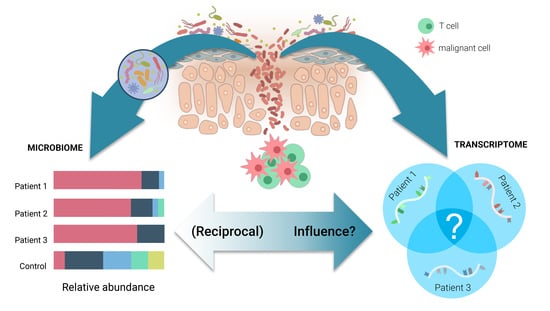
Transcriptional Heterogeneity and the Microbiome of Cutaneous T-Cell Lymphoma
Abstract
1. Introduction
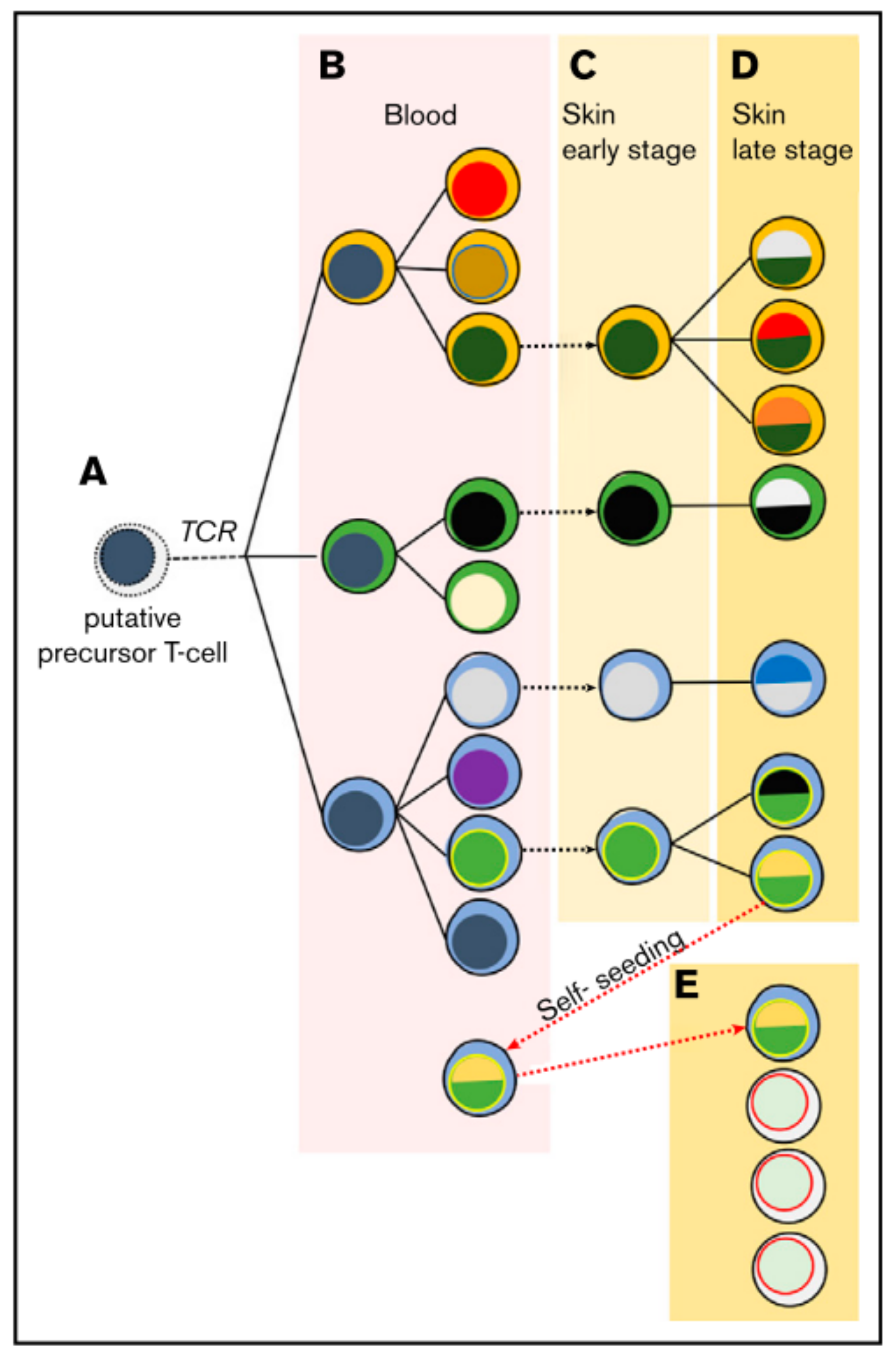
2. T-Cell Receptor Clonality
3. Transcriptional Heterogeneity
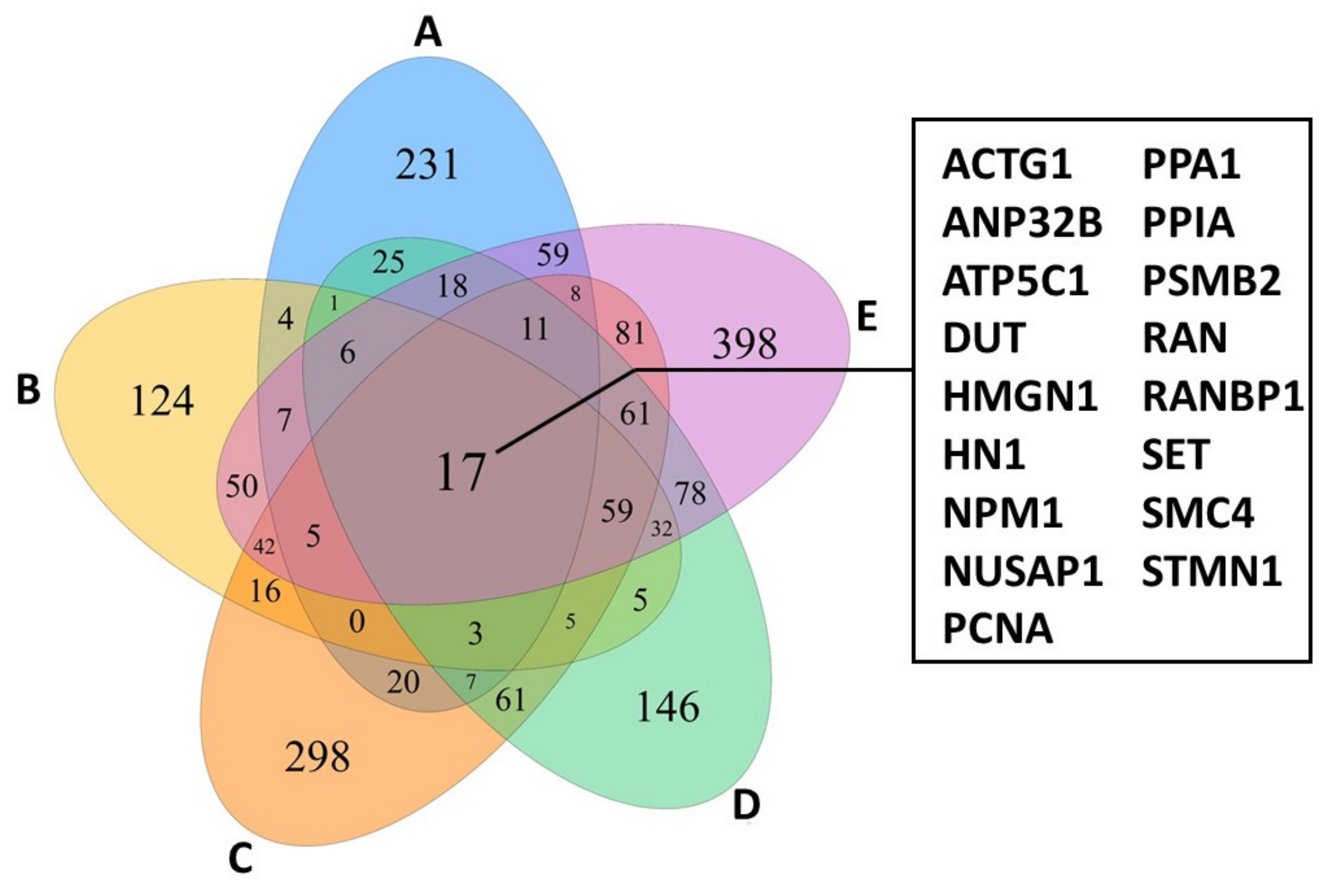
3.1. Single Cell RNA Sequencing Reveals Transcriptional Heterogeneity
3.2. Single Cell Transcriptome Signatures Suggest Microbial Influence on CTCL Heterogeneity
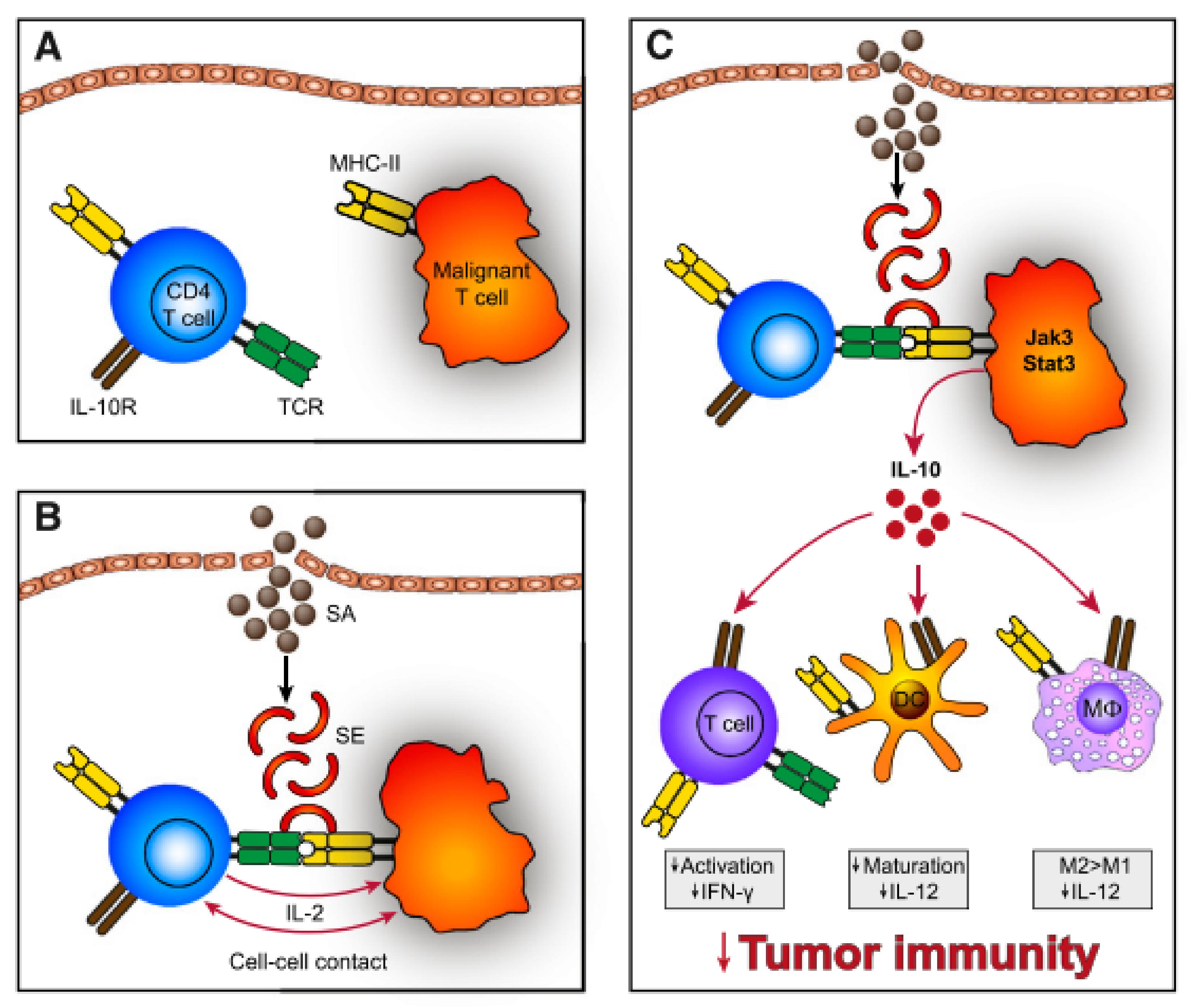
4. Microbiota and CTCL
4.1. Skin Barrier Dysfunction
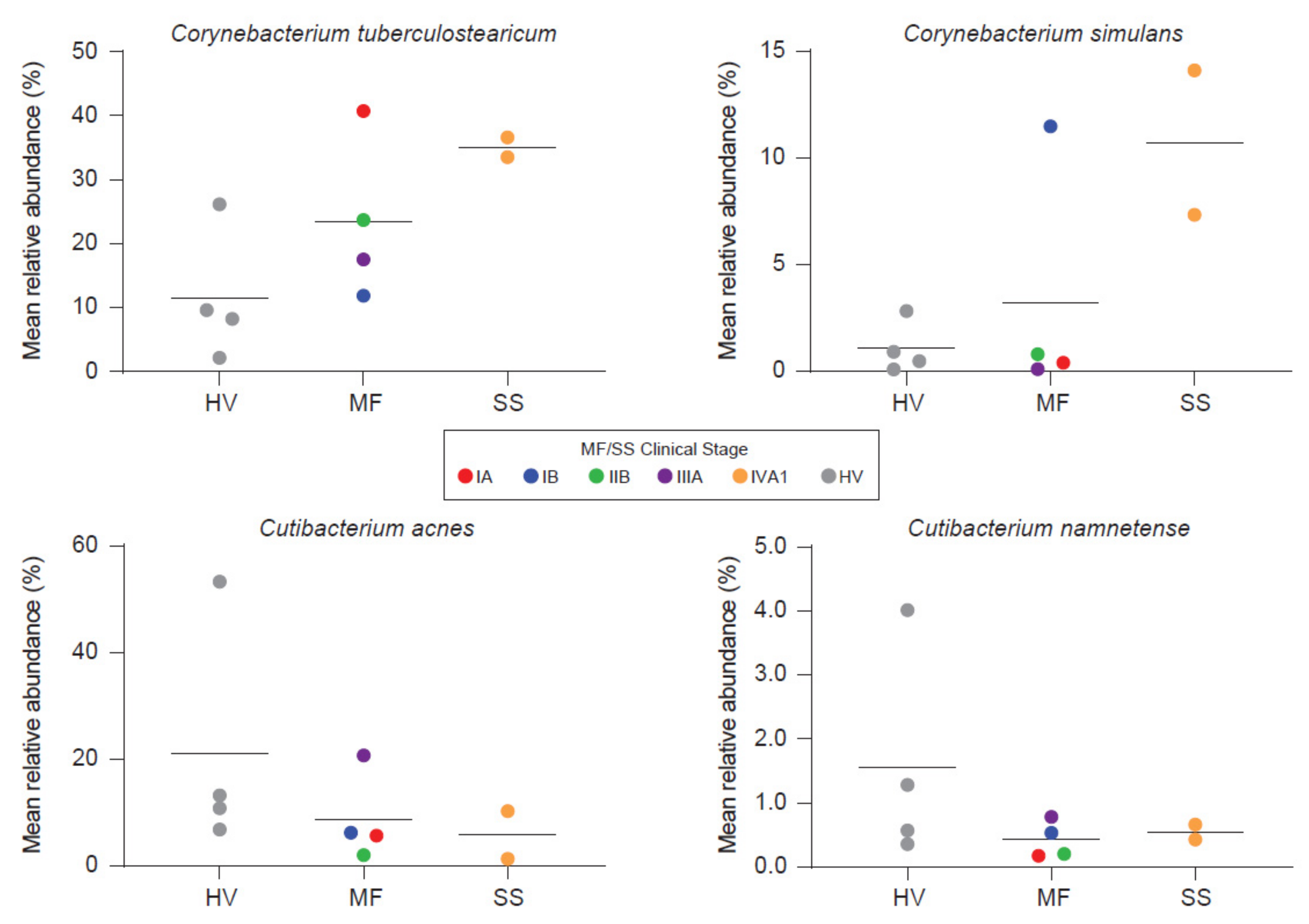
4.2. Microbiome on CTCL Lesions
4.3. Staphylococcus aureus
4.4. S. aureus Eradication
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bradford, P.T.; Devesa, S.S.; Anderson, W.F.; Toro, J.R. Cutaneous lymphoma incidence patterns in the United States: A population-based study of 3884 cases. Blood 2009, 113, 5064–5073. [Google Scholar] [CrossRef] [PubMed]
- Olsen, E.; Vonderheid, E.; Pimpinelli, N.; Willemze, R.; Kim, Y.; Knobler, R.; Zackheim, H.; Duvic, M.; Estrach, T.; Lamberg, S.; et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: A proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Ca. Blood 2007, 110, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Querfeld, C.; Zain, J.; Rosen, S.T. Primary cutaneous T-Cell lymphomas: Mycosis fungoides and Sezary syndrome. In T-Cell and NK-Cell Lymphomas; Springer: Cham, Switzerland, 2019; pp. 225–248. [Google Scholar]
- Agar, N.S.; Wedgeworth, E.; Crichton, S.; Mitchell, T.J.; Cox, M.; Ferreira, S.; Robson, A.; Calonje, E.; Stefanato, C.M.; Wain, E.M.; et al. Survival Outcomes and Prognostic Factors in Mycosis Fungoides/Sézary Syndrome: Validation of the Revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer Staging Proposal. J. Clin. Oncol. 2010, 28, 4730–4739. [Google Scholar] [CrossRef]
- Scarisbrick, J.J.; Quaglino, P.; Prince, H.M.; Papadavid, E.; Hodak, E.; Bagot, M.; Servitje, O.; Berti, E.; Ortiz-Romero, P.; Stadler, R.; et al. The PROCLIPI international registry of early-stage mycosis fungoides identifies substantial diagnostic delay in most patients. Br. J. Dermatol. 2019, 181, 350–357. [Google Scholar] [CrossRef]
- Whittaker, S.; Hoppe, R.; Prince, H.M. How I treat mycosis fungoides and Sezary syndrome. Blood 2016, 127, 3142–3153. [Google Scholar] [CrossRef]
- Kazakov, D.; Burg, G.; Kempf, W. Clinicopathological spectrum of mycosis fungoides. J. Eur. Acad. Dermatol. Venereol. 2004, 18, 397–415. [Google Scholar] [CrossRef] [PubMed]
- De Masson, A.; O’Malley, J.T.; Elco, C.P.; Garcia, S.S.; Divito, S.J.; Lowry, E.L.; Tawa, M.; Fisher, D.C.; Devlin, P.M.; Teague, J.E.; et al. High-throughput sequencing of the T cell receptor β gene identifies aggressive early-stage mycosis fungoides. Sci. Transl. Med. 2018, 10, eaar5894. [Google Scholar] [CrossRef]
- Litvinov, I.V.; Tetzlaff, M.T.; Thibault, P.; Gangar, P.; Moreau, L.; Watters, A.K.; Netchiporouk, E.; Pehr, K.; Prieto, V.G.; Rahme, E.; et al. Gene expression analysis in Cutaneous T-Cell Lymphomas (CTCL) highlights disease heterogeneity and potential diagnostic and prognostic indicators. Oncoimmunology 2017, 6, e1306618. [Google Scholar] [CrossRef]
- Rassek, K.; Iżykowska, K. Single-Cell Heterogeneity of Cutaneous T-Cell Lymphomas Revealed Using RNA-Seq Technologies. Cancers 2020, 12, 2129. [Google Scholar] [CrossRef] [PubMed]
- Lefrançois, P.; Xie, P.; Wang, L.; Tetzlaff, M.T.; Moreau, L.; Watters, A.K.; Netchiporouk, E.; Provost, N.; Gilbert, M.; Ni, X.; et al. Gene expression profiling and immune cell-type deconvolution highlight robust disease progression and survival markers in multiple cohorts of CTCL patients. Oncoimmunology 2018, 7, e1467856. [Google Scholar] [CrossRef]
- Gaydosik, A.M.; Tabib, T.; Geskin, L.J.; Bayan, C.-A.; Conway, J.F.; Lafyatis, R.; Fuschiotti, P. Single-cell Lymphocyte Heterogeneity in Advanced Cutaneous T-cell Lymphoma Skin Tumors. Clin. Cancer Res. 2019, 25, 4443–4454. [Google Scholar] [CrossRef]
