Abstract
Generation of motor neurons (MNs) from human-induced pluripotent stem cells (hiPSCs) overcomes the limited access to human brain tissues and provides an unprecedent approach for modeling MN-related diseases. In this review, we discuss the recent progression in understanding the regulatory mechanisms of MN differentiation and their applications in the generation of MNs from hiPSCs, with a particular focus on two approaches: induction by small molecules and induction by lentiviral delivery of transcription factors. At each induction stage, different culture media and supplements, typical growth conditions and cellular morphology, and specific markers for validation of cell identity and quality control are specifically discussed. Both approaches can generate functional MNs. Currently, the major challenges in modeling neurological diseases using iPSC-derived neurons are: obtaining neurons with high purity and yield; long-term neuron culture to reach full maturation; and how to culture neurons more physiologically to maximize relevance to in vivo conditions.
1. Introduction
Movement disorders are a group of neurological conditions that cause either increased or decreased or slow movements. The movements may be voluntary or involuntary, and implicated in many neurological diseases, such as Dystonia, amyotrophic lateral sclerosis (ALS), Ataxia, Parkinson’s disease [1], and so on. Motor neurons (MNs) are a remarkably powerful cell type in the central nervous system (CNS), and they are involved in both autonomic and voluntary movements. Most prior research of movement disorders was carried out using patient postmortem tissues or rodent models [2,3]. However, some subtle alterations in brain tissues and the dysregulations in specific neuronal subtypes may be transient and therefore difficult to preserve and detect in posthumous patient tissues [4]. While animal models provide insights into disease mechanisms, significant species-dependent differences exist, and animal models only mirror the limited aspects of the pathophysiology of human diseases. It is believed that these species-dependent differences caused the high failure rate in clinical trials that have been derived from successful results in animal models [5,6,7]. Additionally, when using posthumous patient tissues or rodent models, it is difficult to decipher the molecular pathogenesis via biochemical approaches, which require a large number of high-purity living neurons. Human induced pluripotent stem cells (hiPSCs)-derived neurons overcome the limited access to human brain tissues and provide an unprecedented approach to model human neurological diseases [8].
iPSC-based disease modeling starts from Yamanaka and his colleagues’ groundbreaking studies, which demonstrated that somatic cells could be reprogrammed into pluripotent stem cells by ectopic expression of four transcription factors (Oct4 (O), Sox2 (S), Klf4 (K), and c-Myc (M)) under defined culture conditions [9,10]. An essential requirement for fulfilling the potential of hiPSCs is the ability to reliably differentiate into all three germ layers (ectoderm, mesoderm, and endoderm) and generate specific cell types with defined phenotypic traits [11,12,13]. The development of iPSCs offered a new approach for patient specific stem cell-based research, bypassing the reliance on overexpression models, interspecies differences of animal models, and also ethical concerns of using human embryonic stem cells (hESCs) [14]. Given that iPSCs derived from somatic cells can preserve the donor’s genetic background, less transplant rejection will occur when they are used for cell therapy. The in vitro phenotypes of disease-specific iPSC-derived cells hold the most promise to bridge the gap between the clinical phenotype and fundamental molecular and cellular mechanisms, creating new strategies for drug screening and novel therapeutic interventions [15,16]. Additionally, CRISPR engineering of iPSCs enables researchers to make paired patient mutation lines and isogenic control lines, greatly facilitating the research for understanding disease pathogenesis [12,13,17,18,19].
It is also well known that iPSCs and iPSC-derived cells have limitations, especially in the context of culture heterogeneity and dosage variability. The best route of administration and survivability in the hostile inflammatory microenvironment is controversial [20]. Improved ways of making cells, gene-editing technologies, along with patient-derived iPSC cells, have revolutionized the generation of experimental disease models. This provides an unlimited supply of any type of cells from once-inaccessible human tissues for research [21,22]. Neurodevelopmental and neurodegenerative disorders are particularly approachable using iPSC technology as iPSC-derived neurons retain the patient’s genomic context and provide an excellent cellular model system in deciphering the pathophysiology of diseases. Modulating the combination, concentration, and exposure time of crucial signaling molecules has yielded protocols for generating neurons and glia from iPSCs [23]. Thus far, researchers have developed protocols by which several neural and glial cell types could be generated, including glutamatergic neurons [24], GABAergic neurons [25,26], dopaminergic neurons [27], serotonergic neurons [28], MN [29,30,31,32], astrocytes [33], and microglia [34,35]. The improvement techniques and updated protocols in neural differentiation and maturation greatly facilitate the generation of iPSC-derived cells.
On the other hand, iPSC-based disease modelling is relatively new in biomedical research. Substantial progress has been made in developing differentiation protocols for the generation of different cell types. Excitingly, some in vitro models can recapitulate aspects of neuronal synaptic networks, which greatly advances functional modeling of neurodevelopmental and neurodegenerative diseases. Recent studies generating iPSC-derived MNs from movement disorders of ALS [36,37,38] and DYT1 patients [8,31,39] showed great potential to experimentally model molecular events underlying disease pathogenesis. The generation of patient-specific MNs provides an unprecedented approach in modeling MN-related disorders and deciphering cellular and molecular pathogenesis. In this review, we focused on the techniques for the generation of hiPSC-derived cholinergic MNs.
2. Generation of hiPSC-Derived MNs
Many studies are attempting to generate mature MNs from hiPSCs for modeling movement disorders. Developmental biologists have identified the signaling molecules and transcription factors that are involved in MN differentiation and maturation, providing the foundation for the generation of hiPSC-MNs. Currently, scientists are using these extrinsic factors to guide the MN differentiation from stem cells, thereby mimicking regionalization processes during nervous system development. Many protocols have been developed that rely upon core signaling pathways, which could synchronize neuronal induction to MN-specific signaling cascades and upregulate the expression of MN-specific genes.
The generation of hiPSC-MNs is a consecutive process that consists of a variety of induction stages, including iPSC induction, embryoid body (EB) formation, neural rosette growth, neuron progenitor cells (NPCs) differentiation, and MN induction and maturation (Figure 1A). At each induction stage, different culture media with different supplements will be employed to support and drive the cell fate towards MNs. At each differentiation stage, the cells show different growth patterns with unique cellular morphology and express specific protein markers (Figure 1B). These features can be used to verify the cell identity at each induction stage and perform quality controls to obtain highly pure MNs. In the following sections, we will discuss the process of iPSC-MN generation with a focus on the approaches using small molecules (chemicals) and lentiviral delivery of transcription factors. Meanwhile, the regulatory mechanisms of MN differentiation at different stages will be discussed.
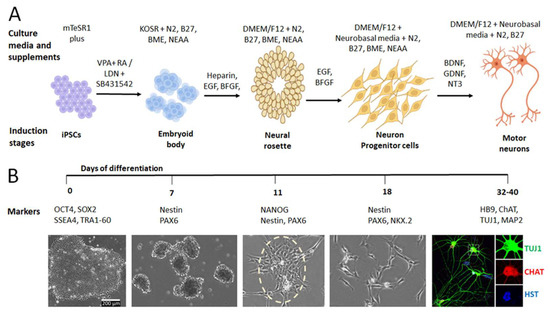
Figure 1.
Generation of iPSC-derived MNs. (A) Schematic shows the process of generating iPSC-MNs. The culture media with different supplements are shown at different induction stages. (B) The timeline of the induction process from hiPSC to MNs. At each stage, specific markers and typical cellular morphology are shown. Image of MNs was adapted from [32].
2.1. Generation of NPCs from hiPSCs
Human nervous system development begins with neural induction converting ectodermal cells into neuroectodermal cells, leading to the formation of the neural plate and finally the neural tube [40,41]. How neural regionalization is precisely controlled to form the forebrain, midbrain, hindbrain, and spinal cord along with the rostral-caudal (R-C) axis remains unknown. Dorso-ventral, rostral, and caudal patterning are very important events in neural tube maturation and specification. The gradients of various morphogens such as the canonical WNT/β-catenin signaling pathway (Wnt), retinoic acid (RA), and Sonic Hedgehog (Shh) signaling are involved in the determination of neuroaxis formation [42,43,44]. Insights from the model animals show that the rostral-caudal (R-C) axis patterning of NPCs is controlled by modulating the Wnt and RA signaling, whereas dorso-ventral patterning is controlled by the modulation of Shh signaling [45,46,47,48]. Most neuronal differentiation schemes mimic embryonic developmental signals by small molecule patterning. This initial study further showed that different combinations of small molecules used as patterning factors could push NPCs toward distinct neuronal fates [49,50,51]. The neuroectoderm is specified by inhibition of mesoderm and endoderm differentiation factors and acquires an initial rostral neural character through the regulation of BMP (bone morphogenetic protein), TGFβ (transforming growth factor-β), FGF (fibroblast growth factor), and Wnt signaling [52,53,54,55,56,57]. The neural fate can be potentially induced by the inhibition of BMP and TGFβ. These rostral neural progenitors are caudalized in response to Wnts, FGFs, and RA during early development [4].
One of the many ways of generating different types of neurons is through the differentiation of NPCs, which are a homogenous, multipotent, undifferentiated, self-renewable cell population. NPCs are committed to become neural lineage and to be differentiated to specific neuronal types in defined culture conditions [58]. iPSCs are induced to become NPCs through neuralization, either by EB formation [59,60,61,62], or by dual SMAD inhibition in monolayers [49,63,64]. EB formation is the most widely used method for neuralization [65,66,67,68,69]. In this process, cells grow in suspension and spontaneously arrange in three-dimensional aggregates upon withdrawal of factors promoting pluripotency. The Dual-SMAD-Inhibition method was developed in 2009 by Chambers and colleagues [49]. This method used a small molecule (Noggin, SB431542) to suppress the TGF-β/Activin/Nodal pathway and the BMP-canonical pathway [70,71]. After a 5-day exposure of the hiPSCs to Noggin/SB431542, the cells became an early-stage neuroepithelial population with expression of SOX1, Paired box protein 6 (PAX6), and Zic family member 1 (ZIC1) markers, and were able to form neural rosette organization [49,72,73]. This protocol showed an 80% efficiency of hESC and hiPSC differentiation into PAX6-positive NPCs. Once the iPSCs cells differentiate into NPCs, the newly formed NPCs continue expressing neuroepithelial markers, such as SOX1 (SRY-Box Transcription Factor 1), SOX3 (SRY-Box Transcription Factor 3), (PSA-NCAM) Polysialylated-neural cell adhesion molecule, and MS1 (Musashi RNA Binding Protein 1) [74,75,76]. Reinhardt and colleagues reported a protocol that NPCs can be generated using only small molecules. Neural induction was introduced through inhibition of both BMP and TGF-β signaling using Dorsomorphin and SB43152. To stimulate the canonical Wnt signaling, CHIR99021, a GSK3β inhibitor, was added to the cell medium, and the Shh pathway was stimulated by using purmorphamine. These neural progenitors are also able to differentiate into different types of neurons, including MNs [70].
Recently, two approaches were reported for generation of NPCs using a combination of small molecules: either RA and VPA (Valproic Acid) or LDN-193189 (SMAD inhibitor) and SB431542 (Activin/BMP/TGF-beta Pathway Inhibitor) [29,77,78]. Both combinations can successfully generate NPCs, which can be spontaneously differentiated into neurons consisting of glutamatergic (~75%), GABAergic (~15%), and dopaminergic (TH+) (~5%) neurons under defined culture conditions. These NPCs can also be differentiated into highly pure (90%) cholinergic MNs via transduction of three transcription factors: NEUROG2 (Neurogenin 2), ISL1 (LIM homeobox 1), and LHX3 (LIM homeobox protein 3) [29,31,32].
2.2. MN Induction via Small Molecules
Small molecules are bioactive compounds that can modulate specific cellular pathways involved in cell signaling, transcription, metabolism, or epigenetics, all of which are modulated during cellular reprogramming. If selective epigenetic modulation can be achieved with chemicals, it could remodel the chromatin structure and activate the gene expression of transcription factors, achieving similar effects to the ectopic expression of reprogramming factors. The advantage of using small molecules in reprogramming is that their biological effects are typically rapid, reversible, and dose-dependent, allowing precise control over specific outcomes by fine-tuning their concentrations and combinations. In Table 1, we summarized the roles of small molecules, including chemicals and peptide growth factors, in MN differentiation. These small molecules target different signaling pathways (Table 2) that promote MN differentiation and maturation. In Table 3, we summarized the chemical cocktails that have been reported in recent publications for the generation of iPSC-MNs.

Table 1.
Roles of small molecules in neuron differentiation.

Table 2.
Signaling pathways participating in MN differentiation.

Table 3.
Chemical cocktails used for MN differentiation from iPSCs.
