Abstract
This review provides a synopsis of transcriptional responses pertaining to interactions between plant viruses and the insect vectors that transmit them in diverse modes. In the process, it attempts to catalog differential gene expression pertinent to virus–vector interactions in vectors such as virus reception, virus cell entry, virus tissue tropism, virus multiplication, and vector immune responses. Whiteflies, leafhoppers, planthoppers, and thrips are the main insect groups reviewed, along with aphids and leaf beetles. Much of the focus on gene expression pertinent to vector–virus interactions has centered around whole-body RNA extraction, whereas data on virus-induced tissue-specific gene expression in vectors is limited. This review compares transcriptional responses in different insect groups following the acquisition of non-persistent, semi-persistent, and persistent (non-propagative and propagative) plant viruses and identifies parallels and divergences in gene expression patterns. Understanding virus-induced changes in vectors at a transcriptional level can aid in the identification of candidate genes for targeting with RNAi and/or CRISPR editing in insect vectors for management approaches.
1. Introduction
Plant viruses cause devastating yield losses in agricultural systems, which result in reduced economic productivity and nutritional insecurity globally [1]. Plant pathogenic viruses are transmitted mechanically, via seeds, and by vectors [2,3,4,5]. Vector-borne viruses are transmitted by insects, mites, nematodes, and fungi [6,7]. Several genera, species, or biotypes from major insect orders have specialized relationships with a wide range of viruses [8]. These relationships have varying degrees of complexity, which is dependent upon the mode of transmission. Ultimately, the specialized dynamics between plant pathogenic viruses and their associated vectors are critical for the successful spread of plant viruses.
Several factors have been attributed to the emergence and successful establishment of plant viruses in a wide array of agricultural systems. These include: extensive monoculture of crops that are susceptible to both viruses and vectors, the movement of virus-infected plant materials and insect vectors from their native to new environments, plant viruses’ ability to rapidly evolve and adapt, and the effect of climate change on the distribution area of hosts and vectors [9,10,11,12,13,14,15,16]. Many plant viruses also possess a broad host range, which in turn aids in their successful establishment in new environments [17].
Among the various insect orders known to transmit plant viruses, insects with piercing and sucking mouthparts comprise the majority [18]. These include aphids (Hemiptera, Aphididea), whiteflies (Hemiptera, Aleyrodidae), leafhoppers (Hemiptera, Cicadellidae), and thrips (Thysanoptera, Thripidae) [19,20]. The geographical distribution of aphids tends to be predominantly in temperate regions, and aphids are major pests of crops such as cereals including wheat and barley [21,22]. Whiteflies are known to be prevalent in sub-tropical and tropical regions and certain members in the cryptic species complex are pests on a variety of vegetable and row crops such as cotton [23,24]. Leaf and planthoppers are pests of host plants such as potato, rice, and soybean in temperate, sub-tropical, and tropical regions of the world [25,26]. Thrips are pervasive in sub-tropical and tropical regions throughout the world and are known pests for a variety of vegetable and row crops, such as tomato and peanut, respectively [27,28].
Insects belonging to the order Hemiptera and Thysanoptera transmit plant viruses in 12 (Begomovirus, Carlavirus, Closterovirus, Crinivirus, Cucumovirus, Ipomovirus, Mastrevirus, Potyvirus, Polerovirus, Phytoreovirus, Tenuivirus, and Torradovirus) and 5 (Orthotospovirus, Ilarvirus, Carmovirus, Sobemovirus, and Machlomovirus) genera, respectively [29,30,31,32]. Additionally, beetles (order Coleoptera) are also reported to transmit over 40 different plant viruses [33]. Details on the specific vector and virus taxonomic classifications from studies discussed in this review are listed in Table 1. Some viruses in the above-listed genera have been ranked in the top ten most agriculturally important plant viruses [34,35]. Plant viruses in the above-mentioned genera diversely interact with their insect vectors, and these interactions influence virus persistence within the insect [36,37,38,39,40].
The interactions between a given vector and the associated virus could differentially impact gene expression in the vectors (Figure 1). To investigate the differential expression patterns in detail, various tools and pipelines have been developed and are detailed in this review. A de novo transcriptome assembly can provide a foundation for determining differentially expressed genes (DEGs) [41]. To attain greater clarity as to the genetic landscape, access to a high-quality genome assembly would be essential. In the case that a reference genome is available, a de novo transcriptome assembly may still be necessary to account for potentially misassembled genes or improper annotations [42,43]. Whole genome assemblies are available for most insect vectors discussed in this review (Table 1). For non-model organisms, a de novo transcriptome or genome assembly is required prior to differential expression analysis.
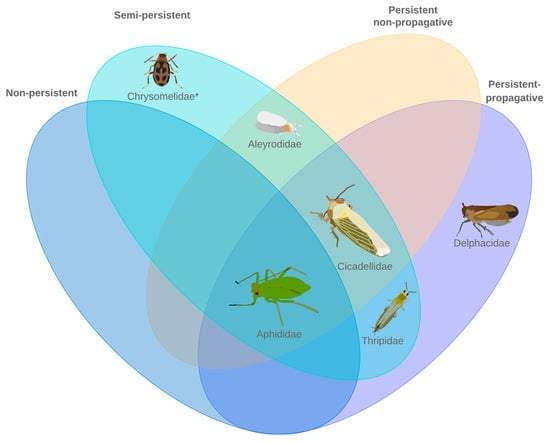
Figure 1.
The six most common plant virus transmitting insect families representing three insect orders. Five of the six families have been investigated for differential gene expression in relation to phytovirus infection. * No differential gene expression studies have been conducted as of December 2021.

Table 1.
Taxonomy of vector–virus associations in which transcriptional responses pertinent to different modes of virus transmission were evaluated. Virus species descriptions are based on the March 2021 International Committee on Taxonomy of Viruses (ICTV) report [44].
Table 1.
Taxonomy of vector–virus associations in which transcriptional responses pertinent to different modes of virus transmission were evaluated. Virus species descriptions are based on the March 2021 International Committee on Taxonomy of Viruses (ICTV) report [44].
| Insect Family | Insect Binomial Name | Insect Common Name | Virus Family | Virus Genus | Virus Species | Reference |
|---|---|---|---|---|---|---|
| Non-persistent | ||||||
| Aphididae | Myzus persicae | Green peach aphid | Bromoviridae | Cucumovirus | Cucumber mosaic virus | [45] |
| Semi-persistent | ||||||
| Aleyrodidae | Bemisia tabaci Middle East-Asia Minor 1 (MEAM1) | Sweetpotato whitefly | Closteroviridae | Crinivirus | Tomato chlorosis virus | [46] |
| Cucurbit yellow stunting disorder virus | [47] | |||||
| Bemisia tabaci Mediterranean (MED) | Tomato chlorosis virus | [48] | ||||
| Cicadellidae | Graminella nigrifrons * | Black-faced leafhopper | Secoviridae | Waikavirus | Maize chlorotic dwarf virus | [49] |
| Persistent non-propagative | ||||||
| Aleyrodidae | Bemisia tabaci (MEAM1) | Sweetpotato whitefly | Geminiviridae | Begomovirus | Tomato yellow leaf curl China virus | [50,51,52,53] |
| Tomato yellow leaf curl virus | [54,55,56] | |||||
| Bemisia tabaci (MED) | [48] | |||||
| Aphididae | Schizaphis graminum | Greenbug | Solemoviridae | Polerovirus | Cereal yellow dwarf virus-RPV | [57] |
| Acyrthosiphon pisum | Pea aphid | Enamovirus | Pea enation mosaic virus 1 | [58] | ||
| Sitobion avenae | English grain aphid | Unassigned species | Barley yellow dwarf virus GAV | [59] | ||
| Persistent-propagative | ||||||
| Thripidae | Frankliniella occidentalis | Western flower thrips | Tospoviridae | Orthotospovirus | Tomato spotted wilt orthotospovirus | [60,61,62,63,64] |
| Frankliniella fusca * | Tobacco thrips | [65] | ||||
| Thrips palmi * | Melon thrips | Capsicum chlorosis orthotospovirus | [66] | |||
| Delphacidae | Nilaparvata lugens | Brown planthopper | Rhabdoviridae | Alphanucleorhabdovirus | Maize mosaic alphanucleorhabdovirus | [67] |
| Laodelphax striatellus | Small brown planthopper | Phenuiviridae | Tenuivirus | Rice stripe tenuivirus | [68,69,70] | |
| Reoviridae | Fijivirus | Southern rice black-streaked dwarf virus | [71] | |||
| Sogatella furcifera | Whitebacked planthopper | [72] | ||||
| Peregrinus maidis | Corn planthopper | Rhabdoviridae | Alphanucleorhabdovirus | Maize mosaic alphanucleorhabdovirus | [73] | |
| Cicadellidae | Graminella nigrifrons * | Blackfaced leafhopper | Gammanucleorhabdovirus | Maize fine streak gammanucleorhabdovirus | [74,75] | |
| Secoviridae | Waikavirus | Maize chlorotic dwarf virus | [49] | |||
* Organisms for which no genome was available as of December 2021.
2. Profiling Transcriptional Responses
2.1. Genetic Foundations
RNA sequencing and bioinformatics analyses are powerful techniques for unraveling the complex interactions between viruses and their vectors. A comparison of transcriptomes between non-viruliferous and viruliferous vectors can aid in understanding qualitative and quantitative differential gene expression modulated by the virus. RNA-Seq methods offer a valuable strategy for sequencing transcripts in rapid, accurate, and informative ways. Studies focused on the six insect families noted in this review have been steadily increasing over the past 10 years (Figure S1). Most of the studies conducted on transcriptomic datasets are based on whole insects rather than specific tissues, mainly due to the limitations of isolating various tissue types [76,77,78] (Figure S2).
For experiments that seek to answer questions about gene regulation, the easiest way to represent how well up or downregulated a gene is to determine the differential expression profile of a viruliferous group against a non-viruliferous group of organisms. In most cases, if not all, this produces an accurate example of the expression levels. The Gene Ontology (GO) database or the Kyoto Encyclopedia of Genes and Genomes (KEGG) database can assign functions to these proteins [79,80]. Having a reference genome provides high levels of certainty as to whether a given gene of interest is up or downregulated [81]. A basic roadmap for the assembly and differential expression analysis is included in Figure 2.
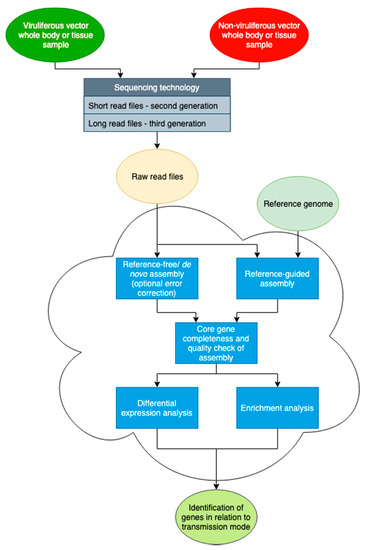
Figure 2.
Diagram detailing a potential pipeline to derive an assembled transcriptome. Initial steps involve the generation of raw data sets from sequencing facilities. Error correction of long read sequences with short reads may be necessary in some cases. The assembly of a transcriptome may be completed de novo if a genome and gene models are not available. The differential expression profile is then determined by mapping the raw read sequences to the assembled genome. Additionally, Gene Ontology term enrichment helps make logical associations between the differentially expressed genes and a given phenotype. See Supplementary File for references.
To attain a concrete resolution on the gene expression profile in each insect/vector, an assembled genome is often an invaluable asset. As sequencing costs are decreasing, more insect genomes are being assembled and projects such as i5k initiative are enabling this knowledge base to grow [82,83]. Some insects, such as the sweetpotato whitefly Middle East-Asia Minor 1 (MEAM1), Bemisia tabaci (Gennadius), have a genome database with a highly curated draft genome, while there is not always a database for other insect vectors [84]. Several aphid genomes have been assembled and annotated, with the first one being the pea aphid, Acyrthosiphon pisum (Harris), and its genome database is known as the AphidBase [85,86]. Additionally, two more aphid genomes have been annotated: the soybean aphid, Aphis glycines (Matsumura), and the wooly apple aphid, Eriosoma lanigerum (Hausmann) [87,88]. Three planthopper genomes have been fully assembled as well; the first was the brown planthopper, Nilaparvata lugens (Stål), followed by the whitebacked planthopper, Sogatella furcifera (Hovárth), and the small brown planthopper, Laodelphax striatellus (Fallén) [89,90,91]. Recently, all three planthoppers’ genomes were revisited with higher resolution from PacBio sequencing [92]. In Thysanoptera, the genome of the western flower thrips, Frankliniella occidentalis (Pergande), was assembled and annotated [93].
