SFRP2 Overexpression Induces an Osteoblast-like Phenotype in Prostate Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line and Cell Culture Conditions
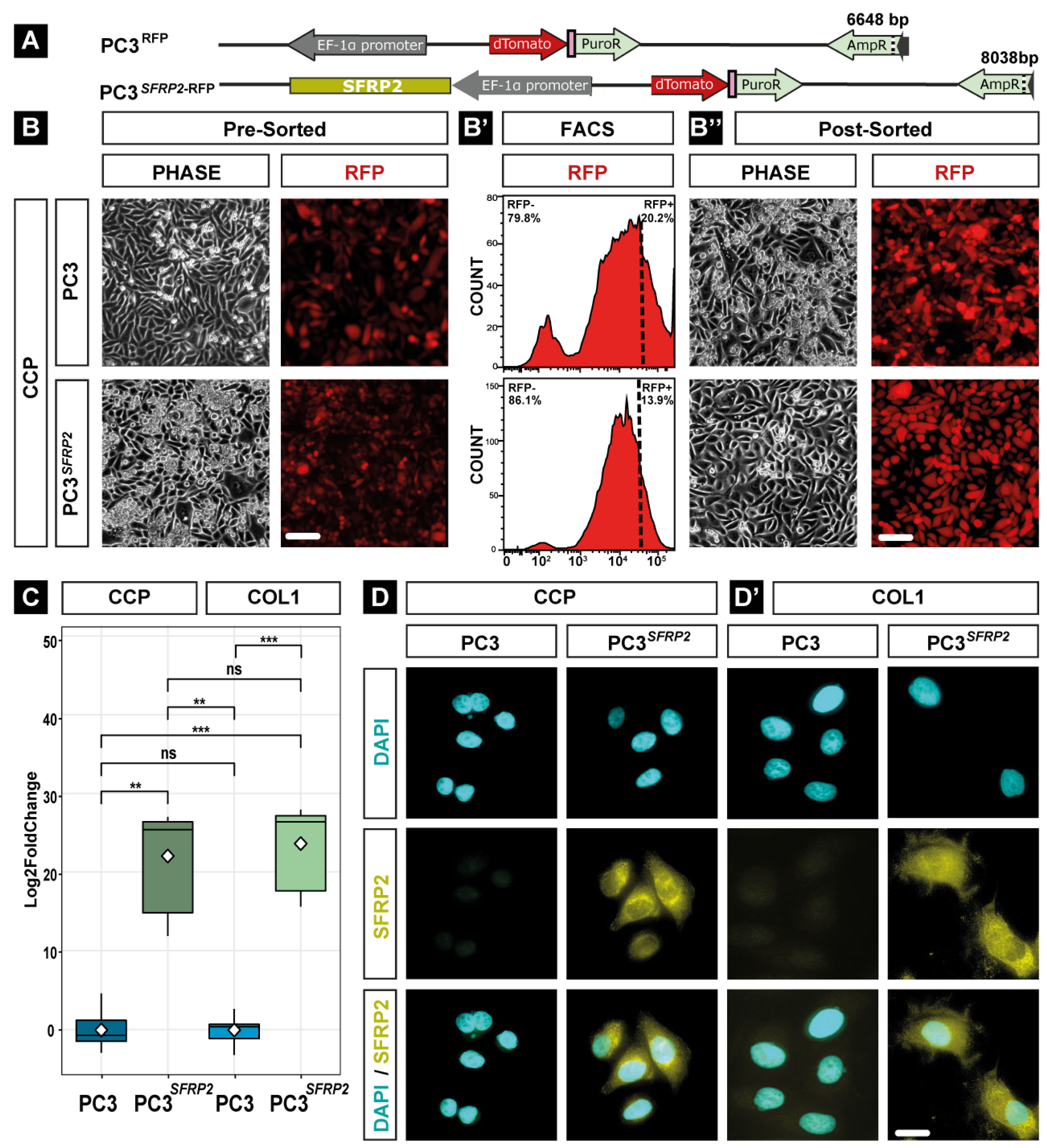
2.2. Generation of PC3 Cells Stably Overexpressing SFRP2
2.3. Surface Coating
2.4. Validation of SFPR2 Overexpression
2.5. Cell Proliferation, Metabolic Activity and Morphology
2.6. Migration, Invasion and Attachment
2.7. Next Generation RNA Sequencing and Bioinformatics Analysis
2.8. Statistical Analysis
3. Results
3.1. Generation and Validation of PC3 Cells Stably Overexpressing SFRP2
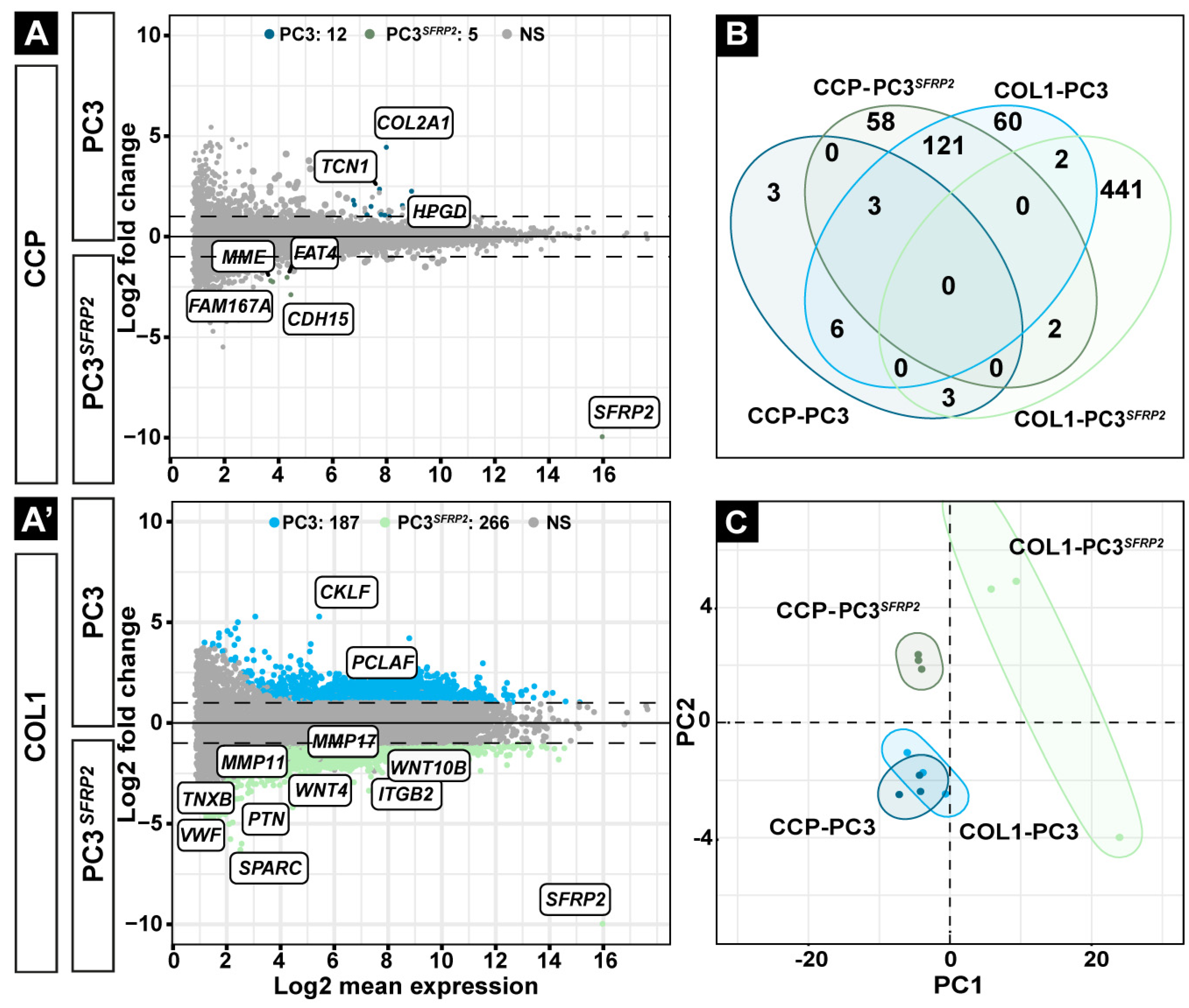
3.2. SFRP2 Overexpression Promotes a Higher Number of Differentially Expressed Genes in PC3 Cells on the COL1-Coated Surface Compared to the CCP Surface
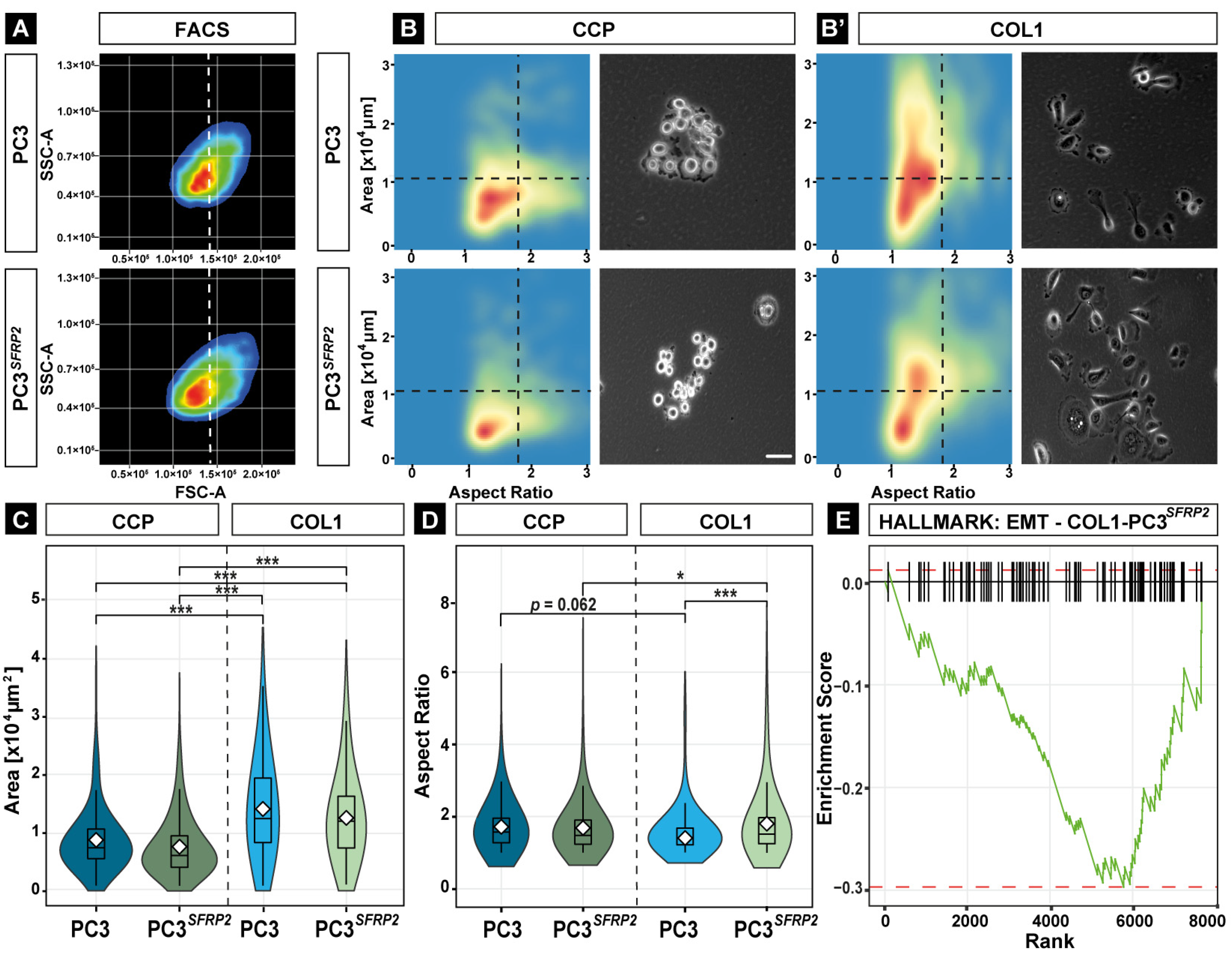
3.3. SFRP2 Overexpression Leads to Osteotropic-like PC3 Cells on the COL1 Surface
3.4. SFRP2 Overexpression Alters the Transcriptome of PC3 Cells towards an Osteoblast-like Phenotype on the COL1 Surface
3.5. SFRP2 Overexpression Induces COL1-Dependent EMT in PC3 Cells
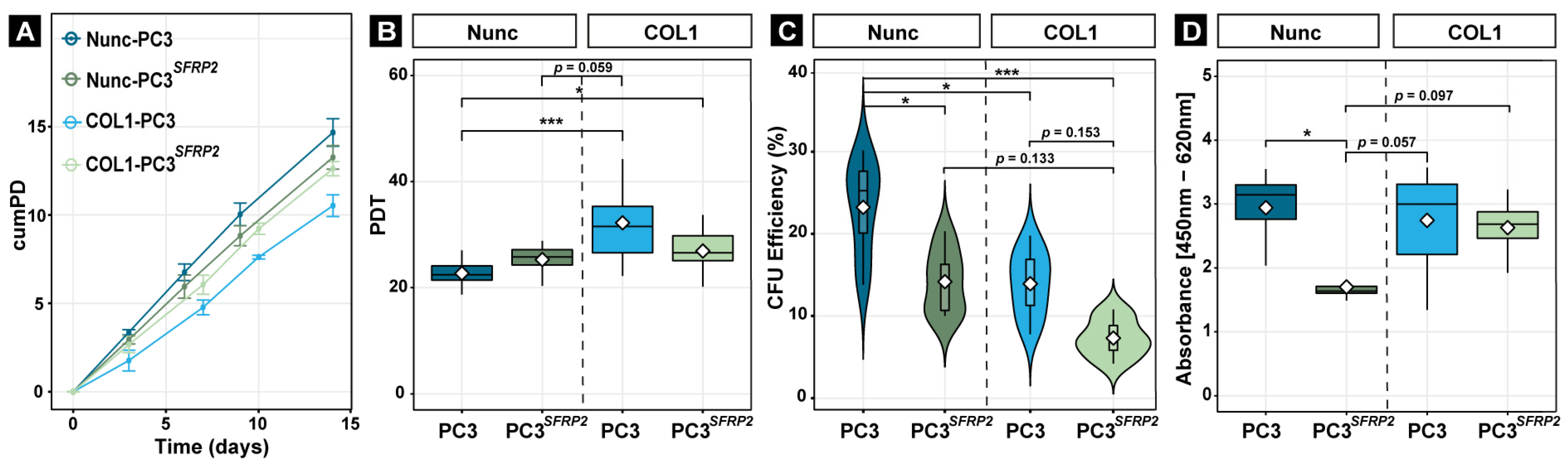
3.6. SFRP2 Overexpression Impedes the Reduction of Proliferation of PC3 Cells on COL1
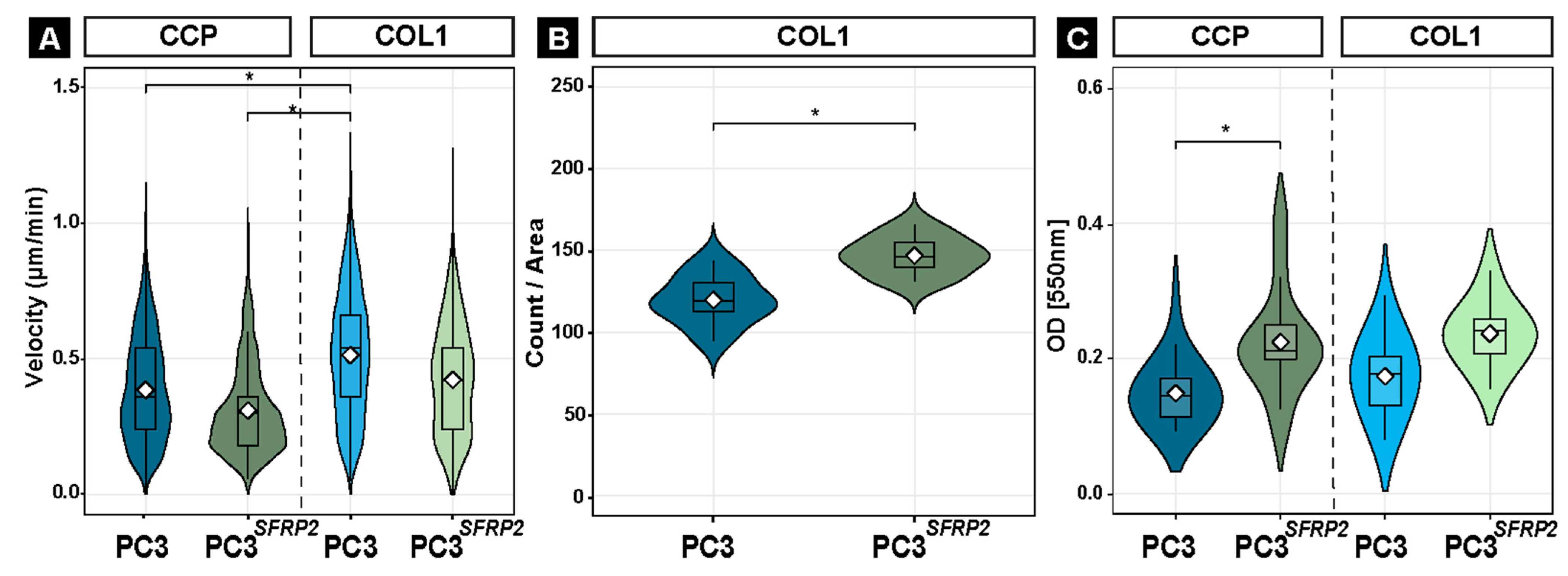
3.7. SFRP2 Overexpression Enhances COL1-Dependent Migration, Invasion, and Metabolic Activity of PC3 Cells
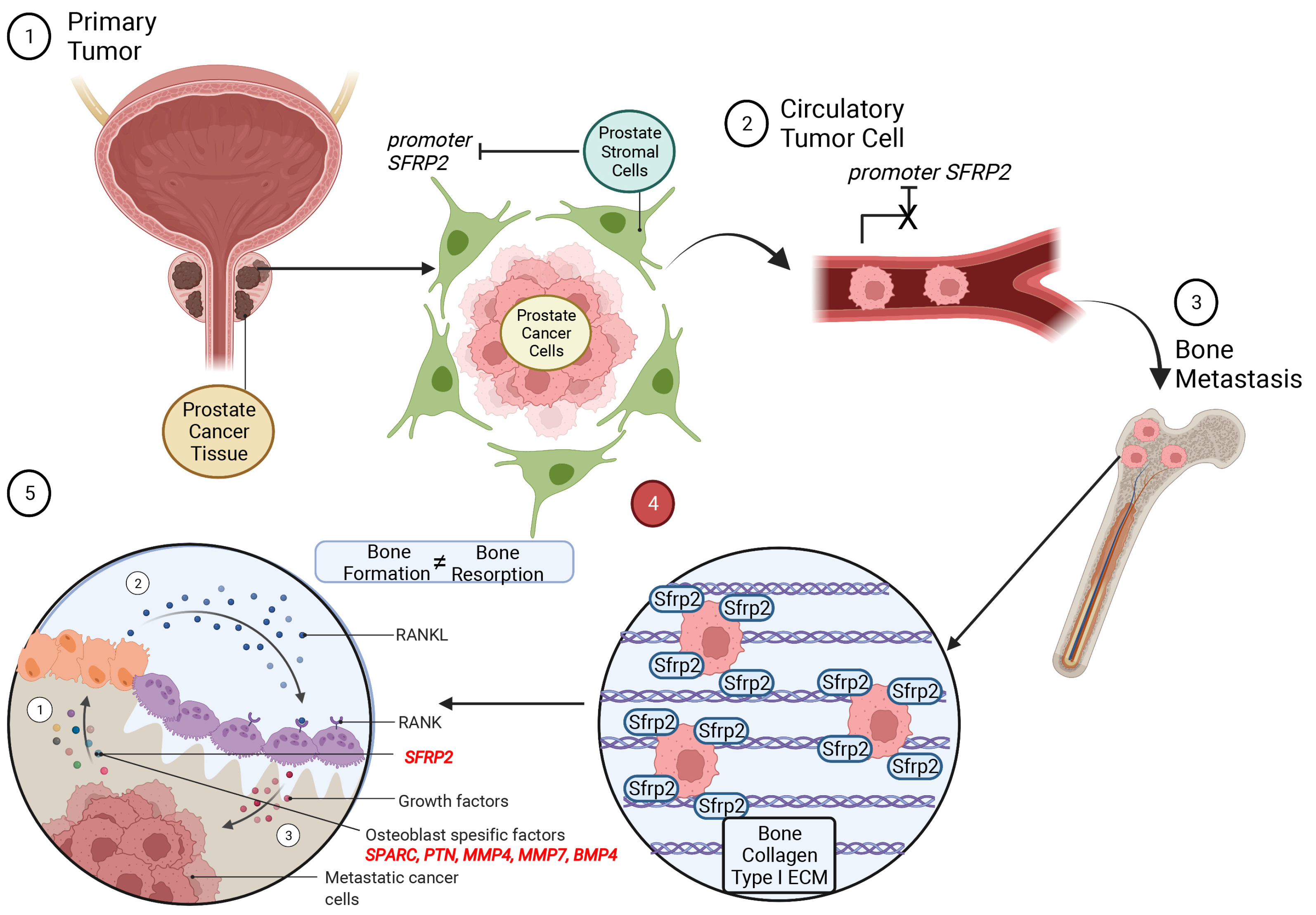
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.-C.; Yu-Lee, L.-Y.; Lin, S.-H. Osteoblastic Factors in Prostate Cancer Bone Metastasis. Curr. Osteoporos. Rep. 2018, 16, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Gandaglia, G.; Abdollah, F.; Schiffmann, J.; Trudeau, V.; Shariat, S.F.; Kim, S.P.; Perrotte, P.; Montorsi, F.; Briganti, A.; Trinh, Q.-D.; et al. Distribution of metastatic sites in patients with prostate cancer: A population-based analysis. Prostate 2014, 74, 210–216. [Google Scholar] [CrossRef]