To acquire the MN progenitor identity, NPCs need to caudalize and ventralize with the action of RA and Shh, respectively [42,109]. Shh induces the expression of homeodomain transcription factors and basic helix-loop-helix (bHLH) [11], the critical intermediaries in the control of cell patterning and neuronal cell fate determination [110,111,112,113]. The combined actions of RA and Shh are thought to establish a spatial and temporal regulation of the expression of transcription factors, such as LHX1 (homeodomain transcription factors) [114,115,116], OLIGO2 (oligodendrocyte transcription factor), MNX1 (MN and pancreas homeobox 1, HB9), and ISL1 [42]. All of these factors are necessary for the subsequent differentiation of NPCs to MNs [117,118,119]. Several studies have identified that OLIG2 is a bHLH protein, which is essential for establishing MN progenitor identity downstream of Shh signaling. OLIG2 also has a key role in specifying the subtype identity and pan-neuronal properties of developing MNs [117,120,121,122]. The expression of downstream transcriptional regulators, particularly HB9, a homeodomain protein expressed in postmitotic MNs, is crucial to consolidate MN identity [123,124,125].
Small molecules have a profound influence on neural induction and promote hiPSC differentiation into MNs combined with the simultaneous inhibition of TGFβ activin, Nodal, and BMP (also known as dual SMAD inhibition) signaling. Dual SMAD inhibition is a well-established method which utilizes small molecules to block endodermal and mesodermal cell fates and promote neuroectoderm conversion. It dramatically enriches neural ectoderm from pluripotent cells with a high expression of PAX6 and sex-determining region Y-box 2 (SOX2). Noggin and SB431542 are the most commonly used dual SMAD inhibitory small molecules. Noggin acts as a BMP inhibitor and SB431542 inhibits the Lefty/Activin/TGFβ pathways by blocking the phosphorylation of ALK4, ALK5, and ALK7 receptors. The GSK-3β inhibitor promotes neural progenitor proliferation by stimulating the canonical Wnt signaling pathway, which contributes to the maintenance of neural precursors. Molecular activation of SHH by Purmorphamine, canonical WNT signaling, and neural patterning by RA have been critical for MN induction. Previous studies have found that Shh induces upregulation of transcription factors OLIG2, NK2 homeobox 2 (NKX2.2), and neurogenin2 (NGN2) to direct the expression of MN fate-consolidating genes such as HB9 and ISL1. Neurotrophic and growth factors, such as BDNF, GDNF, and NT3 are also used as supplements to facilitate MN growth, maturation, and survival [8,29,126]. The entire differentiation process requires from 15 days to up to 2 months to generate fully functional MNs. In 2002, Wichterle and colleagues reported that RA and Shh were used to differentiate mouse ESCs into MNs through EB formation [127,128]. Wada and colleagues differentiated hESCs from the human and monkey ESCs toward MNs through neural rosette formation. They treated ESCs with 1 μM RA and 500 ng/mL Shh, leading to neural precursors becoming Tubulin β III+, Hb9+, Islet1+, and choline acetyltransferase-positive (ChAT+) neurons [129].
Scientists have been working continuously to advance our understanding of MN differentiation and to improve the techniques for generating MNs from iPSCs. The rapid advancement of RNA sequencing technologies contributed to a deep understanding of transcriptome composition and has discovered a large number of non-coding RNAs (ncRNAs) that participate in MN differentiation (Table 4). These ncRNAs have intense regulatory activities in a wide range of biological processes, including neuronal development, subtype diversification, specification, differentiation, and function [130,131,132]. Among the ncRNAs, long non-coding RNAs (lncRNAs) and miRNAs (microRNAs) are especially abundant in the nervous system and have been shown to be implicated in MN development and function.

Table 4.
Noncoding RNAs and their functions in MN differentiation.
Mature miRNAs are ∼22-nucleotide single-stranded RNAs that can recognize the 3′ untranslated region (UTR) of its target mRNAs and negatively regulate gene expression post-transcriptionally [146,147,148]. miRNAs are an integral part of the genetic program controlling MN survival and acquisition of subtype-specific properties [137,138,149]. Several studies also demonstrated that miRNAs mediated post-transcriptional regulation participates in fine-tuning the program of MN progenitor specification [148,150], MN differentiation [151,152,153], and subtype diversification [134,135,136,137,154,155]. miR-9 is involved in fine-tuning the differentiation of MN subtypes. Notably, a recent study revealed that miR-9 is transiently expressed during MN differentiation and regulates the expression of FoxP1 (Forkhead Box P1) and HOX (Homeobox (HOX) transcription factors. These transcription factors play a critical role in coordination of MN subtype identity and connectivity. In mice, overexpression of miR-9 induces neuronal differentiation by inhibiting the nuclear receptor [134,135], suggesting that miR-9 plays a role in fine-tuning the process of specification of MN subtype identity. Other studies showed that ISL1 expression by Onecut transcription factors (OC1) was important to generate LMC MNs [133,156,157]. Studies showed that miR-9 and OC1 are in mutually exclusive patterns in the embryonic spinal cord and miR-9 efficiently represses OC1 expression, demonstrating that regulation of OC1 by miR-9 is a crucial step in the specification of spinal MNs.
miR-218 is the most abundant and highly enriched miRNA in developing and maturing MNs [136,137,158]. miR-218 is decreased in human ALS postmortem spinal cord, and cell-free miR-218 can serve as a marker for MN loss in a rodent model of ALS [159,160]. Studies showed that in the developing spinal cord, the expression of miR-218 is directly upregulated by the Isl1–Lhx3 complex, which drives MN fate. Inhibition of miR-218 suppresses the generation of MNs in both chick neural tube and mouse ESCs, suggesting that miR-218 plays a crucial role in MN differentiation [137]. Previously, it has been found that complete loss of miR-218 results in the breakdown of neuromuscular synaptogenesis, hyperexcitability, post-natal lethality, MN loss, and complete paralysis [136,154,155,161].
Chen and colleagues demonstrated that the repression of Olig2 mRNA (MN progenitor marker) is controlled by mir-17-3p microRNA [138,150]. The expression of miR-17-3p is repressed by Shh, which results in elevated expression of Olig2. Thus, a high amount of Shh will direct neuronal progenitors to differentiate toward MNs [162,163]. Functional studies indicate that miRNA plays a significant role in a broad range of cellular and developmental processes of subsets of MN. By using an in vitro model of human spinal MN development, it has been shown that miR-375 is strongly activated during spinal motor neurogenesis and its expression is specific to MNs [130]. Knockdown of miR-375 significantly impairs MN differentiation, highlighting its essential role in MN development. miR-375 also protects MNs from DNA damage-induced degeneration by inhibiting p53 and therefore preventing apoptosis. Downregulation of the miR-375-3p in patients with spinal muscular atrophy leads to an increase of the p53 protein level and thus to apoptosis [144,145].
Long non-coding RNAs (lncRNAs) are RNAs that exceed 200 nucleotides in length, and they are not translated into proteins. LncRNAs participate in various stages during MNs differentiation, including guiding neural fate choice by driving transcription factor localization [164,165,166], regulating local translation at synapses [167,168], influencing MN development, and contributing to the pathogenic mechanisms underlying MN diseases (MNDs) [164,169].
The lncRNA CAT7 (chromatin-associated transcript 7) is a polyadenylated lncRNA that lies upstream (~400 kb) of MNX1 (MN homobox1). CAT7 has been found to temporally regulate MNX1 expression during the early stages of human ESC-MN differentiation. Loss of CAT7 causes de-repression of MNX1 before committing to motor neuronal lineage [170]. Another lncRNA Meg3 plays a critical role in maintaining postmitotic MN cell fate by repressing progenitor genes that regulate the differentiation of MN identity [171,172,173].
NEAT1 (nuclear-enriched abundant transcript 1) is a well-characterized lncRNA that functions as a chromatin regulator and organizes nuclear structures called ‘paraspeckles’. Paraspeckles contain proteins involved in transcription and RNA processing [174,175]. NEAT1 is highly enriched in neurons of the anterior horn of the spinal cord and in the cortical tissues of ALS patients. Nishimoto and colleagues demonstrated NEAT1 upregulation and increased paraspeckle formation in the MNs during the early phases of ALS pathogenesis [176,177,178]. The exact role of NEAT1 still needs to be resolved. Given their dynamic expression patterns in MNs and emerging roles in MN development and function, it is not surprising that dysregulation of noncoding RNAs has been implicated in MNDs. Understanding the mechanisms of action and functions of lncRNAs may assist the development of new therapies for MNDs.
2.3. MN Induction via Lentiviral Delivery of Transcription Factors
Previous reports have underscored the essential roles of transcription factors in MN development (Table 5). Decades of developmental studies have identified key signaling molecules and cell-intrinsic transcriptional programs [179,180] that specify MN identity during embryonic development. These transcription factors were transduced into neural progenitor cells after differentiation from ESCs/iPSCs, and MNs can be obtained 11 days after the transduction. These transcription factors are generally introduced into the cells via viral transduction (generally the most efficient ones) [29,181]. This transcription factors-mediated differentiation has been shown to produce highly efficient functional MNs with repetitively firing MNs after two weeks post-viral infection (wpi) [31,32].

Table 5.
Roles of transcription factors in MN differentiation.
Transcription factors ISL1 and LHX3 are sufficient to activate a MN gene expression program in other neural progenitors and increase post-mitotic specification by directly reprogramming pluripotent stem cells into MNs [188,189,190]. Specifically, a combinatorial expression of LIM homeodomain transcription factors Lhx3 and Isl-1, together with the expression of the pro-neural gene NGN2, have been shown to be critical to induce MN specification during development [114,191,192,193]. The combinatorial expression of Lhx3 and Isl1 will form an Isl1-Lhx3-hexamer, which will trigger MN specification in a chick spinal cord, ESCs, and iPSCs [114,194,195,196,197]. The binding of the Isl1-Lhx3 complex activates the transcription of genes that are essential for MN specification such as HB9 and promotes the expression of a wide range of terminal differentiation genes, including a battery of cholinergic pathway genes that enable cholinergic neurotransmission [198,199].
The combination of different transcription factors was often used to obtain a high quality of iPSC-MNs. These transcription factors need to be delivered into cells via lentiviral vectors or other vehicles (Table 6). In 2013, the group of Hynek Wichterle demonstrated that overexpression of three transcription factors (Ngn2, Isl1 and Lhx3) was sufficient to rapidly and efficiently program spinal MN identity from the mouse ESCs. Replacement of Lhx3 by Phox2a (Paired Like Homeobox 2A) led to the specification of cranial, rather than spinal MNs. Isl1-Lhx3 and Isl1-Phox2a heterodimers showed different DNA-sequence preferences for the basis of cell reprogramming, indicating that there are synergistic interactions between programming factors underying specification of alternate MN fates [189,200,201,202]. Goto et al. have used a single sendai virus-mediated overexpression of the TF cocktail NGN2, ISL1 and LHX3 in both mice and human iPSCs to promote the expression of MN markers. Notably, after 3 weeks of differentiation, NGN2/ISL1/LHX3-overexpressing neurons were electrophysiologically active and formed neuromuscular junctions (NMJ) with cultured myocytes [203]. The MNs derived via this method from ALS patient’s iPSCs have also shown disease phenotypes. De Santis et al. expressed transcription factors of Ngn2, Isl1, and Phox2a in human iPSCs via Piggy-bac transposable vectors and converted human iPSCs into cranial MNs and upregulated pan-MN genes such as TUBB3, ISL1, and ChAT within the first 3 days of differentiation. HB9 expression was increased when LHX3 was co-expressed, whereas PHOX2B and TBX20 (T-Box Transcription Factor 20) were detected by day 5 [144]. Finally, the authors of this study functionally characterized the cranial MNs obtained after 12 to 13 days to observe that these cells were capable of firing action potentials upon current stimulation, and almost half of all analyzed cells even displayed spontaneous glutamatergic postsynaptic currents.

Table 6.
Transcription factors used in generation of iPSC-MNs.
Goparaju et al. showed that overexpression of NGN1 (Neurogenin 1), NGN2 (Neurogenin 2), NGN3 (Neurogenin 3), NEUROD1 (Neuronal Differentiation 1), and NEUROD2 (Neuronal Differentiation) in human PSCs combined with RA, forskolin, and dual SMAD inhibition via SB431542 and dorsomorphin yields highly pure neuronal cultures expressing the MN markers HB9, ISL1, and ChAT [103]. MNs generated from this method have shown functional activity (repetitive action potentials and calcium-transient) within a week. We have reported that a single lentiviral vector expressing three factors (NGN2, ISL1, and LHX3) is necessary and sufficient to induce iPSC-derived MNs (iPSC-MNs) [29,32]. MNs derived using these methods robustly expressed general neuron markers, such as microtubule-associated protein 2 (MAP2), neurofilament protein (SMI-32), tubulin β-3 class III (TUBB3), and MN-specific markers HB9 and CHAT. These MNs showed electrical maturation within 3 weeks [32].
3. Quality Control: Validation of Neuron Identity and Purity
To ensure that the high quality and purity of MNs can be obtained from hiPSCs, it is necessary to validate the cell identity at each stage during the process of induction and differentiation. Specific markers at different stages could be examined and used to estimate the induction quality and the MN purity (Figure 1B). As the passage number of iPSC may affect the differentiation of iPSC-derived neurons [207], using a lower passage number is recommended in the generation of hiPSC-derived MNs.