2.2. Transmission Modes
The successful transmission of plant viruses is dependent on the nature of the interactions between plant viruses, vectors, and host plants [38,39,40,94,95,96]. Through modulation of plant phenotype by altering plant physiology, such as coloration, nutrients, and volatile organic compounds, plant viruses can influence insect vector behavior and fitness, thereby favoring their transmission [97,98,99]. Currently, there are four described major modes of virus transmission, ranging from non-persistent to persistent-propagative [36,37]. Irrespective of the mode of transmission, many plant viruses infecting their host plants tend to attract their associated vectors [97,100,101]. However, the arrestment duration of their vectors ranging from a brief period (without providing long-term fitness benefits) to a prolonged period (with provision of long-term fitness benefits) is dependent on the mode of transmission [97]. On one end of the spectrum, non-persistent viruses (NPVs), such as cucumber mosaic virus (CMV), briefly arrest their aphid vectors, thereby facilitating rapid virus acquisition and vector dispersal, presumably aided by poor host plant quality [98]. On the opposite end of the spectrum, persistent-propagative viruses, such as tomato spotted wilt orthotospovirus (TSWV), arrest the thrips vectors for longer periods to enable virus acquisition and inoculation thereafter [102,103]. The prolonged feeding by vectors of persistently transmitted viruses is linked to improved plant quality in virus-infected host plants [97,102]. These diverse macro-level effects of plant viruses on their vectors have led researchers to investigate plant viruses-modulated micro-level effects on their vectors using transcriptomic and genomic tools (Table 2).

Table 2.
Summary of insect development stage, number of differentially expressed genes, and sequencing technology used in the reviewed studies.
Non-persistent viruses have short acquisition periods and inoculation periods, lasting only a few seconds, with no latent period [29,104,105,106,107,108,109]. There are currently two documented strategies NPVs use to interact with the receptors of their vectors; these are the “capsid,” and the “helper component” strategies. In the “capsid” strategy, the coat protein (CP) of the virus interacts directly with the vector’s binding sites (Figure 3A) [29,108], while in the “helper component” strategy, the CP interacts indirectly with vector’s binding sites (cell-surface receptor) through the non-structural helper component protein [29]. Viruses that use this mode of transmission often are spread by aphids due to their swift probing behavior associated with assessing host plant suitability [29,104]. NPVs are non-tissue specific and can be transmitted by insects within a short feeding window [19]. NPVs arguably possess the least complicated strategy in terms of virus-vector interaction, followed by the semi-persistent viruses (SPVs). At the time of this review, only one study had examined the transcriptional responses of vector following the acquisition of a NPV (Figure 4A). Liang et al. focused on the impact of CMV on the gene expression profile of the green peach aphid, Myzus persicae (Sulzer) [45].
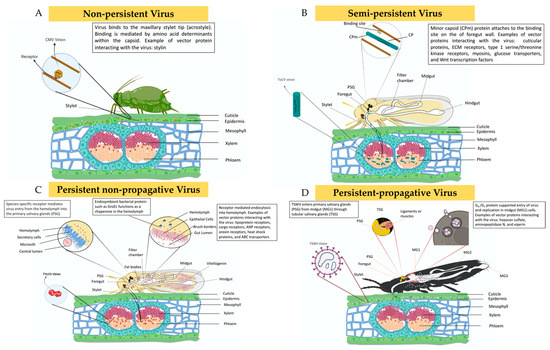
Figure 3.
Schematic diagram explaining the interactions between plant viruses and their vectors with respect to different transmission modes viz., non-persistent, semi-persistent, persistent non-propagative, and persistent-propagative. Modeled after [29,94,95,96,107]. (A) Non-persistent viruses, such as cucumber mosaic virus (CMV), are acquired by aphids from the epidermal cells of infected plants and retained at the tip of its stylet (acrostyle) at the distal end of the common (food/salivary) duct. (B) Semi-persistent viruses, such as tomato chlorosis virus (ToCV), are phloem-limited in infected plants, and the virus attaches to the binding site at the vector’s foregut with the help of the minor capsid protein (CPm). (C) Persistent non-propagative viruses, such as tomato yellow leaf curl virus (TYLCV), are also phloem-limited and are retained in the midgut upon acquisition. Through receptor-mediated endocytosis, the virus traverses the midgut barrier into hemolymph where the endosymbiont protein GroEL interacts with the virion. The virus from the hemolymph reaches primary salivary glands mediated again via species-specific receptors. (D) Thrips acquire persistent propagative viruses, such as tomato spotted wilt virus (TSWV), from epidermal cells of infected plants. Gn/Gc protein supports virus entry into midgut cells, where replication of the virus occurs. The virus TSWV enters primary salivary glands (PSG) from MG1 through tubular salivary glands (TSG).
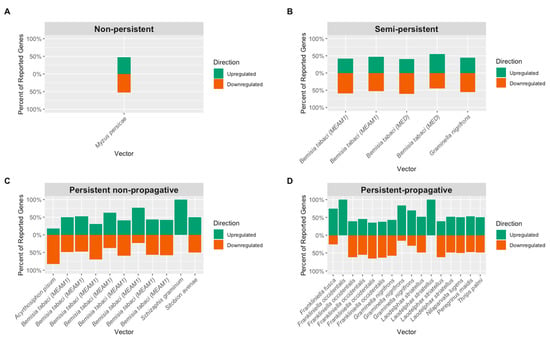
Figure 4.
Comparison of upregulated and downregulated genes reported from studies focusing on (A) non-persistent, (B) semi-persistent, (C) persistent non-propagative, and (D) persistent-propagative transmission. Note that studies with 100% upregulation had very few genes reported due to the method chosen.
Despite the prolonged retention of SPVs by their vectors, these viruses share some similarities with NPVs. For example, SPVs may require helper components to bind to the vector’s foregut (Figure 3B) [29,104]. NPVs may interact with helper components from related viruses to facilitate their transmission, which also may occur with SPVs during co-inoculation [29]. The primary difference between the two transmission strategies is where viruses localize within the vector. Semi-persistently transmitted viruses such as maize chlorotic dwarf virus (MCDV) are phloem-limited in their hosts and typically localize within the foregut of their leafhopper vectors, as opposed to exclusively being stylet-borne as in the case of NPVs [110,111]. Under semi-persistently transmitted viruses, this review focuses on two whitefly-transmitted viruses (tomato chlorosis virus (ToCV) and cucurbit yellow stunting disorder virus (CYSDV)) and one leafhopper-transmitted virus, MCDV. At the micro-level, transcriptional responses relating to virus acquisition have been explored in two whitefly cryptic species, B. tabaci (MEAM1) and Mediterranean (MED), as well in the blackfaced leafhopper, Graminella nigrifrons (Forbes) (Figure 4B). Furthermore, beetles from the family Chrysomelidae can transmit plant viruses, such as the bean pod mottle virus (BPMV), semi-persistently [20,109,112]. However, there are no reported studies on the gene expression level changes induced by viruses in leaf beetles.
Persistent viruses enter their vector through the feeding organs, move through the midgut lumen, translocate across the walls of epithelial cells into the hemolymph, and ultimately travel to the salivary glands; from there, the virions are inoculated into non-infected plants while feeding [19]. This mode of transmission is categorized into non-propagative and propagative, with viruses in the former being incapable of replicating within the insect tissues; while in the latter, the viruses multiply within the host tissues, such as the midgut, salivary gland, and fat body (Figure 3C) [36]. Under the persistent non-propagative category, extensive work has focused on two whitefly-transmitted viruses: tomato yellow leaf curl virus (TYLCV) and tomato yellow leaf curl China virus (TYLCCNV). Additionally, some aphids, such as the greenbug, Schizaphis graminum (Rodani), and the English grain aphid, Sitobion avenae (Fabricius), are known to transmit cereal yellow dwarf virus-RPV (CYDV-RPV) and barley yellow dwarf virus GAV (BYDV-GAV), respectively, in a persistent and non-propagative manner [57,59]. Most studies in this category have focused on the transcriptional responses in B. tabaci (MEAM1) (Figure 4C).
In the persistent-propagative category, extensive work has focused on transcriptional responses in thrips, leafhoppers, and planthoppers (Figure 4D). Persistent-propagative viruses are thought to have originated from animal-infecting viruses, of which several are still capable of infecting insects and are also reported to be transmitted vertically to the next generation [6,111]. Listed below are various pathosystems of persistent-propagative viruses highlighted in this review. In thrips, the virus is acquired during their first instar stage, and it is retained throughout the pupal and adult molts (Figure 3D) [113]. TSWV is one of the most important viruses transmitted by nine thrips species, and gene expression studies have mainly focused on F. occidentalis and tobacco thrips, Frankliniella fusca (Hinds) [60,61,62,63,64,65,114]. Capsicum chlorosis virus (CaCV) is another important virus transmitted by the melon thrips, Thrips palmi (Karny), in Australia and Southeast Asia [115,116]. There are no transcriptional studies on CaCV-infected and non-infected melon thrips. The maize mosaic rhabdovirus (MMV) and maize stripe tenuivirus (MSTV) are viruses transmitted by the corn planthopper, Peregrinus maidis (Ashmead), in a persistent and propagative manner. The rice stripe virus (RSV) and Southern rice black-streaked dwarf virus (SRBSDV) are the other persistent and propagative viruses transmitted by L. striatellus and S. furcifera, respectively [19,117,118,119].
3. Plant-Virus-Induced Responses in Vectors
3.1. Cell-Surface Reception and Virus Tropism
Virion binding to cell-surface receptors facilitates cell entry. Numerous putative cell surface receptors have been identified via transcriptomics, and their diversity seems to vary with virus species and mode of transmission by vectors.
Cuticular proteins present at the stylet tip, such as the stylins of aphids, have been identified as putative receptors of stylet-borne non-persistent viruses [45,120]. Transcripts of aphid cuticular protein genes were associated with the cell entry of zucchini yellow mosaic virus (ZYMV) and with M. persicae and CMV [45,121]. Cuticular protein gene transcripts were also differentially expressed in vectors associated with the transmission of semi-persistent viruses. Five cuticular proteins’ encoding genes were upregulated in B. tabaci (MEAM1) that acquired CYSDV from infected melon, Cucumis melo (L.), plants [47]. In addition to cuticular proteins, extracellular matrix (ECM)-receptors, orphan genes, neuroactive-ligand receptors, and type 1 serine/threonine kinase receptor proteins were identified among the DEGs in B. tabaci (MEAM1) that acquired ToCV—another semi-persistent virus [46]. ECM receptors are transmembrane proteins that are reported as viral receptors favoring viral recognition, attachment, and entry into the cell [122]. Type 1 serine/threonine kinase receptors are ubiquitous transmembrane proteins involved in cell entry of viruses, such as rabies virus [123,124]. The ECM receptors were downregulated, while the neuroactive-ligand and type 1 serine/threonine kinase receptors were upregulated in whiteflies that acquired ToCV [46]. Twenty-one more orphan genes in B. tabaci (MEAM1) that acquired ToCV and CYSDV were downregulated than in non-viruliferous insects [47]. Some of the orphan genes in B. tabaci (MEAM1) that acquired CYSDV and ToCV were speculated to play a role in virus attachment to the vector’s foregut. Orphan genes were also reported in other arthropods to encode surface antigens that are involved in interactions between the viruses and their hosts [125].
In whiteflies that acquired the persistent virus, TYLCV, from infected tomato plants, low-density lipoprotein receptor (LDLR) was upregulated in B. tabaci (MEAM1) [54]. LDLRs are cell-surface proteins involved in the receptor-mediated endocytosis of viruses such as feline leukemia virus (FeLV), hepatitis C virus (HCV), and vesicular stomatitis virus (VSV) [126,127]. Furthermore, in the midgut of B. tabaci (MEAM1) that acquired TYLCV, cargo receptors and epidermal growth factor (EGF)-like repeats, such as laminin subunit alpha-1, LDLR and EGF-like domain 8, were upregulated [55]. Cargo receptors are cell-surface proteins that bind simultaneously to cargo molecules (soluble proteins) and efficiently recruit them to nascent vesicles [128]. Geng et al. speculated that the upregulation of cargo receptors in viruliferous B. tabaci (MEAM1) could facilitate TYLCV endocytosis and crossover from the midgut epithelial cells into the hemolymph [55]. In B. tabaci that acquired another persistent virus from TYLCCNV-infected tobacco (Nicotiana tabacum L.) plants, a putative receptor, heparan sulfate proteoglycan (HSPG), was identified among the upregulated expressed sequence tags [51]. Numerous viruses, such as the herpes simplex virus, were shown to bind to the HSPG receptor, thereby enhancing virus entry into the target cell [129].
In thrips (F. fusca), homologs of heparan sulfate were identified upon the acquisition of a persistent and propagative orthotospovirus (TSWV), but this putative receptor was not identified in the transcriptome of the non-vector flower thrips, Frankliniella tritici (Fitch) [114]. Furthermore, in F. fusca, a homolog of aminopeptidase N, a known virus receptor in aphids, was among the upregulated genes in larvae and adults that fed on TSWV-infected peanut (Arachis hypogaea L.) plants [65].
Transcriptomic studies have revealed the presence of several putative receptors across transmission modes, especially in the persistent virus category. A few receptors appeared to aid in multiple transmission modes. Furthermore, a single virus seems to possess multiple putative receptors within the vector. These receptors are likely present in multiple tissues within a vector. The relevance of receptors’ diversity in the transmission mechanics of most plant viruses remains to be functionally validated. Nevertheless, transcriptomic resources have been of immense importance in establishing and/or speculating virus–vector receptor connections.