- Keller, E.T.; Brown, J. Prostate cancer bone metastases promote both osteolytic and osteoblastic activity. J. Cell. Biochem. 2004, 91, 718–729. [Google Scholar] [CrossRef] [Green Version]
- Croucher, P.I.; McDonald, M.M.; Martin, T.J. Bone metastasis: The importance of the neighbourhood. Nat. Rev. Cancer 2016, 16, 373–386. [Google Scholar] [CrossRef]
- Ban, J.; Fock, V.; Aryee, D.N.T.; Kovar, H. Mechanisms, Diagnosis and Treatment of Bone Metastases. Cells 2021, 10, 2944. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Kuai, Y.; Zhu, R.; Zhou, C.; Tao, Y.; Han, W.; Chen, Q. Prognosis of prostate cancer and bone metastasis pattern of patients: A SEER-based study and a local hospital based study from China. Sci. Rep. 2020, 10, 9104. [Google Scholar] [CrossRef]
- Marchetti, C. Calcium signaling in prostate cancer cells of increasing malignancy. Biomol. Concepts 2022, 13, 156–163. [Google Scholar] [CrossRef]
- Fradet, A.; Sorel, H.; Depalle, B.; Serre, C.M.; Farlay, D.; Turtoi, A.; Bellahcène, A.; Follet, H.; Castronovo, V.; Clézardin, P.; et al. A New Murine Model of Osteoblastic/Osteolytic Lesions from Human Androgen-Resistant Prostate Cancer. PLoS ONE 2013, 8, e75092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, J.S.; Queally, J.M.; Devitt, B.M.; Murray, D.W.; Doran, P.P.; O’Byrne, J.M. Silencing Dkk1 expression rescues dexamethasone-induced suppression of primary human osteoblast differentiation. BMC Musculoskelet. Disord. 2010, 11, 210. [Google Scholar] [CrossRef]
- Hall, C.L.; Bafico, A.; Dai, J.; Aaronson, S.A.; Keller, E.T. Prostate Cancer Cells Promote Osteoblastic Bone Metastases through Wnts. Cancer Res. 2005, 65, 7554–7560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, X. Targeting the Wnt/β-catenin signaling pathway in cancer. J. Hematol. Oncol. 2020, 13, 165. [Google Scholar] [CrossRef]
