CrossRef]
- Beaulieu, J.M.; Zhang, X.; Rodriguiz, R.M.; Sotnikova, T.D.; Cools, M.J.; Wetsel, W.C.; Gainetdinov, R.R.; Caron, M.G. Role of GSK3 beta in behavioral abnormalities induced by serotonin deficiency. Proc. Natl. Acad. Sci. USA 2008, 105, 1333–1338. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, J.; Illiano, P.; Sotnikova, T.D.; Gainetdinov, R.R.; Beaulieu, J.M.; Hébert, M. The electroretinogram as a biomarker of central dopamine and serotonin: Potential relevance to psychiatric disorders. Biol. Psychiatry 2014, 75, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Sachs, B.; Salahi, A.; Caron, M. Congenital brain serotonin deficiency leads to reduced ethanol sensitivity and increased ethanol consumption in mice. Neuropharmacology 2014, 77, 177–184. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lemay, F.; Doré, F.; Beaulieu, J. Increased ethanol consumption despite taste aversion in mice with a human tryptophan hydroxylase 2 loss of function mutation. Neurosci. Lett. 2015, 609, 194–197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Beaulieu, J.M.; Sotnikova, T.D.; Gainetdinov, R.R.; Caron, M.G. Tryptophan hydroxylase-2 controls brain synthesis. Science 2004, 305, 217. [Google Scholar] [CrossRef]
- Elmer, G.; Meisch, R.; George, F. Mouse strain differences in operant self-administration of ethanol. Behav. Genet. 1987, 17, 439–451. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Nebert, D.; Vasiliou, V.; Zhu, H.; Shertzer, H. Genetic differences in alcohol drinking preference between inbred strains of mice. Pharmacogenetics 1997, 7, 223–233. [Google Scholar] [CrossRef]
- Bach, H.; Arango, V.; Huang, Y.; Leong, S.; Mann, J.; Underwood, M. Neuronal tryptophan hydroxylase expression in BALB/cJ and C57Bl/6J mice. J. Neurochem. 2011, 118, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Alenina, N.; Kikic, D.; Todiras, M.; Mosienko, V.; Qadri, F.; Plehm, R.; Boyé, P.; Vilianovitch, L.; Sohr, R.; Tenner, K.; et al. Growth retardation and altered autonomic control in mice lacking brain serotonin. Proc. Natl. Acad. Sci. USA 2009, 106, 10332–10337. [Google Scholar] [CrossRef]
- Savelieva, K.V.; Zhao, S.; Pogorelov, V.M.; Rajan, I.; Yang, Q.; Cullinan, E.; Lanthorn, T.H. Genetic disruption of both tryptophan hydroxylase genes dramatically reduces serotonin and affects behavior in models sensitive to antidepressants. PLoS ONE 2008, 3, e3301. [Google Scholar] [CrossRef] [PubMed]
- Angoa-Pérez, M.; Kane, M.J.; Briggs, D.I.; Herrera-Mundo, N.; Sykes, C.E.; Francescutti, D.M.; Kuhn, D.M. Mice genetically depleted of brain serotonin do not display a depression-like behavioral phenotype. ACS Chem. Neurosci. 2014, 5, 908–919. [Google Scholar] [CrossRef] [PubMed]
- Gutknecht, L.; Popp, S.; Waider, J.; Sommerlandt, F.M.J.; Göppner, C.; Post, A.; Reif, A.; Van Den Hove, D.; Strekalova, T.; Schmitt, A.; et al. Interaction of brain 5-HT synthesis deficiency, chronic stress and sex differentially impact emotional behavior in Tph2 knockout mice. Psychopharmacology 2015, 232, 2429–2441. [Google Scholar] [CrossRef] [PubMed]
- Mosienko, V.; Bert, B.; Beis, D.; Matthes, S.; Fink, H.; Bader, M.; Alenina, N. Exaggerated aggression and decreased anxiety in mice deficient in brain serotonin. Transl. Psychiatry 2012, 2, e122. [Google Scholar] [CrossRef]
- van Lingen, M.; Sidorova, M.; Alenina, N.; Klempin, F. Lack of Brain Serotonin Affects Feeding and Differentiation of Newborn Cells in the Adult Hypothalamus. Front. Cell Dev. Biol. 2019, 7, 65. [Google Scholar] [CrossRef]