Besides receptors, numerous other proteins pertaining to virus tropism within vectors have been identified via transcriptomics. As expected, these proteins seem to vary across vectors and transmission modes. Four myosin genes were upregulated in B. tabaci (MEAM1) following acquisition of the foregut-borne semi-persistent CYSDV [47]. Myosins are molecular motors and have been documented to aid in the intracellular movement of plant viruses, such as tobacco mosaic virus in Nicotiana benthamiana (Domin.) [130]. Their role in interactions with a tissue-specific (foregut borne) virus as its aphid vector remains to be characterized. In another example involving a semi-persistent virus, Ding et al. reported five downregulated facilitative glucose transporters in B. tabaci (MED) adults that acquired ToCV from infected tomato plants [48]. By contrast, in a different study, 10 glucose transporters were upregulated in B. tabaci (MEAM1) that acquired ToCV [46]. Sugar transporters are composed of glucose molecules and are known to transport propagative arthropod viruses, such as Bombyx mori nucleopolyhedrosisvirus (BmNPV), across cells [131]. The role of transporters associated with a non-propagative and exclusively foregut borne virus, such as ToCV, in whiteflies remains to be ascertained.
The atrial natriuretic peptide (ANP) receptors, orexin 2 receptor, and sugar and amino acid transporters were differentially expressed in B. tabaci (MEAM1) that acquired a persistent circulative virus, TYLCV [54]. ANP receptors consist of four distinct domains: ligand-binding domain, transmembrane region, protein kinase-like homology domain and guanylyl cyclase domain [132]. Orexin receptors are natriuretic peptides and affect several circulatory and nervous system functions in higher animals [133,134]. The upregulation of ANP and orexin receptors in B. tabaci (MEAM1) that acquired TYLCV from infected tomato plants was implicated in regulating hemolymph circulation favoring virus movement [54]. Up and downregulation of various transporters in B. tabaci (MEAM1) that acquired TYLCV was linked to facilitating begomovirus movement across the midgut into hemolymph and into primary salivary glands thereafter [48]. In addition, heat shock proteins (HSPs) were upregulated in B. tabaci (MED) following TYLCV acquisition [48]. HSPs are reported to play a role in cell entry, replication and particle assembly, and movement in several animal viruses [135]. HSPs also are molecular chaperones responsible for cellular defense mechanisms against deleterious effects such as protein misfolding, degradation, and insoluble aggregation [136]. HSP 70, upregulated upon TYLCV acquisition in B. tabaci, is also believed to function as a chaperone [137].
Furthermore, in reference to TYLCV, ATP-binding cassette (ABC) transporters and knottin genes were among the identified DEGs in the midguts of B. tabaci (MEAM1) that acquired TYLCV [55]. ABC transporters are membrane proteins with diverse biological functions, including the unidirectional translocation of compounds across cellular membrane [138,139]. Geng et al. speculated that the 16 upregulated and 17 downregulated ABC transporters in viruliferous B. tabaci (MEAM1) could help TYLCV cross the midgut epithelial cells into the hemolymph [55]. Knottins are antimicrobial peptides and have been implicated in the circulative movement of TYLCV in B. tabaci [140,141]. The upregulation of knottin genes in viruliferous B. tabaci (MEAM1) midguts was speculated to be involved in TYLCV and TYLCCNV circulative transmission [51,55,140]. Facilitated trehalose and unknown protein transporters were among the identified upregulated DEGs in B. tabaci (MEAM1) that acquired TYLCV [56]. These genes were speculated to play a crucial role in TYLCV transport in the hemolymph [54,55].
A few transport- and/or tropism-pertinent genes were associated with foregut borne semi-persistent viruses. By contrast, it is logical to identify several genes associated with these functions with persistent circulative viruses that interact with multiple tissue types within the vector.
3.2. Virus Replication, Virus-Induced Metabolism, and Other Cellular Functions
Plant viruses’ entry into vector cells induces a multitude of responses. Virus replication occurs within the cells of insect vectors of persistent-propagative viruses. Consequently, the presence of viruses in cells modulates several metabolic processes.
In whiteflies that acquired a semi-persistent virus, the Wnt and farnesyl pyrophosphate were upregulated in B. tabaci (MEAM1) that acquired ToCV from infected tomato plants [46]. Wnt transcription factors inhibited replication of human T-cell leukemia virus type 1 (HTLV-1), and farnesyl pyrophosphate interacted with HTLV-1 proteins [142,143]. In another whitefly cryptic species (B. tabaci (MED)) that acquired ToCV, eukaryotic translation initiation factor genes were identified among the upregulated DEGs [48]. Those genes were also reported to inhibit the replication of viruses such as influenza A by upregulating the expression level of interferon-induced transmembrane protein 3 [144]. The role of the replication-inhibiting genes indicated above is unclear in the case of semi-persistently transmitted foregut-borne criniviruses, as criniviruses do not replicate within whiteflies. In B. tabaci (MEAM1) that acquired another semi-persistent virus (CYSDV) from infected melon plants, 10 ATPases Associated with cellular Activities (AAA-ATPases) were identified among the downregulated genes [47]. AAA-ATPases are involved in functions such as cell-cycle regulation and vesicle-mediated protein transport [138,145].
The acquisition of a persistent-propagative virus, MMV, in the corn planthopper (P. maidis) resulted in the upregulation of RNA-dependent DNA polymerase (RdRp) genes in the vector, and RdRp is reported to facilitate virus infection and propagation [73]. In the case of another persistent-propagative virus, TSWV, Shrestha et al. found virus inhibitory protein endoplasmic reticulum-associated interferon-inducible (viperin) transcripts in the non-vector F. tritici but not in the vector F. fusca [114]. Although the role of viperin is not functionally validated in thrips, viperin is shown to suppress replication of influenza and rabies viruses as well as lentiviruses [146,147].
The viral activation of cell metabolism provides an increase in the pool of free nucleotides necessary for rapid viral genome replication, as well as increased amino acid production for rapid virion assembly [148]. In the vectors of persistent-propagative viruses, the differential regulation of metabolism-related genes was identified. Han and Rotenberg found that metabolism-associated DEGs, such as putative beta-glucosidase and acyl-CoA desaturase, were mostly downregulated in TSWV-infected F. occidentalis [64]. However, in another congeneric species, F. fusca infected with TSWV, carbohydrate metabolism genes were upregulated [47,65]. In L. striatellus infected with RSV, another persistent-propagative virus, glyoxylate and dicarboxylate metabolism genes were upregulated in both the alimentary canal and salivary glands [68,69]. In G. nigrifrons infected with the maize fine streak virus (MFSV), Cassone et al. found that genes associated with general metabolism were upregulated [49].
The upregulation of virus replication as well as nutrients’ metabolism gene transcripts in the case of persistent-propagative viruses falls in line with the accepted paradigm; however, the differential expression of such genes in the case of non-propagative semi-persistent viruses indicates an anomaly and remains to be investigated.
4. Immune Responses
Upon the detection of the encapsulation proteins of plant viruses, a suite of vectors’ immune responses can be triggered. Below are the different immune genes and pathways that were differentially altered in vectors following the acquisition of plant viruses (Figure 5).
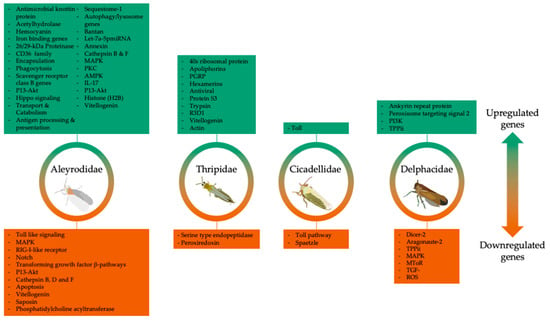
Figure 5.
Major immune-related genes differentially expressed in four insect families. Direction of regulation was variable between insect families, with Aleyrodidae (whiteflies) showing the most identifiable changes across studies.
4.1. Inducible Humoral Response
Humoral response is mediated by antibodies, complement proteins, and antimicrobial peptides. These responses seem to be widespread, regardless of the mode of virus transmission.
In B. tabaci (MEAM1) that acquired a semi-persistent virus from CYSDV-infected melon plants, phosphatidylethanolamine-binding proteins (PEBP) genes were downregulated [47]. PEBPs play an important role in the innate immunity of insects [51,55]. An earlier study reported the downregulation of PEBP genes in B. mori strain resistant to BmNPV. Wang et al. speculated that the downregulation of PEBP could induce enhanced apoptosis, thereby repressing the ability of BmNPV to infect other cells [149]. In B. tabaci (MED) that acquired another semi-persistent virus, ToCV, hemocyanin transcripts were downregulated [48]. Hemocyanins are glycoproteins present in the hemolymph and have been associated with immune responses in insects such as fruit flies, mosquitoes, and psyllids [150,151,152]. Since semi-persistent viruses are limited to the vector’s foregut and do not cross into the hemolymph, the role of PEBPs and hemocyanins in whiteflies that acquire CYSDV and ToCV is rather unclear.
Antimicrobial knottin proteins, platelet-activating factor acetyl hydrolase, and a 26/29-kDa proteinase were other DEGs upregulated in B. tabaci (MEAM1) (whole bodies and midgut tissues) that acquired a persistent circulative virus from TYLCCNV-infected tobacco plants [51,55]. The activation of the knottin protein genes led Luan et al. to speculate that this was a strategy evolved by B. tabaci (MEAM1) to degrade virions [51]. Platelet-activating factor acetyl hydrolase gene was reported as a scavenger of oxidized phospholipids produced by pathogens, including viruses [52,153], while the 26/29-kDa proteinase was speculated to be associated with elimination of xenobiotic proteins in flesh fly, Sarcophaga peregrina (Robineau-Desvoidy) [50,154]. In whiteflies that acquired another persistent circulative virus from TYLCV-infected tomato plants, hemocyanins and iron binding genes were among the identified upregulated DEGs through transcriptomics [48,54]. Iron-binding proteins’ main role in insects is iron transport and immunity [155,156]. The upregulation of hemocyanins and iron binding genes in B. tabaci MEAM1 and MED, respectively, led researchers to speculate about their role in defense response to virus infection [48,54].
In thrips that acquired a persistent-propagative virus, TSWV, from infected plants, several defense and immune response genes, such as apolipophorins, hemocyanins, serine type endopeptidases, peroxiredoxins, peptidoglycan recognition proteins (PGRPs), lysozymes, and trypsin were identified among DEGs [60,63,64]. Schneweis et al. showed that TSWV infection in F. occidentalis increased the expression of phenoloxidase activity in melanogenesis [63]. Medeiros et al. found that 75% of the DEGs identified in TSWV-infected F. occidentalis adults were related to immune function [60]. In the Shrestha et al.’s study, different developmental stages in TSWV-infected F. fusca showed varying regulation patterns, such as the downregulation of genes associated with proteolysis (serine type endopeptidase) and antioxidant defense (peroxiredoxin) exclusively in the larval stage [65]. The few upregulated transcripts identified in the pupal stage were associated with PGRP immune gene homologs [114]. In another example, RSV acquisition in planthoppers caused an upregulation of PGRP and gram-negative bacteria-binding proteins’ genes in the salivary glands [69]. Furthermore, an increase in the expression of PGRP genes in G. nigrifrons that fed on MFSV-infected maize within 4 hr of feeding was observed [49]. By contrast, a noticeable decrease in the expression levels of three of the four PGRP genes in G. nigrifrons designated as transmitters that fed on a MFSV-infected maize was reported [74].
Vectors of persistently-transmitted viruses (non-propagative and propagative) shared only one gene: hemocyanin. As expected, the magnitude of the response was greater in the case of the persistent viruses versus non-persistent viruses, presumably triggered by the persistence of viruses within vectors and increased interactions with multiple tissues.
4.2. Signaling Responses
Signaling responses are initiated upon the detection of virions, and they range from the suppression of immune responses to tumorigenesis. Listed below are the various responses in different vectors upon detecting plant viruses.
In B. tabaci (MEAM1) that acquired ToCV, a semi-persistent virus, several genes involved in the phosphoinositide 3-kinase-protein kinase B (PI3K-Akt), Hippo signaling, transport and catabolism, and antigen processing pathways were identified [46]. The modulation of PI3-Akt in the influenza A and foot-and mouth disease virus and the Hippo pathway in Kaposi sarcoma-associated herpesvirus (KSHV) influenced virus penetration into host cells, the promotion of cell death, and tumorigenesis [157,158]. In the transport and catabolism category, different sets of lysosome DEGs at 24 hr and 72 hr post-acquisition led Kaur et al. to link the upregulation of these genes with virus uptake and downregulation with virus detachment/release post-acquisition [46]. Cathepsins are proteases that perform several roles, including apoptosis, autophagy, and innate immunity [159,160,161,162,163]. In B. tabaci (MED) that acquired ToCV, transcripts of cathepsins B and F were downregulated [48]. On the other hand, the annexin gene was among the identified upregulated genes in B. tabaci (MED) that acquired ToCV [48]. Annexin also was found to function in the apoptosis pathway [164].
