Proteomic Analysis Dissects Molecular Mechanisms Underlying Plant Responses to Phosphorus Deficiency
Abstract
1. Introduction
2. Morphological, Physiological, and Biochemical Responses of Plants to Pi Starvation
3. DAPs Reveal Complex Repones of Plants to P Deficiency
3.1. Identification of DAPs in Plant Leaves/Shoots
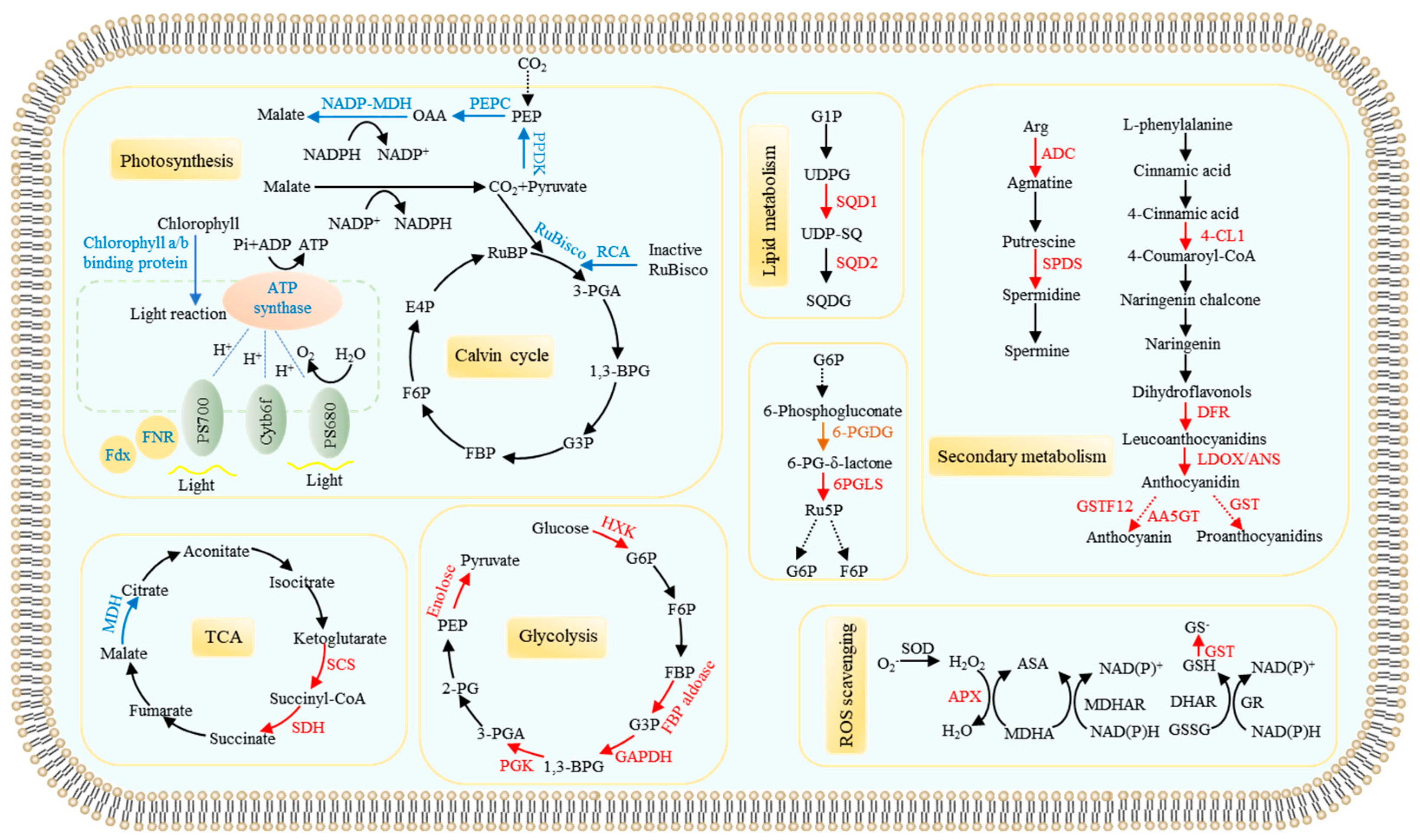
3.1.1. Differential Proteins Related to Photosynthesis and Carbon Metabolism
3.1.2. Pi Starvation Responsive Proteins Related to Remodeling Lipid Membranes
3.1.3. Proteins Involved Anthocyanin, Polyamine, and Reactive Oxygen Species (ROS) Metabolisms
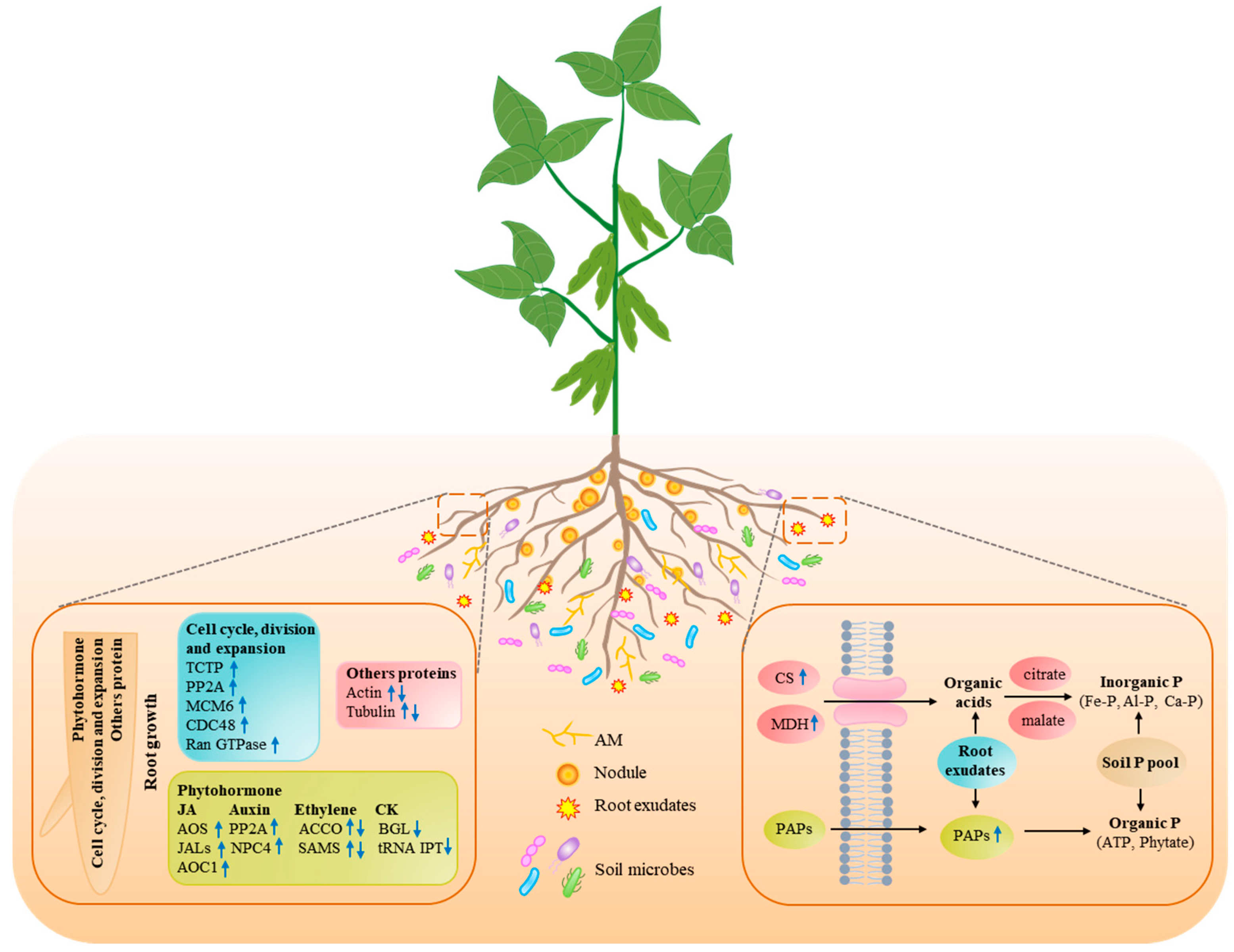
3.2. Identification of DAPs in Plant Roots
3.2.1. Pi Starvation-Responsive Proteins Participated in Root System Remodeling
3.2.2. Pi Starvation-Responsive Proteins Related to Root Exudates
3.2.3. Response of Symbiotic Association to Pi Starvation in Plants
4. Identification of DAPs Exhibiting Post-Transcriptional Modifications
5. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Cashikar, A.G.; Kumaresan, R.; Rao, N.M. Biochemical characterization and subcellular localization of the red kidney bean purple acid phosphatase. Plant Physiol. 1997, 114, 907–915. [Google Scholar] [CrossRef]
- Raghothama, K.G. Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 665–693. [Google Scholar] [CrossRef] [PubMed]
- Plaxton, W.C.; Shane, M.W. The role of post-translational enzyme modifications in the metabolic adaptations of phosphorus-deprived plants. Annu. Plant Rev. 2015, 48, 99–123. [Google Scholar]
- Oldroyd, G.; Leyser, O. A plant’s diet, surviving in a variable nutrient environment. Science 2020, 368, eaba0196. [Google Scholar] [CrossRef] [PubMed]
- Ham, B.K.; Chen, J.; Yan, Y.; Lucas, W.J. Insights into plant phosphate sensing and signaling. Curr. Opin. Biotechnol. 2018, 49, 1–9. [Google Scholar] [CrossRef]
- Hinsinger, P. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: A review. Plant Soil 2001, 237, 173–195. [Google Scholar] [CrossRef]
- Medici, A.; Szponarski, W.; Dangeville, P.; Safi, A.; Dissanayake, I.M.; Saenchai, C.; Emanuel, A.; Rubio, V.; Lacombe, B.; Ruffel, S.; et al. Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. Plant Cell 2019, 31, 1171–1184. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Bouain, N.; Zheng, L.; Rouached, H. Plant resilience to phosphate limitation: Current knowledge and future challenges. Crit. Rev. Biotechnol. 2021, 41, 63–71. [Google Scholar] [CrossRef]
- Lambers, H.; Finnegan, P.M.; Jost, R.; Plaxton, W.C.; Shane, M.W.; Stitt, M. Phosphorus nutrition in Proteaceae and beyond. Nat. Plants 2015, 1, 15109. [Google Scholar] [CrossRef] [PubMed]
- Hallama, M.; Pekrun, C.; Lambers, H.; Kandeler, E. Hidden miners-the roles of cover crops and soil microorganisms in phosphorus cycling through agroecosystems. Plant Soil 2019, 434, 7–45. [Google Scholar] [CrossRef]
- Abdelrahman, M.; El-Sayed, M.A.; Hashem, A.; Abd, A.E.; Alqarawi, A.A.; Burritt, D.J.; Tran, L.P. Metabolomics and transcriptomics in legumes under phosphate deficiency in relation to nitrogen fixation by root nodules. Front. Plant Sci. 2018, 9, 922. [Google Scholar] [CrossRef]
- Deng, Q.; Luo, X.; Chen, Y.; Zhou, Y.; Zhang, F.; Hu, B.; Xie, K. Transcriptome analysis of phosphorus stress responsiveness in the seedlings of Dongxiang wild rice (Oryza rufipogon Griff.). Biol. Res. 2018, 51, 7. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, J.; Trigueros, M.; Rojas-Triana, M.; Fernández, M.; Albar, J.P.; Bustos, R.; Paz-Ares, J.; Rubio, V. Proteomics identifies ubiquitin-proteasome targets and new roles for chromatin-remodeling in the Arabidopsis response to phosphate starvation. J. Proteom. 2013, 94, 1–22. [Google Scholar] [CrossRef]