3.1. Markers of Early Induction from hiPSC to NPC
Patient-specific somatic cells, such as skin fibroblast cells and peripheral blood mononuclear cells (PBMC), can be reprogrammed into the pluripotent state through ectopic expression of Yamanaka factors (Oct3/4, Sox2, Klf4, and c-Myc). The iPSC must be fully characterized to ensure quality before differentiation into neurons or other cell types. The morphology of iPSCs should demonstrate a typical hESC-like appearance composed of tightly packed cells in phase contrast microscopy (Figure 1B). A healthy iPSC line should robustly express the pluripotency markers (SSEA4, TRA-1-81, OCT3/4, and SOX2) and show a normal karyotype [12,13]. There are several techniques that can be used to assess the pluripotency. The pluripotency markers could be examined at protein levels by immunocytochemistry (ICC) and western blot or by using real-time PCR to measure transcription levels. Newly generated iPSC lines need to be validated that they can differentiate into three germ layers: endodermal, mesodermal, and ectodermal cell lineages. This can be verified by two approaches: 1) in vitro differentiation of iPSCs in suspension to form EBs, which highly express trilineage markers [59,208], and 2) EBs injected into immunocompromised NOD/SCID mice will form teratomas, which consist of three germ layers [209,210]. Similarly, three germ layers specific markers can be examined using ICC or western blot at protein level or RT-PCR at transcription level. The iPSCs and differentiated cells need to be confirmed as mycoplasma-negative before establishing a cell line stock.
3.2. Markers of MNs at Early Immature Stages
To achieve more accurate disease modeling and maximize the potential applications, quality controls are critical to verify cell identity and purity. Many methods could be used, including specific marker expression, molecular and functional properties, cellular morphology assay, electrophysiological analysis, and animal transplantation. Many protocols analyze samples within 2 to 5 weeks from the onset of differentiation. Early stages of MNs showed the typical polygonal cell body with few and short dendrites (Weeks 1–2). However, the generation of ISL1/2 and HB9-expressing cells can vary from 3 days to 2 weeks after differentiation. Longer differentiation protocols have an advantage over shorter ones when preparing fully functional mature neurons, which could be used to examine the electrophysiological activity and synaptic network. After treatment with lentivirus-expressing-specific transcription factors or small molecules, usually within a week the cell will become neuron-like with condensed nuclei, long axons, and multiple neurites. Generation of MNs requires the identification of genes that are expressed at the initial stages of MN differentiation. At day 13 of differentiation, studies observed the expression of early MN-specific factors, PAX6, OLIG2, ISL-1 and NEUROD [191]. PAX6 and OLIG2 are required to initiate a general MN fate differentiation [43,117,119]. Studies have also found that ISL1 is the earliest marker involved in the establishment of MN fate. The expression of MN-specific genes becomes evident at week 3 or later, including strong expression of LHX3, ISL1, and HB9. Early MNs are commonly characterized by transient co-expression of HB9 and ISL1/2 [123,211]. Homeobox gene HB9 is required for the consolidation of MN identity, and its expression is restricted to somatic MNs of the hypoglossal nucleus. Transcriptional upregulation of MN markers such as ISL1, HB9, and OLIG2 was shown in the early stage of MN differentiation in several studies.
3.3. Markers of MNs at Late Mature Stages
As HB9 is downregulated over the course of MN maturation, the expression of choline acetyltransferase (ChAT) appeared and temporally increased [212]. As ChAT is an enzyme responsible for the synthesis of neurotransmitter acetylcholine in cholinergic MNs; the expression of ChAT indicates that the cells reach maturation stage. miR-218, abundantly and selectively expressed in maturing MNs, is being recently used as a molecular marker to identify MNs [136,137].
The final maturation can be achieved with the action of neurotrophic factors (BDNF, GDNF, NT3) and demonstrated by increased dendrites arborization and cell-cell connections (weeks 2–6). Mature MNs display larger soma, increased cell shape, complexity of neurite outgrowth, and electrophysiological properties. iPSC-derived MNs are generally considered mature after 3 weeks of differentiation [100]. Higher expression of neuronal markers of TUBB3, MAP2, non-phosphorylated neurofilament heavy chain (SMI32), ChAT, and vesicular acetylcholine transporter (vAChT) indicates the maturation of the MNs. Monitoring the electrophysiological status of MNs in vitro is currently the most comprehensive method to assess their maturation. To demonstrate maturation, MNs are Synapsin-positive and electrophysiologically active. The electrophysiologically mature MNs are able to fire repetitive action potentials and generate spontaneous activity that requires the development of intrinsic (e.g., sufficiently polarized resting membrane potentials) and extrinsic (e.g., synapse formation) properties. Mature MNs are also capable of recreating NMJs when cocultured with myotubes in vitro and expressing acetylcholine receptor (AChR) clusters. All these characteristics indicate that hiPSCs had efficiently differentiated into fully functional MNs.
4. Modeling Neurological Diseases Using hiPSC-Derived MNs
Modeling MN-related diseases using hiPSC-based approaches requires culture conditions in a dish that can recapitulate the events underlying MN differentiation, maturation, aging, and degeneration. Several protocols exist to generate MNs from hiPSCs, and these cells have been used to study the pathophysiology of MN-related diseases, such as ALS, spinal muscular atrophy, and DYT1 dystonia [213,214,215,216,217,218]. The known disease-dependent cellular deficits are excellent features that can be used to validate the hiPSC-MNs in modeling the disease. For example, using patient-specific MNs, we have identified disease-dependent cellular deficits in DYT1 dystonia, including abnormal nuclear envelope morphology, disrupted neurodevelopment, impaired nucleocytoplasmic transport, and mislocalized nuclear Lamin B1 [31,39]. For each preparation of DYT1 iPSC-MNs, we routinely examined these cellular features to ensure that the materials we used are valid in modeling DYT1 dystonia. To identify and characterize functional molecular features of MNs, different technologies have been employed, ranging from fluorescence-based antibody staining of target markers to RNA-seq analysis. We usually perform ICC, Western blot, PCR, and qRT-PCR to verify the expression levels of neural markers at different stages during the induction process. Examination of well-characterized molecular markers is useful for determining the accuracy and efficiency of a protocol. Whole-transcriptome sequencing or RNA-seq, single cell sequencing is also often used to further characterize the expression of these markers in an unbiased manner. Modeling ALS with iPSC-derived MNs has recapitulated several known pathological findings in patient-derived cells, reinforcing the high value of the approach. Compared with healthy controls, patient iPSC-derived cells can be used to identify disease phenotypes at different levels, including molecular profiles, cellular features, and physiological functions. The development of high-throughput single-cell transcriptomics has changed the paradigm, empowering rapid isolation and profiling of MN nuclei, revealing remarkable transcriptional diversity within the skeletal and autonomic nervous systems. In addition, patient-specific iPSCs may also serve as powerful resources for personalized medicine, including drug discovery, genetic testing, and ultimately cell replacement therapy.
5. Future Challenges and Perspectives
Generation of hiPSC-MNs overcomes the limited access to human brain tissues and provides an unprecedented approach for modeling MN-related diseases, offering an excellent platform for developing therapeutic treatments. However, several challenges arise from using this cellular system for disease modeling [32]. First, the purity and yield of iPSC-derived neurons. Current human iPSC-MN induction protocols vary in timescale (ranging from 15 days to more than six weeks) and efficiency, with few protocols achieving both high efficiency and rapid MN generation [219,220]. Some studies require a large number of MNs with high purity, such as transcriptomic studies, to elucidate the alterations of genome-wide gene expression and proteomic studies to identify dysregulated factors in diseased neurons. Second, to obtain fully functional and mature neurons. Because of the lack of simplified and consistent protocols, the generated hiPSC-derived MNs in most studies are often functionally immature and heterogeneous. Some disease-dependent cellular and molecular deficits cannot be noticed until neurons reach full maturation, especially for age-related neurodegenerative diseases such as ALS, Alzheimer’s disease (AD), and Parkinson’s disease (PD). Modelling these late-onset diseases usually requires the long-term culture of neurons from several weeks to a few months, during which the neuron survival and potential contaminations are huge challenges. Third, how to culture the neurons more physiologically? Although chemical or physical modifications of the cell culture plates, such as coating with extracellular matrix (ECM), have been shown to be an efficient method to better mimic in vivo cell behavior [221], the outcomes of some experiments using in vitro cellular systems could be very different from the studies using in vivo models. iPSC-derived neurons cocultured with glial cells and the development of hiPSC to brain organoids under three-dimensional culture conditions could maximize the relevance to in vivo conditions.
To resolve these issues, studies are needed to understand the precise regulatory mechanisms of neural differentiation and maturation using in vivo models. Meanwhile, the protocols for the generation of iPSC-derived neurons need to be updated, simplified, and finally standardized to obtain consistent outcomes for biomedical research. The generation of different neuronal subtypes requires different protocols consisting of different induction factors, specific culture conditions and supplements, and particular treatments and processes. Generally, induction efficiency and neuron survival are paramount to achieve good yield and purity. The combinations of small molecules and/or transcription factors will enhance the induction efficiency and maximize the purity, while the optimized culture conditions will promote neuron maturation and survival, which will lead to a higher yield. One major difficulty in obtaining fully mature iPSC-MNs is the long-term culture issue, in which both neuron survival and potential contaminations are huge challenges. We generate iPSC-MNs using lentiviral delivery transcription factors, and these MNs can reach full maturation at 3 wpi with characterization of high expression of presynaptic proteins, cholinergic markers, and firing action potentials. For setting up experiments that require fully mature neurons, we usually culture iPSC-MN for 4 wpi [29]. For such a long-term culture, culture media supplemented with neurotrophic factors and neurons cocultured with astrocytes are required. For chemical-induced iPSC-MNs, once the MN identity is verified, in theory, the neurons will reach full maturation after culture for a long enough time. However, we have not directly compared the maturation, the culture time, and the detailed characterizations of iPSC-MNs that are generated by different approaches. It is hard to say which approach is better than the other. Excitingly, the techniques of genome-editing and three-dimensional culture of brain organoids greatly expanded the applications of hiPSC in disease modeling, cell therapy, and drug development [222,223].
Author Contributions
Conceptualization, B.D.; Writing, M.A. and B.D.; Funding Acquisition and Supervision, B.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Institute of Health (NIH) National Institute of Neurological Disorders and Stroke (NINDS) (NS112910 to B.D.), Department of Defense (DoD) Peer Reviewed Medical Research Program (PRMRP) Discovery Award (W81XWH2010186 to B.D.), and LSU Health Shreveport Center for Brain Health (CBH) Grant in Aid (Spring 2022 to B.D.).
Institutional Review Board Statement
Note applicable.
