Cannabinoids Reduce Extracellular Vesicle Release from HIV-1 Infected Myeloid Cells and Inhibit Viral Transcription
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Patient Samples
2.3. ZetaView Nanoparticle Tracking Analysis (NTA)
2.4. EV Enrichment Using Nanotrap Particles
2.5. RNA Isolation, Creation of cDNA, and Quantitative Real-Time PCR (RT-qPCR)
2.6. Preparation of Whole-Cell Extracts
2.7. Western Blot
2.8. Cell Viability Assay
2.9. Chromatin Immunoprecipitation (ChIP)
2.10. Kinase Assay
2.11. Neurospheres
2.12. Mass Spectrometry Analysis
2.13. Pathway Enrichment Analysis
2.14. Densitometry Analysis
2.15. Statistical Analysis
3. Results
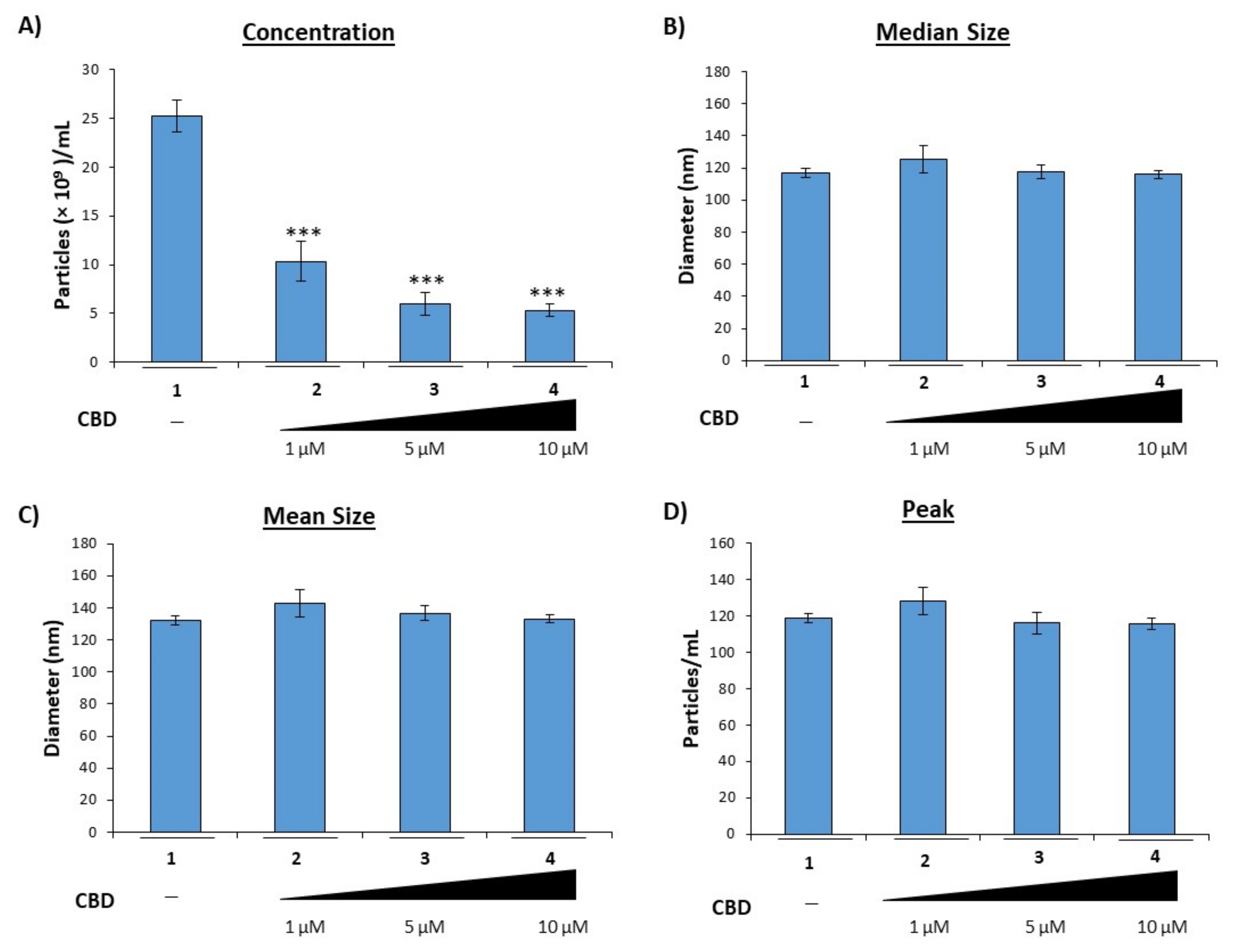
3.1. Cannabidiol Inhibits the Release of EVs from HIV-1 Infected Monocytes
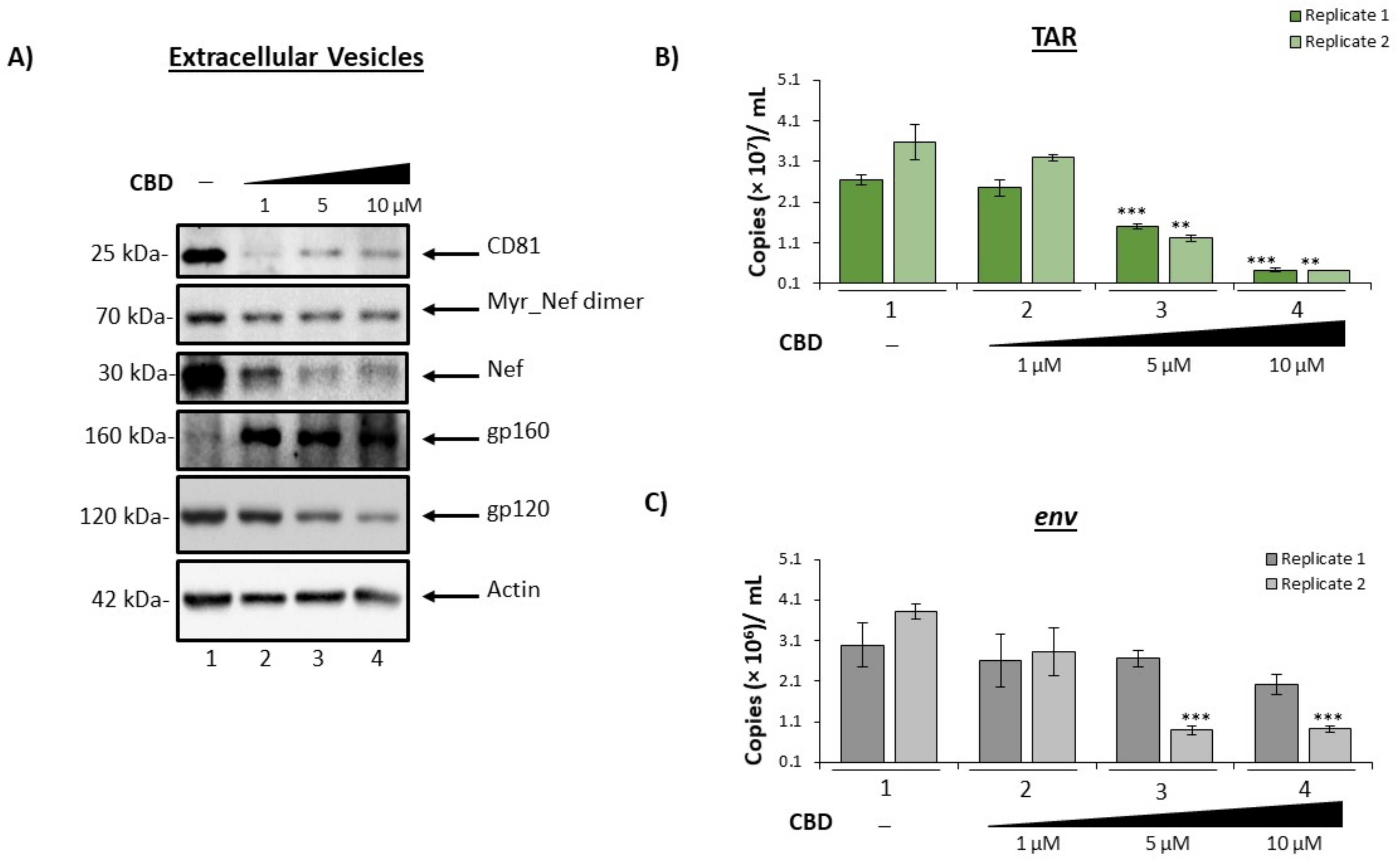
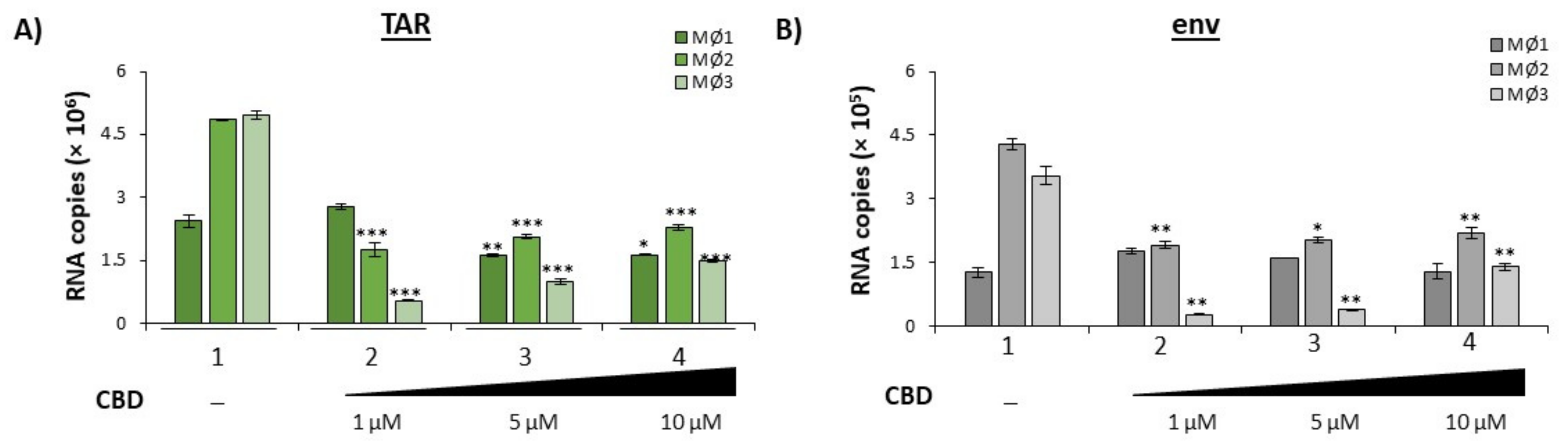
3.2. Cannabidiol Decreases EV-Associated Viral Proteins and Viral RNAs
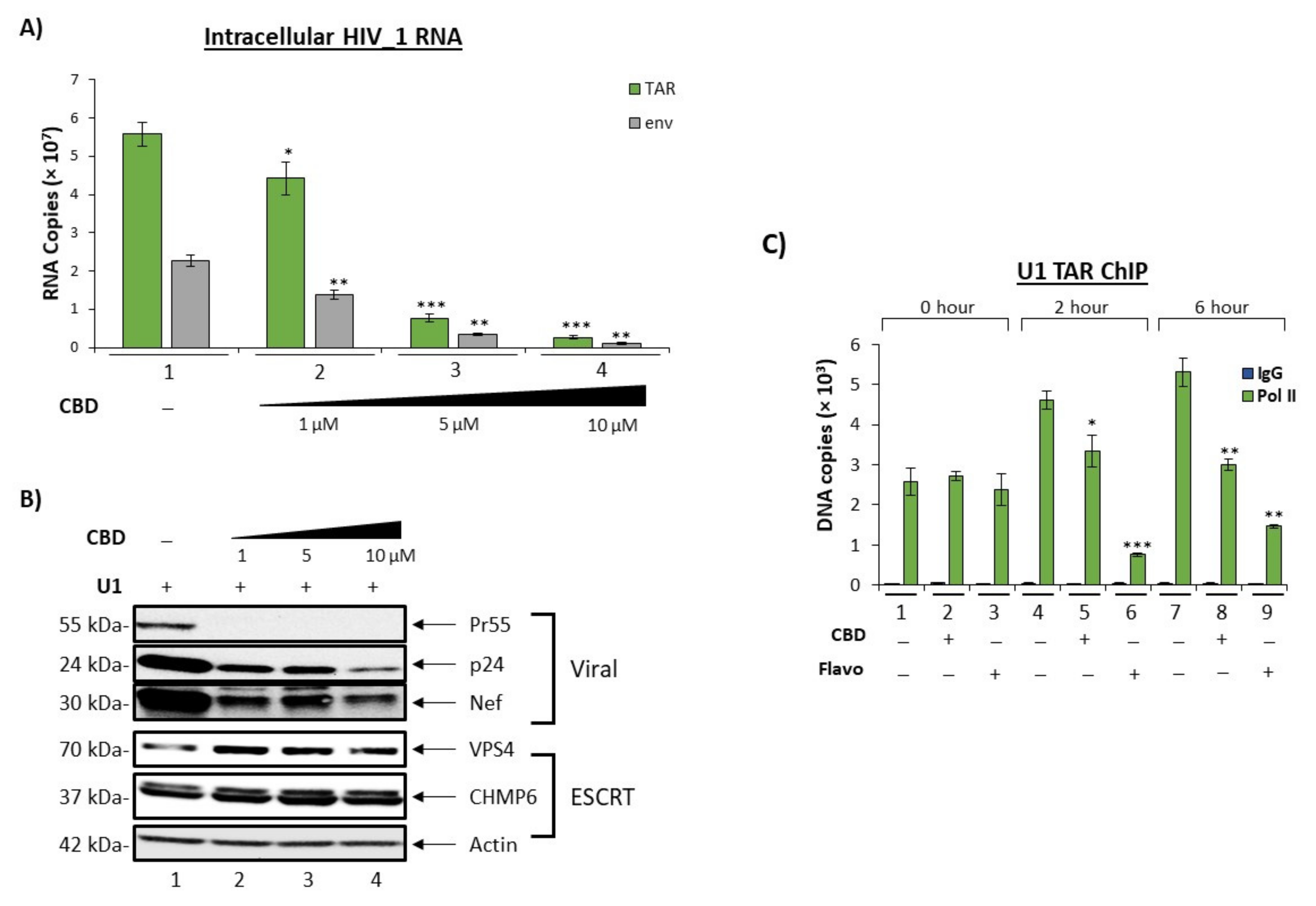
3.3. Cannabidiol Inhibit HIV-1 Transcription in Monocytes
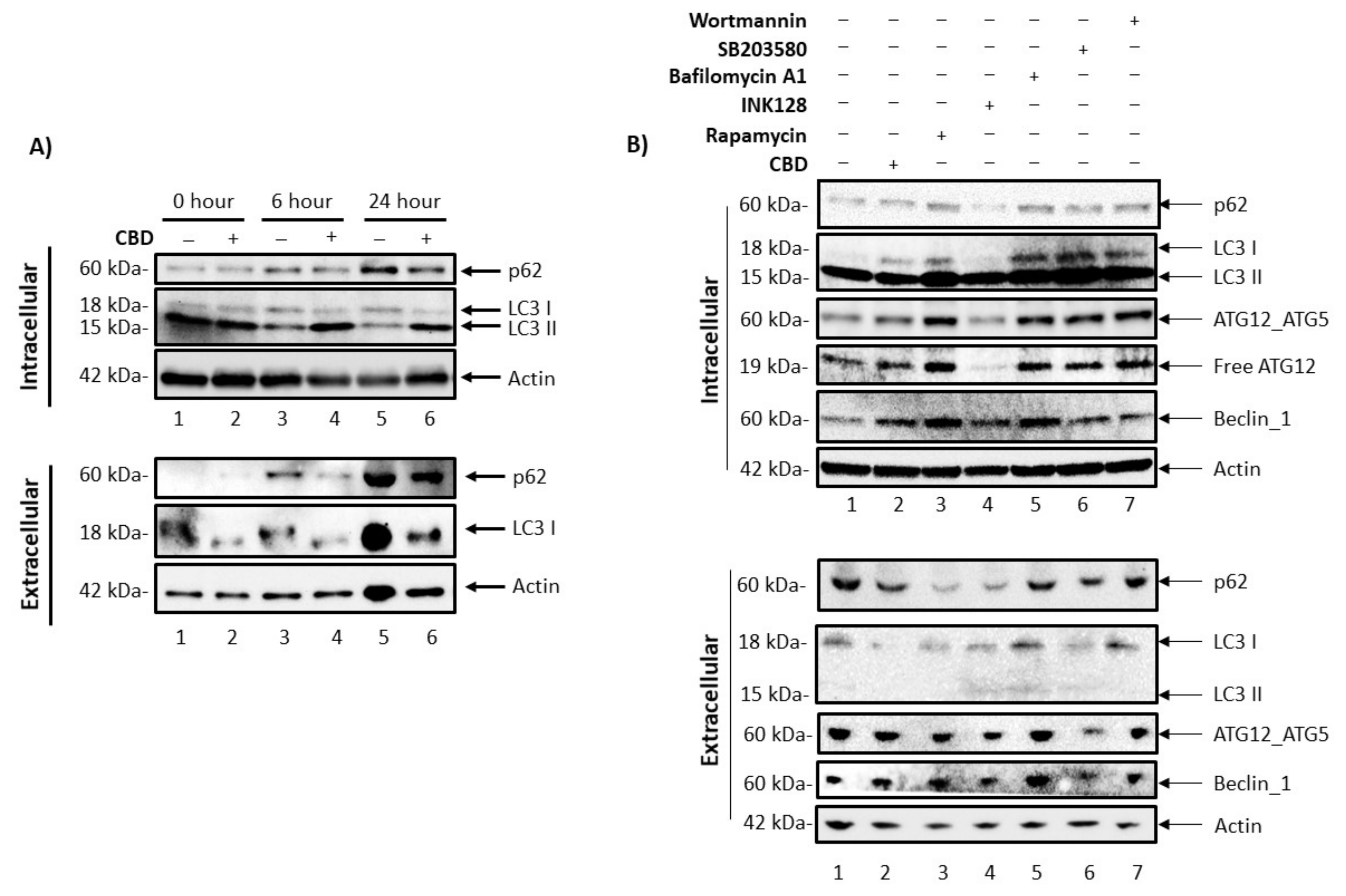
3.4. Cannabidiol Alters Intracellular and Secretory Autophagy Pathways
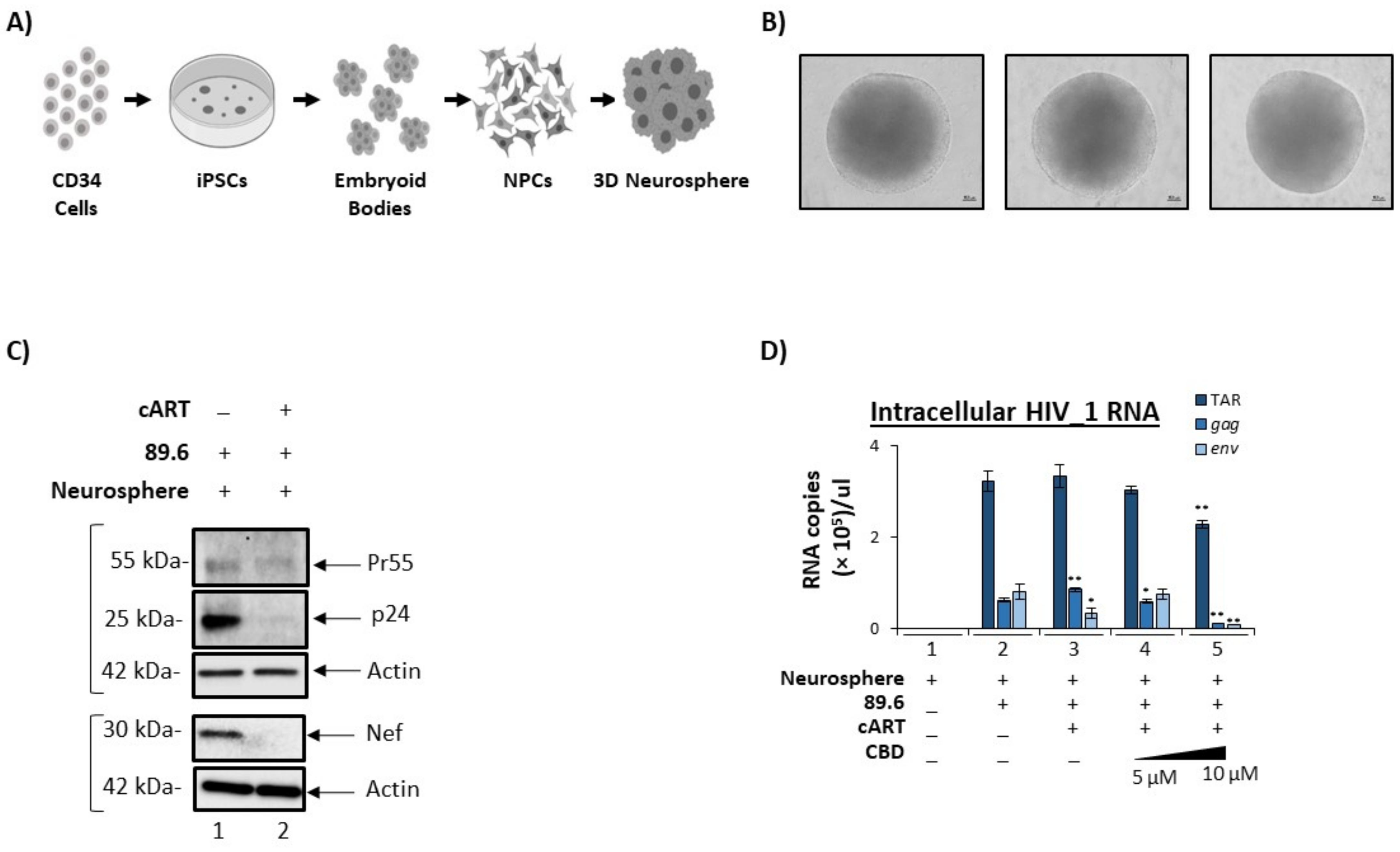
3.5. Cannabidiol Reduces Production of Viral Products in a 3D Neurosphere Model
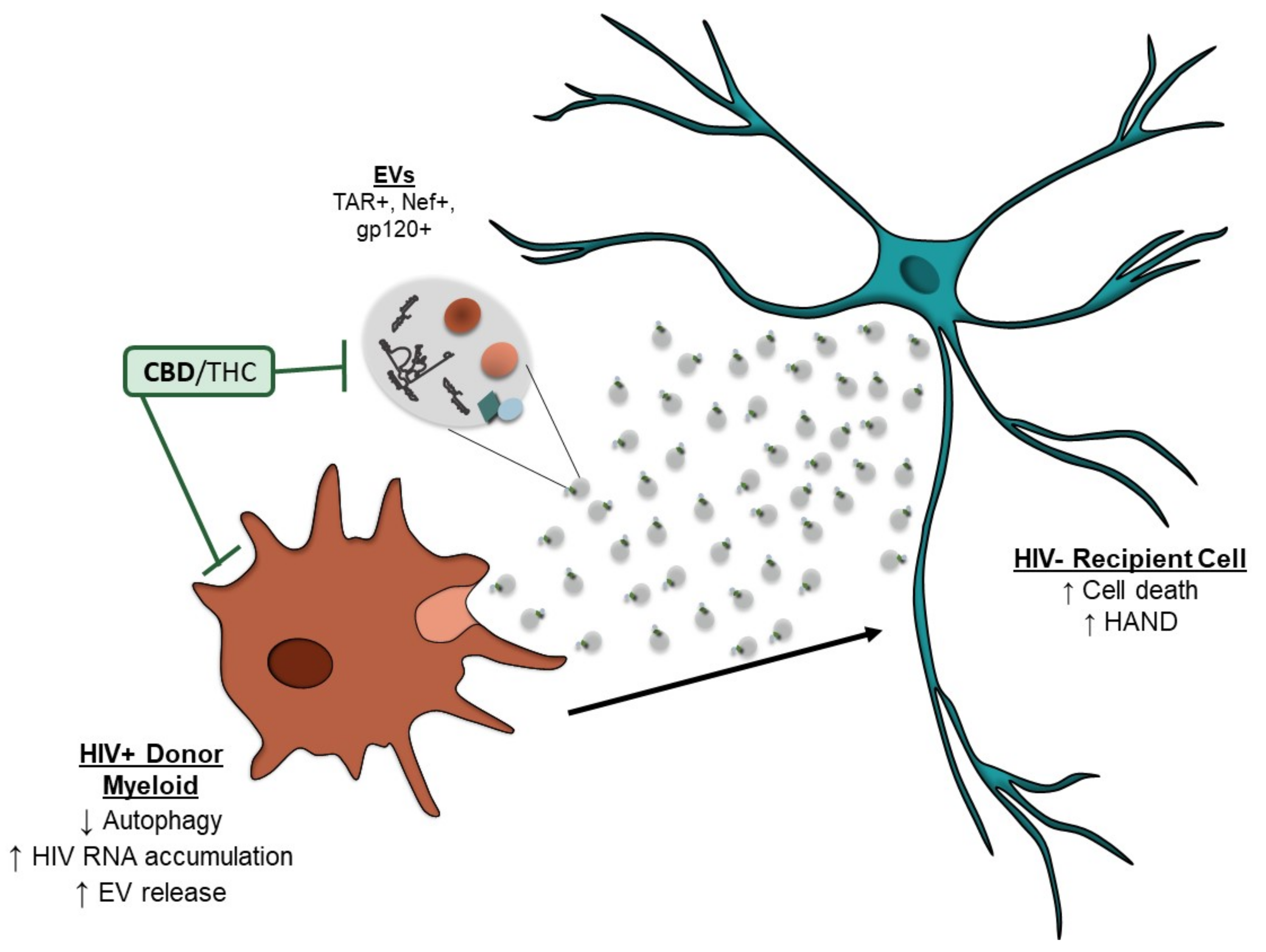
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HAND | HIV-associated neurocognitive disorders |
| HAD | HIV-associated dementia |
| cART | Combined antiretroviral therapy |
| PWH | People with HIV |
| CNS | Central nervous system |
| CSF | Cerebrospinal fluid |
| BBB | Blood–brain barrier |
| THC | Δ9-tetrahydrocannabinol |
| CBD | Cannabidiol |
| TAR | Trans-activation response element |
| ESCRT | Endosomal sorting complexes required for transport |
| iPSCs | induced Pluripotent stem cells |
| pol | Polymerase |
| 3D | Three-dimensional |
References
- Nightingale, S.; Winston, A.; Letendre, S.; Michael, B.; McArthur, J.C.; Khoo, S.; Solomon, T. Controversies in HIV-associated neurocognitive disorders. Lancet. Neurol. 2014, 13, 1139–1151. [Google Scholar] [CrossRef]
- Eggers, C.; For the German Association of Neuro-AIDS und Neuro-Infectiology (DGNANI); Arendt, G.; Hahn, K.; Husstedt, I.W.; Maschke, M.; Neuen-Jacob, E.; Obermann, M.; Rosenkranz, T.; Schielke, E.; et al. HIV-1-associated neurocognitive disorder: Epidemiology, pathogenesis, diagnosis, and treatment. J. Neurol. 2017, 264, 1715–1727. [Google Scholar] [CrossRef] [PubMed]
- Saylor, D.; Dickens, A.; Sacktor, N.; Haughey, N.; Slusher, B.; Pletnikov, M.; Mankowski, J.L.; Brown, A.; Volsky, D.J.; McArthur, J.C. HIV-associated neurocognitive disorder—Pathogenesis and prospects for treatment. Nat. Rev. Neurol. 2016, 12, 234–248. [Google Scholar] [CrossRef] [PubMed]
- McCutchan, J.A.; Wu, J.W.; Robertson, K.; Koletar, S.L.; Ellis, R.J.; Cohn, S.; Taylor, M.; Woods, S.; Heaton, R.; Currier, J.; et al. HIV suppression by HAART preserves cognitive function in advanced, immune-reconstituted AIDS patients. AIDS 2007, 21, 1109–1117. [Google Scholar] [CrossRef]
- DeMarino, C.; Pleet, M.L.; Cowen, M.; Barclay, R.A.; Akpamagbo, Y.; Erickson, J.; Ndembi, N.; Charurat, M.; Jumare, J.; Bwala, S.; et al. Antiretroviral Drugs Alter the Content of Extracellular Vesicles from HIV-1-Infected. Cells Sci. Rep. 2018, 8, 1–20. [Google Scholar] [CrossRef]
- Sampey, G.C.; Meyering, S.S.; Zadeh, M.A.; Saifuddin, M.; Hakami, R.M.; Kashanchi, F. Exosomes and their role in CNS viral infections. J. NeuroVirol. 2014, 20, 199–208. [Google Scholar] [CrossRef]
- Saribas, A.S.; Cicalese, S.; Ahooyi, T.M.; Khalili, K.; Amini, S.; Sariyer, I.K. HIV-1 Nef is released in extracellular vesicles derived from astrocytes: Evidence for Nef-mediated neurotoxicity. Cell Death Dis. 2017, 8, e2542. [Google Scholar] [CrossRef]
- Khan, M.B.; Lang, M.J.; Huang, M.-B.; Raymond, A.; Bond, V.C.; Shiramizu, B.; Powell, M.D. Nef exosomes isolated from the plasma of individuals with HIV-associated dementia (HAD) can induce Aβ1–42 secretion in SH-SY5Y neural cells. J. NeuroVirol. 2015, 22, 179–190. [Google Scholar] [CrossRef]
- Narayanan, A.; Iordanskiy, S.; Das, R.; Van Duyne, R.; Santos, S.; Jaworski, E.; Guendel, I.; Sampey, G.; Dalby, E.; Iglesias-Ussel, M.; et al. Exosomes Derived from HIV-1-infected Cells Contain Trans-activation Response Element RNA. J. Biol. Chem. 2013, 288, 20014–20033. [Google Scholar] [CrossRef]