- Watanabe, T.; Urayama, M.; Shinano, T.; Okada, R.; Osaki, M. Application of ionomics to plant and soil in fields under long-term fertilizer trials. Springerplus 2015, 4, 781. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; He, P.; Zhou, M.; Lu, X.; Chen, K.; Liang, C.; Tian, J. Soybean responds to phosphate starvation through reversible protein phosphorylation. Plant Physiol. Biochem. 2021, 167, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Lan, P.; Li, W.; Schmidt, W. Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell. Proteom. 2012, 11, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Q.; Zhou, X.; Dong, L.; Guo, J.; Chen, Y.; Zhang, Y.; Wu, L.; Xu, M. iTRAQ-based analysis of the Arabidopsis proteome reveals insights into the potential mechanisms of anthocyanin accumulation regulation in response to phosphate deficiency. J. Proteom. 2018, 184, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Liu, H.; Tao, P.; Chen, H. Comparative proteomic analyses provide new insights into low phosphorus stress responses in maize leaves. PLoS ONE 2014, 9, e98215. [Google Scholar]
- Li, K.; Xu, C.; Li, Z.; Zhang, K.; Yang, A.; Zhang, J. Comparative proteome analyses of phosphorus responses in maize (Zea mays L.) roots of wild-type and a low-P-tolerant mutant reveal root characteristics associated with phosphorus efficiency. Plant J. 2008, 55, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Muneer, S.; Jeong, B.R. Proteomic analysis provides new insights in phosphorus homeostasis subjected to Pi (inorganic phosphate) starvation in tomato plants (Solanum lycopersicum L.). PLoS ONE 2015, 10, e0134103. [Google Scholar]
- Sepideh, T.; Matthias, W.; Manzar, H.; Mohammad-Reza, N.; Gilany, K.; Mohammad-Reza, H.; Mansoor, O.; Yazdi-Samadi, B.; Abdelbagi, M.I. A comparative proteome approach to decipher the mechanism of rice adaptation to phosphorous deficiency. Proteomics 2009, 9, 159–170. [Google Scholar]
- Yang, J.; Xie, M.; Yang, X.; Liu, B.; Lin, H. Phosphoproteomic profiling reveals the importance of CK2, MAPKs and CDPKs in response to phosphate starvation in rice. Plant Cell Physiol. 2019, 60, 2785–2796. [Google Scholar] [CrossRef] [PubMed]
- Nadira, U.A.; Ahmed, I.M.; Zeng, J.; Wu, F.; Zhang, G. Identification of the differentially accumulated proteins associated with low phosphorus tolerance in a Tibetan wild barley accession. J. Plant Physiol. 2016, 198, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cui, Q.; Liang, C.; Sun, L.; Tian, J.; Liao, H. Identification of differentially expressed proteins in soybean nodules under phosphorus deficiency through proteomic analysis. Proteomics 2011, 11, 4648–4659. [Google Scholar] [CrossRef]
- Wu, W.; Lin, Y.; Liu, P.; Chen, Q.; Tian, J.; Liang, C. Association of extracellular dNTP utilization with a GmPAP1-like protein identified in cell wall proteomic analysis of soybean roots. J. Exp. Bot. 2018, 69, 603–617. [Google Scholar] [CrossRef]
- Deng, G.; Liu, L.J.; Zhong, X.Y.; Lao, C.Y.; Wang, H.Y.; Wang, B.; Zhu, C.; Shah, F.; Peng, D.X. Comparative proteome analysis of the response of ramie under N, P and K deficiency. Planta 2014, 239, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Liu, H.; Song, J.; Wu, W.; Li, K.; Zhang, J. Physiological and comparative proteome analyses reveal low-phosphate tolerance and enhanced photosynthesis in a maize mutant owing to reinforced inorganic phosphate recycling. BMC Plant Biol. 2016, 16, 129. [Google Scholar] [CrossRef]
- Chu, S.; Li, H.; Zhang, X.; Yu, K.; Chao, M.; Han, S.; Zhang, D. Physiological and proteomics analyses reveal low-phosphorus stress affected the regulation of photosynthesis in soybean. Int. J. Mol. Sci. 2018, 19, 1688. [Google Scholar] [CrossRef]
- Cheng, L.; Min, W.; Li, M.; Zhou, L.; Hsu, C.; Yang, X.; Jiang, X.; Ruan, Z.; Zhong, Y.; Wang, Z.; et al. Quantitative proteomics reveals that GmENO2 proteins are involved in response to phosphate starvation in the leaves of Glycine max L. Int. J. Mol. Sci. 2021, 22, 920. [Google Scholar] [CrossRef]
- Chevalier, F.; Rossignol, M. Proteomic analysis of Arabidopsis thaliana ecotypes with contrasted root architecture in response to phosphate deficiency. J. Plant Physiol. 2011, 168, 1885–1890. [Google Scholar] [CrossRef]
- Li, K.; Xu, C.; Zhang, K.; Yang, A.; Zhang, J. Proteomic analysis of roots growth and metabolic changes under phosphorus deficit in maize (Zea mays L.) plants. Proteomics 2007, 7, 1501–1512. [Google Scholar] [CrossRef]
- Li, K.; Xu, C.; Fan, W.; Zhang, H.; Hou, J.; Yang, A.; Zhang, K. Phosphoproteome and proteome analyses reveal low-phosphate mediated plasticity of root developmental and metabolic regulation in maize (Zea mays L.). Plant Physiol. Biochem. 2014, 83, 232–242. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, J.; Han, Z.; Yang, J.; Ge, C.; Wu, Q. Revealing new insights into different phosphorus-starving responses between two maize (Zea mays) inbred lines by transcriptomic and proteomic studies. Sci. Rep. 2017, 7, 44294. [Google Scholar] [CrossRef]
