Detection of De Novo Dividing Stem Cells In Situ through Double Nucleotide Analogue Labeling
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Delivery of Nucleotide Analogues
2.3. Memantine Injections
2.4. Brain Sample Collection and Tissue Processing
2.5. Visualization of Labels and Cell Type Specific Markers in Series of Brain Slices
2.6. Confocal Microscopy
2.7. Cell Counting
2.8. Data Analysis
3. Results
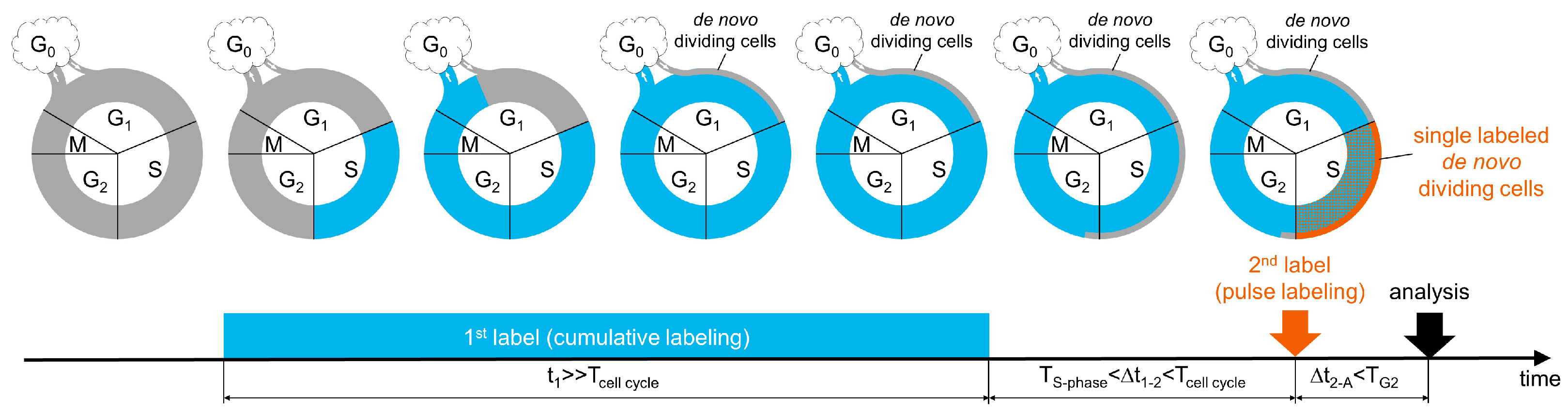
3.1. A New Labeling Scheme for Visualizing De Novo Dividing Cells
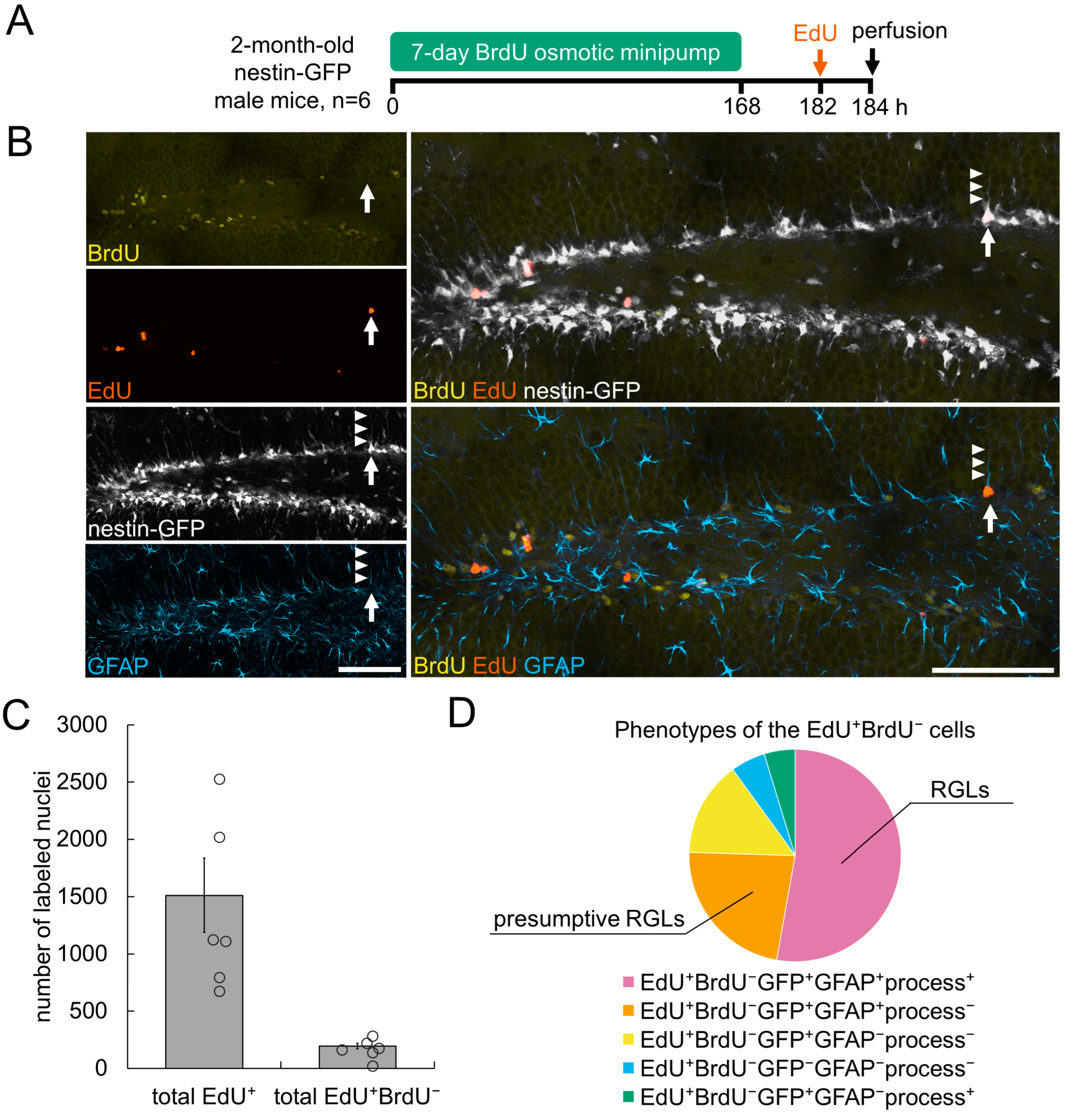
3.2. Detection of De Novo Dividing Stem Cells in the Hippocampal DG
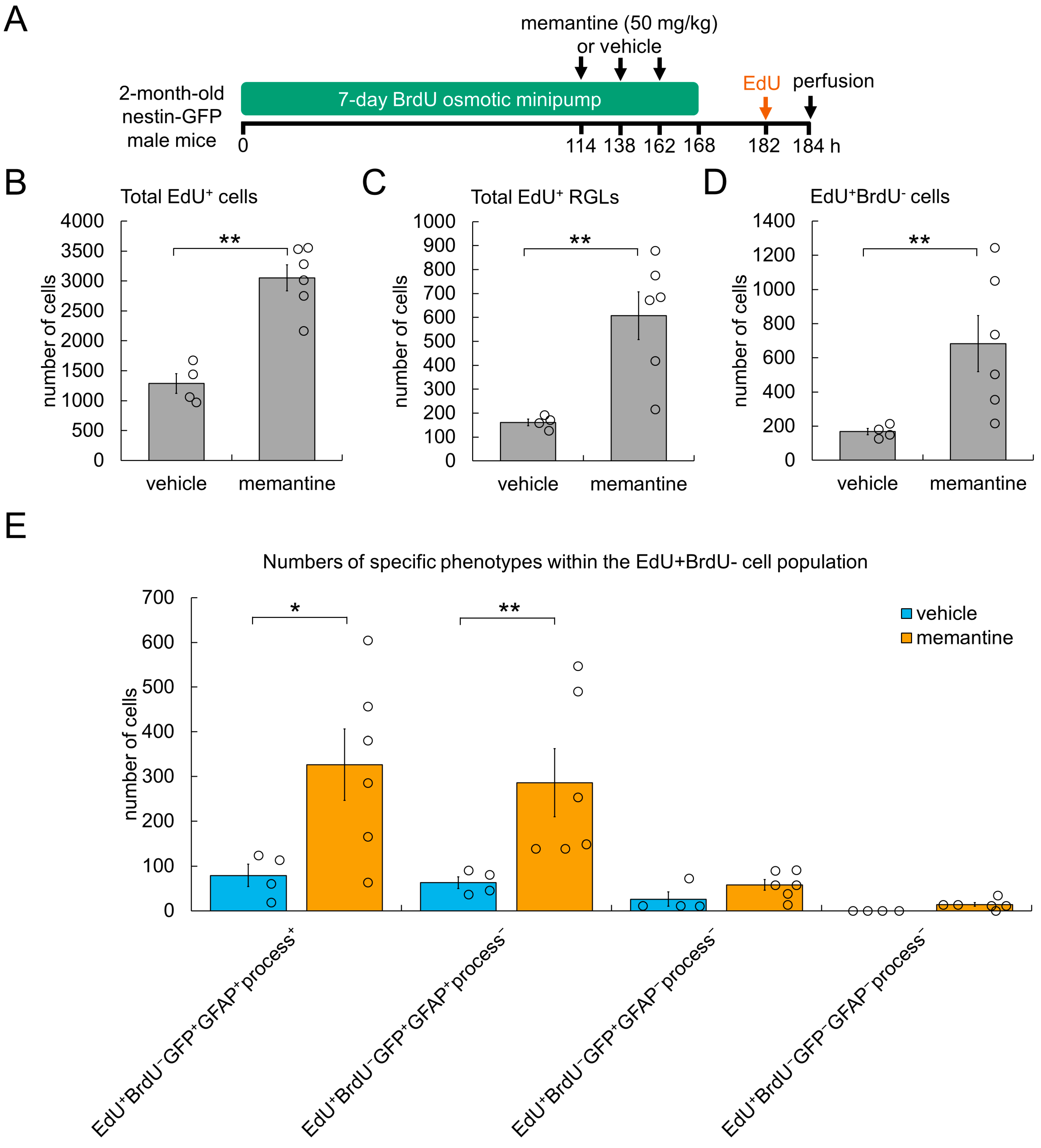
3.3. Cellular Mechanism Underlying AHN Enhancement by Memantine
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Velthoven, C.T.J.; Rando, T.A. Stem Cell Quiescence: Dynamism, Restraint, and Cellular Idling. Cell Stem Cell 2019, 24, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Urbán, N.; Blomfield, I.M.; Guillemot, F. Quiescence of Adult Mammalian Neural Stem Cells: A Highly Regulated Rest. Neuron 2019, 104, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, E.; Blau, H.M. Tissue Stem Cells: Architects of Their Niches. Cell Stem Cell 2020, 27, 532–556. [Google Scholar] [CrossRef]
- Cheung, T.H.; Rando, T.A. Molecular Regulation of Stem Cell Quiescence. Nat. Rev. Mol. Cell Biol. 2013, 14, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Hoggatt, J.; Kfoury, Y.; Scadden, D.T. Hematopoietic Stem Cell Niche in Health and Disease. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 555–581. [Google Scholar] [CrossRef]
- Orford, K.W.; Scadden, D.T. Deconstructing Stem Cell Self-Renewal: Genetic Insights into Cell-Cycle Regulation. Nat. Rev. Genet. 2008, 9, 115–128. [Google Scholar] [CrossRef]
- Oh, J.; Lee, Y.D.; Wagers, A.J. Stem Cell Aging: Mechanisms, Regulators and Therapeutic Opportunities. Nat. Med. 2014, 20, 870–880. [Google Scholar] [CrossRef]
- Sierra, A.; Martín-Suárez, S.; Valcárcel-Martín, R.; Pascual-Brazo, J.; Aelvoet, S.-A.; Abiega, O.; Deudero, J.J.; Brewster, A.L.; Bernales, I.; Anderson, A.E.; et al. Neuronal Hyperactivity Accelerates Depletion of Neural Stem Cells and Impairs Hippocampal Neurogenesis. Cell Stem Cell 2015, 16, 488–503. [Google Scholar] [CrossRef]