- Sampey, G.C.; Saifuddin, M.; Schwab, A.; Barclay, R.; Punya, S.; Chung, M.-C.; Hakami, R.M.; Zadeh, M.A.; Lepene, B.; Klase, Z.A.; et al. Exosomes from HIV-1-infected Cells Stimulate Production of Pro-inflammatory Cytokines through Trans-activating Response (TAR) RNA. J. Biol. Chem. 2016, 291, 1251–1266. [Google Scholar] [CrossRef]
- Kim, Y.; Mensah, G.; Al Sharif, S.; Pinto, D.; Branscome, H.; Yelamanchili, S.; Cowen, M.; Erickson, J.; Khatkar, P.; Mahieux, R.; et al. Extracellular Vesicles from Infected Cells Are Released Prior to Virion Release. Cells 2021, 10, 781. [Google Scholar] [CrossRef] [PubMed]
- Henderson, L.J.; Johnson, T.P.; Smith, B.R.; Reoma, L.; Santamaria, U.A.; Bachani, M.; DeMarino, C.; Barclay, R.A.; Snow, J.; Sacktor, N.; et al. Presence of Tat and transactivation response element in spinal fluid despite antiretroviral therapy. AIDS 2019, 33, S145–S157. [Google Scholar] [CrossRef] [PubMed]
- Maroon, J.; Bost, J. Review of the neurological benefits of phytocannabinoids. Surg. Neurol. Int. 2018, 9, 91. [Google Scholar] [CrossRef]
- Devinsky, O.; Cilio, M.R.; Cross, H.; Fernandez-Ruiz, J.; French, J.; Hill, C.; Katz, R.; Di Marzo, V.; Jutras-Aswad, D.; Notcutt, W.G.; et al. Cannabidiol: Pharmacology and potential therapeutic role in epilepsy and other neuropsychiatric disorders. Epilepsia 2014, 55, 791–802. [Google Scholar] [CrossRef]
- Stampanoni Bassi, M.; Sancesario, A.; Morace, R.; Centonze, D.; Iezzi, E. Cannabinoids in Parkinson’s Disease. Cannabis Cannabinoid Res. 2017, 2, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Notcutt, W.G. Clinical Use of Cannabinoids for Symptom Control in Multiple Sclerosis. Neurotherapeutics 2015, 12, 769–777. [Google Scholar] [CrossRef]
- Kluger, B.; Triolo, P.; Jones, W.; Jankovic, J. The therapeutic potential of cannabinoids for movement disorders. Mov. Disord. 2014, 30, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Thames, A.D.; Mahmood, Z.; Burggren, A.C.; Karimian, A.; Kuhn, T.P. Combined effects of HIV and marijuana use on neurocognitive functioning and immune status. AIDS Care 2015, 28, 628–632. [Google Scholar] [CrossRef]
- Fogarty, A.; Rawstorne, P.; Prestage, G.; Crawford, J.; Grierson, J.; Kippax, S. Marijuana as therapy for people living with HIV/AIDS: Social and health aspects. AIDS Care 2007, 19, 295–301. [Google Scholar] [CrossRef]
- Ellis, R.J.; Peterson, S.; Cherner, M.; Morgan, E.; Schrier, R.; Tang, B.; Hoenigl, M.; Letendre, S.; Iudicello, J. Beneficial Effects of Cannabis on Blood–Brain Barrier Function in Human Immunodeficiency Virus. Clin. Infect. Dis. 2020, 73, 124–129. [Google Scholar] [CrossRef]
- Watson, C.W.-M.; Paolillo, E.W.; Morgan, E.E.; Umlauf, A.; Sundermann, E.E.; Ellis, R.J.; Letendre, S.; Marcotte, T.D.; Heaton, R.K.; Grant, I. Cannabis Exposure is Associated With a Lower Likelihood of Neurocognitive Impairment in People Living With HIV. J. Acquir. Immune Defic. Syndr. 2020, 83, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Raborn, E.S.; Jamerson, M.; Marciano-Cabral, F.; Cabral, G.A. Cannabinoid inhibits HIV-1 Tat-stimulated adhesion of human monocyte-like cells to extracellular matrix proteins. Life Sci. 2014, 104, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Barclay, R.A.; Schwab, A.; DeMarino, C.; Akpamagbo, Y.; Lepene, B.; Kassaye, S.; Iordanskiy, S.; Kashanchi, F. Exosomes from uninfected cells activate transcription of latent HIV-1. J. Biol. Chem. 2017, 292, 11682–11701. [Google Scholar] [CrossRef] [PubMed]
- Pleet, M.L.; Mathiesen, A.; DeMarino, C.; Akpamagbo, Y.A.; Barclay, R.A.; Schwab, A.; Iordanskiy, S.; Sampey, G.C.; Lepene, B.; Ilinykh, P.A.; et al. Ebola VP40 in Exosomes Can Cause Immune Cell Dysfunction. Front. Microbiol. 2016, 7, 1765. [Google Scholar] [CrossRef]
- Pleet, M.L.; Erickson, J.; DeMarino, C.; Barclay, R.A.; Cowen, M.; Lepene, B.; Liang, J.; Kuhn, J.H.; Prugar, L.; Stonier, S.W.; et al. Ebola Virus VP40 Modulates Cell Cycle and Biogenesis of Extracellular Vesicles. J. Infect. Dis. 2018, 218, S365–S387. [Google Scholar] [CrossRef]
- Cotto, B.; Natarajaseenivasan, K.; Ferrero, K.; Wesley, L.; Sayre, M.; Langford, D. Cocaine and HIV-1 Tat disrupt cholesterol homeostasis in astrocytes: Implications for HIV-associated neurocognitive disorders in cocaine user patients. Glia 2018, 66, 889–902. [Google Scholar] [CrossRef]
