Multi-Scope Feature Extraction for Intracranial Aneurysm 3D Point Cloud Completion
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Experimental Dataset
2.3. Proposed Methods
2.3.1. Multi-Scope Aggregate Encoder
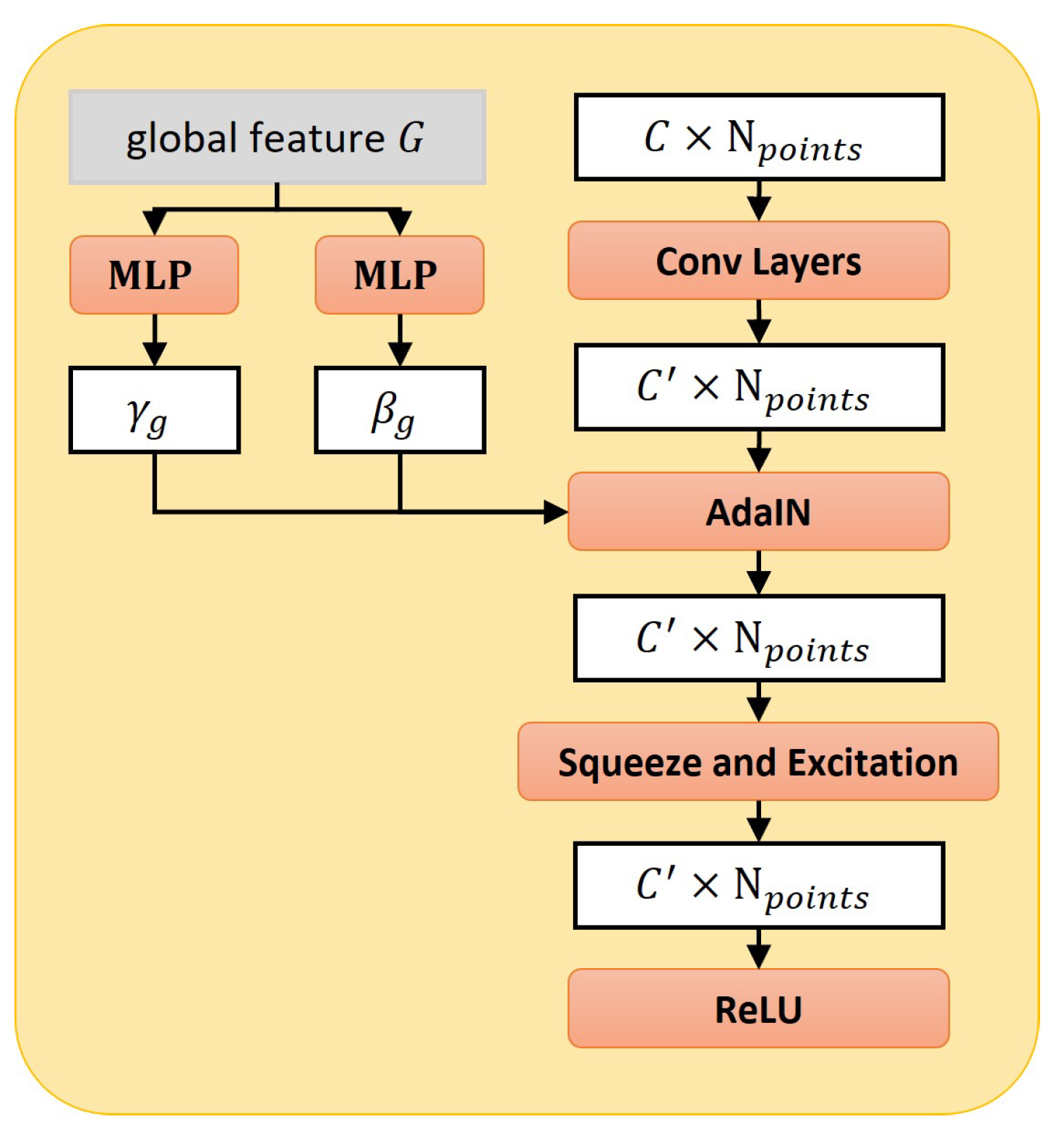
2.3.2. Style-Based Folding Decoder
2.3.3. Refinement Module
2.3.4. Joint Loss Function
2.4. Experimental Setting
2.5. Evaluations
3. Results
3.1. Evaluation on IntrACompletion Dataset
3.2. Evaluation on PCN Dataset
3.3. Ablation Study
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, X.; Xia, D.; Kin, T.; Igarashi, T. INTRA: 3D intracranial aneurysm dataset for deep learning. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 2653–2663. [Google Scholar] [CrossRef]
- Yuan, W.; Khot, T.; Held, D.; Mertz, C.; Hebert, M. PCN: Point completion network. In Proceedings of the 2018 International Conference on 3D Vision, Verona, Italy, 5–8 September 2018; 2018; pp. 728–737. [Google Scholar] [CrossRef]
- Goyal, B.; Dogra, A.; Khoond, R.; Al-Turjman, F. An Efficient Medical Assistive Diagnostic Algorithm for Visualisation of Structural and Tissue Details in CT and MRI Fusion. Cogn. Comput. 2021, 13, 1471–1483. [Google Scholar] [CrossRef]
- Qi, C.R.; Su, H.; Mo, K.; Guibas, L.J. PointNet: Deep learning on point sets for 3D classification and segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 77–85. [Google Scholar] [CrossRef]
- Qi, C.R.; Yi, L.; Su, H.; Guibas, L.J. PointNet++: Deep hierarchical feature learning on point sets in a metric space. Adv. Neural Inf. Process. Syst. 2017, 5100–5109. [Google Scholar]
- Huang, Z.; Yu, Y.; Xu, J.; Ni, F.; Le, X. PF-Net: Point fractal network for 3D point cloud completion. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; Volume 4, pp. 7659–7667. [Google Scholar] [CrossRef]
- Gadelha, M.; Wang, R.; Maji, S. Multiresolution tree networks for 3D point cloud processing. In Computer Vision—ECCV 2018; Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11211, pp. 105–122. [Google Scholar] [CrossRef]
