Proteomic Studies of Roots in Hypoxia-Sensitive and -Tolerant Tomato Accessions Reveal Candidate Proteins Associated with Stress Priming
Abstract
1. Introduction
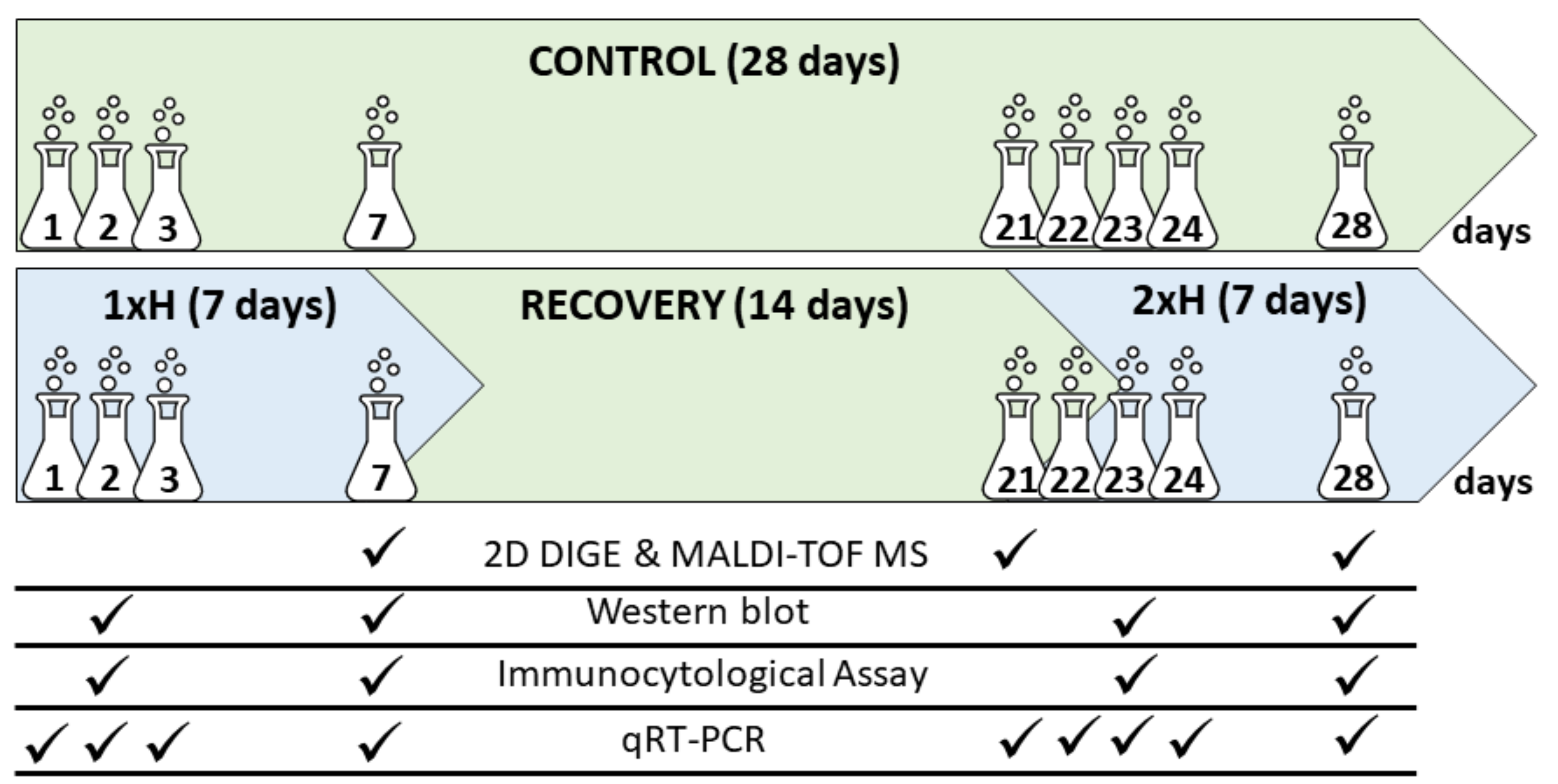
2. Materials and Methods
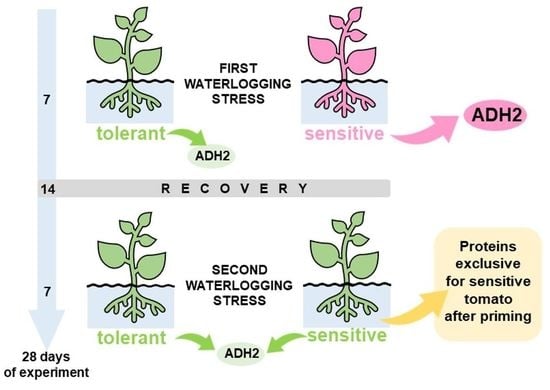
2.1. Plant Material, Cultivation and Stress Treatment
2.2. Soluble Protein Extraction
2.3. D DIGE, Quantitative Protein Analysis, MALDI-TOF MS Analysis
2.4. Validation of Some Key Candidate Proteins by Western-Blot
2.5. Expression of adh Gene by qRT-PCR Assay
2.6. Histological and Immunocytological Analyses
3. Results
3.1. Waterlogging-Induced Changes in the Tomato Root Proteomes
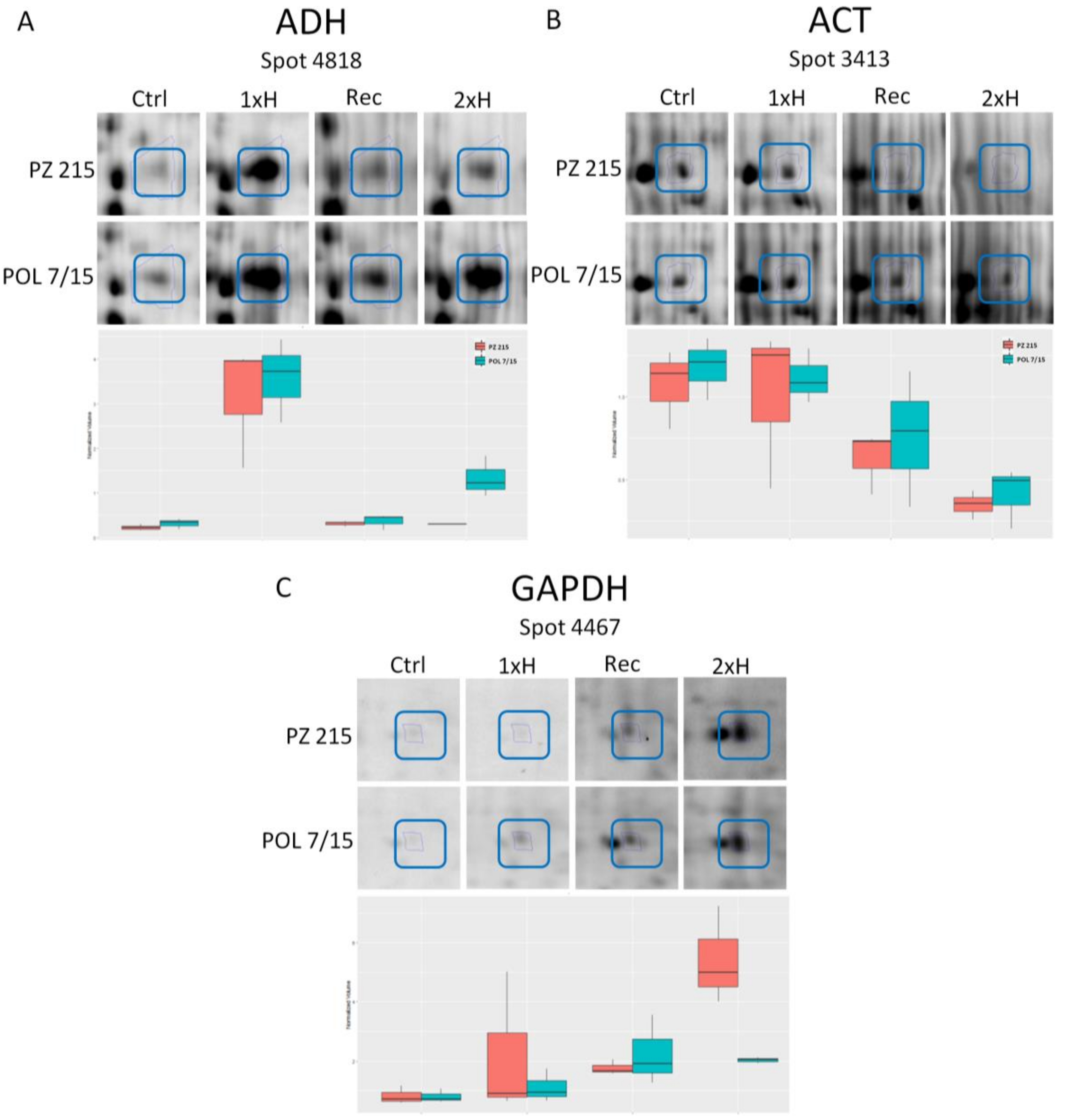
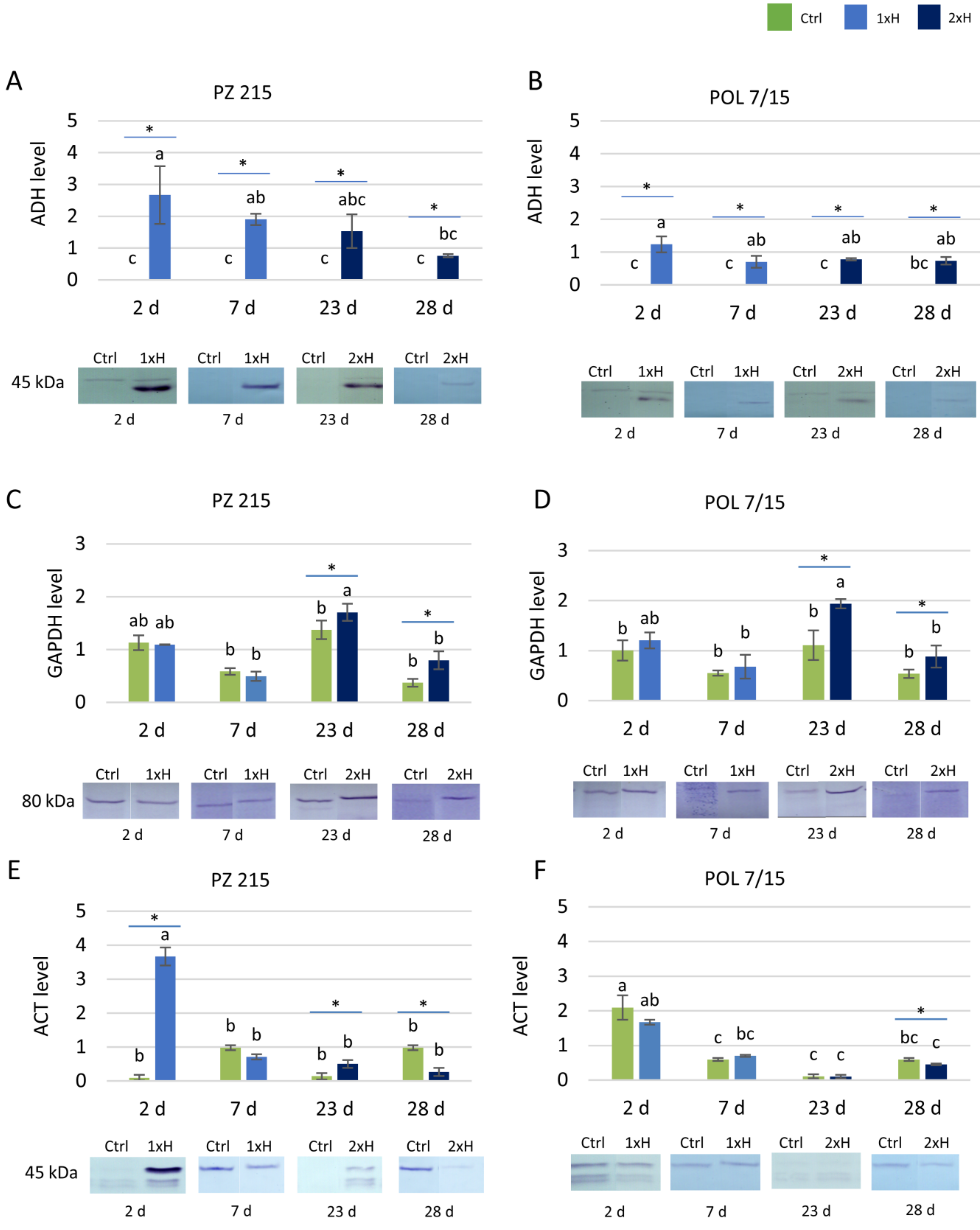
3.2. Validation of Key Proteins by Western Blot
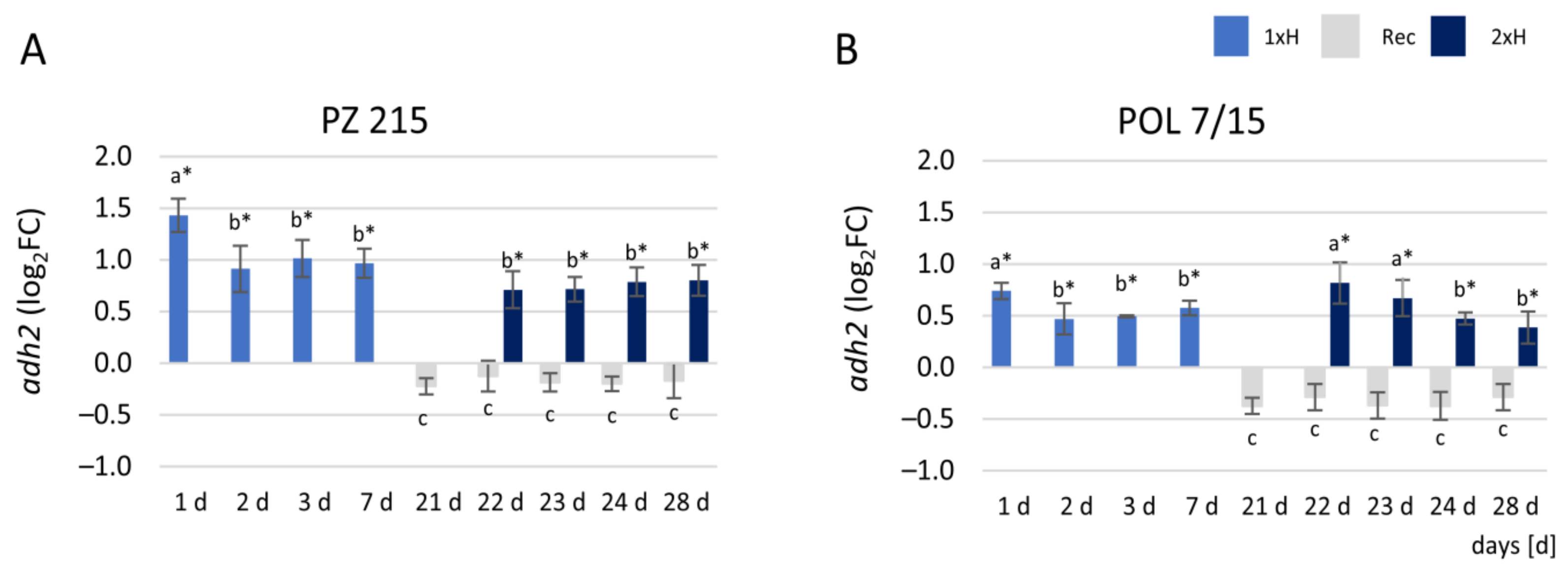
3.3. Expression Profile of adh2 Gene
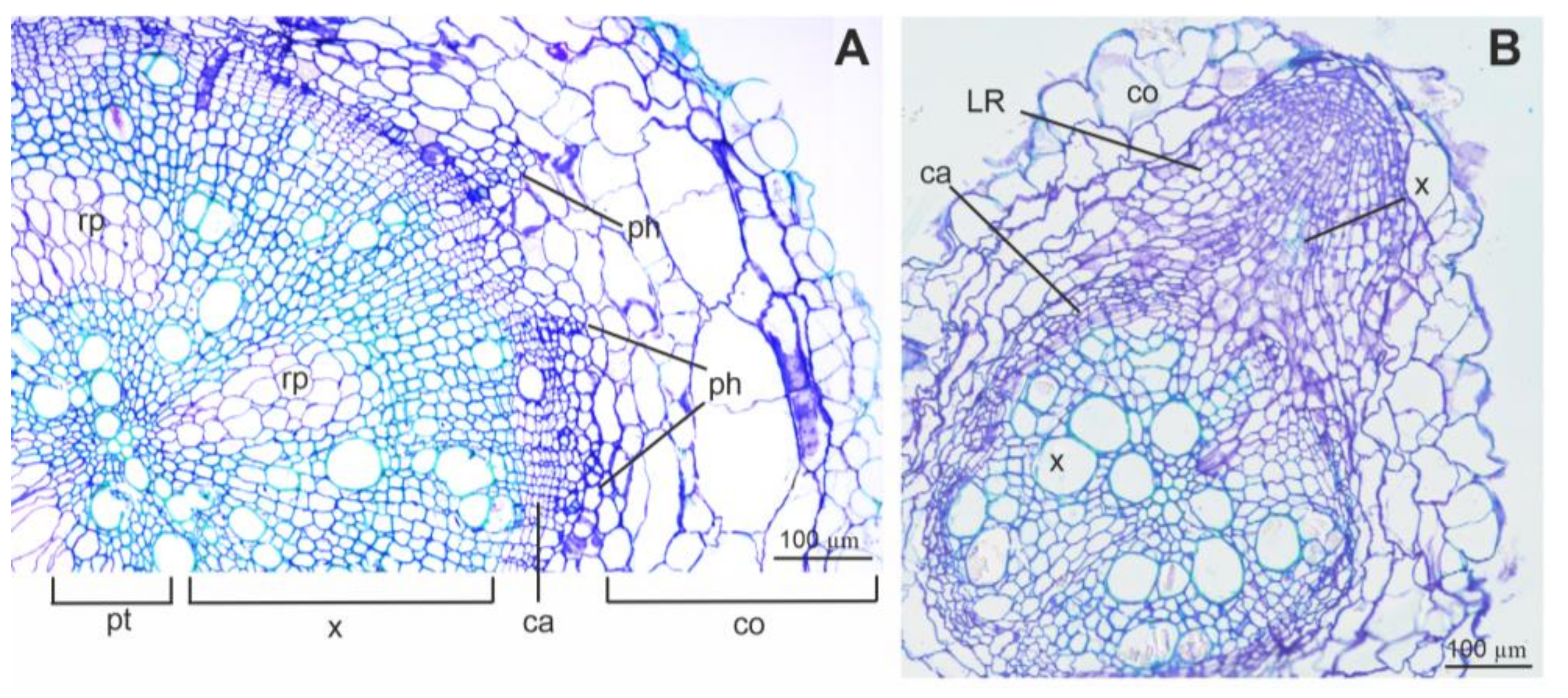
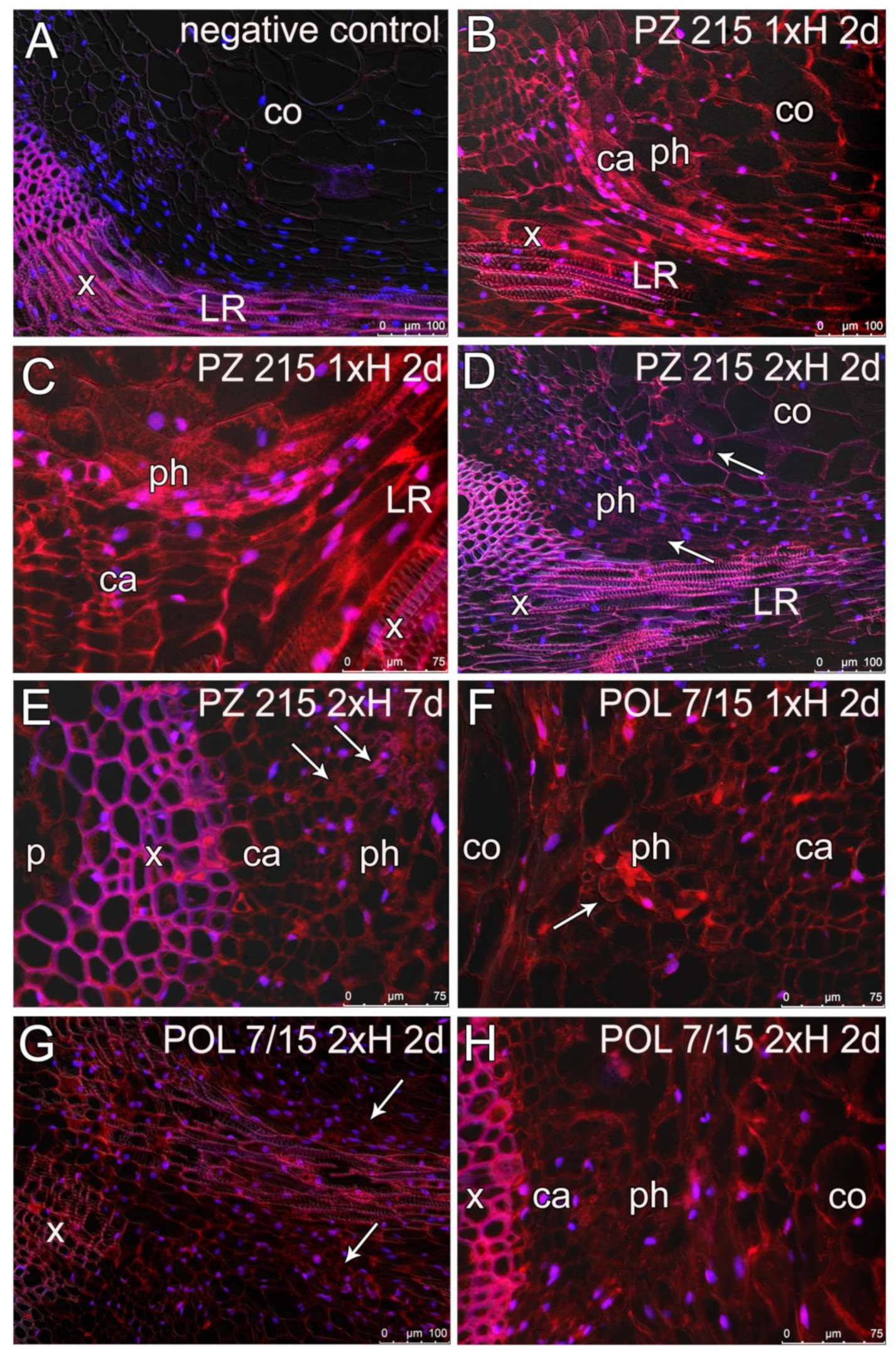
3.4. Root Sections and Immunohistochemical Localization of ADH in Root Tissues
4. Discussion
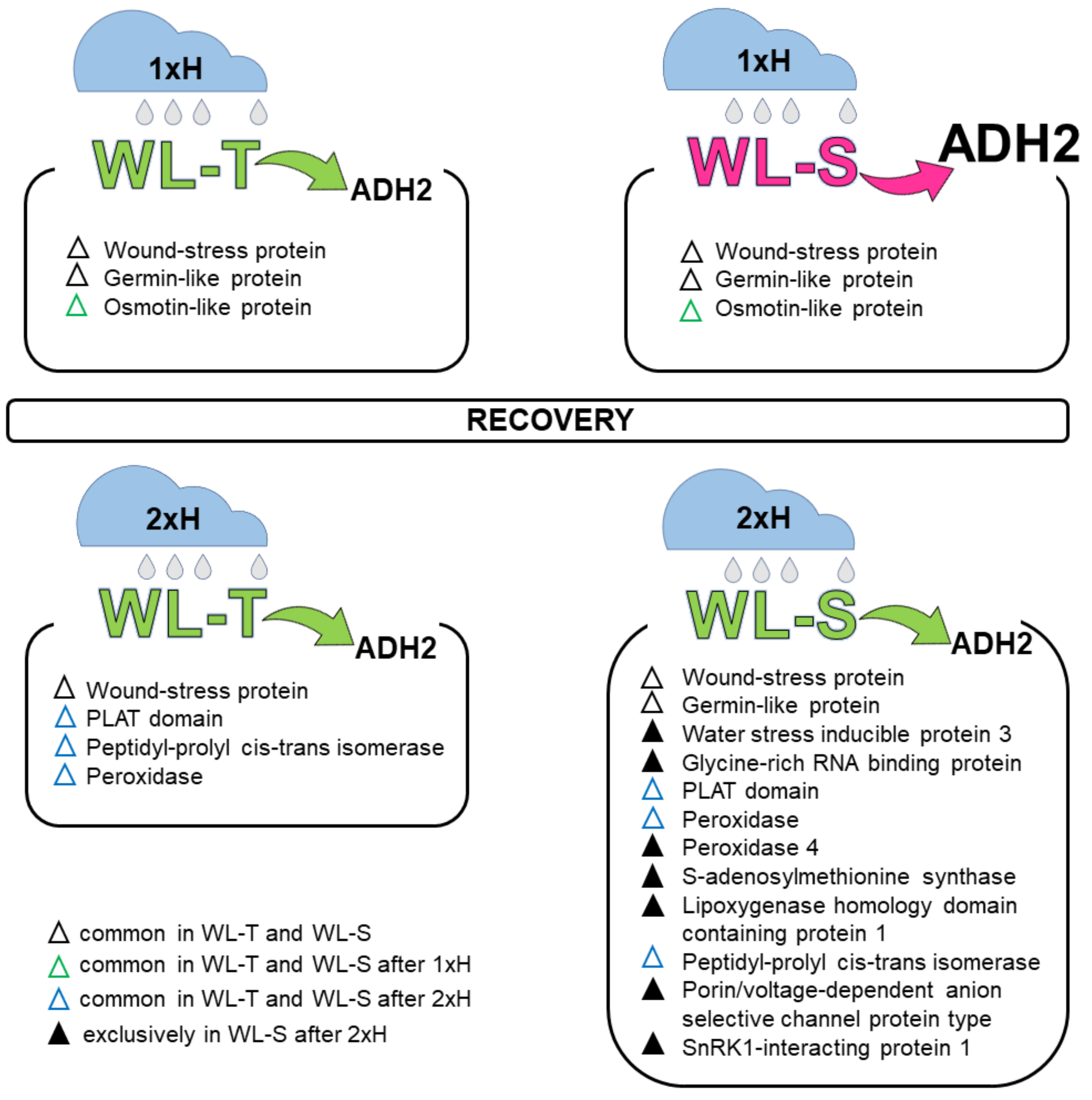
4.1. Global Proteome Analysis after the First Waterlogging (1xH). Different Response of WL-S and WL-T Tomato Accessions
4.2. Global Proteome Analysis after 2xH. Candidates for Priming and Immune Tolerance Acquisition
4.3. Validation of Selected Proteins
4.4. Root Anatomy and ADH Detection Using Immunolabelling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leon, J.; Castillo, M.C.; Beatriz Gayubas, B. The hypoxia-reoxygenation stress in plants. J. Exp. Bot. 2020, 72, 5841–5856. [Google Scholar] [CrossRef] [PubMed]
- Bui, L.T.; Novi, G.; Lombardi, L.; Iannuzzi, C.; Rossi, J.; Santaniello, A.; Mensuali, A.; Corbineau, F.; Giuntoli, B.; Perata, P.; et al. Conservation of ethanol fermentation and its regulation in land plants. J. Exp. Bot. 2019, 70, 1815–1827. [Google Scholar] [CrossRef]
- Diab, H.; Limami, A.M. Reconfiguration of N Metabolism upon Hypoxia Stress and Recovery: Roles of Alanine Aminotransferase (AlaAT) and Glutamate Dehydrogenase (GDH). Plants 2016, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-H.; Yu, S.-i.; Lee, B.-h.; Lee, D.-H. Modulation of Energy Metabolism Is Important for Low-Oxygen Stress Adaptation in Brassicaceae Species. Int. J. Mol. Sci. 2020, 21, 1787. [Google Scholar] [CrossRef] [PubMed]
- Ventura, I.; Brunello, L.; Iacopino, S.; Valeri, M.C.; Novi, G.; Dornbusch, T.; Perata, P.; Loreti, E. Arabidopsis phenotyping reveals the importance of alcohol dehydrogenase and pyruvate decarboxylase for aerobic plant growth. Sci Rep 2020, 10, 16669. [Google Scholar] [CrossRef]
- App, A.A.; Meiss, A.N. Effect of aeration on rice alcohol dehydrogenase. Arch. Biochem. Biophys. 1958, 77, 181–190. [Google Scholar] [CrossRef]
- Safavi-Rizi, V.; Herde, M.; Stöhr, C. RNA-Seq reveals novel genes and pathways associated with hypoxia duration and tolerance in tomato root. Sci. Rep. 2020, 10, 1692. [Google Scholar] [CrossRef]
- Andrzejczak, O.A.; Havelund, J.F.; Wang, W.Q.; Kovalchuk, S.; Hagensen, C.E.; Hasler-Sheetal, H.; Jensen, O.N.; Rogowska-Wrzesinska, A.; Møller, I.M.; Hebelstrup, K.H. The Hypoxic Proteome and Metabolome of Barley (Hordeum vulgare L.) with and without Phytoglobin Priming. Int. J. Mol. Sci. 2020, 21, 1546. [Google Scholar] [CrossRef]
- Cotrozzi, L.; Lorenzini, G.; Nali, C.; Pisuttu, C.; Pampana, S.; Pellegrini, E. Transient Waterlogging Events Impair Shoot and Root Physiology and Reduce Grain Yield of Durum Wheat Cultivars. Plants 2021, 10, 2357. [Google Scholar] [CrossRef]
- Arduini, I.; Baldanzi, M.; Pampana, S. Reduced Growth and Nitrogen Uptake During Waterlogging at Tillering Permanently Affect Yield Components in Late Sown Oats. Front. Plant Sci. 2019, 10, 1087. [Google Scholar] [CrossRef]
- Wang, X.; Liu, F.I.; Dong, J. Priming: A promising strategy for crop production in response to future climate. J. Integr. Agric. 2017, 16, 2709–2716. [Google Scholar] [CrossRef]
- Branco-Price, C.; Kaiser, K.A.; Jang, C.J.H.; Larive, C.K.; Bailey-Serres, J. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabi dopsis thaliana. Plant J. 2008, 56, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Mustafa., G.; Komatsu, S. Quantitative proteomics reveals the effect of protein glycosylation in soybean root under flooding stress. Front Plant Sci. 2014, 5, 627. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, N.; Lee, D.G.; Lee, S.H.; Lee, K.W.; Bahk, J.; Lee, B.H. A proteomic screen and identification of waterlogging-regulated proteins in tomato roots. Plant Soil 2007, 295, 37–51. [Google Scholar] [CrossRef]
- Mignolli, F.; Todaro, J.S.; Vidoz, M.L. Internal aeration and respiration of submerged tomato hypocotyls are enhanced by ethylene-mediated aerenchyma formation and hypertrophy. Physiol. Plant. 2019, 169, 49–63. [Google Scholar] [CrossRef]
- Barik, J.; Panda, D.; Mohanty, S.K.; Lenka, S.K. Leaf photosynthesis and antioxidant response in selected traditional rice landraces of Jeypore tract of Odisha, India to submergence. Physiol. Mol. Biol. Plants 2019, 25, 847–863. [Google Scholar] [CrossRef]