- Guendel, I.; Iordanskiy, S.; Van Duyne, R.; Kehn-Hall, K.; Saifuddin, M.; Das, R.; Jaworski, E.; Sampey, G.C.; Senina, S.; Shultz, L.; et al. Novel Neuroprotective GSK-3β Inhibitor Restricts Tat-Mediated HIV-1 Replication. J. Virol. 2014, 88, 1189–1208. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Chettimada, S.; Lorenz, D.R.; Misra, V.; Wolinsky, S.M.; Gabuzda, D. Small RNA sequencing of extracellular vesicles identifies circulating miRNAs related to inflammation and oxidative stress in HIV patients. BMC Immunol. 2020, 21, 1–20. [Google Scholar] [CrossRef]
- Feliu, A.; Morenomartet, M.; Mecha, M.; Carrillo-Salinas, F.J.; DE Lago, E.; Fernandezruiz, J.; Guaza, C. A Sativex®-like combination of phytocannabinoids as a disease-modifying therapy in a viral model of multiple sclerosis. J. Cereb. Blood Flow Metab. 2015, 172, 3579–3595. [Google Scholar] [CrossRef] [PubMed]
- Jaworski, E.; Saifuddin, M.; Sampey, G.; Shafagati, N.; Van Duyne, R.; Iordanskiy, S.; Kehn-Hall, K.; Liotta, L.; Petricoin, E.; Young, M.; et al. The Use of Nanotrap Particles Technology in Capturing HIV-1 Virions and Viral Proteins from Infected Cells. PLoS ONE 2014, 9, 96778. [Google Scholar] [CrossRef] [PubMed]
- McNamara, R.P.; Costantini, L.M.; Myers, T.A.; Schouest, B.; Maness, N.J.; Griffith, J.D.; Damania, B.A.; MacLean, A.G.; Dittmer, D.P. Nef Secretion into Extracellular Vesicles or Exosomes Is Conserved across Human and Simian Immunodeficiency Viruses. Mbio. 2018, 9, e02344-17. [Google Scholar] [CrossRef]
- Dominkuš, P.P.; Ferdin, J.; Plemenitaš, A.; Peterlin, B.M.; Lenassi, M. Nef is secreted in exosomes from Nef.GFP-expressing and HIV-1-infected human astrocytes. J. NeuroVirol. 2017, 23, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Schierer, S.; Blume, K.; Dindorf, J.; Wittki, S.; Xiang, W.; Ostalecki, C.; Koliha, N.; Wild, S.; Schuler, G.; et al. HIV-Nef and ADAM17-containing plasma extracellular vesicles induce and correlate with immune pathogenesis in chronic HIV infection. EBioMedicine 2016, 6, 103–113. [Google Scholar] [CrossRef]
- Chen, Q.; Wu, H.; Qin, S.; Liu, C.; Chen, Y.; Yang, Y.; Xu, C. The P2X7 Receptor Involved in gp120-Induced Cell Injury in BV2 Microglia. Inflammation 2016, 39, 1814–1826. [Google Scholar] [CrossRef] [PubMed]
- Ivey, N.S.; Renner, N.A.; Moroney-Rasmussen, T.; Mohan, M.; Redmann, R.K.; Didier, P.J.; Alvarez, X.; Lackner, A.A.; MacLean, A.G. Association of FAK activation with lentivirus-induced disruption of blood-brain barrier tight junction–associated ZO-1 protein organization. J. NeuroVirol. 2009, 15, 312–323. [Google Scholar] [CrossRef]
- Liu, J.; Xu, C.; Chen, L.; Xu, P.; Xiong, H. Involvement of Kv1. 3 and p38 MAPK signaling in HIV-1 glycoprotein 120-induced microglia neurotoxicity. Cell Death Dis. 2012, 3, e254. [Google Scholar] [CrossRef]
- Nakamuta, S.; Endo, H.; Higashi, Y.; Kousaka, A.; Yamada, H.; Yano, M.; Kido, H. Human immunodeficiency virus type 1 gp120–mediated disruption of tight junction proteins by induction of proteasome-mediated degradation of zonula occludens-1 and -2 in human brain microvascular endothelial cells. J. NeuroVirol. 2008, 14, 186–195. [Google Scholar] [CrossRef]
- Shrikant, P.; Benos, D.J.; Tang, L.P.; Benveniste, E.N. HIV glycoprotein 120 enhances intercellular adhesion molecule-1 gene expression in glial cells. Involvement of Janus kinase/signal transducer and activator of transcription and protein kinase C signaling pathways. J. Immunol. 1996, 156, 1307–1314. [Google Scholar]
- Song, J.; Wu, C.; Korpos, E.; Zhang, X.; Agrawal, S.M.; Wang, Y.; Faber, C.; Schäfers, M.; Korner, H.; Opdenakker, G.; et al. Focal MMP-2 and MMP-9 Activity at the Blood-Brain Barrier Promotes Chemokine-Induced Leukocyte Migration. Cell Rep. 2015, 10, 1040–1054. [Google Scholar] [CrossRef] [PubMed]
- Ton, H.; Xiong, H. Astrocyte Dysfunctions and HIV-1 Neurotoxicity. J. AIDS Clin. Res. 2013, 4, 255. [Google Scholar] [PubMed]
- Xu, H.; Bae, M.; Tovar-Y.-Romo, L.B.; Patel, N.; Bandaru, V.V.R.; Pomerantz, D.; Steiner, J.P.; Haughey, N.J. The Human Immunodeficiency Virus Coat Protein gp120 Promotes Forward Trafficking and Surface Clustering of NMDA Receptors in Membrane Microdomains. J. Neurosci. 2011, 31, 17074–17090. [Google Scholar] [CrossRef] [PubMed]