- Chakkalakal, J.V.; Christensen, J.; Xiang, W.; Tierney, M.T.; Boscolo, F.S.; Sacco, A.; Brack, A.S. Early Forming Label-Retaining Muscle Stem Cells Require P27kip1 for Maintenance of the Primitive State. Development 2014, 141, 1649–1659. [Google Scholar] [CrossRef]
- Cotsarelis, G.; Sun, T.-T.; Lavker, R.M. Label-Retaining Cells Reside in the Bulge Area of Pilosebaceous Unit: Implications for Follicular Stem Cells, Hair Cycle, and Skin Carcinogenesis. Cell 1990, 61, 1329–1337. [Google Scholar] [CrossRef]
- May, R.; Sureban, S.M.; Hoang, N.; Riehl, T.E.; Lightfoot, S.A.; Ramanujam, R.; Wyche, J.H.; Anant, S.; Houchen, C.W. Doublecortin and CaM Kinase-like-1 and Leucine-Rich-Repeat-Containing G-Protein-Coupled Receptor Mark Quiescent and Cycling Intestinal Stem Cells, Respectively. Stem Cells 2009, 27, 2571–2579. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Laurenti, E.; Oser, G.; van der Wath, R.C.; Blanco-Bose, W.; Jaworski, M.; Offner, S.; Dunant, C.F.; Eshkind, L.; Bockamp, E.; et al. Hematopoietic Stem Cells Reversibly Switch from Dormancy to Self-Renewal during Homeostasis and Repair. Cell 2008, 135, 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Chakkalakal, J.V.; Jones, K.M.; Basson, M.A.; Brack, A.S. The Aged Niche Disrupts Muscle Stem Cell Quiescence. Nature 2012, 490, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Foudi, A.; Hochedlinger, K.; Van Buren, D.; Schindler, J.W.; Jaenisch, R.; Carey, V.; Hock, H. Analysis of Histone 2B-GFP Retention Reveals Slowly Cycling Hematopoietic Stem Cells. Nat. Biotechnol. 2009, 27, 84–90. [Google Scholar] [CrossRef]
- Tumbar, T.; Guasch, G.; Greco, V.; Blanpain, C.; Lowry, W.E.; Rendl, M.; Fuchs, E. Defining the Epithelial Stem Cell Niche in Skin. Science 2004, 303, 359–363. [Google Scholar] [CrossRef]
- Mignone, J.L.; Kukekov, V.; Chiang, A.-S.; Steindler, D.; Enikolopov, G. Neural Stem and Progenitor Cells in Nestin-GFP Transgenic Mice. J. Comp. Neurol. 2004, 469, 311–324. [Google Scholar] [CrossRef]
- Mandyam, C.D.; Harburg, G.C.; Eisch, A.J. Determination of Key Aspects of Precursor Cell Proliferation, Cell Cycle Length and Kinetics in the Adult Mouse Subgranular Zone. Neuroscience 2007, 146, 108–122. [Google Scholar] [CrossRef]