- Kołton, A.; Kęska, K.; Czernicka, M. Selection of Tomato and Cucumber Accessions for Waterlogging Sensitivity through Morpho-Physiological Assessment at an Early Vegetative Stage. Agronomy 2020, 10, 1490. [Google Scholar] [CrossRef]
- Cao, X.; Jiang, X.; Wang, X.; Zang, Y.; Wu, Z. Comprehensive evaluation and screening for chilling-tolerance in tomato lines at the seedling stage. Euphytica 2015, 205, 569–584. [Google Scholar] [CrossRef]
- Kalaji, H.M.; Rastogi, A.; Živˇcák, M.; Brestic, M.; Daszkowska-Golec, A.; Sitko, K.; Alsharafa, K.Y.; Lotfi, R.; Stypiński, P.; Samborska, I.A.; et al. Prompt chlorophyll fluorescence as a tool for crop phenotyping: An example of barley landraces exposed to various abiotic stress factors. Photosynthetica 2018, 56, 953–961. [Google Scholar] [CrossRef]
- Tan, Y.; Qin, Y.; Li, Y.; Li, M.; Ma, F. Overexpression of MpGR-RBP1, a glycine-rich RNA-binding protein gene from Malus prunifolia (Willd.) Borkh, confers salt stress tolerance and protects against oxidative stress in Arabidopsis. PCTOC 2014, 119, 635–646. [Google Scholar] [CrossRef]
- Bohler, S.; Bagard, M.; Oufir, M.; Planchon, S.; Hoffmann, L.; Jolivet, Y.; Hausman, J.F.; Dizengremel, P.; Renaut, J. A DIGE analysis of developing poplar leaves subjected to ozone reveals major changes in carbon metabolism. Proteomics 2007, 7, 1584–1599. [Google Scholar] [CrossRef] [PubMed]
- Alexei, A.; Sharov, D.B.D.; Minoru, S.; Ko, M.S. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics 2005, 21, 2548–2549. [Google Scholar] [CrossRef]
- Shägger, H.; Von Jagow, G. Tricine-sodium dodecyl sulfatepolyacrylamide gel electrophoresis for the separation of proteins in the range for 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- Schneider, C.; Rasband, W.; Eliceiri, K. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Kęska, K.; Szcześniak, M.W.; Makałowska, I.; Czernicka, M. Long-TermWaterlogging as Factor Contributing to Hypoxia Stress Tolerance Enhancement in Cucumber: Comparative Transcriptome Analysis of Waterlogging Sensitive and Tolerant Accessions. Genes 2021, 12, 189. [Google Scholar] [CrossRef] [PubMed]
- Popielarska-Konieczna, M.; Ślesak, H.; Góralski, G. Histological and SEM studies on organogenesis in endosperm-derived callus of kiwifruit (Actinidia deliciosa cv. Hayward). Acta Biol. Crac. Ser. Bot. 2006, 48, 97–104. [Google Scholar]
- Slazak, B.; Kapusta, M.; Malik, S.; Bohdanowicz, J.; Kuta, E.; Malec, P.; Göransson, U. Immunolocalization of cyclotides in plant cells, tissues and organ supports their role in host defense. Planta 2016, 244, 1029–1040. [Google Scholar] [CrossRef]
- Lv, G.Y.; Guo, X.G.; Xie, L.P.; Xie, C.G.; Zhang, X.H.; Yang, Y.; Xiao, L.; Tang, Y.Y.; Pan, X.L.; Guo, A.G.; et al. Molecular Characterization, Gene Evolution, and Expression Analysis of the Fructose-1, 6-bisphosphate Aldolase (FBA) Gene Family in Wheat (Triticum aestivum L.). Front. Plant Sci. 2017, 8, 1030. [Google Scholar] [CrossRef]
- James, D.; Borphukan, B.; Fartyal, D.; Ram, B.; Singh, J.; Manna, M.; Sheri, V.; Panditi, V.; Yadav, R.; Achary, V.M.M.; et al. Concurrent Overexpression of OsGS1;1 and OsGS2 Genes in Transgenic Rice (Oryza sativa L.): Impact on Tolerance to Abiotic Stresses. Front. Plant Sci. 2018, 9, 786. [Google Scholar] [CrossRef]
- Luan, H.; Shen, H.; Pan, Y.; Guo, B.; Lv, C.; Xu, R. Elucidating the hypoxic stress response in barley (Hordeum vulgare L.) during waterlogging: A proteomics approach. Sci. Rep. 2018, 8, 9655. [Google Scholar] [CrossRef]
- Liu, D.; Zhan, J.; Luo, Z.; Zeng, N.; Zhang, W.; Zhang, H.; Li, L. Quantitative Proteomics and Relative Enzymatic Activities Reveal Different Mechanisms in Two Peanut Cultivars (Arachis hypogaea L.) Under Waterlogging Conditions. Front. Plant Sci. 2021, 12, 716114. [Google Scholar] [CrossRef] [PubMed]
- Dunwell, J.M.; Gibbings, J.G.; Mahmood, T.; Saqlan Naqvi, S.M. Germin and germin-like proteins: Evolution, structure, and function. CRC Crit. Rev. Plant Sci. 2008, 27, 342–375. [Google Scholar] [CrossRef]
- Lee, T.G.; Jang, C.S.; Kim, J.Y.; Seong, R.C.; Kim, I.G.; Kim, D.S.; Seo, Y.W. Expressed sequence tags from wheat roots under hypoxia. Russ. J. Plant Physiol. 2007, 54, 659–668. [Google Scholar] [CrossRef]
- Bashir, M.A.; Silvestri, C.; Ahmad, T.; Hafiz, I.A.; Abbasi, N.A.; Manzoor, A.; Cristofori, V.; Rugini, E. Osmotin: A Cationic Protein Leads to Improve Biotic and Abiotic Stress Tolerance in Plants. Plants 2020, 9, 992. [Google Scholar] [CrossRef] [PubMed]
- Reggiani, R.; Brambilla, I.; Bertani, A. Anaerobic protein synthesis and glycosylation in rice seedlings. J. Plant Physiol. 1990, 137, 116–119. [Google Scholar] [CrossRef]
- Do Amaral, M.N.; Arge, L.W.P.; Auler, P.A.; Rosatto, T.; Milech, C.; Magalhães, A.M., Jr.; Braga, E.J.B. Long-term transcriptional memory in rice plants submitted to salt shock. Planta 2020, 251, 111. [Google Scholar] [CrossRef]
- Bittner, A.; van Buer, J.; Baier, M. Cold priming uncouples light- and cold-regulation of gene expression in Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 281. [Google Scholar] [CrossRef]