In B. tabaci (MEAM1) that acquired a persistent circulative virus, TYLCCNV, from infected tomato plants, several genes involved in signaling pathways, such as toll-like signaling, mitogen-activated protein kinase (MAPK), retinoic acid-inducible gene I (RIG-I-like) receptor, Notch, and transforming growth factor beta were downregulated [51]. Signaling transduction pathways can trigger antiviral responses in insects [165,166]. The downregulation of signaling pathways was associated with the suppression of B. tabaci (MEAM1) immunity, thereby enabling the circulative movement of the virus in B. tabaci (MEAM1) [51,165,166]. The upregulation of DEGs in MAPK and protein kinase c (PKC) pathways facilitated immune system activation in whiteflies that acquired TYLCCNV [51]. Furthermore, the MAPK and PKC pathways were implicated in the membrane trafficking of adenovirus type 2 and influenza virus and the blockage of West Nile virus entry into mosquito cell line [167,168,169]. Most autophagy/lysosome genes were upregulated, and genes were reported to combat virus infection in general [170,171,172]. The upregulation of these genes in B. tabaci (MEAM 1) led Luan et al. to conclude that this was an immune response strategy evolved to offset the deleterious effects of the begomovirus [51]. In addition, several genes involved in the apoptosis pathway in viruliferous B. tabaci (MEAM1) were downregulated. Luan et al. associated the downregulation of the apoptosis pathway to TYLCCNV spread or movement within B. tabaci [51]. In another study by Luan et al., only three genes encoding lysosomal proteins, cathepsin D, phosphatidylcholine acyltransferase, and saposin, were downregulated in B. tabaci (MEAM1) that acquired TYLCCNV from infected tobacco plants, while the others, such as AP-1, cathepsin B, iduronate 2-sulfatase, protein tyrosine phosphatase, and vacuolar ATP synthase subunit S1, were upregulated [52].
Cathepsins B and F transcripts were downregulated in another whitefly cryptic species adults, B. tabaci (MED), that acquired TYLCV from infected tomato plants [48]. B. tabaci (MEAM1) that acquired another persistent circulative virus, TYLCV, from infected tomato plants included several DEGs involved in the 5′ adenosine monophosphate-activated protein kinase (AMPK), interleukin 17 (IL-17), and PI3K-Akt pathways [56]. Li et al. speculated that the downregulation of signaling pathways in TYLCV-infected B. tabaci (MEAM1) might aid in the persistence of the virus through the suppression of the vector’s immune system [56]. Further, DEGs associated MAPK and PKC pathways were upregulated in the midgut of B. tabaci (MEAM1) that acquired TYLCV [55].
In F. occidentalis that acquired a persistent-propagative virus (TSWV) from infected tomato plants, 40S ribosomal protein S3, which is involved in the regulation of apoptosis and the regulation of factors in the Toll and Imd pathways, was upregulated [61]. RSV, another persistent-propagative virus, infection in planthopper-repressed genes associated with MAPK, mTOR, and transforming growth factor beta (TGF-β) pathways [69]. In MMV-infected G. nigrifrons, innate immune genes, such as ankyrin repeat protein and peroxisomal targeting signal 2, were upregulated [73]. In G. nigrifrons infected with MFSV, genes associated with the Toll pathway were upregulated [49]. Chen et al. found that G. nigrifrons infected with MFSV reported a significant suppression of transcripts such as Toll and spaetzle [74].
Keeping in line with this trend, more signaling pathways were differentially affected following virus acquisition in the case of persistent viruses as opposed to semi-persistent viruses. The activation of a multitude of pathways, even in the case of non-propagative viruses, suggests robust evidence for an active immune system. The costs of the activation of these pathways in terms of their vectors’ fitness are not clear.
4.3. Cellular Responses
Following the acquisition of ToCV, the histone H2B gene was upregulated in B. tabaci (MED) that acquired ToCV [48]. This gene was reported to play a role in DNA repair inhibition [173].
Following the acquisition of a persistent virus (TYLCV) in B. tabaci (MEAM1), several DEGs associated with the CD36 family (scavenger receptor class B genes), encapsulation, and phagocytosis were upregulated in the midgut [55]. In B. tabaci (MED), scavenger receptor class B genes and sequestosome-1 were among the DEGs identified in viruliferous B. tabaci [48]. The scavenger class B receptor is a regulator of phagocytosis [55,174], and its upregulation in TYLCV-infected B. tabaci (MEAM1) was associated with resistance to the begomovirus [51,53]. The upregulation of sequestome-1, an autophagosome cargo protein in B. tabaci (MEAM1) that acquired TYLCV, was associated with resistance to the begomovirus [51,53].
Major components in insect immune defense mechanisms, pathogen-associated molecular patterns (PAMPs), were found to be involved in the initiation of phagocytotic gene activation [175]. Whitfield et al. found that the expression of autophagy-specific gene 3 (ATG 3), phosphoinositide 3-kinase (PI3K), and tripeptidyl peptidase ii (TPP ii) were elevated in MMV infected P. maidis [67]. Martin et al. found that autophagy-related genes, such as the peroxisomal targeting signal 2 receptor, were upregulated in MMV-infected P. maidis [73]. Evidence for phagocytosis was more commonly detected in the case of persistent viruses, as opposed to the semi-persistent viruses.
4.4. RNAi Responses
The sequence-specific suppression of gene expression by double-stranded RNA is another generic mechanism insect vectors deploy to defend cells against viruses. Wang et al. identified the upregulation of bantam and let-7a-5p microRNAs (miRNA) in B. tabaci (MEAM1) that acquired TYLCCNV from infected tomato plants [53]. The bantam miRNA was reported to simultaneously stimulate cell proliferation and prevent apoptosis in response to patterning cues in Drosophila melanogaster (Meigen) [176]. Wang et al. linked the enhanced expression level of bantam miRNA to its ability to arrest apoptotic response and help maintain homeostasis in the presence of the virus in B. tabaci [53].
The immune response regulation in S. furcifera showed that reactive oxygen species (ROS)-associated genes were suppressed by the presence of a persistent-propagative virus, SRBSDV [72]. A major component that was silenced was the Dicer-2 (DCR2) and Argonaute-2 (AGO2), which may have facilitated SRBSDV propagation in the midgut epithelium. There was a marked reduction of 60 to 70% in the abundance of RNAi-related genes, such as DCR2 and AGO2, in SRBSDV-infected L. striatellus and S. furcifera cells [71]. Similarly, Chen et al. found that G. nigrifrons infected with MFSV reported a significant suppression of transcripts such as DCR-2 and AGO-2 [73].
Comparisons of gene expression levels in non-viruliferous versus viruliferous S. furcifera showed high downregulation of functionally annotated genes associated with oxidative-reduction, response to oxidative stress, and translation [72]. Within the total number of 551 DEGs in RSV-infected L. striatellus, only four genes were predicted to be potential binding sites for the 70 virus-derived small interfering RNAs (vsiRNAs) [70]. Genes such as R3D1 associated with the RNA interference pathway (RNAi) were upregulated in TSWV-infected F. fusca [62]. The downregulation of these RNAi genes was observed in D. melanogaster and coincided with reduction in antivirus defense [165].
5. Vector Biological-Fitness-Related Genes
Vitellogenin-B (Vg-B), a gene associated with fecundity, lifespan, and other housekeeping roles, was the only reported downregulated DEG relating to biological fitness in B. tabaci (MEAM1) that acquired a semi-persistent virus, ToCV [46]. Reproductive vitellogenin (Vg) and development-related genes (juvenile hormone inducible protein genes) were differentially expressed in B. tabaci (MED) that fed on ToCV-infected tomato plants [48]. The upregulation of these genes in B. tabaci (MED) that acquired ToCV pointed to their role in shorter development time on ToCV-infected tomato plants than on non-infected tomato plants [177].
Wang et al. reported the upregulation of let-7a-5p microRNAs (miRNA) and linked it to perturbation of the cell cycle and negative effects on the longevity and fecundity of B. tabaci (MEAM1) that fed on TYLCCNV-infected tomato plants [53]. Vg genes were among the upregulated DEGs identified in viruliferous B. tabaci (MED) that acquired TYLCV, a persistent virus, from infected tomato plants [48]. Su et al. found an upregulation of two Vg genes in TYLCV-infected B. tabaci (MED) and attributed it to an increase in the fecundity of viruliferous B. tabaci (MED) [178]. Several genes associated with amino acid, carbohydrate, and lipid metabolism were differentially expressed in B. tabaci (MEAM1) that fed on TYLCV-infected tomato plants [56]. The downregulation of these genes was linked to deleterious effects in B. tabaci, such as reduced fecundity and longevity [179,180].
Shrestha et al. found that Vg and actin were both upregulated in F. fusca infected with a persistent-propagative virus, TSWV [65]. The majority of the contigs categorized as being involved in reproduction, embryo development, cell differentiation, and growth were upregulated in the viruliferous adults and pupae [65]. Cuticular proteins were among the downregulated transcripts not only in TSWV-infected F. fusca, but also in S. furcifera and L. striatellus that fed on SRBSDV-infected and RSV-infected rice plants, respectively [63,70,72]. Cuticular proteins are integral components of the membrane, and their downregulation implied a slowing-down of T. palmi development to facilitate virus acquisition [66]. Fifty-four transcripts associated with Vg were among the upregulated genes in CaCV-infected thrips [66]. Lee et al. found that Vg in viruliferous L. striatellus was upregulated, indicating that the virus could promote oogenesis to increase the frequency of transovarial transmission [68,181].
While there is substantial evidence of the upregulation of important reproductive hormones, such as vitellogenin, in vectors of persistent viruses, their upregulation in foregut-limiting viruses prompts us to reexamine the correlation. It is not clear how much of this upregulation is direct virus modulation, as opposed to indirect modulation via the host plant. Providing a short acquisition access on infected plants and gut clearing on non-infected plants could be a viable option to exclusively assess persistent virus-modulated effects, even though, in such scenarios, host effects cannot be completely excluded. Therefore, it becomes imperative to point out such caveats before studies attribute these fitness effects to direct virus modulation on their vectors.
6. Discussion
6.1. Vector Expression Profiles
Across a variety of transcriptomic studies considered for this review, 31 studies were selected, all of which met certain criteria for inclusion. These criteria limited the studies to those with transcript expression reported in insect vectors in relation to plant-transmitted viruses. Within all the studies considered in this review, more trends were observed in some DEGs than in others. Conversely, there were some DEGs that were regulated in the opposite direction when considering all the insect vectors.
A limited number of shared DEGs in each of the non-persistent, semi-persistent, and persistent non-propagative, and persistent-propagative categories highlighted in this review were recorded. This limited number of shared DEGs across several studies could have been driven by the duration of the interaction between the virus and vector. Under virus receptor gene families, several DEGs were identified in the different studies reviewed here; however, only the cuticular and transport-related genes were shared across a few transcriptome studies [46,54,55,56,65,70,72]. The orphan genes and glucose transporters associated with virus infection were the only identified DEGs that were shared across two transcriptome studies [46,48]. Among the biological-fitness-related genes, vitellogenin was the only identified DEG shared across four studies, which were focused on whiteflies infected with ToCV or TYLCV and thrips infected with TSWV [46,48,56,65]. Overall, the functional annotations of DEGs in the vectors of plant viruses were based on the gene functions associated with vectors of propagative animal viruses. Their role might be the same in the vectors of plant viruses, especially those that are non-propagative.
The observed similarities and differences in the direction of the expression of a given gene may have a relation to the evolutionary divergence times between the various insect vectors. Some genes may be more highly conserved than others and account for similarities in responses to infection. In some cases, the DEGS in non-persistently transmitted viruses were similar to those found in other modes of transmission. These differences may also be attributed to the limitations of the techniques used at the time of each study. Another factor to consider is the limitations of the respective databases and datasets available at the time of each study. As molecular techniques advance and more curated databases become available, the expression profiles of more vectors increase our understanding of the effects attributed to viral infection.
The DEG profile can be challenging to compare across a wide range of insect species and various methods that are specific to each study. To normalize the comparisons between each study, the main concepts regarding gene regulation were approached with a few key features in mind in this review. The genes described were those that were mainly associated with immune response, development, and reproduction. While the magnitude of expression is important, this was usually inconsistent across studies, which could in part be due to differing methods. However, genes that tended to be upregulated or downregulated were repeatedly mentioned across most of the studies examined in this review. Nevertheless, transcriptomic studies that point toward the overall picture of expression are as good as the baseline from which those studies are built. This baseline is the presence or absence of a well-curated database or genome.
6.2. Implications of the Common Genes/Pathways
A large amount of success has been witnessed in the potential uses of RNAi to disrupt the regulatory elements in insects [182]. In this review, several genes were discussed that were found to be involved in virus transmission that could be potential targets for RNAi. Insects that transmit disease-causing viruses cause significant amounts of damage every year across a wide range of important agricultural crops and their management is crucial. One such application involves the use of exogenous dsRNA to bind to regions of the virus genome and inhibit the production of binding proteins [183]. Techniques or formulations such as BioClay, which are layered double-hydroxide sheets used to coat plant leaves and exogenously deliver dsRNA to insect vectors, such as aphids, upon feeding are gaining traction [183]. Furthermore, exogenous applications are being explored for crop protection [183,184].