- Solius, G.M.; Maltsev, D.I.; Belousov, V.V.; Podgorny, O.V. Recent Advances in Nucleotide Analogue-Based Techniques for Tracking Dividing Stem Cells: An Overview. J. Biol. Chem. 2021, 297, 101345. [Google Scholar] [CrossRef]
- Gould, E.; McEwen, B.S.; Tanapat, P.; Galea, L.A.M.; Fuchs, E. Neurogenesis in the Dentate Gyrus of the Adult Tree Shrew Is Regulated by Psychosocial Stress and NMDA Receptor Activation. J. Neurosci. 1997, 17, 2492–2498. [Google Scholar] [CrossRef]
- Ševc, J.; Matiašová, A.; Smoleková, I.; Jendželovský, R.; Mikeš, J.; Tomášová, L.; Kútna, V.; Daxnerová, Z.; Fedoročko, P. Peroral Administration of 5-Bromo-2-Deoxyuridine in Drinking Water Is Not a Reliable Method for Labeling Proliferating S-Phase Cells in Rats. J. Pharmacol. Toxicol. Methods 2015, 74, 33–39. [Google Scholar] [CrossRef]
- Encinas, J.M.; Enikolopov, G. Identifying and Quantitating Neural Stem and Progenitor Cells in the Adult Brain. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2008; Volume 85, pp. 243–272. ISBN 978-0-12-372558-5. [Google Scholar]
- Maltsev, D.I.; Mellanson, K.A.; Belousov, V.V.; Enikolopov, G.N.; Podgorny, O.V. The Bioavailability Time of Commonly Used Thymidine Analogues after Intraperitoneal Delivery in Mice: Labeling Kinetics in Vivo and Clearance from Blood Serum. Histochem. Cell Biol. 2022, 157, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Podgorny, O.; Peunova, N.; Park, J.-H.; Enikolopov, G. Triple S-Phase Labeling of Dividing Stem Cells. Stem Cell Rep. 2018, 10, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Enikolopov, G.; Overstreet-Wadiche, L.; Ge, S. Viral and Transgenic Reporters and Genetic Analysis of Adult Neurogenesis. Cold Spring Harb. Perspect. Biol. 2015, 7, a018804. [Google Scholar] [CrossRef]
- Mignone, J.; Peunova, N.; Enikolopov, G. Nestin-Based Reporter Transgenic Mouse Lines. In Multipotent Stem Cells of the Hair Follicle; Hoffman, R.M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2016; Volume 1453, pp. 7–14. ISBN 978-1-4939-3784-4. [Google Scholar]