- Patel, S.; Alam, A.; Pant, R.; Chattopadhyay, S. Wnt Signaling and Its Significance Within the Tumor Microenvironment: Novel Therapeutic Insights. Front. Immunol. 2019, 10, 2872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Singhal, U.; Qiao, Y.; Kasputis, T.; Chung, J.-S.; Zhao, H.; Chammaa, F.; Belardo, J.A.; Roth, T.M.; Zhang, H.; et al. Wnt Signaling Drives Prostate Cancer Bone Metastatic Tropism and Invasion. Transl. Oncol. 2020, 13, 100747. [Google Scholar] [CrossRef] [PubMed]
- Agostino, M.; Pohl, S.-G. Wnt Binding Affinity Prediction for Putative Frizzled-Type Cysteine-Rich Domains. Int. J. Mol. Sci. 2019, 20, 4168. [Google Scholar] [CrossRef] [Green Version]
- Gã¶tze, S.; Wolter, M.; Reifenberger, G.; Mã¼Ller, O.; Sievers, S. Frequent promoter hypermethylation of Wnt pathway inhibitor genes in malignant astrocytic gliomas. Int. J. Cancer 2010, 126, 2584–2593. [Google Scholar] [CrossRef]
- Rogan, M.R.; Patterson, L.L.; Byerly, C.D.; Luo, T.; Paessler, S.; Veljkovic, V.; Quade, B.; McBride, J.W. Ehrlichia chaffeensis TRP120 Is a Wnt Ligand Mimetic That Interacts with Wnt Receptors and Contains a Novel Repetitive Short Linear Motif That Activates Wnt Signaling. mSphere 2021, 6, e00216-21. [Google Scholar] [CrossRef]
- Sun, Y.; Zhu, D.; Chen, F.; Qian, M.; Wei, H.; Chen, W.; Xu, J. SFRP2 augments WNT16B signaling to promote therapeutic resistance in the damaged tumor microenvironment. Oncogene 2016, 35, 4321–4334. [Google Scholar] [CrossRef]
- von Marschall, Z.; Fisher, L.W. Secreted Frizzled-related protein-2 (sFRP2) augments canonical Wnt3a-induced signaling. Biochem. Biophys. Res. Commun. 2010, 400, 299–304. [Google Scholar] [CrossRef] [Green Version]
- Perry, A.S.; O’Hurley, G.; Raheem, O.A.; Brennan, K.; Wong, S.; O’Grady, A.; Kennedy, A.-M.; Marignol, L.; Murphy, T.; Sullivan, L.; et al. Gene expression and epigenetic discovery screen reveal methylation of SFRP2 in prostate cancer. Int. J. Cancer 2013, 132, 1771–1780. [Google Scholar] [CrossRef]