- Lebas, E.; Arrese, J.E.; Nikkels, A.F. Risk Factors for Skin Infections in Mycosis Fungoides. Dermatology 2016, 232, 731–737. [Google Scholar] [CrossRef]
- Axelrod, P.I. Infections Complicating Mycosis Fungoides and Sézary Syndrome. JAMA J. Am. Med. Assoc. 1992, 267, 1354. [Google Scholar] [CrossRef]
- Mirvish, E.D.; Pomerantz, R.G.; Geskin, L.J. Infectious agents in cutaneous T-cell lymphoma. J. Am. Acad. Dermatol. 2011, 64, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, I.V.; Shtreis, A.; Kobayashi, K.; Glassman, S.; Tsang, M.; Woetmann, A.; Sasseville, D.; Ødum, N.; Duvic, M. Investigating potential exogenous tumor initiating and promoting factors for Cutaneous T-Cell Lymphomas (CTCL), a rare skin malignancy. Oncoimmunology 2016, 5, e1175799. [Google Scholar] [CrossRef][Green Version]
- Willerslev-Olsen, A.; Krejsgaard, T.; Lindahl, L.M.; Litvinov, I.V.; Fredholm, S.; Petersen, D.L.; Nastasi, C.; Gniadecki, R.; Mongan, N.P.; Sasseville, D.; et al. Staphylococcal enterotoxin A (SEA) stimulates STAT3 activation and IL-17 expression in cutaneous T-cell lymphoma. Blood 2016, 127, 1287–1296. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Harkins, C.P.; MacGibeny, M.A.; Thompson, K.; Bubic, B.; Huang, X.; Brown, I.; Park, J.; Jo, J.-H.; Segre, J.A.; Kong, H.H.; et al. Cutaneous T-Cell Lymphoma Skin Microbiome Is Characterized by Shifts in Certain Commensal Bacteria but not Viruses when Compared with Healthy Controls. J. Investig. Dermatol. 2021, 141, 1604–1608. [Google Scholar] [CrossRef]
- Salava, A.; Deptula, P.; Lyyski, A.; Laine, P.; Paulin, L.; Väkevä, L.; Ranki, A.; Auvinen, P.; Lauerma, A. Skin Microbiome in Cutaneous T-Cell Lymphoma by 16S and Whole-Genome Shotgun Sequencing. J. Investig. Dermatol. 2020, 140, 2304–2308. [Google Scholar] [CrossRef]
- Campbell, J.J.; Clark, R.A.; Watanabe, R.; Kupper, T.S. Sézary syndrome and mycosis fungoides arise from distinct T-cell subsets: A biologic rationale for their distinct clinical behaviors. Blood 2010, 116, 767–771. [Google Scholar] [CrossRef]
- Iyer, A.; Hennessey, D.; O’Keefe, S.; Patterson, J.; Wang, W.; Wong, G.K.-S.; Gniadecki, R. Skin colonization by circulating neoplastic clones in cutaneous T-cell lymphoma. Blood 2019, 134, 1517–1527. [Google Scholar] [CrossRef] [PubMed]
- Yawalkar, N.; Ferenczi, K.; Jones, D.A.; Yamanaka, K.; Suh, K.-Y.; Sadat, S.; Kupper, T.S. Profound loss of T-cell receptor repertoire complexity in cutaneous T-cell lymphoma. Blood 2003, 102, 4059–4066. [Google Scholar] [CrossRef]
- Hamrouni, A.; Fogh, H.; Zak, Z.; Ødum, N.; Gniadecki, R. Clonotypic Diversity of the T-cell Receptor Corroborates the Immature Precursor Origin of Cutaneous T-cell Lymphoma. Clin. Cancer Res. 2019, 25, 3104–3114. [Google Scholar] [CrossRef] [PubMed]
- Iyer, A.; Hennessey, D.; O’Keefe, S.; Patterson, J.; Wang, W.; Salopek, T.; Wong, G.K.-S.; Gniadecki, R. Clonotypic heterogeneity in cutaneous T-cell lymphoma (mycosis fungoides) revealed by comprehensive whole-exome sequencing. Blood Adv. 2019, 3, 1175–1184. [Google Scholar] [CrossRef]
- Iyer, A.; Hennessey, D.; O’Keefe, S.; Patterson, J.; Wang, W.; Wong, G.K.-S.; Gniadecki, R. Branched evolution and genomic intratumor heterogeneity in the pathogenesis of cutaneous T-cell lymphoma. Blood Adv. 2020, 4, 2489–2500. [Google Scholar] [CrossRef]
- Wang, L.; Ni, X.; Covington, K.R.; Yang, B.Y.; Shiu, J.; Zhang, X.; Xi, L.; Meng, Q.; Langridge, T.; Drummond, J.; et al. Genomic profiling of Sézary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat. Genet. 2015, 47, 1426–1434. [Google Scholar] [CrossRef] [PubMed]
- Jackow, C.M.; Cather, J.C.; Hearne, V.; Asano, A.T.; Musser, J.M.; Duvic, M. Association of Erythrodermic Cutaneous T-Cell Lymphoma, Superantigen-Positive Staphylococcus aureus, and Oligoclonal T-Cell Receptor Vβ Gene Expansion. Blood 1997, 89, 32–40. [Google Scholar] [CrossRef]