- Yamakawa, M.; Santosa, S.M.; Chawla, N.; Ivakhnitskaia, E.; del Pino, M.; Giakas, S.; Nadel, A.; Bontu, S.; Tambe, A.; Guo, K.; et al. Transgenic Models for Investigating the Nervous System: Currently Available Neurofluorescent Reporters and Potential Neuronal Markers. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2020, 1864, 129595. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Saunders, T.L.; Enikolopov, G.; Morrison, S.J. Endothelial and Perivascular Cells Maintain Haematopoietic Stem Cells. Nature 2012, 481, 457–462. [Google Scholar] [CrossRef]
- Gleiberman, A.S.; Encinas, J.M.; Mignone, J.L.; Michurina, T.; Rosenfeld, M.G.; Enikolopov, G. Expression of Nestin-Green Fluorescent Protein Transgene Marks Oval Cells in the Adult Liver. Dev. Dyn. 2005, 234, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Gleiberman, A.S.; Michurina, T.; Encinas, J.M.; Roig, J.L.; Krasnov, P.; Balordi, F.; Fishell, G.; Rosenfeld, M.G.; Enikolopov, G. Genetic Approaches Identify Adult Pituitary Stem Cells. Proc. Natl. Acad. Sci. USA 2008, 105, 6332–6337. [Google Scholar] [CrossRef]
- Koning, J.J.; Konijn, T.; Lakeman, K.A.; O’Toole, T.; Kenswil, K.J.G.; Raaijmakers, M.H.G.P.; Michurina, T.V.; Enikolopov, G.; Mebius, R.E. Nestin-Expressing Precursors Give Rise to Both Endothelial as Well as Nonendothelial Lymph Node Stromal Cells. J. Immunol. 2016, 197, 2686–2694. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Ferrer, S.; Michurina, T.V.; Ferraro, F.; Mazloom, A.R.; MacArthur, B.D.; Lira, S.A.; Scadden, D.T.; Ma’ayan, A.; Enikolopov, G.N.; Frenette, P.S. Mesenchymal and Haematopoietic Stem Cells Form a Unique Bone Marrow Niche. Nature 2010, 466, 829–834. [Google Scholar] [CrossRef]
- Mignone, J.L.; Roig-Lopez, J.L.; Fedtsova, N.; Schones, D.E.; Manganas, L.N.; Maletic-Savatic, M.; Keyes, W.M.; Mills, A.A.; Gleiberman, A.; Zhang, M.Q.; et al. Neural Potential of a Stem Cell Population in the Hair Follicle. Cell Cycle 2007, 6, 2161–2170. [Google Scholar] [CrossRef]
- Saboor, F.; Reckmann, A.N.; Tomczyk, C.U.M.; Peters, D.M.; Weissmann, N.; Kaschtanow, A.; Schermuly, R.T.; Michurina, T.V.; Enikolopov, G.; Müller, D.; et al. Nestin-Expressing Vascular Wall Cells Drive Development of Pulmonary Hypertension. Eur. Respir. J. 2016, 47, 876–888. [Google Scholar] [CrossRef] [PubMed]