Informed Consent Statement
Note applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank members of the Ding laboratory for help and discussion.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ALS | Amyotrophic lateral sclerosis |
| ASCL1 | Achaetescute family bHLH transcription factor 1 |
| ATRA | All-trans retinoic acid |
| bHLH | Basic helix-loop-helix |
| bFGF | Basic fibroblast growth factor |
| BMP | Bone morphogenetic protein |
| BDNF | Brain-derived neurotrophic factor |
| CNS | Central nervous system |
| Wnt | Canonical WNT/β-catenin signaling pathway |
| CNTF | Ciliary neurotrophic factor |
| ChAT | Choline acetyltransferase |
| Cpd E | Compound E |
| CAT7 | Chromatin associated transcript 7 |
| EGF | Epidermal growth factor |
| ESCs | Embryonic stem cells |
| FGF | Fibroblast growth factor |
| FOXP1 | Forkhead box protein 1 |
| FGF2 | Fibroblast growth factor 2 |
| GDNF | Glial cell line-derived neurotrophic factor |
| GSK-3 | Glycogen synthase kinase 3 |
| hiPSCs | Human induced pluripotent stem cells |
| HOXC6 | Homeobox C6 |
| HB9 | Homeodomain |
| HST | Hoechst 33342 |
| ISL1 | Insulin gene enhancer 1 |
| LHX3 | LIM/homeobox 3 |
| LHX1 | Lim homeodomain transcription factors |
| LncRNAs | long non-coding RNAs |
| miRNAs | microRNAs |
| MAP2 | Microtubule Associated Protein 2 |
| MNs | Motor neurons |
| MS1 | Musashi RNA Binding Protein |
| MYT1L | Myelin transcription factor 1 like |
| MNX1 | Motor neuron and pancreas homeobox 1 |
| NPCs | Neural progenitor cells |
| NGN2 | Neurogenin-2 |
| NMJs | Neuromuscular junctions |
| DAPT | N-[N- (3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester |
| NKX2.2 | NK2 homeobox 2 |
| NT3 | Neurotrophin-3 |
| ncRNAs | Non-coding RNAs |
| NEAT1 | Nuclear-enriched abundant transcript 1 |
| NGN1 | Neurogenin 1 |
| NGN2 | Neurogenin 2 |
| NGN3 | Neurogenin 3 |
| NEUROD1 | Neuronal Differentiation 1 |
| NEUROD2 | Neuronal Differentiation 2 |
| Olig2 | Oligodendrocyte transcription factor |
| OC1 | Onecut transcription factors |
| PSA-NCAM | Polysialylated-neural cell adhesion molecule |
| POU5F1 | POU class 5 homeobox 1 |
| PHOX2A | Paired like homeobox 2A |
| POU3F2 | POU class 3 homeobox 2 |
| Pax6 | Paired box protein 6 |
| PUR | Purmorphamine |
| qPCR | Quantitative PCR |
| RA | Retinoic acid signaling pathway |
| ROCK | Rho-associated coiled-coil containing protein kinase |
| Shh | Sonic Hedgehog signaling pathway |
| Sox2 | Sex determining region Y-box 2 |
| SAG | Smoothened agonist |
| SOX1 | SRY-Box Transcription Factor 1 |
| SOX3 | SRY-Box Transcription Factor |
| SOX11 | SRY-box transcription factor 11 |
| Synapsins | Regulation of neurotransmitter release at synapses |
| TGFβ | Transforming growth factor-β |
| TUBB3 | Tubulin Beta 3 Class III |
| TBX20 | T-Box Transcription Factor 20 |
| UTR | 3′Untranslated region |
| VPA | Valproic Acid |
| VAChT | Vesicular acetylcholine transporter |
| WPI | Weeks post viral infection |
| ALS | Amyotrophic lateral sclerosis |
| APCDD1 | Adenomatosis polyposis downregulated |
| 1ASCL1 | Achaetescute family bHLH transcription factor 1 |
| ATRA | All-trans retinoic acidbHLH, Basic helix-loop-helix |
| bFGF | Basic fibroblast growth factor |
| BMP | Bone morphogenetic protein |
| BME | β-mercaptoethanol |
| BDNF | Brain-derived neurotrophic factor |
| CNS | Central nervous system |
| Wnt | Canonical WNT/β-catenin signaling pathway |
| CNTF | Ciliary neurotrophic factor |
| ChAT | Choline acetyltransferase |
| Cpd E | Compound E |
| CAT7 | Chromatin associated transcript 7 |
| Dl1 | Delta-like 1 |
| Dl4 | Delta-like 4 |
| DMH1 | a bone morphogenetic protein (BMP) inhibitor |
| EGF | Epidermal growth factor |
| ESCs | Embryonic stem cells |
| FGF | Fibroblast growth factor |
| FOXP1 | Forkhead box protein 1 |
| FGF2 | Fibroblast growth factor 2 |
| GDNF | Glial cell line-derived neurotrophic factor |
| GSK-3 | Glycogen synthase kinase 3 |
| hiPSCs | Human induced pluripotent stem cells |
| HOXC6 | Homeobox C6 |
| HST | Hoechst 33342 |
| ISL1 | Insulin gene enhancer 1 |
| LHX3 | LIM/homeobox 3 |
| LHX1 | Lim homeodomain transcription factors |
| LncRNAs | Long non-coding RNAs |
| miRNAs | microRNAs |
| MAP2 | Microtubule Associated Protein 2 |
| MEG3 | Maternally expressed gene 3 |
| MNs | Motor neurons |
| MS1 | Musashi RNA Binding Protein |
| MYT1L | Myelin transcription factor 1 like |
| MNX1/HB9 | Motor neuron and pancreas homeobox 1 |
| NPCs | Neural progenitor cells |
| NMJs | Neuromuscular junctions |
| DAPT | N-[N- (3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester |
| NKX2.2 | NK2 homeobox 2 |
| NT3 | Neurotrophin-3 |
| ncRNAs | Non-coding RNAs |
| NEAT1 | Nuclear-enriched abundant transcript 1 |
| NGN1 | Neurogenin 1 |
| NGN2 | Neurogenin 2 |
| NGN3 | Neurogenin 3 |
| NEUROD1 | Neuronal Differentiation 1 |
| NEUROD2 | Neuronal Differentiation 2 |
| Olig2 | Oligodendrocyte transcription factor |
| OC1 | Onecut transcription factors |
| PSA-NCAM | Polysialylated-neural cell adhesion molecule |
| POU5F1 | POU class 5 homeobox 1 |
| PHOX2A | Paired like homeobox 2A |
| POU3F2 | POU class 3 homeobox 2 |
| PAX6 | Paired box protein 6 |
| PUR | Purmorphamine |
| qPCR | Quantitative PCR |
| RA | Retinoic acid signaling pathway |
| ROCK | Rho-associated coiled-coil containing protein kinase |
| Shh | Sonic Hedgehog signaling pathway |
| SOX2 | Sex-determining region Y-box 2 |
| SAG | Smoothened agonist |
| SOX1 | SRY-Box Transcription Factor 1 |
| SOX3 | SRY-Box Transcription Factor |
| SOX11 | SRY-box transcription factor 11 |
| Synapsins | Regulation of neurotransmitter release at synapses |
| SYP | Synaptophysin |
| SMI-32 | Neurofilament |
| TGFβ | Transforming growth factor-β |
| TUBB3 | Tubulin Beta 3 Class III |
| TBX20 | T-Box Transcription Factor 20 |
| UTR | 3′Untranslated region |
| VPA | Valproic Acid |
| VAChT | Vesicular acetylcholine transporter |
| WPI | Weeks post-viral infection |
| Waif1/5T4 | Wnt-activated inhibitory factor 1 |
| ZIC1 | Zic family member 1 |
References
- Schugar, R.; Robbins, P.; Deasy, B. Small molecules in stem cell self-renewal and differentiation. Gene Ther. 2008, 15, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Vajkoczy, P.; Faust, K. Morphological Abnormalities in the Basal Ganglia of Dystonia Patients. Stereotact. Funct. Neurosurg. 2021, 99, 351–362. [Google Scholar] [CrossRef]
- Balint, B.; Mencacci, N.E.; Valente, E.M.; Pisani, A.; Rothwell, J.; Jankovic, J.; Vidailhet, M.; Bhatia, K.P. Dystonia. Nat. Rev. Dis. Primers 2018, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Augood, S.J.; Keller-McGandy, C.E.; Siriani, A.; Hewett, J.; Ramesh, V.; Sapp, E.; DiFiglia, M.; Breakefield, X.O.; Standaert, D.G. Distribution and ultrastructural localization of torsinA immunoreactivity in the human brain. Brain Res. 2003, 986, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Dickson, D.W. Parkinson’s disease: Experimental models and reality. Acta. Neuropathol. 2018, 135, 13–32. [Google Scholar] [CrossRef]
- Drummond, E.; Wisniewski, T. Alzheimer’s disease: Experimental models and reality. Acta. Neuropathol. 2017, 133, 155–175. [Google Scholar] [CrossRef] [PubMed]
- Genc, B.; Gozutok, O.; Ozdinler, P.H. Complexity of Generating Mouse Models to Study the Upper Motor Neurons: Let Us Shift Focus from Mice to Neurons. Int. J. Mol. Sci. 2019, 20, 3848. [Google Scholar] [CrossRef] [PubMed]
- Ding, B. Generation of patient-specific motor neurons in modeling movement diseases. Neural. Regen. Res. 2021, 16, 1799. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Shafa, M.; Yang, F.; Fellner, T.; Rao, M.S.; Baghbaderani, B.A. Human-induced pluripotent stem cells manufactured using a current good manufacturing practice-compliant process differentiate into clinically relevant cells from three germ layers. Front. Med. 2018, 5, 69. [Google Scholar] [CrossRef]
- Akter, M.; Cui, H.; Chen, Y.-H.; Ding, B. Generation of two induced pluripotent stem cell lines with heterozygous and homozygous GAG deletion in TOR1A gene from a healthy hiPSC line. Stem Cell Res. 2021, 56, 102536. [Google Scholar] [CrossRef]
- Akter, M.; Cui, H.; Chen, Y.-H.; Ding, B. Generation of gene-corrected isogenic control cell lines from a DYT1 dystonia patient iPSC line carrying a heterozygous GAG mutation in TOR1A gene. Stem Cell Res. 2022, 62, 102807. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Jeong, J.; Choi, D. Small-molecule-mediated reprogramming: A silver lining for regenerative medicine. Exp. Mol. Med. 2020, 52, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Scudellari, M. How iPS cells changed the world. Nature 2016, 534, 310–312. [Google Scholar] [CrossRef]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced pluripotent stem cell technology: A decade of progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef]
- Chang, C.-W.; Lai, Y.-S.; Westin, E.; Khodadadi-Jamayran, A.; Pawlik, K.M.; Lamb, L.S., Jr.; Goldman, F.D.; Townes, T.M. Modeling human severe combined immunodeficiency and correction by CRISPR/Cas9-enhanced gene targeting. Cell Rep. 2015, 12, 1668–1677. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.; Grundmann, A.; Renz, P.; Hänseler, W.; James, W.S.; Cowley, S.A.; Moore, M.D. CRISPR-mediated genotypic and phenotypic correction of a chronic granulomatous disease mutation in human iPS cells. Exp. Hematol. 2015, 43, 838–848.e3. [Google Scholar] [CrossRef]
- Xie, F.; Ye, L.; Chang, J.C.; Beyer, A.I.; Wang, J.; Muench, M.O.; Kan, Y.W. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014, 24, 1526–1533. [Google Scholar] [CrossRef]
- Volpato, V.; Webber, C. Addressing variability in iPSC-derived models of human disease: Guidelines to promote reproducibility. Dis. Model. Mech. 2020, 13, dmm042317. [Google Scholar] [CrossRef]
- Soubannier, V.; Maussion, G.; Chaineau, M.; Sigutova, V.; Rouleau, G.; Durcan, T.M.; Stifani, S. Characterization of human iPSC-derived astrocytes with potential for disease modeling and drug discovery. Neurosci. Lett. 2020, 731, 135028. [Google Scholar] [CrossRef]
- Yamanaka, S. Pluripotent stem cell-based cell therapy-Promise and challenges. Cell Stem Cell 2020, 27, 523–531. [Google Scholar] [CrossRef]
- Sienski, G.; Narayan, P.; Bonner, J.M.; Kory, N.; Boland, S.; Arczewska, A.A.; Ralvenius, W.T.; Akay, L.; Lockshin, E.; He, L. APOE4 disrupts intracellular lipid homeostasis in human iPSC-derived glia. Sci. Transl. Med. 2021, 13, eaaz4564. [Google Scholar] [CrossRef] [PubMed]
- Autar, K.; Guo, X.; Rumsey, J.W.; Long, C.J.; Akanda, N.; Jackson, M.; Narasimhan, N.S.; Caneus, J.; Morgan, D.; Hickman, J.J. A functional hiPSC-cortical neuron differentiation and maturation model and its application to neurological disorders. Stem Cell Rep. 2022, 17, 96–109. [Google Scholar] [CrossRef]
- Gonzalez-Ramos, A.; Waloschková, E.; Mikroulis, A.; Kokaia, Z.; Bengzon, J.; Ledri, M.; Andersson, M.; Kokaia, M. Human stem cell-derived GABAergic neurons functionally integrate into human neuronal networks. Sci. Rep. 2021, 11, 22050. [Google Scholar] [CrossRef]