- Bari, M.; Rapino, C.; Mozetic, P.; Maccarrone, M. The endocannabinoid system in gp120-mediated insults and HIV-associated dementia. Exp. Neurol. 2010, 224, 74–84. [Google Scholar] [CrossRef]
- Moulard, M.; Montagnier, L.; Bahraoui, E. Effects of calcium ions on proteolytic processing of HIV-1 gp160 precursor and on cell fusion. FEBS Lett. 1994, 338, 281–284. [Google Scholar] [CrossRef]
- Purcell, D.F.; Martin, M.A. Alternative Splicing of Human Immunodeficiency Virus Type 1 MRNA Modulates Viral Protein Expression, Replication, and Infectivity. J. Virol. 1993, 67, 6365–6378. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Tkach, M.; Kowal, J.; Théry, C. Why the need and how to approach the functional diversity of extracellular vesicles. Philos. Trans. R. Soc. B Biol. Sci. 2017, 373, 20160479. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT Pathway. Dev. Cell. 2011, 21, 77–91. [Google Scholar] [CrossRef]
- Berro, R.; de la Fuente, C.; Klase, Z.; Kehn-Hall, K.; Parvin, L.; Pumfery, A.; Agbottah, E.; Vertes, A.; Nekhai, S.; Kashanchi, F. Identifying the Membrane Proteome of HIV-1 Latently Infected Cells. J. Biol. Chem. 2007, 282, 8207–8218. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, A.; Sampey, G.; Van Duyne, R.; Guendel, I.; Kehn-Hall, K.; Roman, J.; Currer, R.; Galons, H.; Oumata, N.; Joseph, B.; et al. Use of ATP analogs to inhibit HIV-1 transcription. Virology 2012, 432, 219–231. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Y.; Liu, X.-Y.; De Clercq, E. Role of the HIV-1 positive elongation factor P-TEFb and inhibitors thereof. Mini-Reviews Med. Chem. 2009, 9, 379–385. [Google Scholar] [CrossRef]
- Zhou, M.; Deng, L.; Lacoste, V.; Park, H.U.; Pumfery, A.; Kashanchi, F.; Brady, J.N.; Kumar, A. Coordination of Transcription Factor Phosphorylation and Histone Methylation by the P-TEFb Kinase during Human Immunodeficiency Virus Type 1 Transcription. J. Virol. 2004, 78, 13522–13533. [Google Scholar] [CrossRef]
- Ojha, C.R.; Lapierre, J.; Rodriguez, M.; Dever, S.M.; Zadeh, M.A.; DeMarino, C.; Pleet, M.L.; Kashanchi, F.; El-Hage, N. Interplay between Autophagy, Exosomes and HIV-1 Associated Neurological Disorders: New Insights for Diagnosis and Therapeutic Applications. Viruses 2017, 9, 176. [Google Scholar] [CrossRef] [PubMed]
- Fields, J.; Dumaop, W.; Elueteri, S.; Campos, S.; Serger, E.; Trejo, M.; Kosberg, K.; Adame, A.; Spencer, B.; Rockenstein, E.; et al. HIV-1 Tat Alters Neuronal Autophagy by Modulating Autophagosome Fusion to the Lysosome: Implications for HIV-Associated Neurocognitive Disorders. J. Neurosci. 2015, 35, 1921–1938. [Google Scholar] [CrossRef]
- Saribas, A.S.; Khalili, K.; Sariyer, I.K. Dysregulation of autophagy by HIV-1 Nef in human astrocytes. Cell Cycle 2015, 14, 2899–2904. [Google Scholar] [CrossRef]
- Sarkar, S.; Ravikumar, B.; Floto, R.A.; Rubinsztein, D.C. Rapamycin and MTOR-Independent Autophagy Inducers Ameliorate Toxicity of Polyglutamine-Expanded Huntingtin and Related Proteinopathies. Cell Death Differ. 2009, 16, 46–56. [Google Scholar] [CrossRef]
- Schenone, S.; Brullo, C.; Musumeci, F.M.; Radi, M.; Botta, M. ATP-competitive inhibitors of mTOR: An update. Curr. Med. Chem. 2011, 18, 2995–3014. [Google Scholar] [CrossRef]
- Yamamoto, A.; Tagawa, Y.; Yoshimori, T.; Moriyama, Y.; Masaki, R.; Tashiro, Y. Bafilomycin A1 Prevents Maturation of Autophagic Vacuoles by Inhibiting Fusion between Autophagosomes and Lysosomes in Rat Hepatoma Cell Line, H-4-II-E Cells. Cell Struct. Funct. 1998, 23, 33–42. [Google Scholar] [CrossRef]
- Cicchini, M.; Karantza, V.; Xia, B. Molecular Pathways: Autophagy in Cancer—A Matter of Timing and Context. Clin. Cancer Res. 2015, 21, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Böckmann, S.; Hinz, B. Cannabidiol Promotes Endothelial Cell Survival by Heme Oxygenase-1-Mediated Autophagy. Cells 2020, 9, 1703. [Google Scholar] [CrossRef] [PubMed]
- Bublitz, K.; Böckmann, S.; Peters, K.; Hinz, B. Cannabinoid-Induced Autophagy and Heme Oxygenase-1 Determine the Fate of Adipose Tissue-Derived Mesenchymal Stem Cells under Stressful Conditions. Cells 2020, 9, 2298. [Google Scholar] [CrossRef]
- Gugliandolo, A.; Pollastro, F.; Bramanti, P.; Mazzon, E. Cannabidiol exerts protective effects in an in vitro model of Parkinson’s disease activating AKT/mTOR pathway. Fitoterapia 2020, 143, 104553. [Google Scholar] [CrossRef]