Genome-editing platforms, such as clustered, regularly interspaced, short palindromic repeats (CRISPR)-Cas9 may not be at the point at which developed technologies can be immediately or commercially applied in the field. However, techniques such as RNAi have been studied more, not only in laboratory settings but also in field situations [185]. While it will take time to understand CRISPR in the same sense, its high specificity and ability to edit large or small genomic regions of species of interest could provide a way to increase the effectiveness of RNAi [186,187,188]. Although RNAi technology looks to be promising, it also comes with the challenges of determining the most effective delivery method and screening for off-target effects [182,189,190]. As new genome editing tools are developed, the foundations of fully sequenced genomes and de novo assembled transcriptomes could be used to target specific genetic sites. More investigations into the transcriptomic profiles of insect vectors will help to develop practical applications for controlling vectors’ populations and/or managing devastating plant viruses in production systems. Future applications may result in genetically modified vectors that, by altering or eliminating binding proteins, could prevent the acquisition of viruses.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells11040693/s1. Figure S1: Deposition of gene annotation files uploaded to the National Center for Biotechnology Information (NCBI) gene database over time. Figure S2: Relative frequency of studies using RNA-Seq data derived from various tissue types. References [191,192,193,194,195,196,197] are cited in the supplementary materials.
Author Contributions
Conceptualization, M.A.C., B.K.M., B.D. and R.S.; formal analysis, M.A.C. and H.M.; investigation, M.A.C., H.M., B.K.M. and R.S.; resources, B.D. and R.S.; writing—original draft preparation, M.A.C., H.M., B.K.M. and R.S.; writing—review and editing, M.A.C., H.M., B.K.M., S.P., B.D. and R.S.; visualization, M.A.C., S.P. and H.M.; supervision, B.D. and R.S.; project administration, R.S.; funding acquisition, R.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by USDA-NIFA: UGA-USDA ARS Cooperative Agree.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Strange, R.N.; Scott, P.R. Plant disease: A threat to global food security. Annu. Rev. Phytopathol. 2005, 43, 83–116. [Google Scholar] [CrossRef] [PubMed]
- Dheepa, R.; Paranjothi, S. Transmission of Cucumber Mosaic Virus (CMV) infecting banana by aphid and mechanical methods. Emir. J. Food Agric. 2010, 22, 117–129. [Google Scholar] [CrossRef]
- Sacristan, S.; Diaz, M.; Fraile, A.; Garcia-Arenal, F. Contact transmission of Tobacco mosaic virus: A quantitative analysis of parameters relevant for virus evolution. J. Virol. 2011, 85, 4974–4981. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Li, R.; Shamimuzzaman, M.; Wu, Z.; Ling, K.S. Understanding the Transmissibility of Cucumber Green Mottle Mosaic Virus in Watermelon Seeds and Seed Health Assays. Plant Dis. 2019, 103, 1126–1131. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.M.; Power, A.G. Anthropogenic influences on emergence of vector-borne plant viruses: The persistent problem of Potato virus Y. Curr. Opin. Virol. 2018, 33, 177–183. [Google Scholar] [CrossRef]
- Gray, S.M.; Banerjee, N. Mechanisms of arthropod transmission of plant and animal viruses. Microbiol. Mol. Biol. Rev. 1999, 63, 128–148. [Google Scholar] [CrossRef]
- Campbell, R.N. Fungal transmission of plant viruses. Annu. Rev. Phytopathol. 1996, 34, 87–108. [Google Scholar] [CrossRef]
- Rodhain, F. Insects as vectors: Systematics and biology. Rev. Sci. Tech. l’OIE 2015, 34, 67–96. [Google Scholar] [CrossRef]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of Plant Viruses and Disease Management: Relevance of Genetic Diversity and Evolution. Front. Plant Sci. 2020, 11, 1092. [Google Scholar] [CrossRef]
- Brown, J.K.; Frohlich, D.R.; Rosell, R.C. The Sweetpotato or Silverleaf Whiteflies: Biotypes of Bemisia tabaci or a Species Complex? Annu. Rev. Entomol. 1995, 40, 511–534. [Google Scholar] [CrossRef]
- Rey, M.E.; Ndunguru, J.; Berrie, L.C.; Paximadis, M.; Berry, S.; Cossa, N.; Nuaila, V.N.; Mabasa, K.G.; Abraham, N.; Rybicki, E.P.; et al. Diversity of dicotyledenous-infecting geminiviruses and their associated DNA molecules in southern Africa, including the South-west Indian ocean islands. Viruses 2012, 4, 1753–1791. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.A. Plant virus emergence and evolution: Origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus Res. 2009, 141, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Elena, S.F.; Fraile, A.; Garcia-Arenal, F. Evolution and emergence of plant viruses. Adv. Virus Res. 2014, 88, 161–191. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Kriticos, D.J.; Darnell, R.E.; Yonow, T.; Ota, N.; Sutherst, R.W.; Parry, H.R.; Mugerwa, H.; Maruthi, M.N.; Seal, S.E.; Colvin, J.; et al. Improving climate suitability for Bemisia tabaci in East Africa is correlated with increased prevalence of whiteflies and cassava diseases. Sci. Rep. 2020, 10, 22049. [Google Scholar] [CrossRef]
- Lefeuvre, P.; Martin, D.P.; Elena, S.F.; Shepherd, D.N.; Roumagnac, P.; Varsani, A. Evolution and ecology of plant viruses. Nat. Rev. Microbiol. 2019, 17, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Fermin, G. Host Range, Host–Virus Interactions, and Virus Transmission. Viruses 2018, 101–134. [Google Scholar] [CrossRef]
- Dader, B.; Then, C.; Berthelot, E.; Ducousso, M.; Ng, J.C.K.; Drucker, M. Insect transmission of plant viruses: Multilayered interactions optimize viral propagation. Insect Sci. 2017, 24, 929–946. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; el Ammar, D.; Whitfield, A.E.; Redinbaugh, M.G. Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 2008, 46, 327–359. [Google Scholar] [CrossRef]
- Eigenbrode, S.D.; Bosque-Perez, N.A.; Davis, T.S. Insect-Borne Plant Pathogens and Their Vectors: Ecology, Evolution, and Complex Interactions. Annu. Rev. Entomol. 2018, 63, 169–191. [Google Scholar] [CrossRef]
- Hulle, M.; Coeur d’Acier, A.; Bankhead-Dronnet, S.; Harrington, R. Aphids in the face of global changes. C R Biol. 2010, 333, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, M.W.; Ghani, H.; Ayyub, M.; Ali, Q.; Ahmad, H.M.; Faisal, M.; Ali, A.; Qasim, M.U. Performance of Some Wheat Cultivars against Aphid and Its Damage on Yield and Photosynthesis. J. Glob. Innov. Agric. Soc. Sci. 2019, 105–109. [Google Scholar] [CrossRef]
- Krause-Sakate, R.; Watanabe, L.F.M.; Gorayeb, E.S.; da Silva, F.B.; Alvarez, D.L.; Bello, V.H.; Nogueira, A.M.; de Marchi, B.R.; Vicentin, E.; Ribeiro-Junior, M.R.; et al. Population Dynamics of Whiteflies and Associated Viruses in South America: Research Progress and Perspectives. Insects 2020, 11, 847. [Google Scholar] [CrossRef] [PubMed]
- Mugerwa, H.; Colvin, J.; Alicai, T.; Omongo, C.A.; Kabaalu, R.; Visendi, P.; Sseruwagi, P.; Seal, S.E. Genetic diversity of whitefly (Bemisia spp.) on crop and uncultivated plants in Uganda: Implications for the control of this devastating pest species complex in Africa. J. Pest Sci. 2021, 94, 1307–1330. [Google Scholar] [CrossRef] [PubMed]
- Beanland, L.; Noble, R.; Wolf, T.K. Spatial and Temporal Distribution of North American Grapevine Yellows Disease and of Potential Vectors of the Causal Phytoplasmas in Virginia. Environ. Entomol. 2006, 35, 332–344. [Google Scholar] [CrossRef]
- Shi, L.; Vasseur, L.; Huang, H.; Zeng, Z.; Hu, G.; Liu, X.; You, M. Adult Tea Green Leafhoppers, Empoasca onukii (Matsuda), Change Behaviors under Varying Light Conditions. PLoS ONE 2017, 12, e0168439. [Google Scholar] [CrossRef]
- Riley, D.G.; Joseph, S.V.; Srinivasan, R.; Diffie, S. Thrips Vectors of Tospoviruses. J. Integr. Pest Manag. 2011, 2, I1–I10. [Google Scholar] [CrossRef]
- He, Z.; Guo, J.F.; Reitz, S.R.; Lei, Z.R.; Wu, S.Y. A global invasion by the thrip, Frankliniella occidentalis: Current virus vector status and its management. Insect Sci. 2020, 27, 626–645. [Google Scholar] [CrossRef]
- Ng, J.C.; Falk, B.W. Virus-vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu. Rev. Phytopathol. 2006, 44, 183–212. [Google Scholar] [CrossRef]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A.; Ictv Report, C. ICTV Virus Taxonomy Profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef]
- Ghosh, S.; Kanakala, S.; Lebedev, G.; Kontsedalov, S.; Silverman, D.; Alon, T.; Mor, N.; Sela, N.; Luria, N.; Dombrovsky, A.; et al. Transmission of a New Polerovirus Infecting Pepper by the Whitefly Bemisia tabaci. J. Virol. 2019, 93, e00488-19. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.R. Plant Viruses Transmitted by Thrips. Eur. J. Plant Pathol. 2005, 113, 119–157. [Google Scholar] [CrossRef]
- Nault, L.R. Arthropod Transmission of Plant Viruses: A New Synthesis. Ann. Entomol. Soc. Am. 1997, 90, 521–541. [Google Scholar] [CrossRef]
- Rybicki, E.P. A Top Ten list for economically important plant viruses. Arch. Virol. 2015, 160, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, K.B.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef] [PubMed]
- Dietzgen, R.G.; Mann, K.S.; Johnson, K.N. Plant Virus-Insect Vector Interactions: Current and Potential Future Research Directions. Viruses 2016, 8, 303. [Google Scholar] [CrossRef] [PubMed]
- Gould, F.; Brown, Z.S.; Kuzma, J. Wicked evolution: Can we address the sociobiological dilemma of pesticide resistance? Science 2018, 360, 728–732. [Google Scholar] [CrossRef]
- McElhany, P.; Real, L.A.; Power, A.G. Vector Preference and Disease Dynamics: A Study of Barley Yellow Dwarf Virus. Ecology 1995, 76, 444–457. [Google Scholar] [CrossRef]
- Sisterson, M.S. Effects of Insect-Vector Preference for Healthy or Infected Plants on Pathogen Spread: Insights from a Model. J. Econ. Entomol. 2008, 101, 1–8. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Michalakis, Y.; Munster, M.; Blanc, S.; Biere, A. Plant feeding by insect vectors can affect life cycle, population genetics and evolution of plant viruses. Funct. Ecol. 2013, 27, 610–622. [Google Scholar] [CrossRef]
- Geniza, M.; Jaiswal, P. Tools for building de novo transcriptome assembly. Curr. Plant Biol. 2017, 11–12, 41–45. [Google Scholar] [CrossRef]
- Holzer, M.; Marz, M. De novo transcriptome assembly: A comprehensive cross-species comparison of short-read RNA-Seq assemblers. Gigascience 2019, 8, giz039. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.A.; Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011, 12, 671–682. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Davison, A.J.; Dempsey, D.M.; Dutilh, B.E.; Garcia, M.L.; et al. Changes to virus taxonomy and to the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2021). Arch. Virol. 2021, 166, 2633–2648. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Ma, K.S.; Liang, P.Z.; Yang, L.W.; Zhang, L.; Gao, X.W. Combined Transcriptomic and Proteomic Analysis of Myzus persicae, the Green Peach Aphid, Infected with Cucumber Mosaic Virus. Insects 2021, 12, 372. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Chen, W.; Zheng, Y.; Hasegawa, D.K.; Ling, K.S.; Fei, Z.; Wintermantel, W.M. Transcriptome analysis of the whitefly, Bemisia tabaci MEAM1 during feeding on tomato infected with the crinivirus, Tomato chlorosis virus, identifies a temporal shift in gene expression and differential regulation of novel orphan genes. BMC Genom. 2017, 18, 370. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Chen, W.; Fei, Z.; Wintermantel, W.M. Differences in gene expression in whitefly associated with CYSDV-infected and virus-free melon, and comparison with expression in whiteflies fed on ToCV- and TYLCV-infected tomato. BMC Genom. 2019, 20, 654. [Google Scholar] [CrossRef]