- Balmer, A.; Pastor, V.; Gamir, J.; Flors, V.; Mauch-Mani, B. The ‘prime-ome’: Towards a holistic approach to priming. Trends Plant Sci. 2015, 20, 443–452. [Google Scholar] [CrossRef]
- António, C.; Päpke, C.; Rocha, M.; Diab, H.; Limami, A.; Obata, T.; Fernie, A.R.; van Dongen, J.T. Regulation of Primary Metabolism in Response to Low Oxygen Availability as Revealed by Carbon and Nitrogen Isotope Redistribution. Plant Physiol. 2016, 170, 43–56. [Google Scholar] [CrossRef]
- Wang, K.L.C.; Li, H.; Ecker, J.R. Ethylene biosynthesis and Signaling Networks. The Plant Cell. 2002, 14, S131–S151. [Google Scholar] [CrossRef]
- Wang, F.; Chen, Z.H.; Shabala, S. Hypoxia Sensing in Plants: On a Quest for Ion Channels as Putative Oxygen Sensors. Plant Cell Physiol. 2017, 58, 1126–1142. [Google Scholar] [CrossRef] [PubMed]
- Wurzinger, B.; Nukarinen, E.; Nägele, T.; Weckwerth, W.; Teige, M. The SnRK1 Kinase as Central Mediator of Energy Signaling between Different Organelles. Plant Physiol. 2018, 176, 1085–1094. [Google Scholar] [CrossRef]
- Fukao, T.; Barrera-Figueroa, B.E.; Juntawong, P.; Peña-Castro, J.M. Submergence and Waterlogging Stress in Plants: A Review Highlighting Research Opportunities and Understudied Aspects. Front. Plant Sci. 2019, 10, 340. [Google Scholar] [CrossRef]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.-Y.; Wen, T.N.; Wang, Y.T.; Shih, M.C. Quantitative phosphoproteomics of protein kinase SnRK1 regulated protein phosphorylation in Arabidopsis under submergence. J. Exp. Bot. 2016, 67, 2745–2760. [Google Scholar] [CrossRef] [PubMed]
- Shigeto, J.; Tsutsumi, Y. Diverse functions and reactions of class III peroxidases. New Phytol. 2016, 209, 1395–1402. [Google Scholar] [CrossRef] [PubMed]
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann. Bot. 2003, 91, 179–194. [Google Scholar] [CrossRef]
- Kidwai, M.; Ahmad, I.Z.; Chakrabarty, D. Class III peroxidase: An indispensable enzyme for biotic/abiotic stress tolerance and a potent candidate for crop improvement. Plant Cell Rep. 2020, 39, 1381–1393. [Google Scholar] [CrossRef]
- Yang, C.Y.; Hsu, F.C.; Li, J.P.; Wang, N.N.; Shih, M.C. The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 2011, 156, 202–212. [Google Scholar] [CrossRef]
- Singh, H.; Kaur, K.; Singh, M.; Kaur Gand Singh, P. Plant Cyclophilins: Multifaceted Proteins With Versatile Roles. Front. Plant Sci. 2020, 11, 585212. [Google Scholar] [CrossRef]
- Romano, P.G.N.; Horton, P.; Gray, J.E. The Arabidopsis Cyclophilin Gene Family. Plant Physiol. 2004, 134, 1268–1282. [Google Scholar] [CrossRef] [PubMed]
- Sekhar, K.; Priyanka, B.; Reddy, V.D.; Rao, K.V. Isolation and characterization of a pigeonpea cyclophilin (CcCYP) gene, and its overexpression in Arabidopsis confers multiple abiotic stress tolerance. Plant Cell Environ. 2020, 33, 1324–1338. [Google Scholar] [CrossRef]
- Yoon, D.H.; Lee, S.S.; Park, H.J.; Lyu, J.; Chong, W.S.; Liu, J.R.; Kim, B.G.; Ahn, J.C.; Cho, H.S. Overexpression of OsCYP19-4 increases tolerance to cold stress and enhances grain yield in rice (Oryza sativa). J. Exp. Bot. 2016, 67, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Bissoli, G.; Niñoles, R.; Fresquet, S.; Palombieri, S.; Bueso, E.; Rubio, L.; García-Sánchez, M.J.; Fernández, J.A.; Mulet, J.M.; Serrano, R. Peptidyl-prolyl cis-trans isomerase ROF2 modulates intracellular pH homeostasis in Arabidopsis. Plant J. 2012, 70, 704–716. [Google Scholar] [CrossRef]
- Ricardi, M.M.; Guaimas, F.F.; Gonzalez, R.M.; Burrieza, H.P.; Lopez-Fernandez, M.P.; Jares-Erijman, E.A.; Estévez, J.M.; Iusem, N.D. Nuclear Import and Dimerization of Tomato ASR1, a Water Stress- Inducible Protein Exclusive to Plants. PLoS ONE 2012, 7, e41008. [Google Scholar] [CrossRef]
- Yacoubi, I.; Hamdi, K.; Fourquet, P.; Bignon, C.; Longhi, S. Structural and Functional Characterization of the ABA-Water Deficit Stress Domain from Wheat and Barley: An Intrinsically Disordered Domain behind the Versatile Functions of the Plant Abscissic Acid, Stress and Ripening Protein Family. Int. J. Mol. Sci. 2021, 22, 2314. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, L.; Chandrasekaran, U.; Luo, X.; Zheng, C.; Shu, K. ABA Biosynthesis and Signaling Cascades Under Hypoxia Stress. Front. Plant Sci. 2021, 12, 661228. [Google Scholar] [CrossRef]
- Dombrowski, J.E. Salt Stress Activation of Wound-Related Genes in Tomato Plants. Plant Physiol. 2003, 132, 2098–2107. [Google Scholar] [CrossRef]
- Mirzahosseini, Z.; Shabani, L.; Sabzalian, M.R.; Dayanandan, S. Comparative physiological and proteomic analysis of Arabidopsis thaliana revealed differential wound stress responses following the exposure to different LED light sources. Environ Exp. Bot. 2020, 169, 103895. [Google Scholar] [CrossRef]
- Walper, E.; Weiste, C.; Mueller, M.J.; Hamberg, M.; Dröge-Laser, W. Screen Identifying Arabidopsis Transcription Factors Involved in the Response to 9-Lipoxygenase-Derived Oxylipins. PLoS ONE 2016, 11, e0153216. [Google Scholar] [CrossRef]