- Wang, X.; Ang, M.H.; Lee, G.H. Cascaded Refinement Network for Point Cloud Completion. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 787–796. [Google Scholar] [CrossRef]
- Sarmad, M.; Lee, H.J.; Kim, Y.M. RL-GAN-net: A reinforcement learning agent controlled gan network for real-time point cloud shape completion. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 5891–5900. [Google Scholar] [CrossRef]
- Hu, T.; Han, Z.; Shrivastava, A.; Zwicker, M. Render4Completion: Synthesizing multi-view depth maps for 3D shape completion. In Proceedings of the International Conference on Computer Vision Workshop, Long Beach, CA, USA, 15–20 June 2019; pp. 4114–4122. [Google Scholar] [CrossRef]
- Yang, Y.; Feng, C.; Shen, Y.; Tian, D. FoldingNet: Point Cloud Auto-Encoder via Deep Grid Deformation. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 206–215. [Google Scholar] [CrossRef]
- Groueix, T.; Fisher, M.; Kim, V.G.; Russell, B.C.; Aubry, M. A Papier-Mâché Approach to Learning 3D Surface Generation. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 216–224. [Google Scholar] [CrossRef]
- Ping, Y.; Wei, G.; Yang, L.; Cui, Z.; Wang, W. Self-attention implicit function networks for 3D dental data completion. Comput. Aided Geom. Des. 2021, 90, 102026. [Google Scholar] [CrossRef]
- Kodym, O.; Li, J.; Pepe, A.; Gsaxner, C.; Chilamkurthy, S.; Egger, J.; Španěl, M. SkullBreak/SkullFix—Dataset for automatic cranial implant design and a benchmark for volumetric shape learning tasks. Data Brief 2021, 35, 106902. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Wang, C.; Zhang, B.; Yang, H.; Chen, D.; Wen, F. Style-based Point Generator with Adversarial Rendering for Point Cloud Completion. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021. [Google Scholar] [CrossRef]