- Brandt, M.D.; Hübner, M.; Storch, A. Brief Report: Adult Hippocampal Precursor Cells Shorten S-Phase and Total Cell Cycle Length During Neuronal Differentiation. Stem Cells 2012, 30, 2843–2847. [Google Scholar] [CrossRef] [PubMed]
- Fischer, T.J.; Walker, T.L.; Overall, R.W.; Brandt, M.D.; Kempermann, G. Acute Effects of Wheel Running on Adult Hippocampal Precursor Cells in Mice Are Not Caused by Changes in Cell Cycle Length or S Phase Length. Front. Neurosci. 2014, 8, 314. [Google Scholar] [CrossRef]
- Encinas, J.M.; Michurina, T.V.; Peunova, N.; Park, J.-H.; Tordo, J.; Peterson, D.A.; Fishell, G.; Koulakov, A.; Enikolopov, G. Division-Coupled Astrocytic Differentiation and Age-Related Depletion of Neural Stem Cells in the Adult Hippocampus. Cell Stem Cell 2011, 8, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Lazutkin, A.; Podgorny, O.; Enikolopov, G. Modes of Division and Differentiation of Neural Stem Cells. Behav. Brain Res. 2019, 374, 112118. [Google Scholar] [CrossRef] [PubMed]
- Pilz, G.-A.; Bottes, S.; Betizeau, M.; Jörg, D.J.; Carta, S.; Simons, B.D.; Helmchen, F.; Jessberger, S. Live Imaging of Neurogenesis in the Adult Mouse Hippocampus. Science 2018, 359, 658–662. [Google Scholar] [CrossRef]
- Kronenberg, G.; Reuter, K.; Steiner, B.; Brandt, M.D.; Jessberger, S.; Yamaguchi, M.; Kempermann, G. Subpopulations of Proliferating Cells of the Adult Hippocampus Respond Differently to Physiologic Neurogenic Stimuli. J. Comp. Neurol. 2003, 467, 455–463. [Google Scholar] [CrossRef]
- Moss, J.; Gebara, E.; Bushong, E.A.; Sánchez-Pascual, I.; O’Laoi, R.; El M’Ghari, I.; Kocher-Braissant, J.; Ellisman, M.H.; Toni, N. Fine Processes of Nestin-GFP–Positive Radial Glia-like Stem Cells in the Adult Dentate Gyrus Ensheathe Local Synapses and Vasculature. Proc. Natl. Acad. Sci. USA 2016, 113, E2536–E2545. [Google Scholar] [CrossRef]
- Seri, B.; García-Verdugo, J.M.; Collado-Morente, L.; McEwen, B.S.; Alvarez-Buylla, A. Cell Types, Lineage, and Architecture of the Germinal Zone in the Adult Dentate Gyrus: Organization of the Vertebrate Dentate Gyrus. J. Comp. Neurol. 2004, 478, 359–378. [Google Scholar] [CrossRef]
- Rieskamp, J.; Sarchet, P.; Smith, B.; Kirby, E. Estimation of the Density of Neural, Glial, and Endothelial Lineage Cells in the Adult Mouse Dentate Gyrus. Neural Regen. Res. 2022, 17, 1286. [Google Scholar] [CrossRef]