- Lee, J.-L.; Lin, C.-T.; Chueh, L.-L.; Chang, C.-J. Autocrine/Paracrine Secreted Frizzled-related Protein 2 Induces Cellular Resistance to Apoptosis. J. Biol. Chem. 2004, 279, 14602–14609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murillo-Garzón, V.; Kypta, R. WNT signalling in prostate cancer. Nat. Rev. Urol. 2017, 14, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Ye, Z.; Wan, J.; Liang, J.; Liu, M.; Xu, X.; Li, L. Secreted Frizzled-Related Protein 2 Is Associated with Disease Progression and Poor Prognosis in Breast Cancer. Dis. Markers 2019, 2019, 6149381. [Google Scholar] [CrossRef] [Green Version]
- de Castro, L.F.; Sworder, B.J.; Mui, B.; Futrega, K.; Berendsen, A.; Phillips, M.D.; Burbach, N.J.; Cherman, N.; Kuznetsov, S.; Gabet, Y.; et al. Secreted frizzled related-protein 2 (Sfrp2) deficiency decreases adult skeletal stem cell function in mice. Bone Res. 2021, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- van Loon, K.; Huijbers, E.J.M.; Griffioen, A.W. Secreted frizzled-related protein 2: A key player in noncanonical Wnt signaling and tumor angiogenesis. Cancer Metastasis Rev. 2021, 40, 191–203. [Google Scholar] [CrossRef]
- Schild, T.; Low, V.; Blenis, J.; Gomes, A.P. Unique Metabolic Adaptations Dictate Distal Organ-Specific Metastatic Colonization. Cancer Cell 2018, 33, 347–354. [Google Scholar] [CrossRef] [Green Version]
- Furesi, G.; Rauner, M.; Hofbauer, L.C. Emerging Players in Prostate Cancer–Bone Niche Communication. Trends Cancer 2021, 7, 112–121. [Google Scholar] [CrossRef]
- Jadaan, D.Y.; Jadaan, M.M.; McCabe, J.P. Cellular Plasticity in Prostate Cancer Bone Metastasis. Prostate Cancer 2015, 2015, 651580. [Google Scholar] [CrossRef]
- Gao, D.; Zhang, X.H.F.; Thompson, E.W.; Mittal, V. EMT process in bone metastasis. In Bone Sarcomas and Bone Metastases-From Bench to Bedside; Academic Press: Cambridge, MA, USA, 2022; pp. 359–370. [Google Scholar]
- Monteran, L.; Ershaid, N.; Sabah, I.; Fahoum, I.; Zait, Y.; Shani, O.; Cohen, N.; Eldar-Boock, A.; Satchi-Fainaro, R.; Erez, N. Bone metastasis is associated with acquisition of mesenchymal phenotype and immune suppression in a model of spontaneous breast cancer metastasis. Sci. Rep. 2020, 10, 13838. [Google Scholar] [CrossRef]