- Tegla, C.A.; Herrera, A.M.; Seffens, A.M.; Fanok, M.H.; Dean, G.; Kawaoka, J.; Laird, M.E.; Fulmer, Y.; Willerslev-Olsen, A.; Hymes, K.B.; et al. Skin Associated Staphylococcus aureus Contributes to Disease Progression in CTCL. Blood 2019, 134, 659. [Google Scholar] [CrossRef]
- Williams, M.R.; Costa, S.K.; Zaramela, L.S.; Khalil, S.; Todd, D.A.; Winter, H.L.; Sanford, J.A.; O’Neill, A.M.; Liggins, M.C.; Nakatsuji, T.; et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci. Transl. Med. 2019, 11, eaat8329. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef]
- Choi, J.; Goh, G.; Walradt, T.; Hong, B.S.; Bunick, C.G.; Chen, K.; Bjornson, R.D.; Maman, Y.; Wang, T.; Tordoff, J.; et al. Genomic landscape of cutaneous T cell lymphoma. Nat. Genet. 2015, 47, 1011–1019. [Google Scholar] [CrossRef]
- Fanok, M.H.; Sun, A.; Fogli, L.K.; Narendran, V.; Eckstein, M.; Kannan, K.; Dolgalev, I.; Lazaris, C.; Heguy, A.; Laird, M.E.; et al. Role of Dysregulated Cytokine Signaling and Bacterial Triggers in the Pathogenesis of Cutaneous T-Cell Lymphoma. J. Investig. Dermatol. 2018, 138, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- van Doorn, R.; van Kester, M.S.; Dijkman, R.; Vermeer, M.H.; Mulder, A.A.; Szuhai, K.; Knijnenburg, J.; Boer, J.M.; Willemze, R.; Tensen, C.P. Oncogenomic analysis of mycosis fungoides reveals major differences with Sézary syndrome. Blood 2009, 113, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Vermeer, M.H.; van Doorn, R.; Dijkman, R.; Mao, X.; Whittaker, S.; van Voorst Vader, P.C.; Gerritsen, M.-J.P.; Geerts, M.-L.; Gellrich, S.; Soderberg, O.; et al. Novel and Highly Recurrent Chromosomal Alterations in Sezary Syndrome. Cancer Res. 2008, 68, 2689–2698. [Google Scholar] [CrossRef] [PubMed]
- Damsky, W.E.; Choi, J. Genetics of Cutaneous T Cell Lymphoma: From Bench to Bedside. Curr. Treat. Options Oncol. 2016, 17, 33. [Google Scholar] [CrossRef]
- Litvinov, I.V.; Netchiporouk, E.; Cordeiro, B.; Zargham, H.; Pehr, K.; Gilbert, M.; Zhou, Y.; Moreau, L.; Woetmann, A.; Ødum, N.; et al. Ectopic expression of embryonic stem cell and other developmental genes in cutaneous T-cell lymphoma. Oncoimmunology 2014, 3, e970025. [Google Scholar] [CrossRef]
- Scott, A.C.; Dündar, F.; Zumbo, P.; Chandran, S.S.; Klebanoff, C.A.; Shakiba, M.; Trivedi, P.; Menocal, L.; Appleby, H.; Camara, S.; et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 2019, 571, 270–274. [Google Scholar] [CrossRef]
- Schrader, A.M.R.; Jansen, P.M.; Willemze, R. TOX expression in cutaneous B-cell lymphomas. Arch. Dermatol. Res. 2016, 308, 423–427. [Google Scholar] [CrossRef][Green Version]
- Schrader, A.M.R.; Jansen, P.M.; Willemze, R. TOX expression in cutaneous T-cell lymphomas: An adjunctive diagnostic marker that is not tumour specific and not restricted to the CD4+ CD8− phenotype. Br. J. Dermatol. 2016, 175, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Kuksin, M.; Morel, D.; Aglave, M.; Danlos, F.-X.; Marabelle, A.; Zinovyev, A.; Gautheret, D.; Verlingue, L. Applications of single-cell and bulk RNA sequencing in onco-immunology. Eur. J. Cancer 2021, 149, 193–210. [Google Scholar] [CrossRef] [PubMed]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef]
- Esteve-Codina, A.; Arpi, O.; Martinez-García, M.; Pineda, E.; Mallo, M.; Gut, M.; Carrato, C.; Rovira, A.; Lopez, R.; Tortosa, A.; et al. A Comparison of RNA-Seq results from paired formalin-fixed paraffin-embedded and fresh-frozen glioblastoma tissue samples. PLoS ONE 2017, 12, e0170632. [Google Scholar] [CrossRef]