- Hyun, T.K.; van der Graaff, E.; Albacete, A.; Eom, S.H.; Großkinsky, D.K.; Böhm, H.; Janschek, U.; Rim, Y.; Ali, W.W.; Kim, S.Y.; et al. The Arabidopsis PLAT Domain Protein1 Is Critically Involved in Abiotic Stress Tolerance. PLoS ONE 2014, 9, e112946. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Méndez, K.J.; Sánchez Segura, L.; Chagolla, A.; Lino, B.; González de la Vara, L.E. Voltage-Dependent Anion-Selective Channels and Other Mitochondrial Membrane Proteins Form Diverse Complexes in Beetroots Subjected to Flood-Induced Programmed Cell Death. Front. Plant Sci. 2021, 12, 714847. [Google Scholar] [CrossRef] [PubMed]
- Khatri, N.; Mudgil, Y. Hypothesis: NDL proteins function in stress responses by regulating microtubule organization. Front. Plant Sci. 2015, 6, 947. [Google Scholar] [CrossRef]
- Nick, P. Microtubules, signalling and abiotic stress. Plant J. 2013, 75, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zheng, H.; Wu, J.; Ma, H.P.; Yu, J.; Yiliyaer, M. The key role of microtubules in hypoxia preconditioning-induced nuclear translocation of HIF-1α in rat cardiomyocytes. Peer J. 2017, 5, e3662. [Google Scholar] [CrossRef] [PubMed]
- Strommer, J. The plant ADH gene family. Plant J. 2011, 66, 128–142. [Google Scholar] [CrossRef] [PubMed]
- Mustroph, A.; Albrecht, G. Tolerance of crop plants to oxygen deficiency stress: Fermentative activity and photosynthetic capacity of entire seedlings under hypoxia and anoxia. Physiol. Plant. 2003, 117, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Monk, L.S.; Crawford, R.M.M.; Brändle, R. Fermentation Rates and Ethanol Accumulation in Relation to Flooding Tolerance in Rhizomes of Monocotyledonous Species. J. Exp. Bot. 1984, 35, 738–745. [Google Scholar] [CrossRef]
- Kennedy, R.A.; Rumpho, M.E.; Fox, T.C. Anaerobic metabolism in plants. Plant Physiol. 1992, 100, 1–6. [Google Scholar] [CrossRef]
- Kumar, D.; Das, P.K.; Sarmah, B.K. Reference gene validation for normalization of RT-qPCR assay associated with germination and survival of rice under hypoxic condition. J. Appl. Genet. 2018, 59, 419–430. [Google Scholar] [CrossRef]
- Slajcherová, K.; Fišerová, J.; Fischer, L.; Schwarzerová, K. Multiple actin isotypes in plants: Diverse genes for diverse roles? Front. Plant Sci. 2012, 3, 226. [Google Scholar] [CrossRef]
- Gu, C.; Chen, S.; Liu, Z.; Shan, H.; Luo, H.; Guan, Z.; Chen, F. Reference Gene Selection for Quantitative Real-Time PCR in Chrysanthemum Subjected to Biotic and Abiotic Stress. Mol. Biotechnol. 2011, 49, 192. [Google Scholar] [CrossRef] [PubMed]
- Stocking, C.R. Histology and development of the root. In Pflanze und Wasser/Water Relations of Plants. Handbuch der Pflanzenphysiologie/Encyclopedia of Plant Physiology, vol 3; Adriani, M.J., Aslyng, H.C., Burström, H., Geiger, R., Gessner, F., Härtel, O., Huber, B., Hülsbruch, M., Kalle, K., Kilian, C.H., et al., Eds.; Springer: Berlin, Heidelberg, 1956; pp. 173–187. [Google Scholar] [CrossRef]
- Roberts, J.K.; Callis, J.; Jardetzky, O.; Walbot, V.; Freeling, M. Cytoplasmic acidosis as a determinant of flooding intolerance in plants. Proc. Natl. Acad. Sci. 1984, 81, 6029–6033. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.B.; Ricard, B. Physiology, Biochemistry and Molecular Biology of Plant Root Systems Subjected to Flooding of the Soil. In Root Ecology; de Kroon, H., Visser, E.J.W., Eds.; Springer: Heidelberg/Berlin, Germany, 2003; pp. 193–213. [Google Scholar]
- de Bruxelles, G.L.; Peacock, W.J.; Dennis, E.S.; Dolferus, R. Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiol. 1996, 111, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Kyozuka, J.; Fujimoto, H.; Izawa, T.; Shimamoto, K. Anaerobic induction and tissue-specific expression of maize Adh1 promoter in transgenic rice plants and their progeny. Molec. Gen. Genet. 1991, 228, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Foster Atkinson, E. Alcohol dehydrogenase gene expression in petunia. PhD Thesis, University of Guelph, Guelph, ON, Canada, 1996. [Google Scholar]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and Biological Functions in Plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]
- Notaguchi, M.; Okamoto, S. Dynamics of long-distance signaling via plant vascular tissues. Front. Plant Sci. 2015, 6, 161. [Google Scholar] [CrossRef]
- Morris, H.; Plavcová, L.; Gorai, M.; Klepsch, M.M.; Kotowska, M.; Schenk, H.J.; Jansen, S. Vessel-associated cells in angiosperm xylem: Highly specialized living cells at the symplast–apoplast boundary. Am. J. Bot. 2018, 105, 151–160. [Google Scholar] [CrossRef]


| Spot ID | Protein Name | Funtional Annotation (Gene Ontology) | Protein Tomato Entry | Protein Fold Change (p < 0.01) | |||||
|---|---|---|---|---|---|---|---|---|---|
| PZ 215 (WL-S) | POL 7/15 (WL-T) | ||||||||
| 1xH/Ctrl | Rec/ Ctrl | 2xH/Ctrl | 1xH/Ctrl | Rec/Ctrl | 2xH/ Ctrl | ||||
| Exclusively in PZ 215: | |||||||||
| 3746 | Endochitinase (Chitinase) | cell wall macromolecule catabolic process (GO:0016998) | Solyc10g055810 | 2.3 | - | - | - | - | - |
| 2065 | Fructokinase 2 | sucrose biosynthetic process (GO: 0005986) | Solyc06g073190 | 2.0 | - | - | - | - | - |
| 2215 | Germin | response to cold (GO: 0009409), metal ion binding (GO: 0046872) | Solyc01g102380 | 1.7 | - | - | - | - | - |