- Beis, D.; Holzwarth, K.; Flinders, M.; Bader, M.; Wöhr, M.; Alenina, N. Brain serotonin deficiency leads to social communication deficits in mice. Biol. Lett. 2015, 11, 20150057. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mosienko, V.; Beis, D.; Pasqualetti, M.; Waider, J.; Matthes, S.; Qadri, F.; Bader, M.; Alenina, N. Life without brain serotonin: Reevaluation of serotonin function with mice deficient in brain serotonin synthesis. Behav. Brain Res. 2015, 277, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Fructuoso, M.; Gu, Y.C.; Kassis, N.; de Lagran, M.M.; Dierssen, M.; Janel, N. Ethanol-Induced Changes in Brain of Transgenic Mice Overexpressing DYRK1A. Mol. Neurobiol. 2020, 57, 3195–3205. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, N.; Crabbe, J.C.; Ford, M.M.; Murillo, A.; Finn, D.A. Voluntary ethanol consumption in 22 inbred mouse strains. Alcohol 2008, 42, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.P.; Zou, M.E.; Janak, P.H.; Messing, R.O. Responses to ethanol in C57BL/6 versus C57BL/6 × 129 hybrid mice. Brain Behav. 2012, 2, 22–31. [Google Scholar] [CrossRef]
- Kelaï, S.; Renoir, T.; Chouchana, L.; Saurini, F.; Hanoun, N.; Hamon, M.; Lanfumey, L. Chronic voluntary ethanol intake hypersensitizes 5-HT(1A) autoreceptors in C57BL/6J mice. J. Neurochem. 2008, 107, 1660–1670. [Google Scholar] [CrossRef]
- Mosienko, V.; Matthes, S.; Hirth, N.; Beis, D.; Flinders, M.; Bader, M.; Hansson, A.C.; Alenina, N. Adaptive changes in serotonin metabolism preserve normal behavior in mice with reduced TPH2 activity. Neuropharmacology 2014, 85, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Solich, J.; Pałach, P.; Budziszewska, B.; Dziedzicka-Wasylewska, M. Effect of two behavioral tests on corticosterone level in plasma of mice lacking the noradrenaline transporter. Pharm. Rep. 2008, 60, 1008–1013. [Google Scholar] [CrossRef]
- Costa, A.P.R.; Vieira, C.; Bohner, L.O.; Silva, C.F.; da Silva Santos, E.C.; De Lima, T.C.M.; Lino-de-Oliveira, C. A proposal for refining the forced swim test in Swiss mice. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2013, 45, 150–155. [Google Scholar] [CrossRef] [PubMed]
- McGuier, N.S.; Griffin, W.C.; Gass, J.T.; Padula, A.E.; Chesler, E.J.; Mulholland, P.J. Kv7 channels in the nucleus accumbens are altered by chronic drinking and are targets for reducing alcohol consumption. Addict. Biol. 2016, 21, 1097–1112. [Google Scholar] [CrossRef] [PubMed]
- Paxinos, G.; Franklin, K.B.J. The Mouse Brain in Stereotaxic Coordinates, 2nd ed.; Academic Press: San Diego, CA, USA, 2001. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Yang, W.; Xiu, Z.; Zhang, L.; Ren, X.; Wang, L.; Chen, L.; Asakawa, T. An in vivo explorative study to observe the protective effects of Puerariae flos extract on chronic ethanol exposure and withdrawal male mice. Biomed. Pharmacother. 2021, 137. [Google Scholar] [CrossRef]
- Slattery, D.A.; Desrayaud, S.; Cryan, J.F. GABAB receptor antagonist-mediated antidepressant-like behavior is serotonin-dependent. J. Pharmacol. Exp. Ther. 2005, 312, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Ciccocioppo, R.; Panocka, I.; Froldi, R.; Colombo, G.; Gessa, G.; Massi, M. Antidepressant-like effect of ethanol revealed in the forced swimming test in Sardinian alcohol-preferring rats. Psychopharmacology 1999, 144, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Gutknecht, L.; Araragi, N.; Merker, S.; Waider, J.; Sommerlandt, F.; Mlinar, B.; Baccini, G.; Mayer, U.; Proft, F.; Hamon, M.; et al. Impacts of brain serotonin deficiency following Tph2 inactivation on development and raphe neuron serotonergic specification. PLoS ONE 2012, 7, e43157. [Google Scholar] [CrossRef] [PubMed]