- Lefrançois, P.; Tetzlaff, M.T.; Moreau, L.; Watters, A.K.; Netchiporouk, E.; Provost, N.; Gilbert, M.; Ni, X.; Sasseville, D.; Duvic, M.; et al. TruSeq-Based Gene Expression Analysis of Formalin-Fixed Paraffin-Embedded (FFPE) Cutaneous T-Cell Lymphoma Samples: Subgroup Analysis Results and Elucidation of Biases from FFPE Sample Processing on the TruSeq Platform. Front. Med. 2017, 4, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Slowikowski, K.; Zhang, F. Single-cell transcriptomics in cancer: Computational challenges and opportunities. Exp. Mol. Med. 2020, 52, 1452–1465. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xiong, F.; Wang, Y.; Zhang, S.; Gong, Z.; Li, X.; He, Y.; Shi, L.; Wang, F.; Liao, Q.; et al. What are the applications of single-cell RNA sequencing in cancer research: A systematic review. J. Exp. Clin. Cancer Res. 2021, 40, 163. [Google Scholar] [CrossRef] [PubMed]
- Streets, A.M.; Zhang, X.; Cao, C.; Pang, Y.; Wu, X.; Xiong, L.; Yang, L.; Fu, Y.; Zhao, L.; Tang, F.; et al. Microfluidic single-cell whole-transcriptome sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, 7048–7053. [Google Scholar] [CrossRef]
- Querfeld, C.; Zain, J. T-Cell and NK-Cell Lymphomas; Querfeld, C., Zain, J., Rosen, S.T., Eds.; Cancer Treatment and Research; Springer International Publishing: Cham, Switzerland, 2019; Volume 176. [Google Scholar]
- Krejsgaard, T.; Lindahl, L.M.; Mongan, N.P.; Wasik, M.A.; Litvinov, I.V.; Iversen, L.; Langhoff, E.; Woetmann, A.; Odum, N. Malignant inflammation in cutaneous T-cell lymphoma—A hostile takeover. Semin. Immunopathol. 2017, 39, 269–282. [Google Scholar] [CrossRef]
- Buus, T.B.; Willerslev-Olsen, A.; Fredholm, S.; Blümel, E.; Nastasi, C.; Gluud, M.; Hu, T.; Lindahl, L.M.; Iversen, L.; Fogh, H.; et al. Single-cell heterogeneity in Sézary syndrome. Blood Adv. 2018, 2, 2115–2126. [Google Scholar] [CrossRef]
- Borcherding, N.; Voigt, A.P.; Liu, V.; Link, B.K.; Zhang, W.; Jabbari, A. Single-Cell Profiling of Cutaneous T-Cell Lymphoma Reveals Underlying Heterogeneity Associated with Disease Progression. Clin. Cancer Res. 2019, 25, 2996–3005. [Google Scholar] [CrossRef]
- Herrera, A.; Cheng, A.; Mimitou, E.P.; Seffens, A.; George, D.D.; Bar-Natan, M.; Heguy, A.; Ruggles, K.V.; Scher, J.U.; Hymes, K.; et al. Multimodal single-cell analysis of cutaneous T cell lymphoma reveals distinct sub-clonal tissue-dependent signatures. Blood 2021, 138, 1456–1464. [Google Scholar] [CrossRef]
- Rindler, K.; Bauer, W.M.; Jonak, C.; Wielscher, M.; Shaw, L.E.; Rojahn, T.B.; Thaler, F.M.; Porkert, S.; Simonitsch-Klupp, I.; Weninger, W.; et al. Single-cell RNA sequencing reveals tissue compartment-specific plasticity of mycosis fungoides tumor cells. Front. Immunol. 2021, 12, 666935. [Google Scholar] [CrossRef]
- Wolk, K.; Mitsui, H.; Witte, K.; Gellrich, S.; Gulati, N.; Humme, D.; Witte, E.; Gonsior, M.; Beyer, M.; Kadin, M.E.; et al. Deficient cutaneous antibacterial competence in cutaneous T-Cell lymphomas: Role of Th2-mediated biased Th17 function. Clin. Cancer Res. 2014, 20, 5507–5516. [Google Scholar] [CrossRef]
- Saelens, W.; Cannoodt, R.; Todorov, H.; Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 2019, 37, 547–554. [Google Scholar] [CrossRef]
- Papalexi, E.; Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018, 18, 35–45. [Google Scholar] [CrossRef]
- Fonseca, R.; Beura, L.K.; Quarnstrom, C.F.; Ghoneim, H.E.; Fan, Y.; Zebley, C.C.; Scott, M.C.; Fares-Frederickson, N.J.; Wijeyesinghe, S.; Thompson, E.A.; et al. Developmental plasticity allows outside-in immune responses by resident memory T cells. Nat. Immunol. 2020, 21, 412–421. [Google Scholar] [CrossRef]
- Suga, H.; Sugaya, M.; Miyagaki, T.; Ohmatsu, H.; Kawaguchi, M.; Takahashi, N.; Fujita, H.; Asano, Y.; Tada, Y.; Kadono, T.; et al. Skin barrier dysfunction and low antimicrobial peptide expression in cutaneous T-cell lymphoma. Clin. Cancer Res. 2014, 20, 4339–4348. [Google Scholar] [CrossRef] [PubMed]