| 3768 | Cysteine proteinase cathepsin F | proteolysis (GO:0006508) | Solyc04g080960 | −2.4 | - | - | - | - | - |
| 4667 | Subtilisin-like protease | proteolysis (GO:0006508) | Solyc06g062950 | −1.7 | - | - | - | - | - |
| 1261 | Glutamine synthetase | nitrate assimilation (GO: 0042128) | Solyc04g014510 | 2.0 | - | −2.9 | - | - | - |
| 4117 | Heat shock protein | ATPase activity, coupled (GO:0042623), misfolded protein binding (GO:0051787) | Solyc03g082920 | 2.1 | - | 2.4 | - | - | - |
| 4477 | Pathogenesis-related protein-1A1 | defense response (GO:0006952) | Solyc01g106610 | 2.4 | - | 4.2 | - | - | - |
| 4526 | Wound/stress protein | protein binding (GO:0005515) | Solyc03g096540 | 5.2 | - | 7.1 | - | - | - |
| 3404 | S-adenosylmethionine synthase | S-adenosylmethionine biosynthetic process (GO:0006556) | Solyc12g099000 | −1.5 | - | −3.9 | - | - | - |
| 4272 | Redoxin domain protein | cell redox homeostasis (GO:0045454) | Solyc01g079820 | −1.6 | - | −2.5 | - | - | - |
| 1439 | Peroxidase 1 | oxidoreductase activity (GO:0016491) | Solyc10g076220 | −1.7 | −1.6 | - | - | - | - |
| 3929 | Germin-like protein | nutrient reservoir activity (GO:0045735) | Solyc01g102390 | - | - | −2.1 | - | - | - |
| Exclusively in POL 7/15: | |||||||||
| 467 | 5-methyltetrahydropteroyltriglutamate -homocysteine methyltransferase | methionine biosynthetic process (GO:0009086) | Solyc10g081510 | - | - | - | 3.8 | - | - |
| 145 | Alcohol dehydrogenase 2 | alcohol dehydrogenase activity (GO:0004024) | Solyc06g059740 | - | - | - | 4.5 | - | - |
| 3473 | Fructose-bisphosphate aldolase | glycolytic process (GO:0006096) | Solyc09g009260 | - | - | - | 3.7 | - | - |
| 4212 | Pectinesterase | cell wall modification (GO:0042545) | Solyc02g080220 | - | - | - | −1.6 | - | - |
| 4572 | RNA binding protein-like protein | nucleic acid binding (GO:0003676) | Solyc05g053780 | - | - | - | −1.7 | - | −1.4 |
| Common in PZ 215 & POL 7/15: | |||||||||
| 3439 | Alcohol dehydrogenase 2 | alcohol dehydrogenase activity (GO:0004024) | Solyc06g059740 | 5.0 | - | - | 5.1 | - | - |
| 4134 | Osmotin-like protein | protein binding (GO:0005515) | Solyc08g080670 | 9.4 | - | - | 4.7 | - | - |
| 4043 | Triosephosphate isomerase | glycolytic process (GO:0006096) | Solyc04g011510 | 2.6 | - | - | 3.0 | - | - |
| 4019 | Germin-like protein | nutrient reservoir activity (GO:0045735) | Solyc01g102400 | 2.6 | - | 1.6 | 2.4 | 1.4 | - |
| 4818 | Alcohol dehydrogenase 2 | alcohol dehydrogenase activity, zinc-dependent (GO:0004024) | Solyc06g059740 | 14.0 | - | - | 11.4 | - | 4.2 |
| 3418 | Peroxidase | oxidoreductase activity (GO:0016491) | Solyc07g052510 | −1.9 | - | - | −1.7 | - | - |
| 3567 | RNA-binding protein-like | mRNA binding (GO:0003729) | Solyc07g045240 | −1.5 | - | −3.0 | - | - | −2.8 |
| 3351 | UDP-D-glucose dehydrogenase | oxidation-reduction process (GO:0055114) | Solyc02g067080 | −1.9 | - | −12.3 | - | - | −3.7 |
| 3378 | Nuclear RNA binding protein | RNA binding (GO:0003723) | Solyc01g090190 | - | - | −3.0 | −1.7 | - | −3.0 |
| 4072 | Thaumatin-like protein | defense response (GO:0006952) | Solyc11g066130 | - | - | −1.5 | −2.1 | - | −2.5 |
| Exclusively in PZ 215 2xH: | |||||||||
| 1796 | Actin | ATP binding (GO:0005524) | Solyc03g078400 | - | - | 2.9 | - | - | - |
| 3892 | Alcohol dehydrogenase 2 | alcohol dehydrogenase activity (GO:0004024) | Solyc06g059740 | - | - | 5.3 | - | - | - |
| 3953 | Coatomer subunit delta | endoplasmic reticulum to Golgi vesicle-mediated transport (GO:0006888) | Solyc01g103480 | - | - | 2.1 | - | - | - |
| 4768 | Chitinase | cell wall macromolecule catabolic process (GO:0016998) | Solyc10g055800 | - | - | 3.7 | - | - | - |
| 4804 | Embryo-specific 3 | protein binding (GO:0005515) | Solyc03g116590 | - | - | 3.0 | - | - | - |
| 4760 | Enolase | glycolytic process (GO:0006096) | Solyc09g009020 | - | - | 1.8 | - | - | - |
| 4533 | FK506-binding protein 2 | peptidyl-prolyl cis-trans isomerase activity (GO:0003755) | Solyc09g057670 | - | - | 1.6 | - | - | - |
| 4408 | Glycine-rich RNA-binding protein | nucleic acid binding (GO:0003676) | Solyc10g051380 | - | - | 5.4 | - | - | - |
| 4209 | Kunitz-type protease inhibitor | alpha-amylase inhibitor activity (GO:0015066) | Solyc03g019690 | - | - | 4.1 | - | - | - |
| 2552 | Lipoxygenase homology domain-containing protein 1 | catalase activity (GO:0004096) | Solyc04g054980 | - | - | 2.7 | - | - | - |
| 2474 | Peptidyl-prolyl cis-trans isomerase | brassinosteroid signaling pathway (GO:0009742), response to cytokinin (GO:0009735) | Solyc01g111170 | - | - | 1.7 | - | - | - |
| 1294 | Peroxidase 4 | peroxidase activity (GO:0004601) | Solyc04g071890 | - | - | 1.8 | - | - | - |
| 2944 | Porin/voltage-dependent anion-selective channel protein | transmembrane transport (GO:0055085) | Solyc02g067460 | - | - | 4.0 | - | - | - |
| 1106 | S-adenosylmethionine synthase | S-adenosylmethionine biosynthetic process (GO:0006556) | Solyc09g008280 | - | - | 3.9 | - | - | - |
| 4347 | SnRK1-interacting protein 1 | group II intron splicing (GO:0000373) | Solyc08g005060 | - | - | 2.0 | - | - | - |