- Ding, T.B.; Li, J.; Chen, E.H.; Niu, J.Z.; Chu, D. Transcriptome Profiling of the Whitefly Bemisia tabaci MED in Response to Single Infection of Tomato yellow leaf curl virus, Tomato chlorosis virus, and Their Co-infection. Front. Physiol. 2019, 10, 302. [Google Scholar] [CrossRef]
- Cassone, B.J.; Wijeratne, S.; Michel, A.P.; Stewart, L.R.; Chen, Y.; Yan, P.; Redinbaugh, M.G. Virus-independent and common transcriptome responses of leafhopper vectors feeding on maize infected with semi-persistently and persistent propagatively transmitted viruses. BMC Genom. 2014, 15, 133. [Google Scholar] [CrossRef]
- Li, J.-M.; Ruan, Y.-M.; Li, F.-F.; Liu, S.-S.; Wang, X.-W. Gene expression profiling of the whitefly (Bemisia tabaci) Middle East-Asia Minor 1 feeding on healthy and Tomato yellow leaf curl China virus-infected tobacco. Insect Sci. 2011, 18, 11–22. [Google Scholar] [CrossRef]
- Luan, J.B.; Li, J.M.; Varela, N.; Wang, Y.L.; Li, F.F.; Bao, Y.Y.; Zhang, C.X.; Liu, S.S.; Wang, X.W. Global analysis of the transcriptional response of whitefly to tomato yellow leaf curl China virus reveals the relationship of coevolved adaptations. J. Virol. 2011, 85, 3330–3340. [Google Scholar] [CrossRef] [PubMed]
- Luan, J.B.; Wang, Y.L.; Wang, J.; Wang, X.W.; Liu, S.S. Detoxification activity and energy cost is attenuated in whiteflies feeding on tomato yellow leaf curl China virus-infected tobacco plants. Insect Mol. Biol. 2013, 22, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, L.; Chen, F.; Yang, X.; Ding, M.; Zhang, Z.; Liu, S.S.; Wang, X.W.; Zhou, X. MicroRNA profiling of the whitefly Bemisia tabaci Middle East-Aisa Minor I following the acquisition of Tomato yellow leaf curl China virus. Virol. J. 2016, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, D.K.; Chen, W.; Zheng, Y.; Kaur, N.; Wintermantel, W.M.; Simmons, A.M.; Fei, Z.; Ling, K.S. Comparative transcriptome analysis reveals networks of genes activated in the whitefly, Bemisia tabaci when fed on tomato plants infected with Tomato yellow leaf curl virus. Virology 2018, 513, 52–64. [Google Scholar] [CrossRef]
- Geng, L.; Qian, L.X.; Shao, R.X.; Liu, Y.Q.; Liu, S.S.; Wang, X.W. Transcriptome profiling of whitefly guts in response to Tomato yellow leaf curl virus infection. Virol. J. 2018, 15, 14. [Google Scholar] [CrossRef]
- Li, M.; Zhao, J.; Su, Y.L. Transcriptome Analysis of Gene Expression Profiles of Tomato Yellow Leaf Curl Virus-Infected Whiteflies over Different Viral Acquisition Access Periods. Insects 2020, 11, 297. [Google Scholar] [CrossRef]
- Yang, X.; Thannhauser, T.W.; Burrows, M.; Cox-Foster, D.; Gildow, F.E.; Gray, S.M. Coupling genetics and proteomics to identify aphid proteins associated with vector-specific transmission of polerovirus (luteoviridae). J. Virol. 2008, 82, 291–299. [Google Scholar] [CrossRef]
- Brault, V.; Tanguy, S.; Reinbold, C.; Le Trionnaire, G.; Arneodo, J.; Jaubert-Possamai, S.; Guernec, G.; Tagu, D. Transcriptomic analysis of intestinal genes following acquisition of pea enation mosaic virus by the pea aphid Acyrthosiphon pisum. J. Gen. Virol. 2010, 91, 802–808. [Google Scholar] [CrossRef]
- Li, D.; Su, D.; Tong, Z.; Zhang, C.; Zhang, G.; Zhao, H.; Hu, Z. Virus-Dependent and -Independent Responses of Sitobion avenae (Homoptera: Aphididae) Feeding on Wheat Infected by Transmitted and Nontransmitted Viruses at Transcriptomic Level. J. Econ. Entomol. 2019, 112, 2067–2076. [Google Scholar] [CrossRef]
- Medeiros, R.B.; Resende Rde, O.; de Avila, A.C. The plant virus Tomato Spotted Wilt Tospovirus activates the immune system of its main insect vector, Frankliniella occidentalis. J. Virol. 2004, 78, 4976–4982. [Google Scholar] [CrossRef]
- Badillo-Vargas, I.E.; Rotenberg, D.; Schneweis, D.J.; Hiromasa, Y.; Tomich, J.M.; Whitfield, A.E. Proteomic analysis of Frankliniella occidentalis and differentially expressed proteins in response to tomato spotted wilt virus infection. J. Virol. 2012, 86, 8793–8809. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, P.; Li, W.; Zhang, J.; Huang, F.; Yang, J.; Bei, Y.; Lu, Y. De novo transcriptome sequencing in Frankliniella occidentalis to identify genes involved in plant virus transmission and insecticide resistance. Genomics 2013, 101, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Schneweis, D.J.; Whitfield, A.E.; Rotenberg, D. Thrips developmental stage-specific transcriptome response to tomato spotted wilt virus during the virus infection cycle in Frankliniella occidentalis, the primary vector. Virology 2017, 500, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Rotenberg, D. Integration of transcriptomics and network analysis reveals co-expressed genes in Frankliniella occidentalis larval guts that respond to tomato spotted wilt virus infection. BMC Genom. 2021, 22, 810. [Google Scholar] [CrossRef]
- Shrestha, A.; Champagne, D.E.; Culbreath, A.K.; Rotenberg, D.; Whitfield, A.E.; Srinivasan, R. Transcriptome changes associated with Tomato spotted wilt virus infection in various life stages of its thrips vector, Frankliniella fusca (Hinds). J. Gen. Virol. 2017, 98, 2156–2170. [Google Scholar] [CrossRef]
- Widana Gamage, S.M.K.; Rotenberg, D.; Schneweis, D.J.; Tsai, C.W.; Dietzgen, R.G. Transcriptome-wide responses of adult melon thrips (Thrips palmi) associated with capsicum chlorosis virus infection. PLoS ONE 2018, 13, e0208538. [Google Scholar] [CrossRef]
- Whitfield, A.E.; Rotenberg, D.; Aritua, V.; Hogenhout, S.A. Analysis of expressed sequence tags from Maize mosaic rhabdovirus-infected gut tissues of Peregrinus maidis reveals the presence of key components of insect innate immunity. Insect Mol. Biol. 2011, 20, 225–242. [Google Scholar] [CrossRef]
- Lee, J.H.; Choi, J.Y.; Tao, X.Y.; Kim, J.S.; Kim, W.; Je, Y.H. Transcriptome Analysis of the Small Brown Planthopper, Laodelphax striatellus Carrying Rice stripe virus. Plant Pathol. J. 2013, 29, 330–337. [Google Scholar] [CrossRef]
- Zhao, W.; Lu, L.; Yang, P.; Cui, N.; Kang, L.; Cui, F. Organ-specific transcriptome response of the small brown planthopper toward rice stripe virus. Insect Biochem. Mol. Biol. 2016, 70, 60–72. [Google Scholar] [CrossRef]
- Yang, M.; Xu, Z.; Zhao, W.; Liu, Q.; Li, Q.; Lu, L.; Liu, R.; Zhang, X.; Cui, F. Rice stripe virus-derived siRNAs play different regulatory roles in rice and in the insect vector Laodelphax striatellus. BMC Plant Biol. 2018, 18, 219. [Google Scholar] [CrossRef]
- Lan, H.; Chen, H.; Liu, Y.; Jiang, C.; Mao, Q.; Jia, D.; Chen, Q.; Wei, T. Small Interfering RNA Pathway Modulates Initial Viral Infection in Midgut Epithelium of Insect after Ingestion of Virus. J. Virol. 2016, 90, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, N.; Gao, X.; Guo, D.; Chang, Z.; Fu, Y.; Akinyemi, I.A.; Wu, Q. Understanding the immune system architecture and transcriptome responses to southern rice black-streaked dwarf virus in Sogatella furcifera. Sci. Rep. 2016, 6, 36254. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.M.; Barandoc-Alviar, K.; Schneweis, D.J.; Stewart, C.L.; Rotenberg, D.; Whitfield, A.E. Transcriptomic response of the insect vector, Peregrinus maidis, to Maize mosaic rhabdovirus and identification of conserved responses to propagative viruses in hopper vectors. Virology 2017, 509, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cassone, B.J.; Bai, X.; Redinbaugh, M.G.; Michel, A.P. Transcriptome of the plant virus vector Graminella nigrifrons, and the molecular interactions of Maize fine streak rhabdovirus transmission. PLoS ONE 2012, 7, e40613. [Google Scholar] [CrossRef]
- Chen, Y.; Redinbaugh, M.G.; Michel, A.P. Molecular interactions and immune responses between Maize fine streak virus and the leafhopper vector Graminella nigrifrons through differential expression and RNA interference. Insect Mol. Biol. 2015, 24, 391–401. [Google Scholar] [CrossRef]
- Shen, J.; Chen, X.; Chen, J.; Sun, L. A phloem-limited fijivirus induces the formation of neoplastic phloem tissues that house virus multiplication in the host plant. Sci. Rep. 2016, 6, 29848. [Google Scholar] [CrossRef]
- Bleckmann, M.; Fritz, M.H.; Bhuju, S.; Jarek, M.; Schurig, M.; Geffers, R.; Benes, V.; Besir, H.; van den Heuvel, J. Genomic Analysis and Isolation of RNA Polymerase II Dependent Promoters from Spodoptera frugiperda. PLoS ONE 2015, 10, e0132898. [Google Scholar] [CrossRef]
- Pena-Llopis, S.; Brugarolas, J. Simultaneous isolation of high-quality DNA, RNA, miRNA and proteins from tissues for genomic applications. Nat. Protoc. 2013, 8, 2240–2255. [Google Scholar] [CrossRef]
- The Gene Ontology, C. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Thorpe, P.; Cock, P.J.; Bos, J. Comparative transcriptomics and proteomics of three different aphid species identifies core and diverse effector sets. BMC Genom. 2016, 17, 172. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.E.; Hackett, K.J.; Purcell-Miramontes, M.; Brown, S.J.; Evans, J.D.; Goldsmith, M.R.; Lawson, D.; Okamuro, J.; Robertson, H.M.; Schneider, D.J. Creating a buzz about insect genomes. Science 2011, 331, 1386. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.W.C.; Dohmen, E.; Hughes, D.S.T.; Murali, S.C.; Poelchau, M.; Glastad, K.; Anstead, C.A.; Ayoub, N.A.; Batterham, P.; Bellair, M.; et al. Gene content evolution in the arthropods. Genome Biol. 2020, 21, 15. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Hasegawa, D.K.; Kaur, N.; Kliot, A.; Pinheiro, P.V.; Luan, J.; Stensmyr, M.C.; Zheng, Y.; Liu, W.; Sun, H.; et al. The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 2016, 14, 110. [Google Scholar] [CrossRef]
- International Aphid Genomics, C. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010, 8, e1000313. [Google Scholar] [CrossRef]
- Legeai, F.; Shigenobu, S.; Gauthier, J.P.; Colbourne, J.; Rispe, C.; Collin, O.; Richards, S.; Wilson, A.C.; Murphy, T.; Tagu, D. AphidBase: A centralized bioinformatic resource for annotation of the pea aphid genome. Insect Mol. Biol. 2010, 19 (Suppl. 2), 5–12. [Google Scholar] [CrossRef]
- Mathers, T.C. Improved Genome Assembly and Annotation of the Soybean Aphid (Aphis glycines Matsumura). G3 (Bethesda) 2020, 10, 899–906. [Google Scholar] [CrossRef]
- Biello, R.; Singh, A.; Godfrey, C.J.; Fernandez, F.F.; Mugford, S.T.; Powell, G.; Hogenhout, S.A.; Mathers, T.C. A chromosome-level genome assembly of the woolly apple aphid, Eriosoma lanigerum Hausmann (Hemiptera: Aphididae). Mol. Ecol. Resour. 2021, 21, 316–326. [Google Scholar] [CrossRef]
- Xue, J.; Zhou, X.; Zhang, C.X.; Yu, L.L.; Fan, H.W.; Wang, Z.; Xu, H.J.; Xi, Y.; Zhu, Z.R.; Zhou, W.W.; et al. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 2014, 15, 521. [Google Scholar] [CrossRef]