- Matsunaga, S.; Kishi, T.; Iwata, N. Memantine Monotherapy for Alzheimer’s Disease: A Systematic Review and Meta-Analysis. PLoS ONE 2015, 10, e0123289. [Google Scholar] [CrossRef] [PubMed]
- McShane, R.; Westby, M.J.; Roberts, E.; Minakaran, N.; Schneider, L.; Farrimond, L.E.; Maayan, N.; Ware, J.; Debarros, J. Memantine for Dementia. Cochrane Database Syst. Rev. 2019. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, M.; Namba, T.; Suzuki, E.; Yuasa, S.; Kohsaka, S.; Uchino, S. NMDA Receptor Antagonist Memantine Promotes Cell Proliferation and Production of Mature Granule Neurons in the Adult Hippocampus. Neurosci. Res. 2009, 63, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Namba, T.; Maekawa, M.; Yuasa, S.; Kohsaka, S.; Uchino, S. The Alzheimer’s Disease Drug Memantine Increases the Number of Radial Glia-like Progenitor Cells in Adult Hippocampus. Glia 2009, 57, 1082–1090. [Google Scholar] [CrossRef]
- Bao, H.; Asrican, B.; Li, W.; Gu, B.; Wen, Z.; Lim, S.-A.; Haniff, I.; Ramakrishnan, C.; Deisseroth, K.; Philpot, B.; et al. Long-Range GABAergic Inputs Regulate Neural Stem Cell Quiescence and Control Adult Hippocampal Neurogenesis. Cell Stem Cell 2017, 21, 604–617.e5. [Google Scholar] [CrossRef]
- Unger, M.S.; Marschallinger, J.; Kaindl, J.; Höfling, C.; Rossner, S.; Heneka, M.T.; Van der Linden, A.; Aigner, L. Early Changes in Hippocampal Neurogenesis in Transgenic Mouse Models for Alzheimer’s Disease. Mol. Neurobiol. 2016, 53, 5796–5806. [Google Scholar] [CrossRef]
- Ehm, O.; Göritz, C.; Covic, M.; Schäffner, I.; Schwarz, T.J.; Karaca, E.; Kempkes, B.; Kremmer, E.; Pfrieger, F.W.; Espinosa, L.; et al. RBPJkappa-Dependent Signaling Is Essential for Long-Term Maintenance of Neural Stem Cells in the Adult Hippocampus. J. Neurosci. 2010, 30, 13794–13807. [Google Scholar] [CrossRef]
- Breunig, J.J.; Arellano, J.I.; Macklis, J.D.; Rakic, P. Everything That Glitters Isn’t Gold: A Critical Review of Postnatal Neural Precursor Analyses. Cell Stem Cell 2007, 1, 612–627. [Google Scholar] [CrossRef]
- Martí-Clúa, J. Incorporation of 5-Bromo-2′-Deoxyuridine into DNA and Proliferative Behavior of Cerebellar Neuroblasts: All That Glitters Is Not Gold. Cells 2021, 10, 1453. [Google Scholar] [CrossRef]