- Moins-Teisserenc, H.; Daubord, M.; Clave, E.; Douay, C.; Félix, J.; Marie-Cardine, A.; Ram-Wolff, C.; Maki, G.; Beldjord, K.; Homyrda, L.; et al. CD158k is a reliable marker for diagnosis of Sézary syndrome and reveals an unprecedented heterogeneity of circulating malignant cells. J. Investig. Dermatol. 2015, 135, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Bresnick, A.R.; Weber, D.J.; Zimmer, D.B. S100 proteins in cancer. Nat. Rev. Cancer 2015, 15, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Park, A.J.; Okhovat, J.-P.; Kim, J. Antimicrobial peptides. In Clinical and Basic Immunodermatology; Springer International Publishing: Cham, Switzerland, 2017; pp. 81–95. [Google Scholar]
- Bian, L.; Strzyz, P.; Jonsson, I.-M.; Erlandsson, M.; Hellvard, A.; Brisslert, M.; Ohlsson, C.; Ambartsumian, N.; Grigorian, M.; Bokarewa, M. S100A4 deficiency is associated with efficient bacterial clearance and protects against joint destruction during staphylococcal infection. J. Infect. Dis. 2011, 204, 722–730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jiao, Y.; Hou, S.; Tian, T.; Yuan, Q.; Hao, H.; Wu, Z.; Bao, X. S100A4 contributes to colitis development by increasing the adherence of citrobacter rodentium in intestinal epithelial cells. Sci. Rep. 2017, 7, 12099. [Google Scholar] [CrossRef] [PubMed]
- Corbin, B.D.; Seeley, E.H.; Raab, A.; Feldmann, J.; Miller, M.R.; Torres, V.J.; Anderson, K.L.; Dattilo, B.M.; Dunman, P.M.; Gerads, R.; et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 2008, 319, 962–965. [Google Scholar] [CrossRef]
- Cole, A.M.; Kim, Y.-H.; Tahk, S.; Hong, T.; Weis, P.; Waring, A.J.; Ganz, T. Calcitermin, a novel antimicrobial peptide isolated from human airway secretions. FEBS Lett. 2001, 504, 5–10. [Google Scholar] [CrossRef]
- Nakajima, R.; Miyagaki, T.; Kamijo, H.; Oka, T.; Shishido-Takahashi, N.; Suga, H.; Sugaya, M.; Sato, S. Decreased progranulin expression in Mycosis fungoides: A possible association with the high frequency of skin infections. Eur. J. Dermatol. 2018, 28, 790–794. [Google Scholar] [CrossRef] [PubMed]
- Willerslev-Olsen, A.; Krejsgaard, T.; Lindahl, L.; Bonefeld, C.; Wasik, M.; Koralov, S.; Geisler, C.; Kilian, M.; Iversen, L.; Woetmann, A.; et al. Bacterial toxins fuel disease progression in cutaneous T-cell lymphoma. Toxins 2013, 5, 1402–1421. [Google Scholar] [CrossRef]
- Thode, C.; Woetmann, A.; Wandall, H.H.; Carlsson, M.C.; Qvortrup, K.; Kauczok, C.S.; Wobser, M.; Printzlau, A.; Ødum, N.; Dabelsteen, S. Malignant T cells secrete galectins and induce epidermal hyperproliferation and disorganized stratification in a skin model of cutaneous T-cell lymphoma. J. Investig. Dermatol. 2015, 135, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Tang, J.; Owusu, L.; Sun, M.-Z.; Wu, J.; Zhang, J. Galectin-3 in cancer. Clin. Chim. Acta 2014, 431, 185–191. [Google Scholar] [CrossRef]
- Rabinovich, G.A.; Toscano, M.A. Turning “sweet” on immunity: Galectin–glycan interactions in immune tolerance and inflammation. Nat. Rev. Immunol. 2009, 9, 338–352. [Google Scholar] [CrossRef]
- Larsen, L.; Chen, H.-Y.; Saegusa, J.; Liu, F.-T. Galectin-3 and the skin. J. Dermatol. Sci. 2011, 64, 85–91. [Google Scholar] [CrossRef]
- Stolearenco, V.; Namini, M.R.J.; Hasselager, S.S.; Gluud, M.; Buus, T.B.; Willerslev-Olsen, A.; Ødum, N.; Krejsgaard, T. cellular interactions and inflammation in the pathogenesis of cutaneous T-cell lymphoma. Front. Cell Dev. Biol. 2020, 8, 1–12. [Google Scholar] [CrossRef]
- Howell, M.D.; Boguniewicz, M.; Pastore, S.; Novak, N.; Bieber, T.; Girolomoni, G.; Leung, D.Y.M. Mechanism of HBD-3 deficiency in atopic dermatitis. Clin. Immunol. 2006, 121, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, I.V.; Tetzlaff, M.T.; Rahme, E.; Habel, Y.; Risser, D.R.; Gangar, P.; Jennings, M.A.; Pehr, K.; Prieto, V.G.; Sasseville, D.; et al. Identification of geographic clustering and regions spared by cutaneous T-cell lymphoma in Texas using 2 distinct cancer registries. Cancer 2015, 121, 1993–2003. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, I.V.; Tetzlaff, M.T.; Rahme, E.; Jennings, M.A.; Risser, D.R.; Gangar, P.; Netchiporouk, E.; Moreau, L.; Prieto, V.G.; Sasseville, D.; et al. Demographic patterns of cutaneous T-cell lymphoma incidence in Texas based on two different cancer registries. Cancer Med. 2015, 4, 1440–1447. [Google Scholar] [CrossRef]