- Ivics, Z.; Izsvák, Z. Nonviral Gene Delivery with the Sleeping Beauty Transposon System. Hum. Gene Ther. 2011, 22, 1043–1051. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Meijering, E.; Dzyubachyk, O.; Smal, I. Methods for Cell and Particle Tracking. Methods Enzymol. 2012, 504, 183–200. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Kowarz, E.; Löscher, D.; Marschalek, R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 2015, 10, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Talluri, B.; Amar, K.; Saul, M.; Shireen, T.; Konjufca, V.; Ma, J.; Ha, T.; Chowdhury, F. COL2A1 Is a Novel Biomarker of Melanoma Tumor Repopulating Cells. Biomedicines 2020, 8, 360. [Google Scholar] [CrossRef]
- Donalies, M.; Cramer, M.; Ringwald, M.; Starzinski-Powitz, A. Expression of M-cadherin, a member of the cadherin multigene family, correlates with differentiation of skeletal muscle cells. Proc. Natl. Acad. Sci. USA 1991, 88, 8024–8028. [Google Scholar] [CrossRef] [Green Version]
- Yue, J.; Zhang, K.; Chen, J. Role of Integrins in Regulating Proteases to Mediate Extracellular Matrix Remodeling. Cancer Microenviron. 2012, 5, 275–283. [Google Scholar] [CrossRef] [Green Version]
- Kajitani, N.; Yamada, T.; Kawakami, K.; Matsumoto, K.-I. TNX deficiency results in bone loss due to an increase in multinucleated osteoclasts. Biochem. Biophys. Res. Commun. 2019, 512, 659–664. [Google Scholar] [CrossRef]
- Yang, A.-J.; Wang, M.; Wang, Y.; Cai, W.; Li, Q.; Zhao, T.-T.; Zhang, L.-H.; Houck, K.; Chen, X.; Jin, Y.-L.; et al. Cancer cell-derived von Willebrand factor enhanced metastasis of gastric adenocarcinoma. Oncogenesis 2018, 7, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamprou, M.; Kaspiris, A.; Panagiotopoulos, E.; Giannoudis, P.V.; Papadimitriou, E. The role of pleiotrophin in bone repair. Injury 2014, 45, 1816–1823. [Google Scholar] [CrossRef] [PubMed]