- Zhang, W.; Yan, Q.; Xiao, C. Detail Preserved Point Cloud Completion via Separated Feature Aggregation. In Computer Vision—ECCV 2020; Lecture Notes in Computer Science; Vedaldi, A., Bischof, H., Brox, T., Frahm, J.M., Eds.; Springer: Cham, Switzerland, 2020; Volume 12370, pp. 512–528. [Google Scholar] [CrossRef]
- Mendoza, A.; Apaza, A.; Sipiran, I.; Lopez, C. Refinement of Predicted Missing Parts Enhance Point Cloud Completion. arXiv 2020, arXiv:2010.04278. [Google Scholar]
- Wang, Y.; Sun, Y.; Liu, Z.; Sarma, S.E.; Bronstein, M.M.; Solomon, J.M. Dynamic graph Cnn for learning on point clouds. ACM Trans. Graph. 2019, 38, 1–12. [Google Scholar] [CrossRef]
- Pan, L.; Chen, X.; Cai, Z.; Zhang, J.; Zhao, H.; Yi, S.; Liu, Z. Variational Relational Point Completion Network. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021. [Google Scholar]
- Hong, J.; Kim, K.; Lee, H. Faster dynamic graph CNN: Faster deep learning on 3d point cloud data. IEEE Access 2020, 8, 190529–190538. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Wen, X.; Li, T.; Han, Z.; Liu, Y.S. Point Cloud Completion by Skip-Attention Network with Hierarchical Folding. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 1936–1945. [Google Scholar] [CrossRef]
- Li, R.; Li, X.; Fu, C.W.; Cohen-Or, D.; Heng, P.A. PU-GAN: A point cloud upsampling adversarial network. In Proceedings of the IEEE International Conference on Computer Vision, Long Beach, CA, USA, 15–20 June 2019; pp. 7202–7211. [Google Scholar] [CrossRef]
- Liu, M.; Sheng, L.; Yang, S.; Shao, J.; Hu, S.M. Morphing and sampling network for dense point cloud completion. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; pp. 11596–11603. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Q.; Yue, X.; Lasenby, J.; Kusner, M.J. Unsupervised Point Cloud Pre-training via Occlusion Completion. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021. [Google Scholar] [CrossRef]
- Karras, T.; Laine, S.; Aila, T. A Style-Based Generator Architecture for Generative Adversarial Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 4217–4228. [Google Scholar] [CrossRef] [PubMed]