- Kim, S.G.; Wang, Y.; Lee, C.H.; Mun, B.G.; Kim, P.J.; Lee, S.Y.; Kim, Y.C.; Kang, K.Y.; Rakwal, R.; Agrawal, G.K.; et al. A comparative proteomics survey of proteins responsive to phosphorous starvation in roots of hydroponically-grown rice seedlings. J. Korean Soc. Appl. Biol. Chem. 2011, 54, 667–677. [Google Scholar] [CrossRef]
- Vengavasi, K.; Pandey, R.; Abraham, G.; Yadav, R. Comparative analysis of soybean root proteome reveals molecular basis of differential carboxylate efflux under low phosphorus stress. Genes 2017, 8, 341. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yang, A.; Kong, L.; Xie, F.; Wang, H.; Ao, X. Proteome characterization of two contrasting soybean genotypes in response to different phosphorus treatments. AoB Plants 2021, 13, plab019. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, G.; Li, G.; Yuan, S.; Wang, C.; Xie, Y.; Guo, T.; Kang, G.; Wang, D. TaPHT1;9-4B and its transcriptional regulator TaMYB4-7D contribute to phosphate uptake and plant growth in bread wheat. New Phytol. 2021, 231, 1968–1983. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ma, Z.; Li, C.; Ren, P.; Yao, L.; Li, B.; Meng, Y.; Ma, X.; Si, E.; Yang, K.; et al. Dynamic responses of barley root succinyl-proteome to short-term phosphate starvation and recovery. Front. Plant Sci. 2021, 12, 649147. [Google Scholar] [CrossRef]
- Chen, S.; Luo, Y.; Ding, G.; Xu, F. Comparative analysis of Brassica napus plasma membrane proteins under phosphorus deficiency using label-free and MaxQuant-based proteomics approaches. J. Proteom. 2016, 133, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Huang, L.; Pan, G.; Liu, L.; Wang, X.; Lu, L. Identifying the genes regulated by AtWRKY6 using comparative transcript and proteomic analysis under phosphorus deficiency. Int. J. Mol. Sci. 2017, 18, 1046. [Google Scholar] [CrossRef]
- Yao, Y.; Sun, H.; Xu, F.; Zhang, X.; Liu, S. Comparative proteome analysis of metabolic changes by low phosphorus stress in two Brassica napus genotypes. Planta 2011, 233, 523–537. [Google Scholar] [CrossRef]
- Fan, F.H.; Ding, G.J.; Wen, X.P. Proteomic analyses provide new insights into the responses of Pinus massoniana seedlings to phosphorus deficiency. Proteomics 2016, 16, 504–515. [Google Scholar] [CrossRef]
- Tran, H.T.; Plaxton, W.C. Proteomic analysis of alterations in the secretome of Arabidopsis thaliana suspension cells subjected to nutritional phosphate deficiency. Proteomics 2008, 8, 4317–4326. [Google Scholar] [CrossRef]
- Mehta, D.; Ghahremani, M.; Pérez Fernández, M.; Tan, M.; Schläpfer, P.; Plaxton, W.C.; Uhrig, R.G. Phosphate and phosphite have a differential impact on the proteome and phosphoproteome of Arabidopsis suspension cell cultures. Plant J. 2021, 105, 924–941. [Google Scholar] [CrossRef]
- Uzokwe, V.N.E.; Asafo-Adjei, B.; Fawole, I.; Abaidoo, R.; Odeh, I.O.A.; Ojo, D.K.; Dashiell, K.; Sanginga, N. Generation mean analysis of phosphorus-use efficiency in freely nodulating soybean crosses grown in low-phosphorus soil. Plant Breed. 2017, 136, 139–146. [Google Scholar] [CrossRef]
- Sims, L.; Pastor, J.; Lee, T.; Dewey, B. Nitrogen, phosphorus and light effects on growth and allocation of biomass and nutrients in wild rice. Oecologia 2012, 170, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Wang, C.; Zhang, Q.; He, X.; Whelan, J.; Shou, H. Overexpression of OsPAP10a, a root-associated acid phosphatase, increased extracellular organic phosphorus utilization in rice. J. Integr. Plant Biol. 2012, 54, 631–639. [Google Scholar] [CrossRef]
- Liang, C.; Wang, J.; Zhao, J.; Tian, J.; Liao, H. Control of phosphate homeostasis through gene regulation in crops. Curr. Opin. Plant Biol. 2014, 21, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Dissanayaka, D.M.S.B.; Nishida, S.; Tawaraya, K.; Wasaki, J. Organ-specific allocation pattern of acquired phosphorus and dry matter in two rice genotypes with contrasting tolerance to phosphorus deficiency. Soil Sci. Plant Nutr. 2018, 64, 282–290. [Google Scholar] [CrossRef]