- Grigor’eva, E.V.; Malankhanova, T.B.; Surumbayeva, A.; Pavlova, S.V.; Minina, J.M.; Kizilova, E.A.; Suldina, L.A.; Morozova, K.N.; Kiseleva, E.; Sorokoumov, E.D. Generation of GABAergic striatal neurons by a novel iPSC differentiation protocol enabling scalability and cryopreservation of progenitor cells. Cytotechnology 2020, 72, 649–663. [Google Scholar] [CrossRef]
- Rakovic, A.; Voß, D.; Vulinovic, F.; Meier, B.; Hellberg, A.-K.; Nau, C.; Klein, C.; Leipold, E. Electrophysiological Properties of Induced Pluripotent Stem Cell-Derived Midbrain Dopaminergic Neurons Correlate With Expression of Tyrosine Hydroxylase. Front. Cell. Neurosci. 2022, 121, 817198. [Google Scholar] [CrossRef]
- Valiulahi, P.; Vidyawan, V.; Puspita, L.; Oh, Y.; Juwono, V.B.; Sittipo, P.; Friedlander, G.; Yahalomi, D.; Sohn, J.-W.; Lee, Y.K. Generation of caudal-type serotonin neurons and hindbrain-fate organoids from hPSCs. Stem Cell Rep. 2021, 16, 1938–1952. [Google Scholar] [CrossRef] [PubMed]
- Akter, M.; Cui, H.; Sepehrimanesh, M.; Hosain, M.A.; Ding, B. Generation of highly pure motor neurons from human induced pluripotent stem cells. STAR Protoc. 2022, 3, 101223. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Akter, M.; Zhang, C.-L. Differential influence of sample sex and neuronal maturation on mRNA and protein transport in induced human neurons. Front. Mol. Neurosci. 2020, 13, 46. [Google Scholar] [CrossRef]
- Ding, B.; Tang, Y.; Ma, S.; Akter, M.; Liu, M.-L.; Zang, T.; Zhang, C.-L. Disease modeling with human neurons reveals LMNB1 dysregulation underlying DYT1 dystonia. J. Neurosci. 2021, 41, 2024–2038. [Google Scholar] [CrossRef]
- Sepehrimanesh, M.; Ding, B. Generation and optimization of highly pure motor neurons from human induced pluripotent stem cells via lentiviral delivery of transcription factors. Am. J. Physiol. Cell Physiol. 2020, 319, C771–C780. [Google Scholar] [CrossRef]
- Voulgaris, D.; Nikolakopoulou, P.; Herland, A. Generation of Human iPSC-Derived Astrocytes with a mature star-shaped phenotype for CNS modeling. Stem Cell Rev. Rep. 2022, 18, 2494–2512. [Google Scholar] [CrossRef]
- Xu, R.; Li, X.; Boreland, A.J.; Posyton, A.; Kwan, K.; Hart, R.P.; Jiang, P. Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat. Commun. 2020, 11, 1577. [Google Scholar] [CrossRef]
- Hasselmann, J.; Blurton-Jones, M. Human iPSC-derived microglia: A growing toolset to study the brain’s innate immune cells. Glia 2020, 68, 721–739. [Google Scholar] [CrossRef]
- Ho, R.; Workman, M.J.; Mathkar, P.; Wu, K.; Kim, K.J.; O’Rourke, J.G.; Kellogg, M.; Montel, V.; Banuelos, M.G.; Arogundade, O.A.; et al. Cross-Comparison of Human iPSC Motor Neuron Models of Familial and Sporadic ALS Reveals Early and Convergent Transcriptomic Disease Signatures. Cell Syst. 2021, 12, 159–175.e9. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Zhu, Y.; Hsiao-Nakamoto, J.; Tang, X.; Dugas, J.C.; Moscovitch-Lopatin, M.; Glass, J.D.; Brown, R.H., Jr.; Ladha, S.S.; Lacomis, D. Longitudinal biomarkers in amyotrophic lateral sclerosis. Ann. Clin. Transl. Neurol. 2020, 7, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Ferraiuolo, L.; Maragakis, N.J. Mini-Review: Induced pluripotent stem cells and the search for new cell-specific ALS therapeutic targets. Neurosci. Lett. 2021, 755, 135911. [Google Scholar] [CrossRef] [PubMed]
- Ding, B. Novel insights into the pathogenesis of DYT1 dystonia from induced patient-derived neurons. Neural. Regen. Res. 2022, 17, 561. [Google Scholar] [CrossRef]
- Gilbert, S.F. Developmental Biology; Sinauer Associates, Inc.: Sunderland, MA, USA, 2010. [Google Scholar]
- Leto, K.; Arancillo, M.; Becker, E.B.; Buffo, A.; Chiang, C.; Ding, B.; Dobyns, W.B.; Dusart, I.; Haldipur, P.; Hatten, M.E.; et al. Consensus Paper: Cerebellar Development. Cerebellum 2016, 15, 789–828. [Google Scholar] [CrossRef]
- Briscoe, J.; Ericson, J. Specification of neuronal fates in the ventral neural tube. Curr. Opin. Neurobiol. 2001, 11, 43–49. [Google Scholar] [CrossRef]
- Ericson, J.; Briscoe, J.; Rashbass, P.; Van Heyningen, V.; Jessell, T. Graded sonic hedgehog signaling and the specification of cell fate in the ventral neural tube. In cold Spring Harbor symposia on quantitative biology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997; pp. 451–466. [Google Scholar]
- Muhr, J.; Andersson, E.; Persson, M.; Jessell, T.M.; Ericson, J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell 2001, 104, 861–873. [Google Scholar] [CrossRef]
- Belgacem, Y.H.; Hamilton, A.M.; Shim, S.; Spencer, K.A.; Borodinsky, L.N. The many hats of sonic hedgehog signaling in nervous system development and disease. J. Dev. Biol. 2016, 4, 35. [Google Scholar] [CrossRef]
- Han, Y.-G.; Spassky, N.; Romaguera-Ros, M.; Garcia-Verdugo, J.-M.; Aguilar, A.; Schneider-Maunoury, S.; Alvarez-Buylla, A. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat. Neurosci. 2008, 11, 277–284. [Google Scholar] [CrossRef]
- Lai, K.; Kaspar, B.K.; Gage, F.H.; Schaffer, D.V. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat. Neurosci. 2003, 6, 21–27. [Google Scholar] [CrossRef]
- Wechsler-Reya, R.J.; Scott, M.P. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 1999, 22, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009, 27, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Chambers, S.M.; Qi, Y.; Mica, Y.; Lee, G.; Zhang, X.-J.; Niu, L.; Bilsland, J.; Cao, L.; Stevens, E.; Whiting, P. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat. Biotechnol. 2012, 30, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Wu, T.Y.; Brinker, A.; Peters, E.C.; Hur, W.; Gray, N.S.; Schultz, P.G. Synthetic small molecules that control stem cell fate. Proc. Natl. Acad. Sci. USA 2003, 100, 7632–7637. [Google Scholar] [CrossRef] [PubMed]
- Bragdon, B.; Moseychuk, O.; Saldanha, S.; King, D.; Julian, J.; Nohe, A. Bone morphogenetic proteins: A critical review. Cell. Signal. 2011, 23, 609–620. [Google Scholar] [CrossRef]
- Dhara, S.K.; Stice, S.L. Neural differentiation of human embryonic stem cells. J. Cell. Biochem. 2008, 105, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Dottori, M.; Pera, M.F. Neural differentiation of human embryonic stem cells. In Neural Stem Cells; Springer: Berlin, Germany, 2008; pp. 19–30. [Google Scholar]
- Gaspard, N.; Vanderhaeghen, P. Mechanisms of neural specification from embryonic stem cells. Curr. Opin. Neurobiol. 2010, 20, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J.; Brivanlou, A.H. Proposal of a model of mammalian neural induction. Dev. Biol. 2007, 308, 247–256. [Google Scholar] [CrossRef]
- Pankratz, M.T.; Li, X.-J.; LaVaute, T.M.; Lyons, E.A.; Chen, X.; Zhang, S.-C. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells 2007, 25, 1511–1520. [Google Scholar] [CrossRef]
- Kempermann, G.; Song, H.; Gage, F.H. Neurogenesis in the adult hippocampus. Cold Spring Harb. Perspect. Biol. 2015, 7, a018812. [Google Scholar] [CrossRef] [PubMed]
- Koziol, L.F.; Budding, D.; Andreasen, N.; D’Arrigo, S.; Bulgheroni, S.; Imamizu, H.; Ito, M.; Manto, M.; Marvel, C.; Parker, K. Consensus paper: The cerebellum’s role in movement and cognition. Cerebellum 2014, 13, 151–177. [Google Scholar] [CrossRef] [PubMed]
- Eiraku, M.; Watanabe, K.; Matsuo-Takasaki, M.; Kawada, M.; Yonemura, S.; Matsumura, M.; Wataya, T.; Nishiyama, A.; Muguruma, K.; Sasai, Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008, 3, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Ohgushi, M.; Matsumura, M.; Eiraku, M.; Murakami, K.; Aramaki, T.; Nishiyama, A.; Muguruma, K.; Nakano, T.; Suga, H.; Ueno, M. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell 2010, 7, 225–239. [Google Scholar] [CrossRef]
- Wataya, T.; Ando, S.; Muguruma, K.; Ikeda, H.; Watanabe, K.; Eiraku, M.; Kawada, M.; Takahashi, J.; Hashimoto, N.; Sasai, Y. Minimization of exogenous signals in ES cell culture induces rostral hypothalamic differentiation. Proc. Natl. Acad. Sci. USA 2008, 105, 11796–11801. [Google Scholar] [CrossRef]
- Fedorova, V.; Vanova, T.; Elrefae, L.; Pospisil, J.; Petrasova, M.; Kolajova, V.; Hudacova, Z.; Baniariova, J.; Barak, M.; Peskova, L. Differentiation of neural rosettes from human pluripotent stem cells in vitro is sequentially regulated on a molecular level and accomplished by the mechanism reminiscent of secondary neurulation. Stem Cell Res. 2019, 40, 101563. [Google Scholar] [CrossRef]
- Patani, R.; Compston, A.; Puddifoot, C.A.; Wyllie, D.J.; Hardingham, G.E.; Allen, N.D.; Chandran, S. Activin/Nodal inhibition alone accelerates highly efficient neural conversion from human embryonic stem cells and imposes a caudal positional identity. PLoS ONE 2009, 4, e7327. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, M.W.; Croft, G.F.; Williams, D.J.; O’Keeffe, S.; Carrasco, M.A.; Davis, A.R.; Roybon, L.; Oakley, D.H.; Maniatis, T.; Henderson, C.E. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J. Neurosci. 2013, 33, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; O’Doherty, J.P.; Shimojo, S. Neural computations mediating one-shot learning in the human brain. PLoS Biol. 2015, 13, e1002137. [Google Scholar] [CrossRef]
- Li, Y.; Mao, X.; Zhou, X.; Su, Y.; Zhou, X.; Shi, K.; Zhao, S. An optimized method for neuronal differentiation of embryonic stem cells in vitro. J. Neurosci. Methods 2020, 330, 108486. [Google Scholar] [CrossRef] [PubMed]
- Maury, Y.; Côme, J.; Piskorowski, R.A.; Salah-Mohellibi, N.; Chevaleyre, V.; Peschanski, M.; Martinat, C.; Nedelec, S. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 2015, 33, 89–96. [Google Scholar] [CrossRef]
- Salimi, A.; Nadri, S.; Ghollasi, M.; Khajeh, K.; Soleimani, M. Comparison of different protocols for neural differentiation of human induced pluripotent stem cells. Mol. Biol. Rep. 2014, 41, 1713–1721. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, P.; Glatza, M.; Hemmer, K.; Tsytsyura, Y.; Thiel, C.S.; Höing, S.; Moritz, S.; Parga, J.A.; Wagner, L.; Bruder, J.M.; et al. Correction: Derivation and Expansion Using Only Small Molecules of Human Neural Progenitors for Neurodegenerative Disease Modeling. PLoS ONE 2013, 8, e59252. [Google Scholar] [CrossRef]
- Smith, K.A.; Chocron, S.; von der Hardt, S.; de Pater, E.; Soufan, A.; Bussmann, J.; Schulte-Merker, S.; Hammerschmidt, M.; Bakkers, J. Rotation and Asymmetric Development of the Zebrafish Heart Requires Directed Migration of Cardiac Progenitor Cells. Dev. Cell 2008, 14, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Osumi, N.; Shinohara, H.; Numayama-Tsuruta, K.; Maekawa, M. Concise Review: Pax6 Transcription Factor Contributes to both Embryonic and Adult Neurogenesis as a Multifunctional Regulator. Stem Cells 2008, 26, 1663–1672. [Google Scholar] [CrossRef]
- Zhang, M.; Ngo, J.; Pirozzi, F.; Sun, Y.-P.; Wynshaw-Boris, A. Highly efficient methods to obtain homogeneous dorsal neural progenitor cells from human and mouse embryonic stem cells and induced pluripotent stem cells. Stem Cell Res. Ther. 2018, 9, 67. [Google Scholar] [CrossRef]
- Jiménez-Vaca, A.L.; Benitez-King, G.; Ruiz, V.; Ramírez-Rodríguez, G.B.; Hernández-de la Cruz, B.; Salamanca-Gómez, F.A.; González-Márquez, H.; Ramírez-Sánchez, I.; Ortíz-López, L.; Vélez-del Valle, C. Exfoliated human olfactory neuroepithelium: A source of neural progenitor cells. Mol. Neurobiol. 2018, 55, 2516–2523. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fang, Z.; Wen, J.; Tang, F.; Liao, B.; Jing, N.; Lai, D.; Jin, Y. SOX1 is required for the specification of rostral hindbrain neural progenitor cells from human embryonic stem cells. Iscience 2020, 23, 101475. [Google Scholar] [CrossRef] [PubMed]