- Prim, R.C. Shortest connection networks and some generalizations. Bell Syst. Tech. J. 1957, 36, 1389–1401. [Google Scholar] [CrossRef]
- Fan, H.; Su, H.; Guibas, L. A point set generation network for 3D object reconstruction from a single image. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2463–2471. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019, 32. [Google Scholar]
- Huang, K.; Hussain, A.; Wang, Q.F.; Zhang, R. Deep Learning: Fundamentals, Theory and Applications; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Yu, L.; Li, X.; Fu, C.-W.; Cohen-Or, D.; Heng, P.-A. PU-Net: Point Cloud Upsampling Network Lequan. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 2790–2799. [Google Scholar]
- Xie, H.; Yao, H.; Zhou, S.; Mao, J.; Zhang, S.; Sun, W. GRNet: Gridding Residual Network for Dense Point Cloud Completion. In Computer Vision—ECCV 2020; Lecture Notes in Computer Science; Vedaldi, A., Bischof, H., Brox, T., Frahm, J.M., Eds.; Springer: Cham, Switzerland, 2020; Volume 12354, pp. 365–381. [Google Scholar] [CrossRef]
- Mahmud, M.; Kaiser, M.S.; McGinnity, T.M.; Hussain, A. Deep Learning in Mining Biological Data. Cogn. Comput. 2021, 13, 1–33. [Google Scholar] [CrossRef] [PubMed]


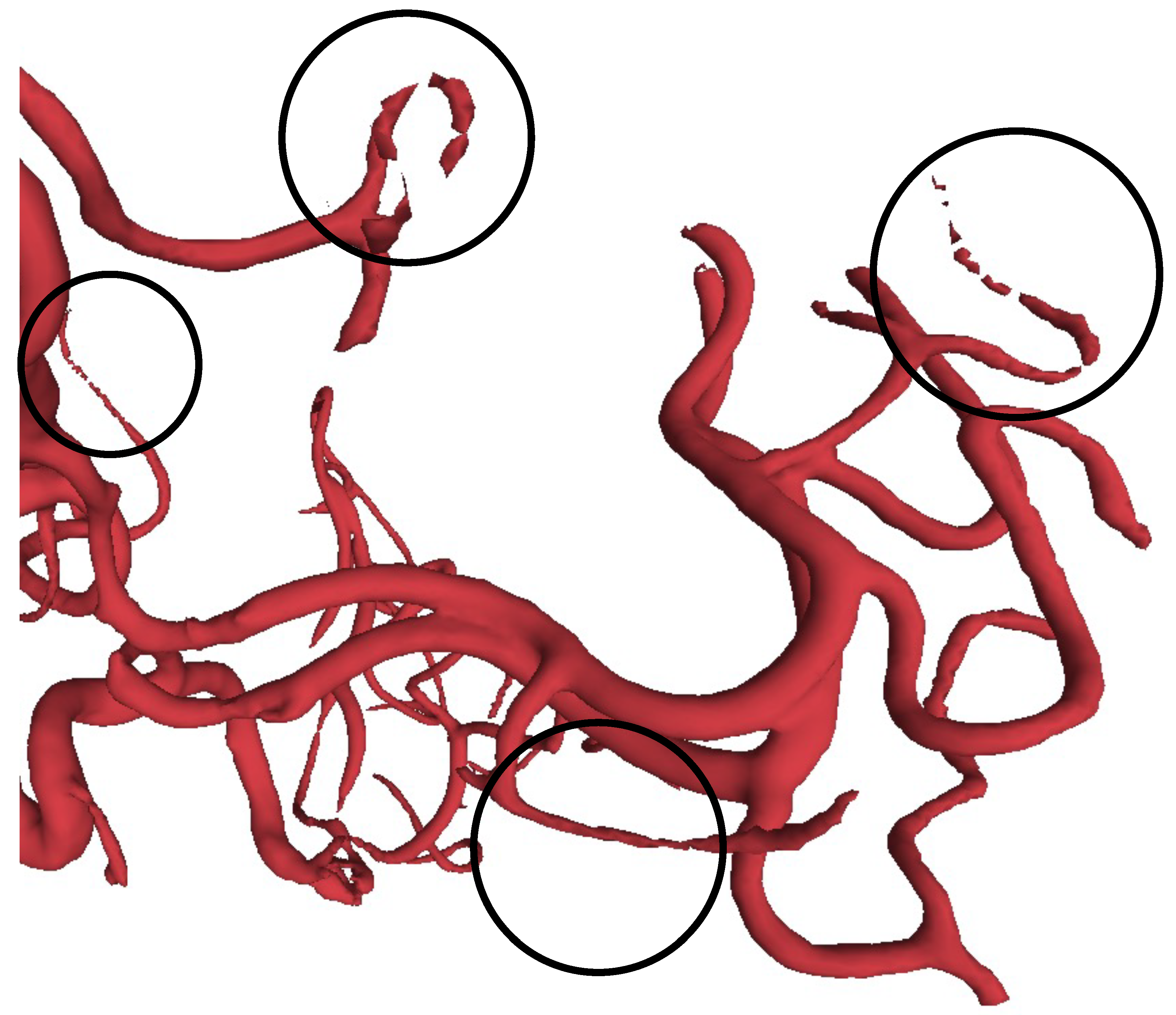
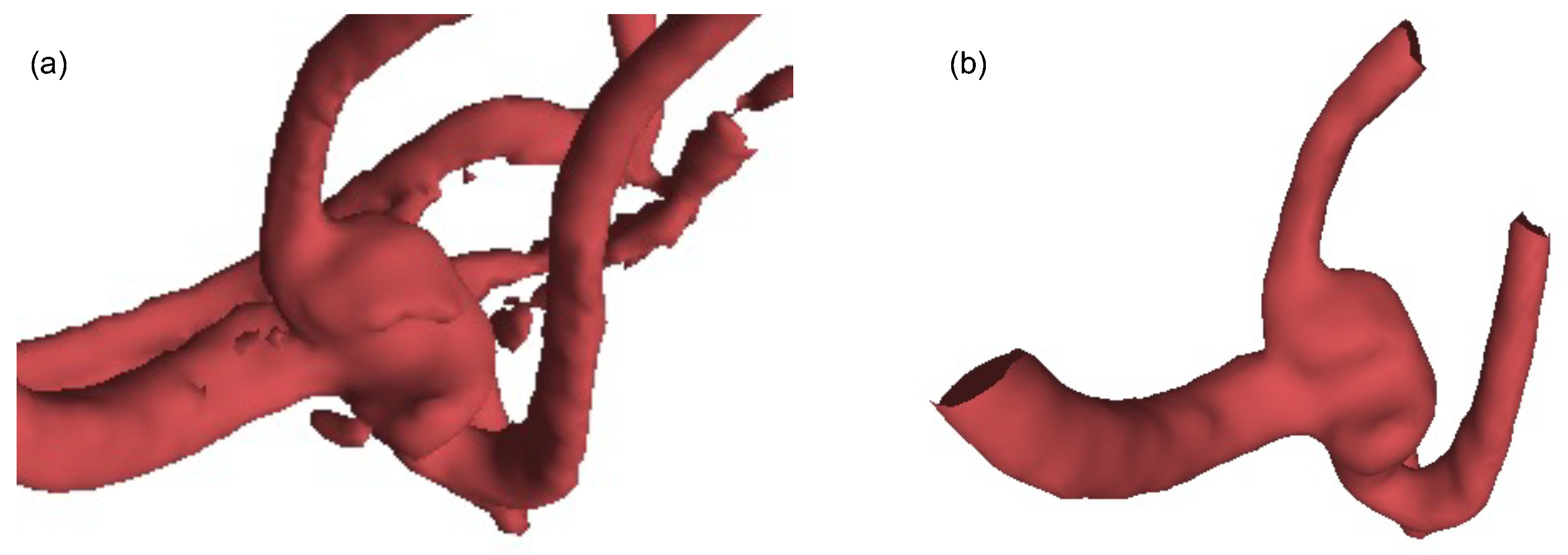






| Methods | Aneurysm | Vessel | Overall |
|---|---|---|---|
| AtlasNet [12] | 0.168 | 0.196 | 0.193 |
| FoldingNet [11] | 0.132 | 0.161 | 0.158 |
| PCN [2] | 0.140 | 0.154 | 0.153 |
| MSN [24] | 0.278 | 0.318 | 0.313 |
| GRNet [33] | 0.224 | 0.247 | 0.245 |
| SpareNet [15] | 0.322 | 0.376 | 0.370 |
| Ours | 0.343 | 0.384 | 0.379 |
| Methods | Aneurysm | Vessel | Overall |
|---|---|---|---|
| AtlasNet [12] | 2.156 | 2.049 | 2.061 |
| FoldingNet [11] | 5.124 | 3.781 | 3.929 |
| PCN [2] | 3.972 | 3.971 | 3.971 |
| MSN [24] | 1.623 | 1.493 | 1.508 |
| GRNet [33] | 2.068 | 2.006 | 2.013 |
| SpareNet [15] | 1.453 | 1.140 | 1.174 |
| Ours | 1.173 | 0.976 | 0.998 |