- Volkow, N.D.; Wang, G.-J.; Fowler, J.S.; Tomasi, D.; Telang, F.; Baler, R. Addiction: Decreased reward sensitivity and increased expectation sensitivity conspire to overwhelm the brain’s control circuit. Bioessays. 2010, 32, 748–788. [Google Scholar] [CrossRef]
- Roy, S. Finding order in slow axonal transport. Curr. Opin. Neurobiol. 2020, 63, 87–94. [Google Scholar] [CrossRef]
- Sahoo, P.K.; Smith, D.S.; Perrone-Bizzozero, N.; Twiss, J.L. Axonal mRNA transport and translation at a glance. J. Cell Sci. 2018, 131, jcs196808. [Google Scholar] [CrossRef]
- Dalla Costa, I.; Buchanan, C.N.; Zdradzinski, M.D.; Sahoo, P.K.; Smith, T.P.; Thames, E.; Kar, A.N.; Twiss, J.L. The functional organization of axonal mRNA transport and translation. Nat. Rev. Neurosci. 2020, 22, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Belmer, A.; Patkar, O.L.; Pitman, K.M.; Bartlett, S.E. Serotonergic Neuroplasticity in Alcohol Addiction. Brain Plast. 2016, 1, 177–206. [Google Scholar] [CrossRef]
- Vilpoux, C.; Warnault, V.; Pierrefiche, O.; Daoust, M.; Naassila, M. Ethanol-sensitive brain regions in rat and mouse: A cartographic review, using immediate early gene expression. Alcohol. Clin. Exp. Res. 2009, 33, 945–969. [Google Scholar] [CrossRef] [PubMed]
- Carkaci-Salli, N.; Salli, U.; Kuntz-Melcavage, K.L.; Pennock, M.M.; Ozgen, H.; Tekin, I.; Freeman, W.M.; Vrana, K.E. TPH2 in the ventral tegmental area of the male rat brain. Brain Res. Bull. 2011, 84, 376–380. [Google Scholar] [CrossRef]
- Popova, N.; Ilchibaeva, T.; Antonov, E.; Pershina, A.; Bazovkina, D.; Naumenko, V. On the interaction between BDNF and serotonin systems: The effects of long-term ethanol consumption in mice. Alcohol 2020, 87, 1–15. [Google Scholar] [CrossRef]
- Richardson, H.; Lee, S.; O’Dell, L.; Koob, G.; Rivier, C. Alcohol self-administration acutely stimulates the hypothalamic-pituitary-adrenal axis, but alcohol dependence leads to a dampened neuroendocrine state. Eur. J. Neurosci. 2008, 28, 1641–1653. [Google Scholar] [CrossRef]
- Heydendael, W.; Jacobson, L. Glucocorticoid status affects antidepressant regulation of locus coeruleus tyrosine hydroxylase and dorsal raphé tryptophan hydroxylase gene expression. Brain Res. 2009, 1288, 69–78. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaniewska, M.; Mosienko, V.; Bader, M.; Alenina, N. Tph2 Gene Expression Defines Ethanol Drinking Behavior in Mice. Cells 2022, 11, 874. https://doi.org/10.3390/cells11050874
Zaniewska M, Mosienko V, Bader M, Alenina N. Tph2 Gene Expression Defines Ethanol Drinking Behavior in Mice. Cells. 2022; 11(5):874. https://doi.org/10.3390/cells11050874
Chicago/Turabian StyleZaniewska, Magdalena, Valentina Mosienko, Michael Bader, and Natalia Alenina. 2022. "Tph2 Gene Expression Defines Ethanol Drinking Behavior in Mice" Cells 11, no. 5: 874. https://doi.org/10.3390/cells11050874
APA StyleZaniewska, M., Mosienko, V., Bader, M., & Alenina, N. (2022). Tph2 Gene Expression Defines Ethanol Drinking Behavior in Mice. Cells, 11(5), 874. https://doi.org/10.3390/cells11050874