- Kricker, A.; Armstrong, B.K.; Hughes, A.M.; Goumas, C.; Smedby, K.E.; Zheng, T.; Spinelli, J.J.; De Sanjosé, S.; Hartge, P.; Melbye, M.; et al. Personal sun exposure and risk of non Hodgkin lymphoma: A pooled analysis from the interlymph consortium. Int. J. Cancer 2008, 122, 144–154. [Google Scholar] [CrossRef]
- Wacker, M.; Holick, M.F. Sunlight and vitamin D. Dermatoendocrinology 2013, 5, 51–108. [Google Scholar] [CrossRef]
- Hattangdi-Haridas, S.R.; Lanham-New, S.A.; Wong, W.H.S.; Ho, M.H.K.; Darling, A.L. Vitamin D deficiency and effects of vitamin D supplementation on disease severity in patients with atopic dermatitis: A systematic review and meta-analysis in adults and children. Nutrients 2019, 11, 1854. [Google Scholar] [CrossRef] [PubMed]
- Barrea, L.; Savanelli, M.C.; Di Somma, C.; Napolitano, M.; Megna, M.; Colao, A.; Savastano, S. Vitamin D and its role in psoriasis: An overview of the dermatologist and nutritionist. Rev. Endocr. Metab. Disord. 2017, 18, 195–205. [Google Scholar] [CrossRef]
- Garland, C.F.; Garland, F.C.; Gorham, E.D.; Lipkin, M.; Newmark, H.; Mohr, S.B.; Holick, M.F. The role of vitamin D in cancer prevention. Am. J. Public Health 2006, 96, 252–261. [Google Scholar] [CrossRef]
- Talpur, R.; Cox, K.M.; Hu, M.; Geddes, E.R.; Parker, M.K.; Yang, B.Y.; Armstrong, P.A.; Liu, P.; Duvic, M. Vitamin D deficiency in mycosis fungoides and sézary syndrome patients is similar to other cancer patients. Clin. Lymphoma Myeloma Leuk. 2014, 14, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Bandurska, K.; Berdowska, A.; Barczyńska-Felusiak, R.; Krupa, P. Unique features of human cathelicidin LL-37. BioFactors 2015, 41, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Talpur, R.; Bassett, R.; Duvic, M. Prevalence and treatment of Staphylococcus aureus colonization in patients with mycosis fungoides and Sézary syndrome. Br. J. Dermatol. 2008, 159, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Locey, K.J.; Lennon, J.T. Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. USA 2016, 113, 5970–5975. [Google Scholar] [CrossRef] [PubMed]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.-F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef]
- Claesson, M.J.; Clooney, A.G.; O’Toole, P.W. A clinician’ s guide to microbiome analysis. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-X.; Qin, Y.; Chen, T.; Lu, M.; Qian, X.; Guo, X.; Bai, Y. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell 2020, 12, 315. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Hsu, T.; Sirota-Madi, A.; Shafquat, A.; Abu-Ali, G.; Morgan, X.C.; Huttenhower, C. Sequencing and beyond: Integrating molecular “omics” for microbial community profiling. Nat. Rev. Microbiol. 2015, 13, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Byrd, A.L.; Deming, C.; Conlan, S.; Kong, H.H.; Segre, J.A. Biogeography and individuality shape function in the human skin metagenome. Nature 2014, 514, 59–64. [Google Scholar] [CrossRef]
- Byrd, A.L.; Deming, C.; Cassidy, S.K.B.; Harrison, O.J.; Ng, W.-I.; Conlan, S.; Belkaid, Y.; Segre, J.A.; Kong, H.H. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 2017, 9, eaal4651. [Google Scholar] [CrossRef]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; Murray, P.R.; et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef]
- Mirvish, J.J.; Pomerantz, R.G.; Falo, L.D.; Geskin, L.J. Role of infectious agents in cutaneous T-cell lymphoma: Facts and controversies. Clin. Dermatol. 2013, 31, 423–431. [Google Scholar] [CrossRef]