- Stevanovic, M.; Drakulic, D.; Lazic, A.; Ninkovic, D.S.; Schwirtlich, M.; Mojsin, M. SOX transcription factors as important regulators of neuronal and glial differentiation during nervous system development and adult neurogenesis. Front. Mol. Neurosci. 2021, 14, 654031. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Sepehrimanesh, M. Nucleocytoplasmic transport: Regulatory mechanisms and the implications in neurodegeneration. Int. J. Mol. Sci. 2021, 22, 4165. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Li, C.; Zhang, M.; Wang, H.; Xie, Y.; Tang, Y. A Step-by-Step Refined Strategy for Highly Efficient Generation of Neural Progenitors and Motor Neurons from Human Pluripotent Stem Cells. Cells 2021, 10, 3087. [Google Scholar] [CrossRef]
- Rolletschek, A.; Wobus, A.M. Induced human pluripotent stem cells: Promises and open questions. Biol. Chem. 2009, 390, 845–849. [Google Scholar] [CrossRef]
- Trawczynski, M.; Liu, G.; David, B.T.; Fessler, R.G. Restoring motor neurons in spinal cord injury with induced pluripotent stem cells. Front. Cell. Neurosci. 2019, 13, 369. [Google Scholar] [CrossRef]
- Valizadeh-Arshad, Z.; Shahbazi, E.; Hashemizadeh, S.; Moradmand, A.; Jangkhah, M.; Kiani, S. In vitro differentiation of neural-like cells from human embryonic stem cells by a combination of dorsomorphin, XAV939, and A8301. Cell J. 2018, 19, 545. [Google Scholar]
- Kamishibahara, Y.; Kawaguchi, H.; Shimizu, N. Rho kinase inhibitor Y-27632 promotes neuronal differentiation in mouse embryonic stem cells via phosphatidylinositol 3-kinase. Neurosci. Lett. 2016, 615, 44–49. [Google Scholar] [CrossRef]
- Dworkin, S.; Mantamadiotis, T. Targeting CREB signalling in neurogenesis. Expert Opin. Ther. Targets 2010, 14, 869–879. [Google Scholar] [CrossRef]
- Du, Z.-W.; Chen, H.; Liu, H.; Lu, J.; Qian, K.; Huang, C.-L.; Zhong, X.; Fan, F.; Zhang, S.-C. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat. Commun. 2015, 6, 6626. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, J.; Noakes, P.G.; Bellingham, M.C. The role of altered BDNF/TrkB signaling in amyotrophic lateral sclerosis. Front. Cell. Neurosci. 2019, 13, 368. [Google Scholar] [CrossRef] [PubMed]
- Cintrón-Colón, A.F.; Almeida-Alves, G.; Boynton, A.M.; Spitsbergen, J.M. GDNF synthesis, signaling, and retrograde transport in motor neurons. Cell Tissue Res. 2020, 382, 47–56. [Google Scholar] [CrossRef]
- Zeng, S.; Zhao, X.; Zhang, L.; Pathak, J.L.; Huang, W.; Li, Y.; Guan, H.; Zhao, W.; Ge, L.; Shu, Y. Effect of ciliary neurotrophic factor on neural differentiation of stem cells of human exfoliated deciduous teeth. J. Biol. Eng. 2020, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Ueno, M.; Kamiya, D.; Nishiyama, A.; Matsumura, M.; Wataya, T.; Takahashi, J.B.; Nishikawa, S.; Nishikawa, S.-i.; Muguruma, K. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007, 25, 681–686. [Google Scholar] [CrossRef]
- Diez del Corral, R.; Morales, A.V. The multiple roles of FGF signaling in the developing spinal cord. Front. Cell Dev. Biol. 2017, 5, 58. [Google Scholar] [CrossRef] [PubMed]
- Scalabrino, G. Epidermal growth factor in the CNS: A beguiling journey from integrated cell biology to multiple sclerosis. an extensive translational overview. Cell. Mol. Neurobiol. 2020, 42, 891–916. [Google Scholar] [CrossRef]
- Colombres, M.; Henríquez, J.P.; Reig, G.F.; Scheu, J.; Calderón, R.; Alvarez, A.; Brandan, E.; Inestrosa, N.C. Heparin activates Wnt signaling for neuronal morphogenesis. J. Cell. Physiol. 2008, 216, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Qi, Y.; Sun, Z. The role of sonic hedgehog pathway in the development of the central nervous system and aging-related neurodegenerative diseases. Front. Mol. Biosci. 2021, 8, 711710. [Google Scholar] [CrossRef]
- Tomishima, M. Neural Induction–Dual SMAD Inhibition; Steam Book: Holland Landing, ON, Canada, 2013. [Google Scholar]
- Mitre, M.; Mariga, A.; Chao, M.V. Neurotrophin signalling: Novel insights into mechanisms and pathophysiology. Clin. Sci. 2017, 131, 13–23. [Google Scholar] [CrossRef]
- Westphal, M.; Panza, P.; Kastenhuber, E.; Wehrle, J.; Driever, W. Wnt/β-catenin signaling promotes neurogenesis in the diencephalospinal dopaminergic system of embryonic zebrafish. Sci. Rep. 2022, 12, 1030. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shushan, E.; Feldman, E.; Reubinoff, B.E. Notch signaling regulates motor neuron differentiation of human embryonic stem cells. Stem Cells 2015, 33, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Allison, R.L.; Welby, E.; Khayrullina, G.; Burnett, B.G.; Ebert, A.D. Viral mediated knockdown of GATA6 in SMA iPSC-derived astrocytes prevents motor neuron loss and microglial activation. Glia 2022, 70, 989–1004. [Google Scholar] [CrossRef] [PubMed]
- Cutarelli, A.; Martínez-Rojas, V.A.; Tata, A.; Battistella, I.; Rossi, D.; Arosio, D.; Musio, C.; Conti, L. A monolayer system for the efficient generation of motor neuron progenitors and functional motor neurons from human pluripotent stem cells. Cells 2021, 10, 1127. [Google Scholar] [CrossRef] [PubMed]
- Solomon, E.; Davis-Anderson, K.; Hovde, B.; Micheva-Viteva, S.; Harris, J.F.; Twary, S.; Iyer, R. Global transcriptome profile of the developmental principles of in vitro iPSC-to-motor neuron differentiation. BMC Mol. Cell Biol. 2021, 22, 13. [Google Scholar] [CrossRef]
- Bianchi, F.; Malboubi, M.; Li, Y.; George, J.H.; Jerusalem, A.; Szele, F.; Thompson, M.S.; Ye, H. Rapid and efficient differentiation of functional motor neurons from human iPSC for neural injury modelling. Stem Cell Res. 2018, 32, 126–134. [Google Scholar] [CrossRef]
- Kiskinis, E.; Kralj, J.M.; Zou, P.; Weinstein, E.N.; Zhang, H.; Tsioras, K.; Wiskow, O.; Ortega, J.A.; Eggan, K.; Cohen, A.E. All-optical electrophysiology for high-throughput functional characterization of a human iPSC-derived motor neuron model of ALS. Stem Cell Rep. 2018, 10, 1991–2004. [Google Scholar] [CrossRef]
- Fujimori, K.; Ishikawa, M.; Otomo, A.; Atsuta, N.; Nakamura, R.; Akiyama, T.; Hadano, S.; Aoki, M.; Saya, H.; Sobue, G. Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat. Med. 2018, 24, 1579–1589. [Google Scholar] [CrossRef]
- Goparaju, S.K.; Kohda, K.; Ibata, K.; Soma, A.; Nakatake, Y.; Akiyama, T.; Wakabayashi, S.; Matsushita, M.; Sakota, M.; Kimura, H. Rapid differentiation of human pluripotent stem cells into functional neurons by mRNAs encoding transcription factors. Sci. Rep. 2017, 7, 42367. [Google Scholar] [CrossRef]
- Guo, W.; Naujock, M.; Fumagalli, L.; Vandoorne, T.; Baatsen, P.; Boon, R.; Ordovás, L.; Patel, A.; Welters, M.; Vanwelden, T. HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat. Commun. 2017, 8, 861. [Google Scholar] [CrossRef]
- Ichiyanagi, N.; Fujimori, K.; Yano, M.; Ishihara-Fujisaki, C.; Sone, T.; Akiyama, T.; Okada, Y.; Akamatsu, W.; Matsumoto, T.; Ishikawa, M. Establishment of in vitro FUS-associated familial amyotrophic lateral sclerosis model using human induced pluripotent stem cells. Stem Cell Rep. 2016, 6, 496–510. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Qian, K.; Du, Z.; Cao, J.; Petersen, A.; Liu, H.; Blackbourn IV, L.W.; Huang, C.-L.; Errigo, A.; Yin, Y. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell 2014, 14, 796–809. [Google Scholar] [CrossRef] [PubMed]
- Qu, Q.; Li, D.; Louis, K.R.; Li, X.; Yang, H.; Sun, Q.; Crandall, S.R.; Tsang, S.; Zhou, J.; Cox, C.L. High-efficiency motor neuron differentiation from human pluripotent stem cells and the function of Islet-1. Nat. Commun. 2014, 5, 3449. [Google Scholar] [CrossRef]
- Dimos, J.T.; Rodolfa, K.T.; Niakan, K.K.; Weisenthal, L.M.; Mitsumoto, H.; Chung, W.; Croft, G.F.; Saphier, G.; Leibel, R.; Goland, R. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 2008, 321, 1218–1221. [Google Scholar] [CrossRef] [PubMed]
- Briscoe, J.; Pierani, A.; Jessell, T.M.; Ericson, J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 2000, 101, 435–445. [Google Scholar] [CrossRef]
- Dasen, J.S.; De Camilli, A.; Wang, B.; Tucker, P.W.; Jessell, T.M. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell 2008, 134, 304–316. [Google Scholar] [CrossRef]
- Dasen, J.S.; Liu, J.-P.; Jessell, T.M. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature 2003, 425, 926–933. [Google Scholar] [CrossRef]
- Dasen, J.S.; Tice, B.C.; Brenner-Morton, S.; Jessell, T.M. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell 2005, 123, 477–491. [Google Scholar] [CrossRef]
- Peljto, M.; Dasen, J.S.; Mazzoni, E.O.; Jessell, T.M.; Wichterle, H. Functional diversity of ESC-derived motor neuron subtypes revealed through intraspinal transplantation. Cell Stem Cell 2010, 7, 355–366. [Google Scholar] [CrossRef]
- Cho, H.-H.; Cargnin, F.; Kim, Y.; Lee, B.; Kwon, R.-J.; Nam, H.; Shen, R.; Barnes, A.P.; Lee, J.W.; Lee, S. Isl1 directly controls a cholinergic neuronal identity in the developing forebrain and spinal cord by forming cell type-specific complexes. PLoS Genet. 2014, 10, e1004280. [Google Scholar] [CrossRef]
- Palmesino, E.; Rousso, D.L.; Kao, T.-J.; Klar, A.; Laufer, E.; Uemura, O.; Okamoto, H.; Novitch, B.G.; Kania, A. Foxp1 and lhx1 coordinate motor neuron migration with axon trajectory choice by gating Reelin signalling. PLoS Biol. 2010, 8, e1000446. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Liu, H.; Zhang, D.; Xu, G.; Liu, J.; Evans, S.M.; Pan, J.; Cui, S. LIM homeodomain transcription factor Isl1 affects urethral epithelium differentiation and apoptosis via Shh. Cell Death Dis. 2019, 10, 713. [Google Scholar] [CrossRef] [PubMed]
- Novitch, B.G.; Chen, A.I.; Jessell, T.M. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 2001, 31, 773–789. [Google Scholar] [CrossRef] [PubMed]
- Novitch, B.G.; Wichterle, H.; Jessell, T.M.; Sockanathan, S. A requirement for retinoic acid-mediated transcriptional activation in ventral neural patterning and motor neuron specification. Neuron 2003, 40, 81–95. [Google Scholar] [CrossRef]
- Vallstedt, A.; Muhr, J.; Pattyn, A.; Pierani, A.; Mendelsohn, M.; Sander, M.; Jessell, T.M.; Ericson, J. Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron 2001, 31, 743–755. [Google Scholar] [CrossRef]
- Lu, Q.R.; Sun, T.; Zhu, Z.; Ma, N.; Garcia, M.; Stiles, C.D.; Rowitch, D.H. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 2002, 109, 75–86. [Google Scholar] [CrossRef]
- Sagner, A.; Gaber, Z.B.; Delile, J.; Kong, J.H.; Rousso, D.L.; Pearson, C.A.; Weicksel, S.E.; Melchionda, M.; Mousavy Gharavy, S.N.; Briscoe, J. Olig2 and Hes regulatory dynamics during motor neuron differentiation revealed by single cell transcriptomics. PLoS Biol. 2018, 16, e2003127. [Google Scholar] [CrossRef] [PubMed]
- Zannino, D.A.; Appel, B. Olig2+ precursors produce abducens motor neurons and oligodendrocytes in the zebrafish hindbrain. J. Neurosci. 2009, 29, 2322–2333. [Google Scholar] [CrossRef]