- Hao, F.; Feng, Y. Cannabidiol (CBD) enhanced the hippocampal immune response and autophagy of APP/PS1 Alzheimer’s mice uncovered by RNA-seq. Life Sci. 2020, 264, 118624. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Gao, Y.; Li, Z.; Zhang, Z.; Lin, X.; Gao, J. Cannabidiol alleviates hemorrhagic shock-induced neural apoptosis in rats by inducing autophagy through activation of the PI3K/AKT pathway. Fundam. Clin. Pharmacol. 2020, 34, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H.K. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef]
- Lanko, K.; Eggermont, K.; Patel, A.; Kaptein, S.; Delang, L.; Verfaillie, C.M.; Neyts, J. Replication of the Zika Virus in Different IPSC-Derived Neuronal Cells and Implications to Assess Efficacy of Antivirals. Antiviral Res. 2017, 145, 82–86. [Google Scholar] [CrossRef]
- Fujimori, K.; Ishikawa, M.; Otomo, A.; Atsuta, N.; Nakamura, R.; Akiyama, T.; Hadano, S.; Aoki, M.; Saya, H.; Sobue, G.; et al. Modeling Sporadic ALS in IPSC-Derived Motor Neurons Identifies a Potential Therapeutic Agent. Nat. Med. 2018, 24, 1579–1589. [Google Scholar] [CrossRef]
- Lebedeva, O.S.; Lagarkova, M.A. Pluripotent Stem Cells for Modelling and Cell Therapy of Parkinson’s Disease. Biochemistry 2018, 83, 1046–1056. [Google Scholar] [CrossRef]
- Monzio Compagnoni, G.; Kleiner, G.; Samarani, M.; Aureli, M.; Faustini, G.; Bellucci, A.; Ronchi, D.; Bordoni, A.; Garbellini, M.; Salani, S.; et al. Mitochondrial Dysregulation and Impaired Autophagy in IPSC-Derived Dopaminergic Neurons of Multiple System Atrophy. Stem Cell Rep. 2018, 11, 1185–1198. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; West, N.; Vider, J.; Zhang, P.; Griffiths, R.E.; Wolvetang, E.; Burtonclay, P.; Warrilow, D. Inflammatory responses to a pathogenic West Nile virus strain. BMC Infect. Dis. 2019, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Penney, J.; Ralvenius, W.T.; Tsai, L.-H. Modeling Alzheimer’s Disease with IPSC-Derived Brain Cells. Mol. Psychiatry 2020, 25, 148–167. [Google Scholar] [CrossRef] [PubMed]
- Teque, F.; Ye, L.; Xie, F.; Wang, J.; Morvan, M.G.; Kan, Y.W.; Levy, J.A. Genetically-edited induced pluripotent stem cells derived from HIV-1-infected patients on therapy can give rise to immune cells resistant to HIV-1 infection. AIDS 2020, 34, 1141–1149. [Google Scholar] [CrossRef]
- Csobonyeiova, M.; Polak, S.; Danisovic, L. Recent Overview of the Use of iPSCs Huntington’s Disease Modeling and Therapy. Int. J. Mol. Sci. 2020, 21, 2239. [Google Scholar] [CrossRef]
- Garcez, P.P.; Loiola, E.C.; Madeiro Da Costa, R.; Higa, L.M.; Trindade, P.; DelVecchio, R.; Nascimento, J.M.; Brindeiro, R.; Tanuri, A.; Rehen, S.K. Zika virus impairs growth in human neurospheres and brain organoids. Science 2016, 352, 816–818. [Google Scholar] [CrossRef]
- Garcez, P.P.; Nascimento, J.; De Vasconcelos, J.M.; Da Costa, R.M.; DelVecchio, R.; Trindade, P.; Loiola, E.; Higa, L.M.; Cassoli, J.S.; Vitória, G.; et al. Zika virus disrupts molecular fingerprinting of human neurospheres. Sci. Rep. 2017, 7, srep40780. [Google Scholar] [CrossRef]
- D’Aiuto, L.; Naciri, J.; Radio, N.; Tekur, S.; Clayton, D.; Apodaca, G.; Di Maio, R.; Zhi, Y.; Dimitrion, P.; Piazza, P.; et al. Generation of three-dimensional human neuronal cultures: Application to modeling CNS viral infections. Stem Cell Res. Ther. 2018, 9, 134. [Google Scholar] [CrossRef]
- Branscome, H.; Khatkar, P.; Al Sharif, S.; Yin, D.; Jacob, S.; Cowen, M.; Kim, Y.; Erickson, J.; Brantner, C.A.; El-Hage, N.; et al. Retroviral infection of human neurospheres and use of stem Cell EVs to repair cellular damage. Sci. Rep. 2022, 12, 1–27. [Google Scholar] [CrossRef]
- Pertwee, R.G.; Howlett, A.C.; Abood, M.E.; Alexander, S.P.H.; Di Marzo, V.; Elphick, M.R.; Greasley, P.J.; Hansen, H.S.; Kunos, G.; Mackie, K.; et al. International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid Receptors and Their Ligands: Beyond CB1 and CB2. Pharmacol. Rev. 2010, 62, 588–631. [Google Scholar] [CrossRef]
- Ahsan, N.A.; Sampey, G.C.; Lepene, B.; Akpamagbo, Y.; Barclay, R.A.; Iordanskiy, S.; Hakami, R.M.; Kashanchi, F. Presence of Viral RNA and Proteins in Exosomes from Cellular Clones Resistant to Rift Valley Fever Virus Infection. Front. Microbiol. 2016, 7, 139. [Google Scholar] [CrossRef]