- Baird, A.; Lindsay, T.; Everett, A.; Iyemere, V.; Paterson, Y.Z.; McClellan, A.; Henson, F.M.D.; Guest, D.J. Osteoblast differentiation of equine induced pluripotent stem cells. Biol. Open 2018, 7, bio033514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Pan, Y.; Ji, J.; Xu, Y.; Zhang, Q.; Qin, L. Roles and action mechanisms of WNT4 in cell differentiation and human diseases: A review. Cell Death Discov. 2021, 7, 287. [Google Scholar] [CrossRef]
- Bennett, C.N.; Longo, K.A.; Wright, W.S.; Suva, L.J.; Lane, T.F.; Hankenson, K.D.; MacDougald, O.A. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc. Natl. Acad. Sci. USA 2005, 102, 3324–3329. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef]
- Chang, S.-F.; Chang, T.-K.; Peng, H.-H.; Yeh, Y.-T.; Lee, D.-Y.; Yeh, C.-R.; Zhou, J.; Cheng, C.-K.; Chang, C.A.; Chiu, J.-J. BMP-4 Induction of Arrest and Differentiation of Osteoblast-Like Cells via p21CIP1 and p27KIP1 Regulation. Mol. Endocrinol. 2009, 23, 1827–1838. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.-C.; Cheng, C.-J.; Bilen, M.A.; Lu, J.-F.; Satcher, R.L.; Yu-Lee, L.-Y.; Gallick, G.E.; Maity, S.N.; Lin, S.-H. BMP4 Promotes Prostate Tumor Growth in Bone through Osteogenesis. Cancer Res. 2011, 71, 5194–5203. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- The Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Vergara, D.; Simeone, P.; Franck, J.; Trerotola, M.; Giudetti, A.; Capobianco, L.; Tinelli, A.; Bellomo, C.; Fournier, I.; Gaballo, A.; et al. Translating epithelial mesenchymal transition markers into the clinic: Novel insights from proteomics. EuPA Open Proteom. 2016, 10, 31–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teti, A. Osteomimicry how tumor cells try to deceive the bone. Front. Biosci. 2010, 2, 907–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, A.C.; Ramaswamy, S. Mechanisms of Cancer Cell Dormancy—Another Hallmark of Cancer? Cancer Res. 2015, 75, 5014–5022. [Google Scholar] [CrossRef] [Green Version]