| 2511 | Tubulin alpha-3 chain | microtubule cytoskeleton organization (GO:0000226) | Solyc02g091870 | - | - | 3.2 | - | - | - |
| 4482 | Water-stress inducible protein 3 | response to water deprivation (GO:0009414) | Solyc04g071610 | - | - | 6.3 | - | - | - |
| 4406 | Wound/stress protein | protein binding (GO:0005515) | Solyc03g096540 | - | - | 3.1 | - | - | - |
| 4722 | 2 3-bisphosphoglycerate-independent phosphoglycerate mutase | carbohydrate metabolic process (GO:0044262) | Solyc07g044840 | - | - | −10.1 | - | - | - |
| 3413 | Actin | ATP binding (GO:0005524) | Solyc10g080500 | - | - | −3.1 | - | - | - |
| 922 | Adenosylhomocysteinase | S-adenosylmethionine cycle (GO:0033353) | Solyc09g092380 | - | - | −3.3 | - | - | - |
| 954 | Alanine aminotransferase | pyridoxal phosphate binding (GO:0030170) | Solyc03g123610 | - | - | −2.2 | - | - | - |
| 980 | Alcohol dehydrogenase 2 | alcohol dehydrogenase activity (GO:0004024) | Solyc06g059740 | - | - | −2.7 | - | - | - |
| 3949 | Asparagine synthetase B | asparagine metabolic process (GO:0006528) | Solyc09g082780 | - | - | −2.0 | - | - | - |
| 4766 | ATP synthase subunit 1 | ATP biosynthetic process (GO:0006754) | Solyc11g039980 | - | - | −3.9 | - | - | - |
| 3287 | ATP synthase subunit beta | ATP biosynthetic process (GO:0006754) | Solyc05g008460 | - | - | −5.0 | - | - | - |
| 3529 | Cathepsin B-like cysteine proteinase | proteolysis (GO:0006508) | Solyc12g088670 | - | - | −1.6 | - | - | - |
| 3212 | Chaperone DnaK | protein folding (GO:0006457) | Solyc01g106210 | - | - | −3.0 | - | - | - |
| 4705 | Chaperonin | protein folding (GO:0006457) | Solyc01g028810 | - | - | −2.1 | - | - | - |
| 931 | Acetyltransferase component of pyruvate dehydrogenase complex | pyruvate metabolic process (GO:0006090) | Solyc07g006790 | - | - | −3.6 | - | - | - |
| 3583 | Ferredoxin--NADP reductase | oxidation-reduction process (GO:0055114) | Solyc02g024050 | - | - | −5.0 | - | - | - |
| 3601 | Fructokinase 2 | sucrose biosynthetic process (GO: 0005986) | Solyc06g073190 | - | - | −3.6 | - | - | - |
| 3535 | Fructose-bisphosphate aldolase | glycolytic process (GO:0006096) | Solyc05g008600 | - | - | −2.9 | - | - | - |
| 3477 | GDSL esterase/lipase | hydrolase activity (GO:0016788) | Solyc02g071700 | - | - | −2.6 | - | - | - |
| 3423 | Glutamate dehydrogenase | glutamate catabolic process (GO: 0006538) | Solyc10g078550 | - | - | −2.9 | - | - | - |
| 3452 | Glyceraldehyde 3-phosphate dehydrogenase | glycolytic process (GO:0006096) | Solyc05g014470 | - | - | −4.6 | - | - | - |
| 1499 | Guanine nucleotide-binding protein beta subunit-like protein | positive regulation of protein phosphorylation (GO:0001934) | Solyc12g040510 | - | - | −2.2 | - | - | - |
| 3270 | Leucyl aminopeptidase | proteolysis (GO:0006508) | Solyc12g010040 | - | - | −3.8 | - | - | - |
| 3539 | Malate dehydrogenase | carbohydrate metabolic process (GO:0005975) | Solyc09g090140 | - | - | −3.8 | - | - | - |
| 3305 | Mitochondrial processing peptidase alpha subunit | proteolysis (GO:0006508) | Solyc12g008630 | - | - | −6.5 | - | - | - |
| 3604 | Phenazine biosynthesis protein PhzF family | biosynthetic process (GO:0009058) | Solyc09g064940 | - | - | −2.6 | - | - | - |
| 4702 | Subtilisin-like protease | proteolysis (GO:0006508) | Solyc02g092670 | - | - | −1.8 | - | - | - |
| 3208 | Succinate dehydrogenase flavoprotein subunit | anaerobic respiration (GO: 0009061) | Solyc02g085350 | - | - | −5.4 | - | - | - |
| 1023 | Tubulin alpha-3 chain | microtubule cytoskeleton organization (GO:0000226) | Solyc04g077020 | - | - | −7.2 | - | - | - |
| 3664 | Tubulin beta chain | microtubule cytoskeleton organization (GO:0000226) | Solyc03g025730 | - | - | −2.3 | - | - | - |
| 3291 | UDP-D-glucose dehydrogenase | oxidation-reduction process (GO:0055114) | Solyc02g067080 | - | - | −3.9 | - | - | - |
| 3341 | UDP-glucosyltransferase | secondary metabolite biosynthetic process (GO:0044550) | Solyc11g066670 | - | - | −3.4 | - | - | - |
| 991 | UV excision repair protein RAD23 | proteasome-mediated ubiquitin-dependent protein catabolic process (GO:0043161) | Solyc04g007120 | - | - | −2.5 | - | - | - |
| 3321 | V-type ATP synthase beta chain | ATP metabolic process (GO:0046034) | Solyc01g111760 | - | - | −4.6 | - | - | - |
| 968 | Xylose isomerase | carbohydrate metabolic process (GO:0005975) | Solyc07g006650 | - | - | −2.7 | - | - | - |
| 4272 | Redoxin domain protein | cell redox homeostasis (GO:0045454) | Solyc01g079820 | −1.6 | - | −2.5 | - | - | - |
| 3404 | S-adenosylmethionine synthase | S-adenosylmethionine biosynthetic process (GO:0006556) | Solyc12g099000 | −1.5 | - | −3.9 | - | - | - |
| 3528 | Adenosine kinase | purine ribonucleoside salvage (GO:0006166) | Solyc10g086190 | - | −1.3 | −2.8 | - | - | - |
| 4364 | Class II heat shock protein | response to reactive oxygen species (GO:0000302) | Solyc08g062450 | - | −1.5 | −2.4 | - | - | - |
| 3590 | Cysteine synthase | cysteine biosynthetic process from serine (GO:0006535) | Solyc09g082060 | - | −2.4 | −6.7 | - | - | - |
| 3298 | Acetyltransferase component of pyruvate dehydrogenase complex | pyruvate metabolic process (GO:0006090) | Solyc07g006790 | - | −2.3 | −2.7 | - | - | - |