- Caobi, A.; Nair, M.; Raymond, A.D. Extracellular Vesicles in the Pathogenesis of Viral Infections in Humans. Viruses 2020, 12, 1200. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; DeMarino, C.; Pleet, M.L.; Cowen, M.; Branscome, H.; Al Sharif, S.; Jones, J.; Dutartre, H.; Lepene, B.; Liotta, L.A.; et al. HTLV-1 Extracellular Vesicles Promote Cell-to-Cell Contact. Front. Microbiol. 2019, 10, 2147. [Google Scholar] [CrossRef] [PubMed]
- Pleet, M.L.; Branscome, H.; DeMarino, C.; Pinto, D.; Zadeh, M.A.; Rodriguez, M.; Sariyer, I.K.; El-Hage, N.; Kashanchi, F. Autophagy, EVs, and Infections: A Perfect Question for a Perfect Time. Front. Cell. Infect. Microbiol. 2018, 8, 362. [Google Scholar] [CrossRef] [PubMed]
- Nath, A. Human Immunodeficiency Virus (HIV) Proteins in Neuropathogenesis of HIV Dementia. J. Infect. Dis. 2002, 186, S193–S198. [Google Scholar] [CrossRef] [PubMed]
- Imamichi, H.; Dewar, R.L.; Adelsberger, J.W.; Rehm, C.A.; O’Doherty, U.; Paxinos, E.E.; Fauci, A.S.; Lane, H.C. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc. Natl. Acad. Sci. USA 2016, 113, 8783–8788. [Google Scholar] [CrossRef]
- Hatano, H.; Jain, V.; Hunt, P.W.; Lee, T.-H.; Sinclair, E.; Do, T.D.; Hoh, R.; Martin, J.N.; McCune, J.M.; Hecht, F.; et al. Cell-Based Measures of Viral Persistence Are Associated With Immune Activation and Programmed Cell Death Protein 1 (PD-1)–Expressing CD4+ T cells. J. Infect. Dis. 2012, 208, 50–56. [Google Scholar] [CrossRef]
- Hladnik, A.; Ferdin, J.; Goričar, K.; Deeks, S.; Peterlin, B.M.; Plemenitaš, A.; Vita, D.; Metka, L.; Dolžan, V.; Lenassi, M. Trans-Activation Response Element RNA is Detectable in the Plasma of a Subset of Aviremic HIV-1–Infected Patients. Acta Chim. Slov. 2017, 64, 530–536. [Google Scholar] [CrossRef]
- Chen, L.; Feng, Z.; Yue, H.; Bazdar, D.; Mbonye, U.; Zender, C.; Harding, C.V.; Bruggeman, L.; Karn, J.; Sieg, S.F.; et al. Exosomes derived from HIV-1-infected cells promote growth and progression of cancer via HIV TAR RNA. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Kallianpur, K.J.; Birn, R.; Ndhlovu, L.C.; Souza, S.A.; Mitchell, B.; Paul, R.; Chow, D.C.; Kohorn, L.; Shikuma, C.M. Impact of Cannabis Use on Brain Structure and Function in Suppressed HIV Infection. J. Behav. Brain Sci. 2020, 10, 344–370. [Google Scholar] [CrossRef]
- Rizzo, M.D.; Crawford, R.B.; Henriquez, J.E.; Aldhamen, Y.; Gulick, P.; Amalfitano, A.; Kaminski, N.E. HIV-Infected Cannabis Users Have Lower Circulating CD16+ Monocytes and IP-10 Levels Compared to Non-Using HIV Patients. AIDS 2018, 32, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, M.D.; Henriquez, J.E.; Blevins, L.K.; Bach, A.; Crawford, R.B.; Kaminski, N.E. Targeting Cannabinoid Receptor 2 on Peripheral Leukocytes to Attenuate Inflammatory Mechanisms Implicated in HIV-Associated Neurocognitive Disorder. J. Neuroimmune Pharmacol. 2020, 15, 780–793. [Google Scholar] [CrossRef] [PubMed]
- Juknat, A.; Pietr, M.; Kozela, E.; Rimmerman, N.; Levy, R.; Coppola, G.; Geschwind, D.; Vogel, Z. Differential transcriptional profiles mediated by exposure to the cannabinoids cannabidiol and Δ9-tetrahydrocannabinol in BV-2 microglial cells. J. Cereb. Blood Flow Metab. 2012, 165, 2512–2528. [Google Scholar] [CrossRef] [PubMed]
- Juknat, A.; Pietr, M.; Kozela, E.; Rimmerman, N.; Levy, R.; Gao, F.; Coppola, G.; Geschwind, D.; Vogel, Z. Microarray and Pathway Analysis Reveal Distinct Mechanisms Underlying Cannabinoid-Mediated Modulation of LPS-Induced Activation of BV-2 Microglial Cells. PLoS ONE 2013, 8, e61462. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.M.; Kaplan, B.L. Immune Responses Regulated by Cannabidiol. Cannabis Cannabinoid Res. 2020, 5, 12–31. [Google Scholar] [CrossRef] [PubMed]
- Leymarie, O.; Lepont, L.; Berlioz-Torrent, C. Canonical and Non-Canonical Autophagy in HIV-1 Replication Cycle. Viruses 2017, 9, 270. [Google Scholar] [CrossRef] [PubMed]
- Vrechi, T.A.M.; Leão, A.H.F.F.; Morais, I.B.M.; Abílio, V.C.; Zuardi, A.W.; Hallak, J.E.C.; Crippa, J.A.; Bincoletto, C.; Ureshino, R.P.; Smaili, S.S.; et al. Cannabidiol Induces Autophagy via ERK1/2 Activation in Neural Cells. Sci. Rep. 2021, 11, 5434. [Google Scholar] [CrossRef]