- Le Bras, G.F.; Taubenslag, K.J.; Andl, C.D. The regulation of cell-cell adhesion during epithelial-mesenchymal transition, motility and tumor progression. Cell Adhes. Migr. 2012, 6, 365–373. [Google Scholar] [CrossRef] [Green Version]
- Lau, W.M.; Weber, K.L.; Doucet, M.; Chou, Y.-T.; Brady, K.; Kowalski, J.; Tsai, H.-L.; Yang, J.; Kominsky, S.L. Identification of prospective factors promoting osteotropism in breast cancer: A potential role for CITED2. Int. J. Cancer 2010, 126, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.F.; Jenkinson, A.; Pohl, K.; O’Brien, F.J.; Morgan, M.P. Osteomimicry of Mammary Adenocarcinoma Cells In Vitro; Increased Expression of Bone Matrix Proteins and Proliferation within a 3D Collagen Environment. PLoS ONE 2012, 7, e41679. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.-K.; Dayyani, F.; Gallick, G.E. Steps in prostate cancer progression that lead to bone metastasis. Int. J. Cancer 2011, 128, 2545–2561. [Google Scholar] [CrossRef] [Green Version]
- Rosset, E.M.; Bradshaw, A.D. SPARC/osteonectin in mineralized tissue. Matrix Biol. 2016, 52–54, 78–87. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Xing, F.; Liu, Y.; Wu, K.; Said, N.; Pochampally, R.; Shiozawa, Y.; Lin, H.-K.; Balaji, K.; Watabe, K. Secreted Protein Acidic and Rich in Cysteine (SPARC) Mediates Metastatic Dormancy of Prostate Cancer in Bone. J. Biol. Chem. 2016, 291, 19351–19363. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Shen, M.; Hsu, E.-C.; Zhang, C.A.; Garcia-Marques, F.; Nolley, R.; Koul, K.; Rice, M.A.; Aslan, M.; Pitteri, S.J.; et al. Discovery of PTN as a serum-based biomarker of pro-metastatic prostate cancer. Br. J. Cancer 2021, 124, 896–900. [Google Scholar] [CrossRef]
- Tare, R.S.; Oreffo, R.O.C.; Clarke, N.M.P.; Roach, H.I. Pleiotrophin/Osteoblast-Stimulating Factor 1: Dissecting Its Diverse Functions in Bone Formation. J. Bone Miner. Res. 2002, 17, 2009–2020. [Google Scholar] [CrossRef] [PubMed]