- Lopez-Bucio, J.; Cruz-Ramirez, A.; Herrera-Estrella, L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 2003, 6, 280–287. [Google Scholar] [CrossRef]
- Peret, B.; Desnos, T.; Jost, R.; Kanno, S.; Berkowitz, O.; Nussaume, L. Root architecture responses: In search of phosphate. Plant Physiol. 2014, 166, 1713–1723. [Google Scholar] [CrossRef]
- Horsham, V. Proteoid root morphology and function in lupinus albus. Plant Soil 1981, 60, 143–147. [Google Scholar]
- Neumann, G.; Massonneau, A.; Langlade, N.; Dinkelaker, B.; Hengeler, C.; Roemheld, V.; Martinoia, E. Physiological aspects of cluster root function and development in phosphorus-deficient white lupin (Lupinus albus L.). Ann. Bot. 2000, 85, 909–919. [Google Scholar] [CrossRef]
- Lambers, H.; Finnegan, P.M.; Laliberte, E.; Pearse, S.J.; Ryan, M.H.; Shane, M.W.; Veneklaas, E.J. Update on phosphorus nutrition in proteaceae. Phosphorus nutrition of proteaceae in severely phosphorus-impoverished soils: Are there lessons to be learned for future crops? Plant Physiol. 2011, 156, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.C.; Liao, H. Organic acid anions: An effective defensive weapon for plants against aluminum toxicity and phosphorus deficiency in acidic soils. J. Genet. Genom. 2016, 43, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Dissanayaka, D.; Plaxton, W.C.; Lambers, H.; Siebers, M.; Marambe, B.; Wasaki, J. Molecular mechanisms underpinning phosphorus-use efficiency in rice. Plant Cell Environ. 2018, 41, 1483–1496. [Google Scholar] [CrossRef] [PubMed]
- Tarafdar, J.C.; Marchner, H. Efficiency of VAM hyphae in utilisation of organic phosphorus by wheat plants. Soil Sci. Plant Nutr. 1994, 40, 593–600. [Google Scholar] [CrossRef]
- Sa, T.M.; Israel, D.W. Energy status and functioning of phosphorus-deficient soybean nodules. Plant Physiol. 1991, 97, 928–935. [Google Scholar] [CrossRef]
- Drevon, J.; Hartwig, U.A. Phosphorus deficiency increases the argon-induced decline of nodule nitrogenase activity in soybean and alfalfa. Planta 1997, 201, 463–469. [Google Scholar] [CrossRef]
- Hodge, A.; Berta, G.; Doussan, C.; Merchan, F.; Crespi, M. Plant root growth, architecture and function. Plant Soil 2009, 321, 153–187. [Google Scholar] [CrossRef]
- Hiruma, K.; Gerlach, N.; Sacristan, S.; Nakano, R.T.; Hacquard, S.; Kracher, B.; Neumann, U.; Ramirez, D.; Bucher, M.; O’Connell, R.J.; et al. Root endophyte colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 2016, 165, 464–474. [Google Scholar] [CrossRef]
- Umehara, M.; Hanada, A.; Magome, H.; Takeda-Kamiya, N.; Yamaguchi, S. Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 2010, 51, 1118–1126. [Google Scholar] [CrossRef]
- Mghase, J.J.; Shiwachi, H.; Takahashi, H.; Irie, K. Nutrient deficiencies and their symptoms in upland rice. J. ISSAAS 2011, 17, 59–67. [Google Scholar]
- Ruan, W.; Guo, M.; Xu, L.; Wang, X.; Zhao, H.; Wang, J.; Yi, K. An SPX-RLI1 module regulates leaf inclination in response to phosphate availability in rice. Plant Cell 2018, 30, 853–870. [Google Scholar] [CrossRef]
- Mo, X.; Zhang, M.; Zhang, Z.; Lu, X.; Liang, C.; Tian, J. Phosphate (Pi) starvation up-regulated GmCSN5A/B participates in anthocyanin synthesis in Soybean (Glycine max) dependent on Pi availability. Int. J. Mol. Sci. 2021, 22, 12348. [Google Scholar] [CrossRef] [PubMed]
- Ticconi, C.A.; Abel, S. Short on phosphate: Plant surveillance and countermeasures. Trends Plant Sci. 2004, 9, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Liu, D. Signaling components involved in plant responses to phosphate starvation. J. Integr. Plant Biol. 2008, 50, 849–859. [Google Scholar] [CrossRef]
- Liang, C.; Tian, J.; Liao, H. Proteomics dissection of plant responses to mineral nutrient deficiency. Proteomics 2013, 13, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Motte, H.; Vanneste, S.; Beeckman, T. Molecular and environmental regulation of root development. Annu. Rev. Plant Biol. 2019, 70, 465–488. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. Homeostasis of adenylate status during photosynthesis in a fluctuating environment. J. Exp. Bot. 2000, 51, 347–356. [Google Scholar] [CrossRef]
- Rochaix, J.D. Role of thylakoid protein kinases in photosynthetic acclimation. FEBS Lett. 2007, 581, 2768–2775. [Google Scholar] [CrossRef]