| Methods | Aneurysm | Vessel | Overall |
|---|---|---|---|
| AtlasNet [12] | 4.158 | 3.983 | 4.002 |
| FoldingNet [11] | 5.401 | 4.724 | 4.799 |
| PCN [2] | 5.466 | 5.381 | 5.391 |
| MSN [24] | 3.524 | 3.323 | 3.345 |
| GRNet [33] | 4.164 | 4.048 | 4.061 |
| SpareNet [15] | 3.379 | 2.885 | 2.940 |
| Ours | 3.000 | 2.719 | 2.750 |
| Methods | Airplane | Cabinet | Car | Chair | Lamp | Sofa | Table | Vessel | Overall |
|---|---|---|---|---|---|---|---|---|---|
| AtlasNet [12] | 0.590 | 0.096 | 0.160 | 0.149 | 0.211 | 0.106 | 0.199 | 0.263 | 0.222 |
| FoldingNet [11] | 0.509 | 0.134 | 0.159 | 0.161 | 0.247 | 0.117 | 0.229 | 0.279 | 0.229 |
| PCN [2] | 0.611 | 0.148 | 0.200 | 0.185 | 0.293 | 0.128 | 0.240 | 0.312 | 0.265 |
| MSN [24] | 0.670 | 0.117 | 0.202 | 0.229 | 0.380 | 0.148 | 0.276 | 0.357 | 0.297 |
| GRNet [33] | 0.554 | 0.133 | 0.176 | 0.197 | 0.365 | 0.132 | 0.233 | 0.315 | 0.263 |
| SpareNet [15] | 0.664 | 0.162 | 0.204 | 0.240 | 0.436 | 0.153 | 0.275 | 0.378 | 0.314 |
| Ours | 0.661 | 0.164 | 0.206 | 0.249 | 0.439 | 0.167 | 0.297 | 0.380 | 0.320 |
| Methods | Airplane | Cabinet | Car | Chair | Lamp | Sofa | Table | Vessel | Overall |
|---|---|---|---|---|---|---|---|---|---|
| AtlasNet [12] | 1.926 | 3.809 | 3.090 | 3.581 | 3.920 | 4.001 | 3.244 | 2.879 | 3.307 |
| FoldingNet [11] | 2.090 | 3.458 | 3.229 | 3.711 | 3.609 | 3.940 | 3.188 | 2.913 | 3.267 |
| PCN [2] | 1.867 | 3.261 | 2.720 | 3.275 | 3.268 | 3.552 | 2.935 | 2.664 | 2.943 |
| MSN [24] | 1.704 | 3.668 | 2.844 | 3.049 | 2.968 | 3.600 | 2.727 | 2.486 | 2.881 |
| GRNet [33] | 2.013 | 3.474 | 2.954 | 3.188 | 2.836 | 3.705 | 2.977 | 2.583 | 2.966 |
| SpareNet [15] | 1.762 | 3.442 | 2.913 | 3.231 | 2.691 | 3.659 | 2.937 | 2.529 | 2.895 |
| Ours | 1.778 | 3.399 | 2.889 | 3.176 | 2.697 | 3.600 | 2.854 | 2.472 | 2.858 |
| Methods | Airplane | Cabinet | Car | Chair | Lamp | Sofa | Table | Vessel | Overall |
|---|---|---|---|---|---|---|---|---|---|