- Wang, L.; Tang, N.; Gao, X.; Chang, Z.; Zhang, L.; Zhou, G.; Guo, D.; Zeng, Z.; Li, W.; Akinyemi, I.A.; et al. Genome sequence of a rice pest, the white-backed planthopper (Sogatella furcifera). Gigascience 2017, 6, 1–9. [Google Scholar] [CrossRef]
- Zhu, J.; Jiang, F.; Wang, X.; Yang, P.; Bao, Y.; Zhao, W.; Wang, W.; Lu, H.; Wang, Q.; Cui, N.; et al. Genome sequence of the small brown planthopper, Laodelphax striatellus. Gigascience 2017, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Xu, L.; Hua, H.; Chen, M.; Guo, M.; He, K.; Zhao, J.; Li, F. Chromosomal-level genomes of three rice planthoppers provide new insights into sex chromosome evolution. Mol. Ecol. Resour. 2021, 21, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Rotenberg, D.; Baumann, A.A.; Ben-Mahmoud, S.; Christiaens, O.; Dermauw, W.; Ioannidis, P.; Jacobs, C.G.C.; Vargas Jentzsch, I.M.; Oliver, J.E.; Poelchau, M.F.; et al. Genome-enabled insights into the biology of thrips as crop pests. BMC Biol. 2020, 18, 142. [Google Scholar] [CrossRef]
- Ghosh, S.; Ghanim, M. Factors Determining Transmission of Persistent Viruses by Bemisia tabaci and Emergence of New Virus-Vector Relationships. Viruses 2021, 13, 1808. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, A.E.; Falk, B.W.; Rotenberg, D. Insect vector-mediated transmission of plant viruses. Virology 2015, 479–480, 278–289. [Google Scholar] [CrossRef]
- Montero-Astua, M.; Ullman, D.E.; Whitfield, A.E. Salivary gland morphology, tissue tropism and the progression of tospovirus infection in Frankliniella occidentalis. Virology 2016, 493, 39–51. [Google Scholar] [CrossRef]
- Mauck, K.; Bosque-Pérez, N.A.; Eigenbrode, S.D.; De Moraes, C.M.; Mescher, M.C.; Fox, C. Transmission mechanisms shape pathogen effects on host-vector interactions: Evidence from plant viruses. Funct. Ecol. 2012, 26, 1162–1175. [Google Scholar] [CrossRef]
- Mauck, K.E.; De Moraes, C.M.; Mescher, M.C. Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proc. Natl. Acad. Sci. USA 2010, 107, 3600–3605. [Google Scholar] [CrossRef]
- Bosque-Perez, N.A.; Eigenbrode, S.D. The influence of virus-induced changes in plants on aphid vectors: Insights from luteovirus pathosystems. Virus Res. 2011, 159, 201–205. [Google Scholar] [CrossRef]
- Legarrea, S.; Barman, A.; Marchant, W.; Diffie, S.; Srinivasan, R. Temporal Effects of a Begomovirus Infection and Host Plant Resistance on the Preference and Development of an Insect Vector, Bemisia tabaci, and Implications for Epidemics. PLoS ONE 2015, 10, e0142114. [Google Scholar] [CrossRef]
- Gautam, S.; Mugerwa, H.; Sundaraj, S.; Gadhave, K.R.; Murphy, J.F.; Dutta, B.; Srinivasan, R. Specific and Spillover Effects on Vectors Following Infection of Two RNA Viruses in Pepper Plants. Insects 2020, 11, 602. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Srinivasan, R.; Riley, D.G.; Culbreath, A.K. Direct and indirect effects of a thrips-transmitted Tospoviruson the preference and fitness of its vector, Frankliniella fusca. Entomol. Exp. Appl. 2012, 145, 260–271. [Google Scholar] [CrossRef]
- Maris, P.C.; Joosten, N.N.; Goldbach, R.W.; Peters, D. Tomato spotted wilt virus Infection Improves Host Suitability for Its Vector Frankliniella occidentalis. Phytopathology 2004, 94, 706–711. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Tjallingii, W.F.; Fernandez-Mata, G.; Fereres, A. Differences in the mechanism of inoculation between a semi-persistent and a non-persistent aphid-transmitted plant virus. J. Gen. Virol. 2012, 93, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Pirone, T.P.; Harris, K.F. Nonpersistent Transmission of Plant Viruses by Aphids. Annu. Rev. Phytopathol. 1977, 15, 55–73. [Google Scholar] [CrossRef]
- Froissart, R.; Michalakis, Y.; Blanc, S. Helper component-transcomplementation in the vector transmission of plant viruse. Phytopathology 2002, 92, 576–579. [Google Scholar] [CrossRef]
- Gadhave, K.R.; Gautam, S.; Rasmussen, D.A.; Srinivasan, R. Aphid Transmission of Potyvirus: The Largest Plant-Infecting RNA Virus Genus. Viruses 2020, 12, 773. [Google Scholar] [CrossRef]
- Uzest, M.; Gargani, D.; Drucker, M.; Hebrard, E.; Garzo, E.; Candresse, T.; Fereres, A.; Blanc, S. A protein key to plant virus transmission at the tip of the insect vector stylet. Proc. Natl. Acad. Sci. USA 2007, 104, 17959–17964. [Google Scholar] [CrossRef]
- Fereres, A.; Raccah, B. Plant Virus Transmission by Insects; John Wiley & Sons, Ltd: Chichester, UK, 2015; pp. 1–12. [Google Scholar] [CrossRef]
- Childress, S.A.; Harris, K.F. Localization of Virus-like Particles in the Foreguts of Viruliferous Graminella nigrifrons Leafhoppers Carrying the Semi-persistent Maize Chlorotic Dwarf Virus. J. Gen. Virol. 1989, 70, 247–251. [Google Scholar] [CrossRef]
- Huo, Y.; Liu, W.; Zhang, F.; Chen, X.; Li, L.; Liu, Q.; Zhou, Y.; Wei, T.; Fang, R.; Wang, X. Transovarial transmission of a plant virus is mediated by vitellogenin of its insect vector. PLoS Pathog. 2014, 10, e1003949. [Google Scholar] [CrossRef]
- Smith, C.M.; Gedling, C.R.; Wiebe, K.F.; Cassone, B.J. A sweet story: Bean pod mottle virus transmission dynamics by Mexican bean beetles (Epilachna varivestis). Genome Biol. Evol. 2017, 9, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Stafford-Banks, C.A.; Rotenberg, D.; Johnson, B.R.; Whitfield, A.E.; Ullman, D.E. Analysis of the salivary gland transcriptome of Frankliniella occidentalis. PLoS ONE 2014, 9, e94447. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Champagne, D.E.; Culbreath, A.K.; Abney, M.R.; Srinivasan, R. Comparison of transcriptomes of an orthotospovirus vector and non-vector thrips species. PLoS ONE 2019, 14, e0223438. [Google Scholar] [CrossRef] [PubMed]
- Sharman, M.; Persley, D.M. Field isolates of Tomato spotted wilt virusovercoming resistance in capsicum in Australia. Australas. Plant Pathol. 2006, 35, 123–128. [Google Scholar] [CrossRef]
- Akella, S.V.; Kirk, W.D.; Lu, Y.B.; Murai, T.; Walters, K.F.; Hamilton, J.G. Identification of the aggregation pheromone of the melon thrips, Thrips palmi. PLoS ONE 2014, 9, e103315. [Google Scholar] [CrossRef]
- Ramirez, B.C.; Haenni, A.L. Molecular biology of tenuiviruses, a remarkable group of plant viruses. J. Gen. Virol. 1994, 75 Pt 3, 467–475. [Google Scholar] [CrossRef]
- el Ammar, D.; Tsai, C.W.; Whitfield, A.E.; Redinbaugh, M.G.; Hogenhout, S.A. Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu. Rev. Entomol. 2009, 54, 447–468. [Google Scholar] [CrossRef]
- Rotenberg, D.; Jacobson, A.L.; Schneweis, D.J.; Whitfield, A.E. Thrips transmission of tospoviruses. Curr. Opin. Virol. 2015, 15, 80–89. [Google Scholar] [CrossRef]
- Deshoux, M.; Monsion, B.; Uzest, M. Insect cuticular proteins and their role in transmission of phytoviruses. Curr. Opin. Virol. 2018, 33, 137–143. [Google Scholar] [CrossRef]
- Liang, Y.; Gao, X.W. The Cuticle Protein Gene MPCP4 of Myzus persicae (Homoptera: Aphididae) Plays a Critical Role in Cucumber Mosaic Virus Acquisition. J. Econ. Entomol. 2017, 110, 848–853. [Google Scholar] [CrossRef]
- Stavolone, L.; Lionetti, V. Extracellular Matrix in Plants and Animals: Hooks and Locks for Viruses. Front. MicroBiol. 2017, 8, 1760. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, J.; Shuai, L.; Ma, X.; Zhang, H.; Liu, R.; Chen, W.; Wang, X.; Ge, J.; Wen, Z.; et al. The Serine/Threonine Kinase AP2-Associated Kinase 1 Plays an Important Role in Rabies Virus Entry. Viruses 2019, 12, 45. [Google Scholar] [CrossRef]
- Botello-Smith, W.M.; Alsamarah, A.; Chatterjee, P.; Xie, C.; Lacroix, J.J.; Hao, J.; Luo, Y. Polymodal allosteric regulation of Type 1 Serine/Threonine Kinase Receptors via a conserved electrostatic lock. PLoS Comput. Biol. 2017, 13, e1005711. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.H.; Kissinger, J.C. Consistent and contrasting properties of lineage-specific genes in the apicomplexan parasites Plasmodium and Theileria. BMC Evol. Biol. 2008, 8, 108. [Google Scholar] [CrossRef]
- Agnello, V.; Abel, G.; Elfahal, M.; Knight, G.B.; Zhang, Q.X. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 1999, 96, 12766–12771. [Google Scholar] [CrossRef] [PubMed]
- Finkelshtein, D.; Werman, A.; Novick, D.; Barak, S.; Rubinstein, M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 2013, 110, 7306–7311. [Google Scholar] [CrossRef]
- Mitrovic, S.; Ben-Tekaya, H.; Koegler, E.; Gruenberg, J.; Hauri, H.P. The cargo receptors Surf4, endoplasmic reticulum-Golgi intermediate compartment (ERGIC)-53, and p25 are required to maintain the architecture of ERGIC and Golgi. Mol. Biol. Cell 2008, 19, 1976–1990. [Google Scholar] [CrossRef] [PubMed]
- Feyzi, E.; Trybala, E.; Bergstrom, T.; Lindahl, U.; Spillmann, D. Structural requirement of heparan sulfate for interaction with herpes simplex virus type 1 virions and isolated glycoprotein C. J. Biol. Chem. 1997, 272, 24850–24857. [Google Scholar] [CrossRef][Green Version]
- Amari, K.; Di Donato, M.; Dolja, V.V.; Heinlein, M. Myosins VIII and XI play distinct roles in reproduction and transport of tobacco mosaic virus. PLoS Pathog. 2014, 10, e1004448. [Google Scholar] [CrossRef]
- Govindaraj, L.; Gupta, T.; Esvaran, V.G.; Awasthi, A.K.; Ponnuvel, K.M. Genome-wide identification, characterization of sugar transporter genes in the silkworm Bombyx mori and role in Bombyx mori nucleopolyhedrovirus (BmNPV) infection. Gene 2016, 579, 162–171. [Google Scholar] [CrossRef]
- Potter, L.R.; Yoder, A.R.; Flora, D.R.; Antos, L.K.; Dickey, D.M. Natriuretic peptides: Their structures, receptors, physiologic functions and therapeutic applications. Handb. Exp. Pharmacol. 2009, 341–366. [Google Scholar] [CrossRef]
- Tsunematsu, T.; Yamanaka, A. The role of orexin/hypocretin in the central nervous system and peripheral tissues. Vitam Horm 2012, 89, 19–33. [Google Scholar] [CrossRef]
- Kakava-Georgiadou, N.; Bullich-Vilarrubias, C.; Zwartkruis, M.M.; Luijendijk, M.C.M.; Garner, K.M.; Adan, R.A.H. Considerations related to the use of short neuropeptide promoters in viral vectors targeting hypothalamic neurons. Sci. Rep. 2019, 9, 11146. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.H.; Huang, L.M.; Tarn, W.Y. The Host Heat Shock Protein MRJ/DNAJB6 Modulates Virus Infection. Front. MicroBiol. 2019, 10, 2885. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.G.; Amici, C.; Rossi, A. Role of Heat Shock Proteins in Viral Infection. In Prokaryotic and Eukaryotic Heat Shock Proteins in Infectious Disease; Pockley, A.G., Calderwood, S.K., Santoro, M.G., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 51–84. [Google Scholar] [CrossRef]
- Gorovits, R.; Czosnek, H. The Involvement of Heat Shock Proteins in the Establishment of Tomato Yellow Leaf Curl Virus Infection. Front. Plant Sci. 2017, 8, 355. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Q.; Hu, N.; Dong, F.F.; Chen, T.T.; Jiang, Y.M.; Chen, P.; Lu, C.; Pan, M.H. Baculovirus LEF-11 Hijack Host ATPase ATAD3A to Promote Virus Multiplication in Bombyx mori cells. Sci. Rep. 2017, 7, 46187. [Google Scholar] [CrossRef]
- Lewis, V.G.; Ween, M.P.; McDevitt, C.A. The role of ATP-binding cassette transporters in bacterial pathogenicity. Protoplasma 2012, 249, 919–942. [Google Scholar] [CrossRef] [PubMed]
- Hariton Shalev, A.; Sobol, I.; Ghanim, M.; Liu, S.S.; Czosnek, H. The Whitefly Bemisia tabaci Knottin-1 Gene Is Implicated in Regulating the Quantity of Tomato Yellow Leaf Curl Virus Ingested and Transmitted by the Insect. Viruses 2016, 8, 205. [Google Scholar] [CrossRef] [PubMed]