- Altonsy, M.O.; Kurwa, H.A.; Lauzon, G.J.; Amrein, M.; Gerber, A.N.; Almishri, W.; Mydlarski, P.R. Corynebacterium tuberculostearicum, a human skin colonizer, induces the canonical nuclear factor-κB inflammatory signaling pathway in human skin cells. Immun. Inflamm. Dis. 2020, 8, 62–79. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, M.M.; Freire, M.O.; Gabrilska, R.A.; Rumbaugh, K.P.; Lemon, K.P. Staphylococcus aureus shifts toward commensalism in response to corynebacterium species. Front. Microbiol. 2016, 7, 1230. [Google Scholar] [CrossRef]
- Cogen, A.L.; Yamasaki, K.; Sanchez, K.M.; Dorschner, R.A.; Lai, Y.; MacLeod, D.T.; Torpey, J.W.; Otto, M.; Nizet, V.; Kim, J.E.; et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Investig. Dermatol. 2010, 130, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Lina, G.; Boutite, F.; Tristan, A.; Bes, M.; Etienne, J.; Vandenesch, F. Bacterial competition for human nasal cavity colonization: Role of Staphylococcal agr alleles. Appl. Environ. Microbiol. 2003, 69, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcus aureus toxins. Curr. Opin. Microbiol. 2014, 17, 32–37. [Google Scholar] [CrossRef]
- Blümel, E.; Willerslev-Olsen, A.; Gluud, M.; Lindahl, L.M.; Fredholm, S.; Nastasi, C.; Krejsgaard, T.; Surewaard, B.G.J.; Koralov, S.B.; Hu, T.; et al. Staphylococcal alpha-toxin tilts the balance between malignant and non-malignant CD4+ T cells in cutaneous T-cell lymphoma. Oncoimmunology 2019, 8, e1641387. [Google Scholar] [CrossRef]
- Blümel, E.; Munir Ahmad, S.; Nastasi, C.; Willerslev-Olsen, A.; Gluud, M.; Fredholm, S.; Hu, T.; Surewaard, B.G.J.; Lindahl, L.M.; Fogh, H.; et al. Staphylococcus aureus alpha-toxin inhibits CD8+ T cell-mediated killing of cancer cells in cutaneous T-cell lymphoma. Oncoimmunology 2020, 9, 1751561. [Google Scholar] [CrossRef]
- Spaulding, A.R.; Salgado-Pabon, W.; Kohler, P.L.; Horswill, A.R.; Leung, D.Y.M.; Schlievert, P.M. Staphylococcal and streptococcal superantigen exotoxins. Clin. Microbiol. Rev. 2013, 26, 422–447. [Google Scholar] [CrossRef] [PubMed]
- Macias, E.S.; Pereira, F.A.; Rietkerk, W.; Safai, B. Superantigens in dermatology. J. Am. Acad. Dermatol. 2011, 64, 455–472. [Google Scholar] [CrossRef]
- Fraser, J.D.; Proft, T. The bacterial superantigen and superantigen-like proteins. Immunol. Rev. 2008, 225, 226–243. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Tamoutounour, S. The influence of skin microorganisms on cutaneous immunity. Nat. Rev. Immunol. 2016, 16, 353–366. [Google Scholar] [CrossRef]
- Krejsgaard, T.; Willerslev-Olsen, A.; Lindahl, L.M.; Bonefeld, C.M.; Koralov, S.B.; Geisler, C.; Wasik, M.A.; Gniadecki, R.; Kilian, M.; Iversen, L.; et al. Staphylococcal enterotoxins stimulate lymphoma-associated immune dysregulation. Blood 2014, 124, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Nisini, R.; Matricardi, P.M.; Fattorossi, A.; Biselli, R.; D’amelio, R. Presentation of superantigen by human T cell clones: A model of T-T cell interaction. Eur. J. Immunol. 1992, 22, 2033–2039. [Google Scholar] [CrossRef]
- Helft, J.; Jacquet, A.; Joncker, N.T.; Grandjean, I.; Dorothée, G.; Kissenpfennig, A.; Malissen, B.; Matzinger, P.; Lantz, O. Antigen-specific T-T interactions regulate CD4 T-cell expansion. Blood 2008, 112, 1249–1258. [Google Scholar] [CrossRef]
- Willerslev-Olsen, A.; Gjerdrum, L.M.R.; Lindahl, L.M.; Buus, T.B.; Pallesen, E.M.H.; Gluud, M.; Bzorek, M.; Nielsen, B.S.; Kamstrup, M.R.; Rittig, A.H.; et al. Staphylococcus aureus Induces Signal Transducer and Activator of Transcription 5—Dependent miR-155 Expression in Cutaneous T-Cell Lymphoma. J. Invest. Dermatol. 2021, 141, 2449–2458. [Google Scholar] [CrossRef]
- Willerslev-Olsen, A.; Buus, T.B.; Nastasi, C.; Blümel, E.; Gluud, M.; Bonefeld, C.M.; Geisler, C.; Lindahl, L.M.; Vermeer, M.; Wasik, M.A.; et al. Staphylococcus aureus enterotoxins induce FOXP3 in neoplastic T cells in Sézary syndrome. Blood Cancer J. 2020, 10, 57. [Google Scholar] [CrossRef]