- Pinto, D.O.; DeMarino, C.; Vo, T.T.; Cowen, M.; Kim, Y.; Pleet, M.L.; Barclay, R.A.; Hooten, N.N.; Evans, M.K.; Heredia, A.; et al. Low-Level Ionizing Radiation Induces Selective Killing of HIV-1-Infected Cells with Reversal of Cytokine Induction Using mTOR Inhibitors. Viruses 2020, 12, 885. [Google Scholar] [CrossRef]
- Mehrbod, P.; Ande, S.R.; Alizadeh, J.; Rahimizadeh, S.; Shariati, A.; Malek, H.; Hashemi, M.; Glover, K.K.M.; Sher, A.A.; Coombs, K.M.; et al. The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence 2019, 10, 376–413. [Google Scholar] [CrossRef]
- Reggio, P.H. Endocannabinoid Binding to the Cannabinoid Receptors: What Is Known and What Remains Unknown. Curr. Med. Chem. 2010, 17, 1468–1486. [Google Scholar] [CrossRef]
- Scuderi, C.; Steardo, L.; Esposito, G. Cannabidiol Promotes Amyloid Precursor Protein Ubiquitination and Reduction of Beta Amyloid Expression in SHSY5YAPP+ Cells through PPARγ Involvement. Phytother. Res. 2014, 28, 1007–1013. [Google Scholar] [CrossRef] [PubMed]
- Hind, W.H.; England, T.J.; O’Sullivan, S.E. Cannabidiol protects an in vitro model of the blood-brain barrier from oxygen-glucose deprivation via PPARγ and 5-HT1A receptors. J. Cereb. Blood Flow Metab. 2015, 173, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Vallée, A.; Lecarpentier, Y.; Guillevin, R.; Vallée, J.-N. Effects of cannabidiol interactions with Wnt/β-catenin pathway and PPARγ on oxidative stress and neuroinflammation in Alzheimer’s disease. Acta Biochim. Biophys. Sin. 2017, 49, 853–866. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Scuderi, C.; Valenza, M.; Togna, G.I.; Latina, V.; De Filippis, D.; Cipriano, M.; Carratù, M.R.; Iuvone, T.; Steardo, L. Cannabidiol Reduces Aβ-Induced Neuroinflammation and Promotes Hippocampal Neurogenesis through PPARγ Involvement. PLoS ONE 2011, 6, e28668. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, S.; Kendall, D. Cannabinoid activation of peroxisome proliferator-activated receptors: Potential for modulation of inflammatory disease. Immunobiology 2010, 215, 611–616. [Google Scholar] [CrossRef]
- Sartim, A.; Guimarães, F.; Joca, S. Antidepressant-like effect of cannabidiol injection into the ventral medial prefrontal cortex—Possible involvement of 5-HT1A and CB1 receptors. Behav. Brain Res. 2016, 303, 218–227. [Google Scholar] [CrossRef]
- Linge, R.; Jiménez-Sánchez, L.; Campa, L.; Pilar-Cuellar, F.; Vidal, R.; Pazos, A.; Adell, A.; Díaz, Á. Cannabidiol induces rapid-acting antidepressant-like effects and enhances cortical 5-HT/glutamate neurotransmission: Role of 5-HT1A receptors. Neuropharmacology 2016, 103, 16–26. [Google Scholar] [CrossRef]
- Pazos, M.R.; Mohammed, N.; Lafuente, H.; Santos, M.; Martinez-Pinilla, E.; Moreno, E.; Valdizan, E.; Romero, J.; Pazos, A.; Franco, R.; et al. Mechanisms of cannabidiol neuroprotection in hypoxic–ischemic newborn pigs: Role of 5HT1A and CB2 receptors. Neuropharmacology 2013, 71, 282–291. [Google Scholar] [CrossRef]
- Russo, E.; Burnett, A.; Hall, B.; Parker, K.K. Agonistic Properties of Cannabidiol at 5-HT1a Receptors. Neurochem. Res. 2005, 30, 1037–1043. [Google Scholar] [CrossRef]
- Mecha, M.; Feliu, A.; Iñigo, P.; Mestre, L.; Carrillo-Salinas, F.J.; Guaza, C. Cannabidiol provides long-lasting protection against the deleterious effects of inflammation in a viral model of multiple sclerosis: A role for A2A receptors. Neurobiol. Dis. 2013, 59, 141–150. [Google Scholar] [CrossRef]
- Carrier, E.J.; Auchampach, J.A.; Hillard, C.J. Inhibition of an equilibrative nucleoside transporter by cannabidiol: A mechanism of cannabinoid immunosuppression. Proc. Natl. Acad. Sci. USA 2006, 103, 7895–7900. [Google Scholar] [CrossRef] [PubMed]
- Sagredo, O.; Ramos, J.A.; Decio, A.; Mechoulam, R.; Fernández-Ruiz, J. Cannabidiol Reduced the Striatal Atrophy Caused 3-Nitropropionic Acid in Vivo by Mechanisms Independent of the Activation of Cannabinoid, Vanilloid TRPV1 and Adenosine A2A Receptors. Eur. J. Neurosci. 2007, 26, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.S.; Stella, N.; Catterall, W.A.; Westenbroek, R.E. Cannabidiol attenuates seizures and social deficits in a mouse model of Dravet syndrome. Proc. Natl. Acad. Sci. USA 2017, 114, 11229–11234. [Google Scholar] [CrossRef] [PubMed]