- Arber, S.; Han, B.; Mendelsohn, M.; Smith, M.; Jessell, T.M.; Sockanathan, S. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron 1999, 23, 659–674. [Google Scholar] [CrossRef]
- Odden, J.P.; Holbrook, S.; Doe, C.Q. DrosophilaHB9 is expressed in a subset of motoneurons and interneurons, where it regulates gene expression and axon pathfinding. J. Neurosci. 2002, 22, 9143–9149. [Google Scholar] [CrossRef]
- Stifani, N. Motor neurons and the generation of spinal motor neuron diversity. Front. Cell. Neurosci. 2014, 8, 293. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Dobner, P.R.; Mullikin-Kilpatrick, D.; Wang, W.; Zhu, H.; Chow, C.W.; Cave, J.W.; Gronostajski, R.M.; Kilpatrick, D.L. BDNF activates an NFI-dependent neurodevelopmental timing program by sequestering NFATc4. Mol. Biol. Cell. 2018, 29, 975–987. [Google Scholar] [CrossRef] [PubMed]
- Frank-Kamenetsky, M.; Zhang, X.M.; Bottega, S.; Guicherit, O.; Wichterle, H.; Dudek, H.; Bumcrot, D.; Wang, F.Y.; Jones, S.; Shulok, J. Small-molecule modulators of Hedgehog signaling: Identification and characterization of Smoothened agonists and antagonists. J. Biol. 2002, 1, 10. [Google Scholar] [CrossRef]
- Wichterle, H.; Lieberam, I.; Porter, J.A.; Jessell, T.M. Directed differentiation of embryonic stem cells into motor neurons. Cell 2002, 110, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Wada, T.; Honda, M.; Minami, I.; Tooi, N.; Amagai, Y.; Nakatsuji, N.; Aiba, K. Highly efficient differentiation and enrichment of spinal motor neurons derived from human and monkey embryonic stem cells. PLoS ONE 2009, 4, e6722. [Google Scholar] [CrossRef]
- Bhinge, A.; Namboori, S.C.; Bithell, A.; Soldati, C.; Buckley, N.J.; Stanton, L.W. MiR-375 is essential for human spinal motor neuron development and may be involved in motor neuron degeneration. Stem Cells 2016, 34, 124–134. [Google Scholar] [CrossRef]
- Dajas-Bailador, F.; Bonev, B.; Garcez, P.; Stanley, P.; Guillemot, F.; Papalopulu, N. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat. Neurosci. 2012, 15, 697. [Google Scholar] [CrossRef]
- Haramati, S.; Chapnik, E.; Sztainberg, Y.; Eilam, R.; Zwang, R.; Gershoni, N.; McGlinn, E.; Heiser, P.W.; Wills, A.-M.; Wirguin, I. miRNA malfunction causes spinal motor neuron disease. Proc. Natl. Acad. Sci. USA 2010, 107, 13111–13116. [Google Scholar] [CrossRef]
- Luxenhofer, G.; Helmbrecht, M.S.; Langhoff, J.; Giusti, S.A.; Refojo, D.; Huber, A.B. MicroRNA-9 promotes the switch from early-born to late-born motor neuron populations by regulating Onecut transcription factor expression. Dev. Biol. 2014, 386, 358–370. [Google Scholar] [CrossRef]
- Otaegi, G.; Pollock, A.; Hong, J.; Sun, T. MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J. Neurosci. 2011, 31, 809–818. [Google Scholar] [CrossRef]
- Otaegi, G.; Pollock, A.; Sun, T. An optimized sponge for microRNA miR-9 affects spinal motor neuron development in vivo. Front. Neurosci. 2012, 5, 146. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Amin, N.D.; Bai, G.; Klug, J.R.; Bonanomi, D.; Pankratz, M.T.; Gifford, W.D.; Hinckley, C.A.; Sternfeld, M.J.; Driscoll, S.P.; Dominguez, B. Loss of motoneuron-specific microRNA-218 causes systemic neuromuscular failure. Science 2015, 350, 1525–1529. [Google Scholar] [CrossRef] [PubMed]
- Thiebes, K.P.; Nam, H.; Cambronne, X.A.; Shen, R.; Glasgow, S.M.; Cho, H.-H.; Kwon, J.-S.; Goodman, R.H.; Lee, J.W.; Lee, S. miR-218 is essential to establish motor neuron fate as a downstream effector of Isl1–Lhx3. Nat. Commun. 2015, 6, 7718. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.-T.; Lu, Y.-L.; Peng, K.-C.; Yen, Y.-P.; Chang, M.; Li, J.; Jung, H.; Thams, S.; Huang, Y.-P.; Hung, J.-H. Mir-17∼ 92 governs motor neuron subtype survival by mediating nuclear PTEN. Cell Rep. 2015, 11, 1305–1318. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.-T.; Peng, K.-C.; Chen, Y.-C.; Yen, Y.-P.; Chang, M.; Thams, S.; Chen, J.-A. Mir-17∼ 92 confers motor neuron subtype differential resistance to ALS-associated degeneration. Cell Stem Cell 2019, 25, 193–209.e7. [Google Scholar] [CrossRef]
- Li, L.; Luo, Z. Dysregulated miR-27a-3p promotes nasopharyngeal carcinoma cell proliferation and migration by targeting Mapk10. Oncol. Rep. 2017, 37, 2679–2687. [Google Scholar] [CrossRef]
- Chen, Y.; Wei, Q.; Chen, X.; Li, C.; Cao, B.; Ou, R.; Hadano, S.; Shang, H.-F. Aberration of miRNAs expression in leukocytes from sporadic amyotrophic lateral sclerosis. Front. Mol. Neurosci. 2016, 9, 69. [Google Scholar] [CrossRef]
- Li, C.; Chen, Y.; Chen, X.; Wei, Q.; Ou, R.; Gu, X.; Cao, B.; Shang, H. MicroRNA-183-5p is stress-inducible and protects neurons against cell death in amyotrophic lateral sclerosis. J. Cell. Mol. Med. 2020, 24, 8614–8622. [Google Scholar] [CrossRef]
- Asli, N.S.; Kessel, M. Spatiotemporally restricted regulation of generic motor neuron programs by miR-196-mediated repression of Hoxb8. Dev. Biol. 2010, 344, 857–868. [Google Scholar] [CrossRef] [PubMed]
- De Santis, R.; Garone, M.G.; Pagani, F.; de Turris, V.; Di Angelantonio, S.; Rosa, A. Direct conversion of human pluripotent stem cells into cranial motor neurons using a piggyBac vector. Stem Cell Res. 2018, 29, 189–196. [Google Scholar] [CrossRef]
- Rohm, M.; May, C.; Marcus, K.; Steinbach, S.; Theis, V.; Theiß, C.; Matschke, V. The microRNA miR-375-3p and the tumor suppressor NDRG2 are involved in sporadic amyotrophic lateral sclerosis. Cell Physiol. Biochem. 2019, 52, 1412–1426. [Google Scholar] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- DING, B. Gene expression in maturing neurons: Regulatory mechanisms and related neurodevelopmental disorders. Acta Physiol. Sin. 2015, 67, 113–133. [Google Scholar]
- Williams, A.H.; Valdez, G.; Moresi, V.; Qi, X.; McAnally, J.; Elliott, J.L.; Bassel-Duby, R.; Sanes, J.R.; Olson, E.N. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 2009, 326, 1549–1554. [Google Scholar] [CrossRef]
- Chen, J.-A.; Huang, Y.-P.; Mazzoni, E.O.; Tan, G.C.; Zavadil, J.; Wichterle, H. Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron 2011, 69, 721–735. [Google Scholar] [CrossRef]
- Cao, X.; Pfaff, S.L.; Gage, F.H. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007, 21, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, J.; Lee, S.; Lee, B.; Lee, J.W.; Lee, S.-K. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007, 21, 744–749. [Google Scholar] [CrossRef]
- Visvanathan, J.; Lee, S.; Lee, B.; Lee, S.-K. MIR-124 antagonizes the anti-neural rest/scp1 pathway during embryonic development. Dev. Biol. 2008, 2, 574. [Google Scholar] [CrossRef]
- Amin, N.D.; Senturk, G.; Costaguta, G.; Driscoll, S.; O’Leary, B.; Bonanomi, D.; Pfaff, S.L. A hidden threshold in motor neuron gene networks revealed by modulation of miR-218 dose. Neuron 2021, 109, 3252–3267.e6. [Google Scholar] [CrossRef]
- Amin, N.D.; Senturk, G.; Hayashi, M.; Driscoll, S.P.; Pfaff, S.L. Detecting microRNA-mediated gene regulatory effects in murine neuronal subpopulations. STAR Protoc. 2022, 3, 101130. [Google Scholar] [CrossRef] [PubMed]
- Francius, C.; Clotman, F. Generating spinal motor neuron diversity: A long quest for neuronal identity. Cell. Mol. Life Sci. 2014, 71, 813–829. [Google Scholar] [CrossRef] [PubMed]
- Toch, M.; Harris, A.; Schakman, O.; Kondratskaya, E.; Boulland, J.-L.; Dauguet, N.; Debrulle, S.; Baudouin, C.; Hidalgo-Figueroa, M.; Mu, X. Onecut-dependent Nkx6. 2 transcription factor expression is required for proper formation and activity of spinal locomotor circuits. Sci. Rep. 2020, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Shu, P.; Wu, C.; Liu, W.; Ruan, X.; Liu, C.; Hou, L.; Zeng, Y.; Fu, H.; Wang, M.; Chen, P. The spatiotemporal expression pattern of microRNAs in the developing mouse nervous system. J. Biol. Chem. 2019, 294, 3444–3453. [Google Scholar] [CrossRef] [PubMed]
- Emde, A.; Eitan, C.; Liou, L.L.; Libby, R.T.; Rivkin, N.; Magen, I.; Reichenstein, I.; Oppenheim, H.; Eilam, R.; Silvestroni, A. Dysregulated mi RNA biogenesis downstream of cellular stress and ALS-causing mutations: A new mechanism for ALS. EMBO J. 2015, 34, 2633–2651. [Google Scholar] [CrossRef] [PubMed]
- Hoye, M.L.; Koval, E.D.; Wegener, A.J.; Hyman, T.S.; Yang, C.; O’Brien, D.R.; Miller, R.L.; Cole, T.; Schoch, K.M.; Shen, T. MicroRNA profiling reveals marker of motor neuron disease in ALS models. J. Neurosci. 2017, 37, 5574–5586. [Google Scholar] [CrossRef]
- Church, V.A.; Yoo, A.S. MiR-218 steps down to a threshold of motor impairment. Neuron 2021, 109, 3233–3235. [Google Scholar] [CrossRef]
- Emery, B.; Lu, Q.R. Transcriptional and epigenetic regulation of oligodendrocyte development and myelination in the central nervous system. Cold Spring Harb. Perspect. Biol. 2015, 7, a020461. [Google Scholar] [CrossRef]
- Kye, M.J.; Goncalves, I.d.C.G. The role of miRNA in motor neuron disease. Front. Cell. Neurosci. 2014, 8, 15. [Google Scholar] [CrossRef]
- Chalei, V.; Sansom, S.N.; Kong, L.; Lee, S.; Montiel, J.F.; Vance, K.W.; Ponting, C.P. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife 2014, 3, e04530. [Google Scholar] [CrossRef]
- Ng, S.-Y.; Bogu, G.K.; Soh, B.S.; Stanton, L.W. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 2013, 51, 349–359. [Google Scholar] [CrossRef]
- Vance, K.W.; Sansom, S.N.; Lee, S.; Chalei, V.; Kong, L.; Cooper, S.E.; Oliver, P.L.; Ponting, C.P. The long non-coding RNA P aupar regulates the expression of both local and distal genes. EMBO J. 2014, 33, 296–311. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-G.; Liao, Z.; Xiao, H.; Liu, H.; Hu, Y.-H.; Liao, Q.-D.; Zhong, D. LncRNA KCNQ1OT1 promoted BMP2 expression to regulate osteogenic differentiation by sponging miRNA-214. Exp. Mol. 2019, 107, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Biscarini, S.; Capauto, D.; Peruzzi, G.; Lu, L.; Colantoni, A.; Santini, T.; Shneider, N.A.; Caffarelli, E.; Laneve, P.; Bozzoni, I. Characterization of the lncRNA transcriptome in mESC-derived motor neurons: Implications for FUS-ALS. Stem Cell Res. 2018, 27, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Ray, M.K.; Wiskow, O.; King, M.J.; Ismail, N.; Ergun, A.; Wang, Y.; Plys, A.J.; Davis, C.P.; Kathrein, K.; Sadreyev, R. CAT7 and cat7l long non-coding RNAs tune polycomb repressive complex 1 function during human and zebrafish development. J. Biol. Chem. 2016, 291, 19558–19572. [Google Scholar] [CrossRef]
- Quan, Y.; Wang, J.; Wang, S.; Zhao, J. Association of the plasma long non-coding RNA MEG3 with Parkinson’s disease. Front. Neurol. 2020, 11, 532891. [Google Scholar] [CrossRef]
- Yan, H.; Rao, J.; Yuan, J.; Gao, L.; Huang, W.; Zhao, L.; Ren, J. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate ischemic neuronal death by targeting miR-21/PDCD4 signaling pathway. Cell Death Dis. 2017, 8, 3211. [Google Scholar] [CrossRef]
- Yen, Y.-P.; Hsieh, W.-F.; Tsai, Y.-Y.; Lu, Y.-L.; Liau, E.S.; Hsu, H.-C.; Chen, Y.-C.; Liu, T.-C.; Chang, M.; Li, J. Dlk1-Dio3 locus-derived lncRNAs perpetuate postmitotic motor neuron cell fate and subtype identity. Elife 2018, 7, e38080. [Google Scholar] [CrossRef]
- Naganuma, T.; Nakagawa, S.; Tanigawa, A.; Sasaki, Y.F.; Goshima, N.; Hirose, T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012, 31, 4020–4034. [Google Scholar] [CrossRef]