| 3242 | Protein disulfide isomerase | proteolysis (GO:0006508) | Solyc06g005940 | - | −1.8 | −3.7 | - | - | - |
| 3337 | Tubulin alpha-7 chain | microtubule cytoskeleton organization (GO:0000226) | Solyc02g087880 | - | −2.7 | −4.6 | - | - | - |
| Exclusively in POL 7/15 2xH: | |||||||||
| 1849 | Alcohol dehydrogenase 2 | alcohol dehydrogenase activity (GO:0004024) | Solyc06g059740 | - | - | - | - | - | 1.8 |
| 4839 | Peptidyl-prolyl cis-trans isomerase | brassinosteroid signaling pathway (GO:0009742), response to cytokinin (GO:0009735) | Solyc01g111170 | - | - | - | - | 2.0 | 1.8 |
| 1185 | Actin | ATP binding (GO:0005524) | Solyc11g005330 | - | - | - | - | - | −2.8 |
| 4079 | Adenylate kinase | pyrimidine nucleotide biosynthetic process (GO:0006221) | Solyc01g088480 | - | - | - | - | - | −1.9 |
| 3959 | Asparagine synthetase B | asparagine metabolic process (GO:0006528) | Solyc09g082780 | - | - | - | - | - | −2.3 |
| 4197 | Cold shock protein-1 | heat shock protein binding (GO:0031072) | Solyc01g111300 | - | - | - | - | - | −2.8 |
| 631 | Heat shock protein | ATPase activity, coupled (GO:0042623), misfolded protein binding (GO:0051787) | Solyc09g010630 | - | - | - | - | - | −2.1 |
| 908 | Methylmalonate-semialdehyde dehydrogenase | oxidation-reduction process (GO:0055114) | Solyc01g106080 | - | - | - | - | - | −1.7 |
| 3422 | Patatin | phospholipase activity (GO:0004620) | Solyc08g006860 | - | - | - | - | - | −2 |
| 4599 | Peptidyl-prolyl cis-trans isomerase | chaperone-mediated protein folding (GO:0061077) | Solyc01g105710 | - | - | - | - | - | −1.8 |
| 3576 | Peroxidase 57 | oxidation-reduction process (GO:0055114) | Solyc09g072700 | - | - | - | - | - | −1.6 |
| 1575 | Proteasome subunit alpha type | ubiquitin-dependent protein catabolic process (GO:0006511) | Solyc04g080590 | - | - | - | - | - | −1.9 |
| 1188 | Protein phosphatase 2C | phosphatase activity (GO:0016791) | Solyc03g007270 | - | - | - | - | - | −1.9 |
| 3344 | Reductase | oxidation-reduction process (GO:0055114) | Solyc08g081530 | - | - | - | - | - | −2.8 |
| 3367 | SGT1 | protein binding (GO:0005515) | Solyc03g007670 | - | - | - | - | - | −1.7 |
| 4821 | Succinate dehydrogenase iron-sulfur | aerobic respiration (GO:0009060) | Solyc02g093680 | - | - | - | - | - | −1.7 |
| 4355 | Ubiquitin-conjugating enzyme family protein-like | ubiquitin conjugating enzyme binding (GO:0031624) | Solyc10g083120 | - | - | - | - | - | −1.6 |
| 3748 | V-type ATP synthase alpha chain | ATP metabolic process (GO:0046034) | Solyc12g055800 | - | - | - | - | - | −2.2 |
| Common in PZ 215 & POL 7/15 (2xH): | |||||||||
| 530 | Subtilisin-like protease | proteolysis (GO:0006508) | Solyc08g079870 | - | - | 6.0 | - | - | 4.4 |
| 3652 | Peroxidase | oxidation-reduction process (GO:0055114) | Solyc01g105070 | - | - | 3.4 | - | - | 4.2 |
| 2662 | Glyceraldehyde 3-phosphate dehydrogenase dehydrogenase | glycolytic process (GO:0006096) | Solyc05g014470 | - | - | 5.9 | - | - | 2.7 |
| 4467 | Glyceraldehyde-3-phosphate dehydrogenase dehydrogenase | glycolytic process (GO:0006096) | Solyc06g071920 | - | - | 6.6 | - | - | 2.5 |
| 4559 | Thiosulfate sulfurtransferase rhodanese domain protein | thiosulfate sulfurtransferase activity (GO:0004792) | Solyc02g083730 | - | - | 4.9 | - | - | 3.2 |
| 3934 | Alcohol dehydrogenase 2 | alcohol dehydrogenase activity (GO:0004024) | Solyc06g059740 | 3.7 | 1.8 | 8.0 | - | - | 6.7 |
| 2602 | LH2 PLAT domain-containing protein | protein binding (GO:0005515) | Solyc03g096550 | 4.0 | - | 6.2 | - | - | 4.6 |
| 4514 | Wound/stress protein | protein binding (GO:0005515) | Solyc03g096540 | - | - | 3.7 | 2.4 | - | 3.7 |
| 1138 | Activator of heat shock protein ATPase homolog 1 | chaperone binding (GO:0051087) | Solyc10g078930 | - | - | −6.1 | - | - | −3.2 |
| 669 | Chaperone DnaK | protein folding (GO:0006457) | Solyc01g106210 | - | - | −6.3 | - | - | −3.8 |
| 3392 | Cinnamyl alcohol dehydrogenase | oxidation-reduction process (GO:0055114) | Solyc01g107590 | - | - | −6.5 | - | - | −3.5 |
| 3340 | Acetyltransferase component of pyruvate dehydrogenase complex | pyruvate metabolic process (GO:0006090) | Solyc07g006790 | - | - | −3.4 | - | - | −1.6 |
| 4723 | Dihydroxy-acid dehydratase | branched-chain amino acid biosynthetic process (GO:0009082) | Solyc12g043020 | - | - | −9.4 | - | - | −3.4 |
| 4846 | Glyceraldehyde 3-phosphate dehydrogenase | glycolytic process (GO:0006096) | Solyc05g014470 | - | - | −2.8 | - | - | −1.8 |
| 1024 | Heterogeneous nuclear ribonucleoprotein A3 | RNA binding (GO:0003723) | Solyc09g090520 | - | - | −3.8 | - | - | −2.2 |
| 3244 | Ketol-acid reductoisomerase | branched-chain amino acid biosynthetic process (GO:0009082) | Solyc07g053280 | - | - | −3.8 | - | - | −2.6 |
| 3543 | Malate dehydrogenase | carbohydrate metabolic process (GO:0005975) | Solyc09g090140 | - | - | −4.3 | - | - | −3.2 |
| 4728 | Insulinase | proteolysis (GO:0006508) | Solyc02g088700 | - | - | −4.5 | - | - | −1.8 |
| 489 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 1 | cellular respiration (GO:0045333) | Solyc11g011470 | - | - | −6.0 | - | - | −2.7 |