- Carmo-Silva, E.; Scales, J.C.; Madgwick, P.J.; Parry, M.A. Optimizing Rubisco and its regulation for greater resource use efficiency. Plant Cell Environ. 2015, 38, 1817–1832. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y. Phosphate starvation and membrane lipid remodeling in seed plants. Prog. Lipid Res. 2013, 52, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Preiss, J. Bacterial glycogen synthesis and its regulation. Annu. Rev. Microbiol. 1984, 38, 419–458. [Google Scholar] [CrossRef] [PubMed]
- Carrera, D.N.Ï.; Oddsson, S.; Grossmann, J.; Trachsel, C.; Streb, S. Comparative proteomic analysis of plant acclimation to six different long-term environmental changes. Plant Cell Physiol. 2018, 59, 510–526. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Gao, X.; Liao, L.; Harberd, N.P.; Fu, X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the Gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007, 145, 1460–1470. [Google Scholar] [CrossRef]
- Yin, Y.; Borges, G.; Sakuta, M.; Crozier, A.; Ashihara, H. Effect of phosphate deficiency on the content and biosynthesis of anthocyanins and the expression of related genes in suspension-cultured grape (Vitis sp.) cells. Plant Physiol. Biochem. 2012, 55, 77–84. [Google Scholar] [CrossRef]
- Manna, M.; Islam, T.; Kaul, T.; Reddy, C.S.; Fartyal, D.; James, D.; Reddy, M.K. A comparative study of effects of increasing concentrations of phosphate and phosphite on rice seedlings. Acta Physiol. Plant 2015, 37, 258. [Google Scholar] [CrossRef]
- Tominaga-Wada, R.; Masakane, A.; Wada, T. Effect of phosphate deficiency-induced anthocyanin accumulation on the expression of Solanum lycopersicum GLABRA3 (SlGL3) in tomato. Plant Signal. Behav. 2018, 13, e1477907. [Google Scholar] [CrossRef]
- Sakuta, M. Diversity in plant red pigments: Anthocyanins and betacyanins. Plant Biotechnol. Rep. 2014, 8, 37–48. [Google Scholar] [CrossRef]
- Lutts, S.; Hausman, J.F.; Quinet, M.; Lefèvre, I. Polyamines and their roles in the alleviation of ion toxicities in Plants. In Ecophysiology and Responses of Plants under Salt Stress; Parvaiz, A., Azooz, M.M., Prasad, M.N.V., Eds.; Springer: New York, NY, USA, 2013; pp. 315–353. [Google Scholar]
- Niu, Y.F.; Chai, R.S.; Jin, G.L.; Wang, H.; Tang, C.X.; Zhang, Y.S. Responses of root architecture development to low phosphorus availability: A review. Ann. Bot. 2013, 112, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Borch, K.; Bouma, T.J.; Lynch, J.P.; Brown, K.M. Ethylene: A regulator of root architectural responses to soil phosphorus availability. Plant Cell Environ. 1999, 22, 425–431. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Lynch, J.P.; Brown, K.M. Ethylene and phosphorus availability have interacting yet distinct effects on root hair development. J. Exp. Bot. 2003, 54, 2351–2361. [Google Scholar] [CrossRef] [PubMed]
- Talboys, P.J.; Healey, J.R.; Withers, P.J.; Jones, D.L. Phosphate depletion modulates auxin transport in Triticum aestivum leading to altered root branching. J. Exp. Bot. 2014, 65, 5023–5032. [Google Scholar] [CrossRef] [PubMed]
- Khan, G.A.; Vogiatzaki, E.; Glauser, G.; Poirier, Y. Phosphate deficiency induces the jasmonate pathway and enhances resistance to insect herbivory. Plant Physiol. 2016, 171, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Yu, H.; Dong, J.; Che, X.; Jiao, Y.; Liu, D. The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genet. 2016, 12, e1006194. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Zhu, C.Q.; Zhao, X.S.; Zheng, S.J.; Shen, R.F. Ethylene is involved in root phosphorus remobilization in rice (Oryza sativa) by regulating cell-wall pectin and enhancing phosphate translocation to shoots. Ann. Bot. 2016, 118, 645–653. [Google Scholar] [CrossRef]
- Park, C.H.; Roh, J.; Youn, J.; Son, S.; Park, J.H.; Kim, S.Y.; Kim, T.; Kim, S. Arabidopsis ACC oxidase 1 coordinated by multiple signals mediates ethylene biosynthesis and is involved in root development. Mol. Cells 2018, 41, 923–932. [Google Scholar] [PubMed]
- Bhosale, R.; Giri, J.; Pandey, B.K.; Giehl, R.F.H.; Hartmann, A.; Trainil, R.; Truskina, J.; Leftley, N.; Hanlon, M.; Swarup, K.; et al. A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nat. Commun. 2018, 9, 1409. [Google Scholar] [CrossRef]