- Beissert, S.; Schwarz, A.; Schwarz, T. Regulatory T Cells. J. Investig. Dermatol. 2006, 126, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Tett, A.; Pasolli, E.; Farina, S.; Truong, D.T.; Asnicar, F.; Zolfo, M.; Beghini, F.; Armanini, F.; Jousson, O.; De Sanctis, V.; et al. Unexplored diversity and strain-level structure of the skin microbiome associated with psoriasis. NPJ Biofilms Microbiomes 2017, 3, 14. [Google Scholar] [CrossRef]
- Tokura, Y.; Yagi, H.; Ohshima, A.; Kurokawa, S.; Wakita, H.; Yokote, R.; Shirahama, S.; Flirukawa, F.; Takigawa, M. Cutaneous colonization with staphylococci influences the disease activity of Sézary syndrome: A potential role for bacterial superantigens. Br. J. Dermatol. 1995, 133, 6–12. [Google Scholar] [CrossRef]
- Lindahl, L.M.; Willerslev-Olsen, A.; Gjerdrum, L.M.R.; Nielsen, P.R.; Blümel, E.; Rittig, A.H.; Celis, P.; Herpers, B.; Becker, J.C.; Stausbøl-Grøn, B.; et al. Antibiotics inhibit tumor and disease activity in cutaneous T-cell lymphoma. Blood 2019, 134, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, L.M.; Iversen, L.; Ødum, N.; Kilian, M. Staphylococcus aureus and Antibiotics in Cutaneous T-Cell Lymphoma. Dermatology 2021, 7, 1–3. [Google Scholar] [CrossRef]
- Emge, D.A.; Bassett, R.L.; Duvic, M.; Huen, A.O. Methicillin-resistant Staphylococcus aureus (MRSA) is an important pathogen in erythrodermic cutaneous T-cell lymphoma (CTCL) patients. Arch. Dermatol. Res. 2020, 312, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.J.; Holder, B.B.; Duvic, M. The “Duvic regimen” for erythrodermic flares secondary to Staphylococcus aureus in mycosis fungoides and Sézary syndrome. Int. J. Dermatol. 2018, 57, 123–124. [Google Scholar] [CrossRef]
- Ansell, S.M. Non-Hodgkin lymphoma: Diagnosis and treatment. Mayo Clin. Proc. 2015, 90, 1152–1163. [Google Scholar] [CrossRef] [PubMed]
- Stadler, R.; Scarisbrick, J.J. Maintenance therapy in patients with mycosis fungoides or Sézary syndrome: A neglected topic. Eur. J. Cancer 2021, 142, 38–47. [Google Scholar] [CrossRef]
- Molloy, K.; Jonak, C.; Woei-A-Jin, F.J.S.H.; Guenova, E.; Busschots, A.M.; Bervoets, A.; Hauben, E.; Knobler, R.; Porkert, S.; Fassnacht, C.; et al. Characteristics associated with significantly worse quality of life in mycosis fungoides/Sézary syndrome from the prospective cutaneous lymphoma international prognostic index (PROCLIPI) study. Br. J. Dermatol. 2020, 182, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Grice, E. The skin microbiome: Potential for novel diagnostic and therapeutic approaches to cutaneous disease. Semin. Cutan. Med. Surg. 2014, 33, 98–103. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Hata, T.R.; Tong, Y.; Cheng, J.Y.; Shafiq, F.; Butcher, A.M.; Salem, S.S.; Brinton, S.L.; Rudman Spergel, A.K.; Johnson, K.; et al. Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat. Med. 2021, 27, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Byrd, A.L.; Park, M.; Kong, H.H.; Segre, J.A. Temporal stability of the human skin microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; Green, E.D.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial community variation in human body habitats across space and time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef] [PubMed]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Licht, P.; Mailänder, V. Transcriptional Heterogeneity and the Microbiome of Cutaneous T-Cell Lymphoma. Cells 2022, 11, 328. https://doi.org/10.3390/cells11030328
Licht P, Mailänder V. Transcriptional Heterogeneity and the Microbiome of Cutaneous T-Cell Lymphoma. Cells. 2022; 11(3):328. https://doi.org/10.3390/cells11030328
Chicago/Turabian StyleLicht, Philipp, and Volker Mailänder. 2022. "Transcriptional Heterogeneity and the Microbiome of Cutaneous T-Cell Lymphoma" Cells 11, no. 3: 328. https://doi.org/10.3390/cells11030328
APA StyleLicht, P., & Mailänder, V. (2022). Transcriptional Heterogeneity and the Microbiome of Cutaneous T-Cell Lymphoma. Cells, 11(3), 328. https://doi.org/10.3390/cells11030328