- Vashisht, S.; Bagler, G. An Approach for the Identification of Targets Specific to Bone Metastasis Using Cancer Genes Interactome and Gene Ontology Analysis. PLoS ONE 2012, 7, e49401. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, S.; Guo, J.; Zhou, L.; You, L.; Zhang, T.; Zhao, Y. Insights into the distinct roles of MMP-11 in tumor biology and future therapeutics (Review). Int. J. Oncol. 2016, 48, 1783–1793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, X.; Gong, K.; Zhang, X.; Wu, S.; Cui, Y.; Qian, B.-Z. Osteopontin as a multifaceted driver of bone metastasis and drug resistance. Pharmacol. Res. 2019, 144, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Papke, C.L.; Yamashiro, Y.; Yanagisawa, H. MMP17/MT4-MMP and Thoracic Aortic Aneurysms. Circ. Res. 2015, 117, 109–112. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akova Ölken, E.; Aszodi, A.; Taipaleenmäki, H.; Saito, H.; Schönitzer, V.; Chaloupka, M.; Apfelbeck, M.; Böcker, W.; Saller, M.M. SFRP2 Overexpression Induces an Osteoblast-like Phenotype in Prostate Cancer Cells. Cells 2022, 11, 4081. https://doi.org/10.3390/cells11244081
Akova Ölken E, Aszodi A, Taipaleenmäki H, Saito H, Schönitzer V, Chaloupka M, Apfelbeck M, Böcker W, Saller MM. SFRP2 Overexpression Induces an Osteoblast-like Phenotype in Prostate Cancer Cells. Cells. 2022; 11(24):4081. https://doi.org/10.3390/cells11244081
Chicago/Turabian StyleAkova Ölken, Elif, Attila Aszodi, Hanna Taipaleenmäki, Hiroaki Saito, Veronika Schönitzer, Michael Chaloupka, Maria Apfelbeck, Wolfgang Böcker, and Maximilian Michael Saller. 2022. "SFRP2 Overexpression Induces an Osteoblast-like Phenotype in Prostate Cancer Cells" Cells 11, no. 24: 4081. https://doi.org/10.3390/cells11244081
APA StyleAkova Ölken, E., Aszodi, A., Taipaleenmäki, H., Saito, H., Schönitzer, V., Chaloupka, M., Apfelbeck, M., Böcker, W., & Saller, M. M. (2022). SFRP2 Overexpression Induces an Osteoblast-like Phenotype in Prostate Cancer Cells. Cells, 11(24), 4081. https://doi.org/10.3390/cells11244081