- Su, Y.; Li, M.; Guo, L.; Wang, X. Different effects of phospholipase Dζ2 and non-specific phospholipase C4 on lipid remodeling and root hair growth in Arabidopsis response to phosphate deficiency. Plant J. 2018, 94, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pei, L.; Jin, Z.; Zhang, K.; Zhang, J. Overexpression of the protein phosphatase 2A regulatory subunit a gene ZmPP2AA1 improves low phosphate tolerance by remodeling the root system architecture of maize. PLoS ONE 2017, 12, e0176538. [Google Scholar] [CrossRef][Green Version]
- Chen, Y.F.; Li, L.Q.; Xu, Q.; Kong, Y.H.; Wang, H.; Wu, W.H. The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 2009, 21, 3554–3566. [Google Scholar] [CrossRef]
- Nordstrom, A.; Tarkowski, P.; Tarkowska, D.; Norbaek, R.; Astot, C.; Dolezal, K.; Sandberg, G. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 2004, 101, 8039–8044. [Google Scholar] [CrossRef] [PubMed]
- Laplaze, L.; Benkova, E.; Casimiro, I.; Maes, L.; Vanneste, S.; Swarup, R.; Weijers, D.; Calvo, V.; Parizot, B.; Herrera-Rodriguez, M.B.; et al. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 2007, 19, 3889–3900. [Google Scholar] [CrossRef] [PubMed]
- Ruzicka, K.; Ljung, K.; Vanneste, S.; Podhorska, R.; Beeckman, T.; Friml, J.; Benkova, E. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 2007, 19, 2197–2212. [Google Scholar] [CrossRef]
- Swarup, R.; Perry, P.; Hagenbeek, D.; Straeten, D.V.D.; Beemster, G.T.S.; Sandberg, G.; Bhalerao, R.; Ljung, K.; Bennett, M.J. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 2007, 19, 2186–2196. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Yun, J.; Likhacheva, A.V.; Alonso, J.M. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 2007, 19, 2169–2185. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhao, J.; Li, X.; Qin, L.; Yan, X.; Liao, H. A soybean beta-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011, 66, 541–552. [Google Scholar] [CrossRef]
- Lopez-Arredondo, D.L.; Leyva-Gonzalez, M.A.; Gonzalez-Morales, S.I.; Lopez-Bucio, J.; Herrera-Estrella, L. Phosphate nutrition: Improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef] [PubMed]
- Bommer, U.A.; Thiele, B.J. The translationally controlled tumour protein (TCTP). Int. J. Biochem. Cell Biol. 2004, 36, 379–385. [Google Scholar] [CrossRef]
- Tao, J.J.; Cao, Y.R.; Chen, H.W.; Wei, W.; Li, Q.T.; Ma, B.; Zhang, W.K.; Chen, S.Y.; Zhang, J.S. Tobacco translationally controlled tumor protein interacts with ethylene receptor tobacco histidine kinase1 and enhances plant growth through promotion of cell proliferation. Plant Physiol. 2015, 169, 96–114. [Google Scholar] [CrossRef] [PubMed]
- Branco, R.; Masle, J. Systemic signalling through translationally controlled tumour protein controls lateral root formation in Arabidopsis. J. Exp. Bot. 2019, 70, 3927–3940. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Liang, W.; Sturrock, C.J.; Pandey, B.K.; Giri, J.; Mairhofer, S.; Wang, D.; Muller, L.; Tan, H.; York, L.M.; et al. Rice actin binding protein RMD controls crown root angle in response to external phosphate. Nat. Commun. 2018, 9, 2346. [Google Scholar] [CrossRef]
- Chai, Y.N.; Schachtman, D.P. Root exudates impact plant performance under abiotic stress. Trends Plant Sci. 2021, 27, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Wang, X.; Tong, Y.; Chen, X.; Liao, H. Bioengineering and management for efficient phosphorus utilization in crops and pastures. Curr. Opin. Biotechnol. 2012, 23, 866–871. [Google Scholar] [CrossRef]
- Koyama, H.; Kawamura, A.; Kihara, T.; Hara, T.; Takita, E.; Shibata, D. Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus-limited soil. Plant Cell Physiol. 2000, 41, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, D. Functions and regulation of phosphate starvation-induced secreted acid phosphatases in higher plants. Plant Sci. 2018, 271, 108–116. [Google Scholar] [CrossRef]
- Liu, P.; Cai, Z.; Chen, Z.; Mo, X.; Ding, X.; Liang, C.; Liu, G.; Tian, J. A root-associated purple acid phosphatase, SgPAP23, mediates extracellular phytate-P utilization in Stylosanthes guianensis. Plant Cell Environ. 2018, 41, 2821–2834. [Google Scholar] [CrossRef]