| AtlasNet [12] | 0.431 | 1.836 | 0.977 | 1.470 | 1.952 | 2.014 | 1.405 | 0.948 | 1.379 |
| FoldingNet [11] | 0.582 | 1.746 | 1.208 | 1.946 | 2.203 | 2.151 | 1.689 | 1.182 | 1.589 |
| PCN [2] | 0.408 | 1.420 | 0.775 | 1.291 | 1.234 | 1.527 | 1.223 | 0.816 | 1.087 |
| MSN [24] | 0.315 | 1.691 | 0.828 | 1.153 | 1.094 | 1.607 | 0.959 | 0.684 | 1.041 |
| GRNet [33] | 0.441 | 1.360 | 0.874 | 1.189 | 0.994 | 1.826 | 1.101 | 0.691 | 1.059 |
| SpareNet [15] | 0.320 | 1.374 | 0.880 | 1.192 | 0.891 | 1.501 | 0.973 | 0.649 | 0.972 |
| Ours | 0.331 | 1.374 | 0.874 | 1.198 | 0.945 | 1.485 | 0.951 | 0.633 | 0.974 |
| Multi-Scope | Partial Input | EMD (Lower Is Better) | CD (Lower Is Better) | F-Score (Higher Is Better) |
|---|---|---|---|---|
| 3.064 | 1.267 | 0.354 | ||
| ✓ | 2.973 | 1.181 | 0.351 | |
| ✓ | 2.931 | 1.118 | 0.358 | |
| ✓ | ✓ | 2.750 | 0.998 | 0.379 |
| Multi-Scope | Partial Input | EMD (Lower Is Better) | CD (Lower Is Better) | F-Score (Higher Is Better) |
|---|---|---|---|---|
| 2.890 | 0.955 | 0.313 | ||
| ✓ | 2.878 | 0.977 | 0.312 | |
| ✓ | 2.862 | 1.011 | 0.314 | |
| ✓ | ✓ | 2.858 | 0.974 | 0.320 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, W.; Yang, X.; Wang, Q.; Huang, K.; Huang, X. Multi-Scope Feature Extraction for Intracranial Aneurysm 3D Point Cloud Completion. Cells 2022, 11, 4107. https://doi.org/10.3390/cells11244107
Ma W, Yang X, Wang Q, Huang K, Huang X. Multi-Scope Feature Extraction for Intracranial Aneurysm 3D Point Cloud Completion. Cells. 2022; 11(24):4107. https://doi.org/10.3390/cells11244107
Chicago/Turabian StyleMa, Wuwei, Xi Yang, Qiufeng Wang, Kaizhu Huang, and Xiaowei Huang. 2022. "Multi-Scope Feature Extraction for Intracranial Aneurysm 3D Point Cloud Completion" Cells 11, no. 24: 4107. https://doi.org/10.3390/cells11244107
APA StyleMa, W., Yang, X., Wang, Q., Huang, K., & Huang, X. (2022). Multi-Scope Feature Extraction for Intracranial Aneurysm 3D Point Cloud Completion. Cells, 11(24), 4107. https://doi.org/10.3390/cells11244107