| 1030 | Reductase | oxidation-reduction process (GO:0055114) | Solyc08g081530 | - | - | −3.6 | - | - | −1.8 |
| 3567 | RNA-binding protein-like | mRNA binding (GO:0003729) | Solyc07g045240 | −1.5 | - | −3.0 | - | - | −2.8 |
| 3412 | S-(hydroxymethyl)glutathione dehydrogenase | ethanol oxidation (GO:0006069) | Solyc09g064370 | - | - | −4.6 | - | - | −2.1 |
| 622 | Stress-induced protein sti1-like protein | protein binding (GO:0005515) | Solyc08g079170 | - | - | −6.0 | - | - | −2.5 |
| 714 | Succinate dehydrogenase flavoprotein subunit | anaerobic respiration (GO: 0009061) | Solyc02g085350 | - | - | −5.3 | - | - | −2.6 |
| 3202 | Transketolase 1 | transketolase activity (GO:0004802) | Solyc05g050970 | - | - | −15.0 | - | - | −4.2 |
| 959 | Tubulin beta chain | microtubule cytoskeleton organization (GO:0000226) | Solyc10g080940 | - | - | −5.2 | - | - | −3.3 |
| 3317 | Tubulin beta-1 chain | microtubule cytoskeleton organization (GO:0000226) | Solyc04g081490 | - | - | −5.6 | - | - | −4.3 |
| 1063 | UDP-glucose 6-dehydrogenase | oxidation-reduction process (GO:0055114) | Solyc02g088690 | - | - | −5.0 | - | - | −2.5 |
| 1022 | UV excision repair protein RAD23 | proteasome-mediated ubiquitin-dependent protein catabolic process (GO:0043161) | Solyc02g063130 | - | - | −2.1 | - | - | −2.2 |
| 3215 | V-type ATP synthase alpha chain | ATP metabolic process (GO:0046034) | Solyc12g055800 | - | - | −4.3 | - | - | −3.0 |
| 2460 | Eukaryotic translation IF 5A | translation (GO:0006412) | Solyc07g005560 | - | - | −1.2 | - | - | −2.4 |
| 4072 | Thaumatin-like protein | defense response (GO:0006952) | Solyc11g066130 | - | - | −1.5 | −2.1 | - | −2.5 |
| Exclusively in PZ 215 Rec: | |||||||||
| 1429 | Fructokinase 2 | sucrose biosynthetic process (GO: 0005986) | Solyc06g073190 | - | −1.9 | - | - | - | - |
| 4364 | Class II heat shock protein | response to reactive oxygen species (GO:0000302) | Solyc08g062450 | - | −1.5 | −2.4 | - | - | - |
| 3590 | Cysteine synthase | cysteine biosynthetic process from serine (GO:0006535) | Solyc09g082060 | - | −2.3 | −6.7 | - | - | - |
| 3298 | Acetyltransferase component of pyruvate dehydrogenase complex | pyruvate metabolic process (GO:0006090) | Solyc07g006790 | - | −2.3 | −2.7 | - | - | - |
| 3242 | Protein disulfide isomerase | proteolysis (GO:0006508) | Solyc06g005940 | - | −1.8 | −3.7 | - | - | - |
| 3337 | Tubulin alpha-7 chain | microtubule cytoskeleton organization (GO:0000226) | Solyc02g087880 | - | −2.7 | −4.6 | - | - | - |
| 3236 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | carbohydrate metabolic process (GO:0044262) | Solyc07g044840 | - | −1.6 | - | - | - | −2.2 |
| 3351 | UDP-D-glucose dehydrogenase | oxidation-reduction process (GO:0055114) | Solyc02g067080 | - | −1.9 | −12.3 | - | - | −3.7 |
| Exclusively in POL 7/15 Rec: | |||||||||
| 3825 | Actin | ATP binding (GO:0005524) | Solyc02g067080 | - | - | - | - | 1.1 | - |
| 4130 | Osmotin-like protein | protein binding (GO:0005515) | Solyc08g080660 | - | - | - | - | 1.5 | - |
| 4260 | Tubulin alpha-3 chain | microtubule cytoskeleton organization (GO:0000226) | Solyc04g077020 | - | - | - | - | 1.6 | - |
| 4839 | Peptidyl-prolyl cis-trans isomerase | brassinosteroid signaling pathway (GO:0009742), response to cytokinin (GO:0009735) | Solyc01g111170 | - | - | - | - | 2.0 | 1.8 |
| 1213 | Peroxidase | oxidoreductase activity (GO:0016491) | Solyc07g052510 | - | - | - | - | −1.8 | - |
| 4697 | Peroxidase 4 | oxidoreductase activity (GO:0016491) | Solyc04g071890 | - | - | - | - | −2.2 | - |
| 726 | Pyruvate decarboxylase | pyruvate decarboxylase activity (GO:0004737) | Solyc10g076510 | - | - | - | - | −1.5 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czernicka, M.; Kęska, K.; Planchon, S.; Kapusta, M.; Popielarska-Konieczna, M.; Wesołowski, W.; Szklarczyk, M.; Renaut, J. Proteomic Studies of Roots in Hypoxia-Sensitive and -Tolerant Tomato Accessions Reveal Candidate Proteins Associated with Stress Priming. Cells 2022, 11, 500. https://doi.org/10.3390/cells11030500
Czernicka M, Kęska K, Planchon S, Kapusta M, Popielarska-Konieczna M, Wesołowski W, Szklarczyk M, Renaut J. Proteomic Studies of Roots in Hypoxia-Sensitive and -Tolerant Tomato Accessions Reveal Candidate Proteins Associated with Stress Priming. Cells. 2022; 11(3):500. https://doi.org/10.3390/cells11030500
Chicago/Turabian StyleCzernicka, Małgorzata, Kinga Kęska, Sébastien Planchon, Małgorzata Kapusta, Marzena Popielarska-Konieczna, Wojciech Wesołowski, Marek Szklarczyk, and Jenny Renaut. 2022. "Proteomic Studies of Roots in Hypoxia-Sensitive and -Tolerant Tomato Accessions Reveal Candidate Proteins Associated with Stress Priming" Cells 11, no. 3: 500. https://doi.org/10.3390/cells11030500
APA StyleCzernicka, M., Kęska, K., Planchon, S., Kapusta, M., Popielarska-Konieczna, M., Wesołowski, W., Szklarczyk, M., & Renaut, J. (2022). Proteomic Studies of Roots in Hypoxia-Sensitive and -Tolerant Tomato Accessions Reveal Candidate Proteins Associated with Stress Priming. Cells, 11(3), 500. https://doi.org/10.3390/cells11030500