- Zhu, S.; Chen, M.; Liang, C.; Xue, Y.; Lin, S.; Tian, J. Characterization of purple acid phosphatase family and functional analysis of GmPAP7a/7b involved in extracellular ATP utilization in soybean. Front. Plant Sci. 2020, 11, 661. [Google Scholar] [CrossRef] [PubMed]
- Bozzo, G.G.; Dunn, E.L.; Plaxton, W.C. Differential synthesis of phosphate-starvation inducible purple acid phosphatase isozymes in tomato (Lycopersicon esculentum) suspension cells and seedlings. Plant Cell Environ. 2006, 29, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Qian, W.; Hurley, B.A.; She, Y.M.; Wang, D.; Plaxton, W.C. Biochemical and molecular characterization of AtPAP12 and AtPAP26: The predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant Cell Environ. 2010, 33, 1789–1803. [Google Scholar] [CrossRef] [PubMed]
- Robinson, W.D.; Park, J.; Tran, H.T.; Del, V.H.; Ying, S.; Zins, J.L.; Patel, K.; McKnight, T.D.; Plaxton, W.C. The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate-scavenging by Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 6531–6542. [Google Scholar] [CrossRef]
- Wu, W.; Zhu, S.; Chen, Q.; Lin, Y.; Tian, J.; Liang, C. Cell wall proteins play critical roles in plant adaptation to phosphorus deficiency. Int. J. Mol. Sci. 2019, 20, 5259. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.K.; George, T.S.; Thompson, J.A.; Wright, G.; Lyon, J.; Dupuy, L.; Hubbard, S.F.; White, P.J. What are the implications of variation in root hair length on tolerance to phosphorus deficiency in combination with water stress in barley (Hordeum vulgare)? Ann. Bot. 2012, 110, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Jiang, H.; Tian, J.; Zhao, J.; Liao, H. Rhizobia enhance acquisition of phosphorus from different sources by soybean plants. Plant Soil 2011, 349, 25–36. [Google Scholar] [CrossRef]
- Chen, L.; Qin, L.; Zhou, L.; Li, X.; Chen, Z.; Sun, L.; Wang, W.; Lin, Z.; Zhao, J.; Yamaji, N.; et al. A nodule-localized phosphate transporter GmPT7 plays an important role in enhancing symbiotic N2 fixation and yield in soybean. New Phytol. 2018, 221, 2013–2025. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Zhao, J.; Tian, J.; Chen, L.; Sun, Z.; Guo, Y.; Lu, X.; Gu, M.; Xu, G.; Liao, H. The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol. 2012, 159, 1634–1643. [Google Scholar] [CrossRef]
- Zhu, S.; Chen, Z.; Xie, B.; Guo, Q.; Chen, M.; Liang, C.; Bai, Z.; Wang, X.; Wang, H.; Liao, H.; et al. A phosphate starvation responsive malate dehydrogenase, GmMDH12 mediates malate synthesis and nodule size in soybean (Glycine max). Environ. Exp. Bot. 2021, 189, 104560. [Google Scholar] [CrossRef]
- Pan, W.; Wu, Y.; Xie, Q. Regulation of ubiquitination is central to the phosphate starvation response. Trends Plant Sci. 2019, 24, 755–769. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Y.; Wang, F.; Yang, J.; Gao, M.; Li, C.; Liu, Y.; Liu, Y.; Yamaji, N.; Ma, J.F.; et al. The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell 2015, 27, 711–723. [Google Scholar] [CrossRef]
- Gelova, Z.; Gallei, M.; Pernisova, M.; Brunoud, G.; Zhang, X.; Glanc, M.; Li, L.; Michalko, J.; Pavlovicova, Z.; Verstraeten, I.; et al. Developmental roles of auxin binding protein 1 in Arabidopsis thaliana. Plant Sci. 2021, 303, 110750. [Google Scholar] [CrossRef] [PubMed]
- Washburn, M.; Wolters, D.; Yates, J. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001, 19, 242–247. [Google Scholar] [CrossRef]
- Unwin, R.; Pierce, A.; Watson, R.; Sternberg, D.; Whetton, A. Quantitative proteomic analysis using isobaric protein tags enables rapid comparison of changes in transcript and protein levels in transformed cells. Mol. Cell. Proteom. 2005, 4, 924–935. [Google Scholar] [CrossRef]
- Thompson, A.; Schafer, J.; Kuhn, K.; Kienle, S.; Schwarz, J.; Schmidt, G.; Neumann, T.; Hamon, C. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003, 75, 1895–1904. [Google Scholar] [CrossRef]
- Ong, S.; Blagoev, B.; Kratchmarova, I.; Kristensen, D.; Steen, H.; Pandey, A.; Mann, M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteom. 2002, 1, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Messner, C.; Demichev, V.; Bloomfield, N.; Yu, J.; White, M.; Kreidl, M.; Egger, A.; Freiwald, A.; Ivosev, G.; Wasim, F.; et al. Ultra-fast proteomics with scanning SWATH. Nat. Biotechnol. 2021, 39, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, P.; Guo, T.; Zhao, H.; Bensaddek, D.; Aebersold, R.; Xiong, L. Arabidopsis proteome and the mass spectral assay library. Sci. Data 2019, 6, 278. [Google Scholar] [CrossRef]