- Nishimoto, Y.; Nakagawa, S.; Hirose, T.; Okano, H.J.; Takao, M.; Shibata, S.; Suyama, S.; Kuwako, K.-I.; Imai, T.; Murayama, S. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain 2013, 6, 31. [Google Scholar] [CrossRef]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Shelkovnikova, T.A.; Kukharsky, M.S.; An, H.; Dimasi, P.; Alexeeva, S.; Shabir, O.; Heath, P.R.; Buchman, V.L. Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 2018, 13, 30. [Google Scholar] [CrossRef]
- Suzuki, H.; Shibagaki, Y.; Hattori, S.; Matsuoka, M. C9-ALS/FTD-linked proline–arginine dipeptide repeat protein associates with paraspeckle components and increases paraspeckle formation. Cell Death Dis. 2019, 10, 746. [Google Scholar] [CrossRef]
- Ding, B.; Cave, J.W.; Dobner, P.R.; Mullikin-Kilpatrick, D.; Bartzokis, M.; Zhu, H.; Chow, C.W.; Gronostajski, R.M.; Kilpatrick, D.L. Reciprocal autoregulation by NFI occupancy and ETV1 promotes the developmental expression of dendrite-synapse genes in cerebellar granule neurons. Mol. Biol. Cell. 2016, 27, 1488–1499. [Google Scholar] [CrossRef]
- Ding, B.; Wang, W.; Selvakumar, T.; Xi, H.S.; Zhu, H.; Chow, C.W.; Horton, J.D.; Gronostajski, R.M.; Kilpatrick, D.L. Temporal regulation of nuclear factor one occupancy by calcineurin/NFAT governs a voltage-sensitive developmental switch in late maturing neurons. J. Neurosci. 2013, 33, 2860–2872. [Google Scholar] [CrossRef]
- Ding, B.; Kilpatrick, D.L. Lentiviral vector production, titration, and transduction of primary neurons. Methods Mol. Biol. 2013, 1018, 119–131. [Google Scholar] [CrossRef]
- Hulme, A.J.; Maksour, S.; Glover, M.S.-C.; Miellet, S.; Dottori, M. Making neurons, made easy: The use of Neurogenin-2 in neuronal differentiation. Stem Cell Rep. 2021, 17, 13–14. [Google Scholar] [CrossRef]
- Gong, J.; Hu, S.; Huang, Z.; Hu, Y.; Wang, X.; Zhao, J.; Qian, P.; Wang, C.; Sheng, J.; Lu, X. The requirement of Sox2 for the spinal cord motor neuron development of Zebrafish. Front. Mol. Neurosci. 2020, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Bhati, M.; Llamosas, E.; Jacques, D.A.; Jeffries, C.M.; Dastmalchi, S.; Ripin, N.; Nicholas, H.R.; Matthews, J.M. Interactions between LHX3-and ISL1-family LIM-homeodomain transcription factors are conserved in Caenorhabditis elegans. Sci. Rep. 2017, 7, 4579. [Google Scholar] [CrossRef] [PubMed]
- Rao, Z.; Wang, R.; Li, S.; Shi, Y.; Mo, L.; Yuan, J.; Jing, N.; Cheng, L. Molecular mechanisms underlying ascl1-mediated astrocyte-to-neuron conversion. Stem Cell Rep. 2021, 16, 534–547. [Google Scholar] [CrossRef] [PubMed]
- El Wazan, L.; Urrutia-Cabrera, D.; Wong, R.C.-B. Using transcription factors for direct reprogramming of neurons in vitro. World J. Stem Cells 2019, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Son, E.Y.; Ichida, J.K.; Wainger, B.J.; Toma, J.S.; Rafuse, V.F.; Woolf, C.J.; Eggan, K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 2011, 9, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-K.; Pfaff, S.L. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron 2003, 38, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, E.O.; Mahony, S.; Closser, M.; Morrison, C.A.; Nedelec, S.; Williams, D.J.; An, D.; Gifford, D.K.; Wichterle, H. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 2013, 16, 1219–1227. [Google Scholar] [CrossRef]
- Pfaff, S.L.; Mendelsohn, M.; Stewart, C.L.; Edlund, T.; Jessell, T.M. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell 1996, 84, 309–320. [Google Scholar] [CrossRef]
- Hester, M.E.; Murtha, M.J.; Song, S.; Rao, M.; Miranda, C.J.; Meyer, K.; Tian, J.; Boulting, G.; Schaffer, D.V.; Zhu, M.X. Rapid and efficient generation of functional motor neurons from human pluripotent stem cells using gene delivered transcription factor codes. Mol. Ther. 2011, 19, 1905–1912. [Google Scholar] [CrossRef]
- Lee, S.; Cuvillier, J.M.; Lee, B.; Shen, R.; Lee, J.W.; Lee, S.-K. Fusion protein Isl1-Lhx3 specifies motor neuron fate by inducing motor neuron genes and concomitantly suppressing the interneuron programs. Proc. Natl. Acad. Sci. USA 2012, 109, 3383–3388. [Google Scholar] [CrossRef]
- Lee, S.; Shen, R.; Cho, H.-H.; Kwon, R.-J.; Seo, S.Y.; Lee, J.W.; Lee, S.-K. STAT3 promotes motor neuron differentiation by collaborating with motor neuron-specific LIM complex. Proc. Natl. Acad. Sci. USA 2013, 110, 11445–11450. [Google Scholar] [CrossRef]
- Erb, M.; Lee, B.; Seo, S.Y.; Lee, J.W.; Lee, S.; Lee, S.-K. The Isl1-Lhx3 complex promotes motor neuron specification by activating transcriptional pathways that enhance its own expression and formation. eNeuro 2017, 4, ENEURO.0349-16.2017. [Google Scholar] [CrossRef]
- Kim, N.; Park, C.; Jeong, Y.; Song, M.-R. Functional diversification of motor neuron-specific Isl1 enhancers during evolution. PLoS Genet. 2015, 11, e1005560. [Google Scholar] [CrossRef]
- Lee, S.; Lee, B.; Joshi, K.; Pfaff, S.L.; Lee, J.W.; Lee, S.-K. A regulatory network to segregate the identity of neuronal subtypes. Dev. Cell 2008, 14, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Thaler, J.P.; Lee, S.-K.; Jurata, L.W.; Gill, G.N.; Pfaff, S.L. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell 2002, 110, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Kania, A. Concocting cholinergy. PLoS Genet. 2014, 10, e1004313. [Google Scholar] [CrossRef]
- Patel, T.; Hammelman, J.; Closser, M.; Gifford, D.K.; Wichterle, H. General and cell-type-specific aspects of the motor neuron maturation transcriptional program. bioRxiv 2021. [Google Scholar] [CrossRef]
- Coppola, E.; Pattyn, A.; Guthrie, S.C.; Goridis, C.; Studer, M. Reciprocal gene replacements reveal unique functions for Phox2 genes during neural differentiation. EMBO J. 2005, 24, 4392–4403. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, M.-R.; Glover, J.C.; Dufour, H.D.; Brunet, J.-F.; Goridis, C. Forced expression of Phox2 homeodomain transcription factors induces a branchio-visceromotor axonal phenotype. Dev. Biol. 2007, 303, 687–702. [Google Scholar] [CrossRef]
- Pattyn, A.; Morin, X.; Cremer, H.; Goridis, C.; Brunet, J.-F. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development 1997, 124, 4065–4075. [Google Scholar] [CrossRef]
- Goto, K.; Imamura, K.; Komatsu, K.; Mitani, K.; Aiba, K.; Nakatsuji, N.; Inoue, M.; Kawata, A.; Yamashita, H.; Takahashi, R. Simple derivation of spinal motor neurons from ESCs/iPSCs using sendai virus vectors. Mol. Ther. Methods Clin. Dev. 2017, 4, 115–125. [Google Scholar] [CrossRef]
- Limone, F.; Mitchell, J.M.; San Juan, I.G.; Smith, J.L.; Raghunathan, K.; Couto, A.; Ghosh, S.D.; Meyer, D.; Mello, C.J.; Nemesh, J. Efficient generation of lower induced Motor Neurons by coupling Ngn2 expression with developmental cues. bioRxiv 2022. [Google Scholar] [CrossRef]
- Lee, H.; Lee, H.Y.; Lee, B.E.; Gerovska, D.; Park, S.Y.; Zaehres, H.; Araúzo-Bravo, M.J.; Kim, J.-I.; Ha, Y.; Schöler, H.R. Sequentially induced motor neurons from human fibroblasts facilitate locomotor recovery in a rodent spinal cord injury model. Elife 2020, 9, e52069. [Google Scholar] [CrossRef] [PubMed]
- Garone, M.G.; de Turris, V.; Soloperto, A.; Brighi, C.; De Santis, R.; Pagani, F.; Di Angelantonio, S.; Rosa, A. Conversion of human induced pluripotent stem cells (iPSCs) into functional spinal and cranial motor neurons using PiggyBac vectors. JoVE J. Vis. Exp. 2019, 1, e59321. [Google Scholar] [CrossRef] [PubMed]
- Cantor, E.L.; Shen, F.; Jiang, G.; Tan, Z.; Cunningham, G.M.; Wu, X.; Philips, S.; Schneider, B.P. Passage number affects differentiation of sensory neurons from human induced pluripotent stem cells. Sci. Rep. 2022, 12, 15869. [Google Scholar] [CrossRef] [PubMed]
- Baghbaderani, B.A.; Syama, A.; Sivapatham, R.; Pei, Y.; Mukherjee, O.; Fellner, T.; Zeng, X.; Rao, M.S. Detailed characterization of human induced pluripotent stem cells manufactured for therapeutic applications. Stem Cell Rev. Rep. 2016, 12, 394–420. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.; Wanczyk, H.; Jensen, T.; Finck, C. Assessment of iPSC teratogenicity throughout directed differentiation toward an alveolar-like phenotype. Differentiation 2019, 105, 45–53. [Google Scholar] [CrossRef]
- Nelakanti, R.V.; Kooreman, N.G.; Wu, J.C. Teratoma formation: A tool for monitoring pluripotency in stem cell research. Curr. Protoc. Stem Cell Biol. 2015, 32, 4A.8.1–4A.8.17. [Google Scholar] [CrossRef]
- Thaler, J.; Harrison, K.; Sharma, K.; Lettieri, K.; Kehrl, J.; Pfaff, S.L. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron 1999, 23, 675–687. [Google Scholar] [CrossRef]
- Geula, C.; Schatz, C.; Mesulam, M.-M. Differential localization of NADPH-diaphorase and calbindin-D28k within the cholinergic neurons of the basal forebrain, striatum and brainstem in the rat, monkey, baboon and human. Neuroscience 1993, 54, 461–476. [Google Scholar] [CrossRef]
- Egawa, N.; Kitaoka, S.; Tsukita, K.; Naitoh, M.; Takahashi, K.; Yamamoto, T.; Adachi, F.; Kondo, T.; Okita, K.; Asaka, I. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci. Transl. Med. 2012, 4, ra104–ra145. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Kanning, K.C.; Kaplan, A.; Henderson, C.E. Motor neuron diversity in development and disease. Annu. Rev. Neurosci. 2010, 33, 409–440. [Google Scholar] [CrossRef]
- Kiskinis, E.; Sandoe, J.; Williams, L.A.; Boulting, G.L.; Moccia, R.; Wainger, B.J.; Han, S.; Peng, T.; Thams, S.; Mikkilineni, S. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell 2014, 14, 781–795. [Google Scholar] [CrossRef]
- Li, X.-J.; Du, Z.-W.; Zarnowska, E.D.; Pankratz, M.; Hansen, L.O.; Pearce, R.A.; Zhang, S.-C. Specification of motoneurons from human embryonic stem cells. Nat. Biotechnol. 2005, 23, 215–221. [Google Scholar] [CrossRef]
- Nijssen, J.; Comley, L.H.; Hedlund, E. Motor neuron vulnerability and resistance in amyotrophic lateral sclerosis. Acta Neuropathol. 2017, 133, 863–885. [Google Scholar] [CrossRef]
- Faravelli, I.; Bucchia, M.; Rinchetti, P.; Nizzardo, M.; Simone, C.; Frattini, E.; Corti, S. Motor neuron derivation from human embryonic and induced pluripotent stem cells: Experimental approaches and clinical perspectives. Stem Cell Res. Ther. 2014, 5, 87. [Google Scholar] [CrossRef] [PubMed]
- Sances, S.; Bruijn, L.I.; Chandran, S.; Eggan, K.; Ho, R.; Klim, J.R.; Livesey, M.R.; Lowry, E.; Macklis, J.D.; Rushton, D. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat. Neurosci. 2016, 19, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Lam, D.; Enright, H.A.; Cadena, J.; Peters, S.K.; Sales, A.P.; Osburn, J.J.; Soscia, D.A.; Kulp, K.S.; Wheeler, E.K.; Fischer, N.O. Tissue-specific extracellular matrix accelerates the formation of neural networks and communities in a neuron-glia co-culture on a multi-electrode array. Sci. Rep. 2019, 9, 4159. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Meng, T.; Yang, J.; Hu, N.; Zhao, H.; Tian, T. Three-dimensional in vitro tissue culture models of brain organoids. Exp. Neurol. 2021, 339, 113619. [Google Scholar] [CrossRef]
- Kampmann, M. CRISPR-based functional genomics for neurological disease. Nat. Rev. Neurol. 2020, 16, 465–480. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).