- McHugh, D.; Hu, S.S.; Rimmerman, N.; Juknat, A.; Vogel, Z.; Walker, J.M.; Bradshaw, H.B. N-arachidonoyl glycine, an abundant endogenous lipid, potently drives directed cellular migration through GPR18, the putative abnormal cannabidiol receptor. BMC Neurosci. 2010, 11, 44. [Google Scholar] [CrossRef]
- Hassan, S.; Eldeeb, K.; Millns, P.J.; Bennett, A.J.; Alexander, S.P.H.; Kendall, D.A. Cannabidiol enhances microglial phagocytosis via transient receptor potential (TRP) channel activation. Br. J. Pharmacol. 2014, 171, 2426–2439. [Google Scholar] [CrossRef] [PubMed]
- Iannotti, F.A.; Hill, C.L.; Leo, A.; Alhusaini, A.; Soubrane, C.; Mazzarella, E.; Russo, E.; Whalley, B.J.; Di Marzo, V.; Stephens, G.J. Nonpsychotropic Plant Cannabinoids, Cannabidivarin (CBDV) and Cannabidiol (CBD), Activate and Desensitize Transient Receptor Potential Vanilloid 1 (TRPV1) Channels in Vitro: Potential for the Treatment of Neuronal Hyperexcitability. ACS Chem. Neurosci. 2014, 5, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Simeoli, R.; Montague, K.; Jones, H.R.; Castaldi, L.; Chambers, D.; Kelleher, J.H.; Vacca, V.; Pitcher, T.; Grist, J.; Al-Ahdal, H.; et al. Exosomal cargo including microRNA regulates sensory neuron to macrophage communication after nerve trauma. Nat. Commun. 2017, 8, 1–17. [Google Scholar] [CrossRef]
- Nagarkatti, P.; Pandey, R.; Rieder, S.A.; Hegde, V.L.; Nagarkatti, M. Cannabinoids as novel anti-inflammatory drugs. Futur. Med. Chem. 2009, 1, 1333–1349. [Google Scholar] [CrossRef]
- Fitzgerald, W.; Freeman, M.L.; Lederman, M.M.; Vasilieva, E.; Romero, R.; Margolis, L. A System of Cytokines Encapsulated in ExtraCellular Vesicles. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Haque, S.; Kodidela, S.; Sinha, N.; Kumar, P.; Cory, T.J.; Kumar, S. Differential packaging of inflammatory cytokines/ chemokines and oxidative stress modulators in U937 and U1 macrophages-derived extracellular vesicles upon exposure to tobacco constituents. PLoS ONE 2020, 15, e0233054. [Google Scholar] [CrossRef]
- Tokarz, A.; Szuścik, I.; Kuśnierz-Cabala, B.; Kapusta, M.; Konkolewska, M.; Żurakowski, A.; Georgescu, A.; Stępień, E. Extracellular vesicles participate in the transport of cytokines and angiogenic factors in diabetic patients with ocular complications. Folia Med. Cracov. 2015, 55, 35–48. [Google Scholar] [PubMed]
| % Change | |||
|---|---|---|---|
| Extracellular Vesicles (EVs) Release | EVs Characteristics | Concentration Size | ↓79% − |
| EV-Associated Viral Proteins | Nef (30 kDa) Myristoylated Nef dimer (70 kDa) Gp120 | ↓79% ↓31% ↓58% | |
| EV-Associated Viral RNAs | TAR env | ↓55–86% ↓33–75% | |
| EV-Associated Autophagy Proteins | p62 LC3 I | ↓50% ↓46% | |
| Intracellular Effects | HIV-1 Transcription * | TAR env | ↓95.4% ↓95.7% |
| HIV-1 Proteins | Pr55 p24 Nef | ↓99% ↓76% ↓62% | |
| ESCRT (Endosomal Sorting Complexes Required for Transport Pathway) Proteins | VPS4 CHMP6 | − − | |
| Autophagy Proteins | p62 LC3 I LC3 II | ↓53% ↓72% ↑53% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
DeMarino, C.; Cowen, M.; Khatkar, P.; Cotto, B.; Branscome, H.; Kim, Y.; Sharif, S.A.; Agbottah, E.T.; Zhou, W.; Costiniuk, C.T.; et al. Cannabinoids Reduce Extracellular Vesicle Release from HIV-1 Infected Myeloid Cells and Inhibit Viral Transcription. Cells 2022, 11, 723. https://doi.org/10.3390/cells11040723
DeMarino C, Cowen M, Khatkar P, Cotto B, Branscome H, Kim Y, Sharif SA, Agbottah ET, Zhou W, Costiniuk CT, et al. Cannabinoids Reduce Extracellular Vesicle Release from HIV-1 Infected Myeloid Cells and Inhibit Viral Transcription. Cells. 2022; 11(4):723. https://doi.org/10.3390/cells11040723
Chicago/Turabian StyleDeMarino, Catherine, Maria Cowen, Pooja Khatkar, Bianca Cotto, Heather Branscome, Yuriy Kim, Sarah Al Sharif, Emmanuel T. Agbottah, Weidong Zhou, Cecilia T. Costiniuk, and et al. 2022. "Cannabinoids Reduce Extracellular Vesicle Release from HIV-1 Infected Myeloid Cells and Inhibit Viral Transcription" Cells 11, no. 4: 723. https://doi.org/10.3390/cells11040723
APA StyleDeMarino, C., Cowen, M., Khatkar, P., Cotto, B., Branscome, H., Kim, Y., Sharif, S. A., Agbottah, E. T., Zhou, W., Costiniuk, C. T., Jenabian, M.-A., Gelber, C., Liotta, L. A., Langford, D., & Kashanchi, F. (2022). Cannabinoids Reduce Extracellular Vesicle Release from HIV-1 Infected Myeloid Cells and Inhibit Viral Transcription. Cells, 11(4), 723. https://doi.org/10.3390/cells11040723