- Miller, M.W.; Nowakowski, R.S. Use of Bromodeoxyuridine-Immunohistochemistry to Examine the Proliferation, Migration and Time of Origin of Cells in the Central Nervous System. Brain Res. 1988, 457, 44–52. [Google Scholar] [CrossRef]
- Ivanova, A.; Gruzova, O.; Ermolaeva, E.; Astakhova, O.; Itaman, S.; Enikolopov, G.; Lazutkin, A. Synthetic Thymidine Analog Labeling without Misconceptions. Cells 2022, 11, 1888. [Google Scholar] [CrossRef] [PubMed]
- Ponti, G.; Obernier, K.; Guinto, C.; Jose, L.; Bonfanti, L.; Alvarez-Buylla, A. Cell Cycle and Lineage Progression of Neural Progenitors in the Ventricular-Subventricular Zones of Adult Mice. Proc. Natl. Acad. Sci. USA 2013, 110, E1045–E1054. [Google Scholar] [CrossRef] [PubMed]
- Kohlmeier, F.; Maya-Mendoza, A.; Jackson, D.A. EdU Induces DNA Damage Response and Cell Death in MESC in Culture. Chromosome Res. 2013, 21, 87–100. [Google Scholar] [CrossRef]
- Ligasová, A.; Strunin, D.; Friedecký, D.; Adam, T.; Koberna, K. A Fatal Combination: A Thymidylate Synthase Inhibitor with DNA Damaging Activity. PLoS ONE 2015, 10, e0117459. [Google Scholar] [CrossRef]
- Neef, A.B.; Luedtke, N.W. Dynamic Metabolic Labeling of DNA in Vivo with Arabinosyl Nucleosides. Proc. Natl. Acad. Sci. USA 2011, 108, 20404–20409. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Itaman, S.; Enikolopov, G.; Podgorny, O.V. Detection of De Novo Dividing Stem Cells In Situ through Double Nucleotide Analogue Labeling. Cells 2022, 11, 4001. https://doi.org/10.3390/cells11244001
Itaman S, Enikolopov G, Podgorny OV. Detection of De Novo Dividing Stem Cells In Situ through Double Nucleotide Analogue Labeling. Cells. 2022; 11(24):4001. https://doi.org/10.3390/cells11244001
Chicago/Turabian StyleItaman, Sheed, Grigori Enikolopov, and Oleg V. Podgorny. 2022. "Detection of De Novo Dividing Stem Cells In Situ through Double Nucleotide Analogue Labeling" Cells 11, no. 24: 4001. https://doi.org/10.3390/cells11244001
APA StyleItaman, S., Enikolopov, G., & Podgorny, O. V. (2022). Detection of De Novo Dividing Stem Cells In Situ through Double Nucleotide Analogue Labeling. Cells, 11(24), 4001. https://doi.org/10.3390/cells11244001