- Gelly, J.C.; Gracy, J.; Kaas, Q.; Le-Nguyen, D.; Heitz, A.; Chiche, L. The KNOTTIN website and database: A new information system dedicated to the knottin scaffold. Nucleic Acids Res. 2004, 32, D156–D159. [Google Scholar] [CrossRef]
- Ma, G.; Yasunaga, J.; Akari, H.; Matsuoka, M. TCF1 and LEF1 act as T-cell intrinsic HTLV-1 antagonists by targeting Tax. Proc. Natl. Acad. Sci. USA 2015, 112, 2216–2221. [Google Scholar] [CrossRef]
- Lefebvre, L.; Vanderplasschen, A.; Ciminale, V.; Heremans, H.; Dangoisse, O.; Jauniaux, J.C.; Toussaint, J.F.; Zelnik, V.; Burny, A.; Kettmann, R.; et al. Oncoviral bovine leukemia virus G4 and human T-cell leukemia virus type 1 p13(II) accessory proteins interact with farnesyl pyrophosphate synthetase. J. Virol. 2002, 76, 1400–1414. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chi, X.; Wei, H.; Chen, Y.; Chen, Z.; Huang, S.; Chen, J.L. Influenza A virus-induced degradation of eukaryotic translation initiation factor 4B contributes to viral replication by suppressing IFITM3 protein expression. J. Virol. 2014, 88, 8375–8385. [Google Scholar] [CrossRef] [PubMed]
- Bar-Nun, S.; Glickman, M.H. Proteasomal AAA-ATPases: Structure and function. Biochim. Biophys. Acta 2012, 1823, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Dukhovny, A.; Shlomai, A.; Sklan, E.H. The antiviral protein Viperin suppresses T7 promoter dependent RNA synthesis-possible implications for its antiviral activity. Sci. Rep. 2018, 8, 8100. [Google Scholar] [CrossRef]
- Seo, J.Y.; Yaneva, R.; Cresswell, P. Viperin: A multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 2011, 10, 534–539. [Google Scholar] [CrossRef]
- Sanchez, E.L.; Lagunoff, M. Viral activation of cellular metabolism. Virology 2015, 479–480, 609–618. [Google Scholar] [CrossRef]
- Wang, X.Y.; Yu, H.Z.; Xu, J.P.; Zhang, S.Z.; Yu, D.; Liu, M.H.; Wang, L.L. Comparative Subcellular Proteomics Analysis of Susceptible and Near-isogenic Resistant Bombyx mori (Lepidoptera) Larval Midgut Response to BmNPV infection. Sci. Rep. 2017, 7, 45690. [Google Scholar] [CrossRef]
- Wang, Z.; Wilhelmsson, C.; Hyrsl, P.; Loof, T.G.; Dobes, P.; Klupp, M.; Loseva, O.; Morgelin, M.; Ikle, J.; Cripps, R.M.; et al. Pathogen entrapment by transglutaminase--a conserved early innate immune mechanism. PLoS Pathog. 2010, 6, e1000763. [Google Scholar] [CrossRef]
- Eliautout, R.; Dubrana, M.P.; Vincent-Monegat, C.; Vallier, A.; Braquart-Varnier, C.; Poirie, M.; Saillard, C.; Heddi, A.; Arricau-Bouvery, N. Immune response and survival of Circulifer haematoceps to Spiroplasma citri infection requiRes. expression of the gene hexamerin. Dev. Comp. Immunol. 2016, 54, 7–19. [Google Scholar] [CrossRef]
- Ramsey, J.S.; Chavez, J.D.; Johnson, R.; Hosseinzadeh, S.; Mahoney, J.E.; Mohr, J.P.; Robison, F.; Zhong, X.; Hall, D.G.; MacCoss, M.; et al. Protein interaction networks at the host-microbe interface in Diaphorina citri, the insect vector of the citrus greening pathogen. R. Soc. Open Sci. 2017, 4, 160545. [Google Scholar] [CrossRef]
- Arai, H.; Koizumi, H.; Aoki, J.; Inoue, K. Platelet-activating factor acetylhydrolase (PAF-AH). J. BioChem. 2002, 131, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Kurata, S.; Natori, S. Purification and characterization of a hemocyte proteinase of Sarcophaga, possibly participating in elimination of foreign substances. Eur. J. BioChem. 1992, 209, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Yoshiga, T.; Georgieva, T.; Dunkov, B.C.; Harizanova, N.; Ralchev, K.; Law, J.H. Drosophila melanogaster transferrin. Cloning, deduced protein sequence, expression during the life cycle, gene localization and up-regulation on bacterial infection. Eur. J. BioChem. 1999, 260, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Yoshiga, T.; Hernandez, V.P.; Fallon, A.M.; Law, J.H. Mosquito transferrin, an acute-phase protein that is up-regulated upon infection. Proc. Natl. Acad. Sci. USA 1997, 94, 12337–12342. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.T.; Liu, H.J. PI3K-Akt signaling and viral infection. Recent Pat. Biotechnol. 2008, 2, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Yu, F.X.; Kim, Y.C.; Meng, Z.; Naipauer, J.; Looney, D.J.; Liu, X.; Gutkind, J.S.; Mesri, E.A.; Guan, K.L. Kaposi sarcoma-associated herpesvirus promotes tumorigenesis by modulating the Hippo pathway. Oncogene 2015, 34, 3536–3546. [Google Scholar] [CrossRef] [PubMed]
- Kubo, Y.; Hayashi, H.; Matsuyama, T.; Sato, H.; Yamamoto, N. Retrovirus entry by endocytosis and cathepsin proteases. Adv. Virol. 2012, 2012, 640894. [Google Scholar] [CrossRef]
- Sim, S.; Ramirez, J.L.; Dimopoulos, G. Dengue virus infection of the Aedes aegypti salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012, 8, e1002631. [Google Scholar] [CrossRef]
- Saikhedkar, N.; Summanwar, A.; Joshi, R.; Giri, A. Cathepsins of lepidopteran insects: Aspects and prospects. Insect BioChem. Mol. Biol. 2015, 64, 51–59. [Google Scholar] [CrossRef]
- Yadati, T.; Houben, T.; Bitorina, A.; Shiri-Sverdlov, R. The Ins and Outs of Cathepsins: Physiological Function and Role in Disease Management. Cells 2020, 9, 1679. [Google Scholar] [CrossRef]
- Patel, S.; Homaei, A.; El-Seedi, H.R.; Akhtar, N. Cathepsins: Proteases that are vital for survival but can also be fatal. Biomed. Pharmacother. 2018, 105, 526–532. [Google Scholar] [CrossRef]
- Patel, D.M.; Ahmad, S.F.; Weiss, D.G.; Gerke, V.; Kuznetsov, S.A. Annexin A1 is a new functional linker between actin filaments and phagosomes during phagocytosis. J. Cell Sci. 2011, 124, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Zambon, R.A.; Vakharia, V.N.; Wu, L.P. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell MicroBiol. 2006, 8, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Ramirez, J.L.; Dimopoulos, G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog 2008, 4, e1000098. [Google Scholar] [CrossRef]
- Nakano, M.Y.; Boucke, K.; Suomalainen, M.; Stidwill, R.P.; Greber, U.F. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J. Virol. 2000, 74, 7085–7095. [Google Scholar] [CrossRef]
- Sieczkarski, S.B.; Brown, H.A.; Whittaker, G.R. Role of protein kinase C betaII in influenza virus entry via late endosomes. J. Virol. 2003, 77, 460–469. [Google Scholar] [CrossRef]
- Chu, J.J.; Leong, P.W.; Ng, M.L. Analysis of the endocytic pathway mediating the infectious entry of mosquito-borne flavivirus West Nile into Aedes albopictus mosquito (C6/36) cells. Virology 2006, 349, 463–475. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Kriventseva, E.V.; Meister, S.; Xi, Z.; Alvarez, K.S.; Bartholomay, L.C.; Barillas-Mury, C.; Bian, G.; Blandin, S.; Christensen, B.M.; et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 2007, 316, 1738–1743. [Google Scholar] [CrossRef]
- Christophides, G.K.; Zdobnov, E.; Barillas-Mury, C.; Birney, E.; Blandin, S.; Blass, C.; Brey, P.T.; Collins, F.H.; Danielli, A.; Dimopoulos, G.; et al. Immunity-related genes and gene families in Anopheles gambiae. Science 2002, 298, 159–165. [Google Scholar] [CrossRef]
- Jiravanichpaisal, P.; Lee, B.L.; Soderhall, K. Cell-mediated immunity in arthropods: Hematopoiesis, coagulation, melanization and opsonization. Immunobiology 2006, 211, 213–236. [Google Scholar] [CrossRef]
- Ronningen, T.; Shah, A.; Oldenburg, A.R.; Vekterud, K.; Delbarre, E.; Moskaug, J.O.; Collas, P. Prepatterning of differentiation-driven nuclear lamin A/C-associated chromatin domains by GlcNAcylated histone H2B. Genome Res. 2015, 25, 1825–1835. [Google Scholar] [CrossRef] [PubMed]
- Franc, N.C.; Heitzler, P.; Ezekowitz, R.A.; White, K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science 1999, 284, 1991–1994. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Wang, J.L.; Cheng, Y.; Wang, J.X.; Zou, Z. Pattern recognition receptors from lepidopteran insects and their biological functions. Dev. Comp. Immunol. 2020, 108, 103688. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. bantam Encodes a Developmentally Regulated microRNA that Controls Cell Proliferation and Regulates the Proapoptotic Gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef]
- Shi, X.; Tang, X.; Zhang, X.; Zhang, D.; Li, F.; Yan, F.; Zhang, Y.; Zhou, X.; Liu, Y. Transmission Efficiency, Preference and Behavior of Bemisia tabaci MEAM1 and MED under the Influence of Tomato Chlorosis Virus. Front. Plant Sci. 2017, 8, 2271. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Preisser, E.L.; Zhou, X.M.; Xie, W.; Liu, B.M.; Wang, S.L.; Wu, Q.J.; Zhang, Y.J. Manipulation of Host Quality and Defense by a Plant Virus Improves Performance of Whitefly Vectors. J. Econ. Entomol. 2015, 108, 11–19. [Google Scholar] [CrossRef]
- Rubinstein, G.; Czosnek, H. Long-term association of tomato yellow leaf curl virus with its whitefly vector Bemisia tabaci: Effect on the insect transmission capacity, longevity and fecundity. J. Gen. Virol. 1997, 78 Pt 10, 2683–2689. [Google Scholar] [CrossRef]
- Pusag, J.C.; Hemayet Jahan, S.M.; Lee, K.S.; Lee, S.; Lee, K.Y. Upregulation of temperature susceptibility in Bemisia tabaci upon acquisition of Tomato yellow leaf curl virus (TYLCV). J. Insect Physiol. 2012, 58, 1343–1348. [Google Scholar] [CrossRef]
- Zhai, Y.; Zhang, J.; Sun, Z.; Dong, X.; He, Y.; Kang, K.; Liu, Z.; Zhang, W. Proteomic and transcriptomic analyses of fecundity in the brown planthopper Nilaparvata lugens (Stal). J. Proteome Res. 2013, 12, 5199–5212. [Google Scholar] [CrossRef]
- Mamta, B.; Rajam, M.V. RNAi technology: A new platform for crop pest control. Physiol. Mol. Biol. Plants 2017, 23, 487–501. [Google Scholar] [CrossRef]
- Worrall, E.A.; Bravo-Cazar, A.; Nilon, A.T.; Fletcher, S.J.; Robinson, K.E.; Carr, J.P.; Mitter, N. Exogenous Application of RNAi-Inducing Double-Stranded RNA Inhibits Aphid-Mediated Transmission of a Plant Virus. Front. Plant Sci. 2019, 10, 265. [Google Scholar] [CrossRef]
- Das, P.R.; Sherif, S.M. Application of Exogenous dsRNAs-induced RNAi in Agriculture: Challenges and Triumphs. Front. Plant Sci. 2020, 11, 946. [Google Scholar] [CrossRef]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.W.; Broeck, J.V. RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front. Physiol. 2018, 9, 1912. [Google Scholar] [CrossRef]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Fajardo-Ortiz, D.; Shattuck, A.; Hornbostel, S. Mapping the coevolution, leadership and financing of research on viral vectors, RNAi, CRISPR/Cas9 and other genomic editing technologies. PLoS ONE 2020, 15, e0227593. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liao, Z.; Chen, Y.; Han, L.; Yin, Q.; Xiao, H. Application of Various Delivery Methods for CRISPR/dCas9. Mol. Biotechnol. 2020, 62, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.Y.; Palli, S.R. Mechanisms, Applications, and Challenges of Insect RNA Interference. Annu. Rev. Entomol. 2020, 65, 293–311. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 15, 2114–2120. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 19, 3210–3212. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNAseq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 1, 139–140. [Google Scholar] [CrossRef]
- Conesa, A.; Göz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).