- Liu, S.; Li, Z.; Yu, B.; Wang, S.; Shen, Y.; Cong, H. Recent advances on protein separation and purification methods. Adv. Colloid Interface Sci. 2020, 284, 102254. [Google Scholar] [CrossRef] [PubMed]
| Plant Species | Organ/Tissues | Culture Time before Treatment (d) | Treatment Time (d) | Protein Separation Method | Total Protein Number (#) | Number of DAPs (#) | Protein Number Identified by MS Analysis | References | |
|---|---|---|---|---|---|---|---|---|---|
| Up-Regulated | Down-Regulated | ||||||||
| Arabidopsis thaliana | Leaves | 10 | 7 | SCX iTRAQ LC-MS/MS | 5106 | 156 | 106 | 50 | [17] |
| Zea mays | Leaves | 4 | 25 | 2-DE MALDI TOF MS/TOF | 1342 | 200 | nd | nd | [26] |
| Zea mays | Leaves a | 18 | 25 | 2-DE MALDI-TOF/MALDI-TOF-TOF MS | 680/592 | 29/71 | 9/20 | 55/16 | [27] |
| Glycine max | Leaves | 5 | 14 | 2D-IEF/SDS-PAGE MALDI-TOF MS | 55 | 17 | 7 | 10 | [28] |
| Glycine max | Leaves | 3 | 14 | SDS-PAGE Gel Digestion LC-MS/MS | 4219 | 707 | 267 | 440 | [29] |
| Solanum lycopersicum | Leaves | nd | 10 | 2-DE MALDI-TOF MS/MS/MS | 600 | 46 | 31 | 15 | [20] |
| Arabidopsis thaliana | Roots | 10 | 3 | 2-DE MALDI TOF MS | 456 | 30 | nd | nd | [30] |
| Arabidopsis thaliana | Roots | 10 | 3 | 2-DE iTARQ LC-MS | 13,298 | 356 | 199 | 157 | [16] |
| Arabidopsis thaliana | Roots | nd | 14 | 2-DIGE MALDI-TOF/TOF | 1420 | 30 | 14 | 16 | [13] |
| Zea mays | Roots | 24 | 17 | 2-DE MALDI TOF MS | 1300 | 254 | 76 | 30 | [31] |
| Zea mays | Roots | 10 | 1/3/7/11 | 2-DE MALDI-TOF-MS | 850 | 91 | nd | nd | [32] |
| Zea mays | Roots a | 24 | 17 | 2-DE MALDI TOF MS | 2822 | 73/95 | 25/24 | 12/6 | [19] |
| Zea mays | Roots a | nd | 10 | 2-DE MALDI-TOF | nd | 83/325 | 30/246 | 53/79 | [33] |
| Oryza sativa | Roots | 3 | 80 | 2-DE MALDI TOF MS | 669 | 34 | nd | nd | [21] |
| Oryza sativa | Roots | 7 | 21 | 2-DE MALDI-TOF MS | 140 | 10 | 2 | 8 | [34] |
| Glycine max | Roots | nd | 20 | 2-DIGE | 325 | 105 | 61 | 44 | [35] |
| Glycine max | Roots a | 3 | 9 | SDS-PAGE TMT | 4216 | 660/133 | 656/127 | 4/6 | [36] |
| Glycine max | Roots | nd | 10 | iTRAQ LC-MS/MS | nd | 71 | 30 | 41 | [25] |
| Glycine max | Roots | 5 | 14 | iTRAQ | nd | 427 | 213 | 214 | [15] |
| Triticum aestivum | Roots | 14 | 8 | iTRAQ | 6842 | 323 | nd | nd | [37] |
| Hordeum vulgare | Roots | 10 | 0.25/2 | SDS PAGE LC-MS/MS | nd | 697 | nd | nd | [38] |
| Brassica napus | Roots a | 20 | 3 | 2-phase LC/MS-MS | 828 | 31/40 | 8/28 | 23/12 | [39] |
| Arabidopsis thaliana | Leaves/roots | 7 | 3 | 2-DIGE MALDI TOF/TOF MS | 88 | nd | nd | nd | [40] |
| Hordeum vulgare | Leaves/roots | 17 | 21 | 2-DE MALDI-TOF/TOF-MS | nd | 31 | nd | nd | [23] |
| Brassica napus | Leaves/roots | 20 | 26 | 2-DE MALDI TOF MS | 1000 | 32 | 4/12 | 13/3 | [41] |
| Pinus massoniana | Seedlings | 10 | 58 | 2-DE MALDI-TOF/TOF MS | nd | 98 | 44 | 54 | [42] |
| Arabidopsis thaliana | Suspension cells | 7 | 7 | 2-DE MALDI TOF MS | 110 | 46 | 26 | 6 | [43] |
| Arabidopsis thaliana | Suspension cells | 9 | 2 | SDS-PAGE LFQ | 5013/1881 | 1169/994 | nd | nd | [44] |
| Glycine max | Nodules | 5 | 25 | 2-DE MALDI TOF MS | nd | 44 | 17 | 27 | [24] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, M.; Zhu, S.; Mo, X.; Guo, Q.; Li, Y.; Tian, J.; Liang, C. Proteomic Analysis Dissects Molecular Mechanisms Underlying Plant Responses to Phosphorus Deficiency. Cells 2022, 11, 651. https://doi.org/10.3390/cells11040651
Zhou M, Zhu S, Mo X, Guo Q, Li Y, Tian J, Liang C. Proteomic Analysis Dissects Molecular Mechanisms Underlying Plant Responses to Phosphorus Deficiency. Cells. 2022; 11(4):651. https://doi.org/10.3390/cells11040651
Chicago/Turabian StyleZhou, Ming, Shengnan Zhu, Xiaohui Mo, Qi Guo, Yaxue Li, Jiang Tian, and Cuiyue Liang. 2022. "Proteomic Analysis Dissects Molecular Mechanisms Underlying Plant Responses to Phosphorus Deficiency" Cells 11, no. 4: 651. https://doi.org/10.3390/cells11040651
APA StyleZhou, M., Zhu, S., Mo, X., Guo, Q., Li, Y., Tian, J., & Liang, C. (2022). Proteomic Analysis Dissects Molecular Mechanisms Underlying Plant Responses to Phosphorus Deficiency. Cells, 11(4), 651. https://doi.org/10.3390/cells11040651







