The Predicted Splicing Variant c.11+5G>A in RPE65 Leads to a Reduction in mRNA Expression in a Cell-Specific Manner
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Cell Lines
2.3. In Silico Studies
2.4. Generation of Vectors
2.5. Midigene Splicing Assays
2.6. Luciferase Assays
2.7. Induced Pluripotent Stem Cell (iPSC) Differentiation into Photoreceptor Precursor Cells (PPCs)
2.8. Induced Pluripotent Stem Cell (iPSC) Differentiation into RPE Cells
2.9. Quantitative PCR (qPCR)
2.10. Immunochemistry Assays
2.11. Cell-Size Measurement
2.12. Western Blot
2.13. Immunoprecipitation
2.14. In-Gel Digestion and Proteomic Analysis
2.15. Analysis of Proteomic Data
2.16. PACBIO Long-Read Genome Sequencing and Analysis
2.17. Statistical Studies
3. Results
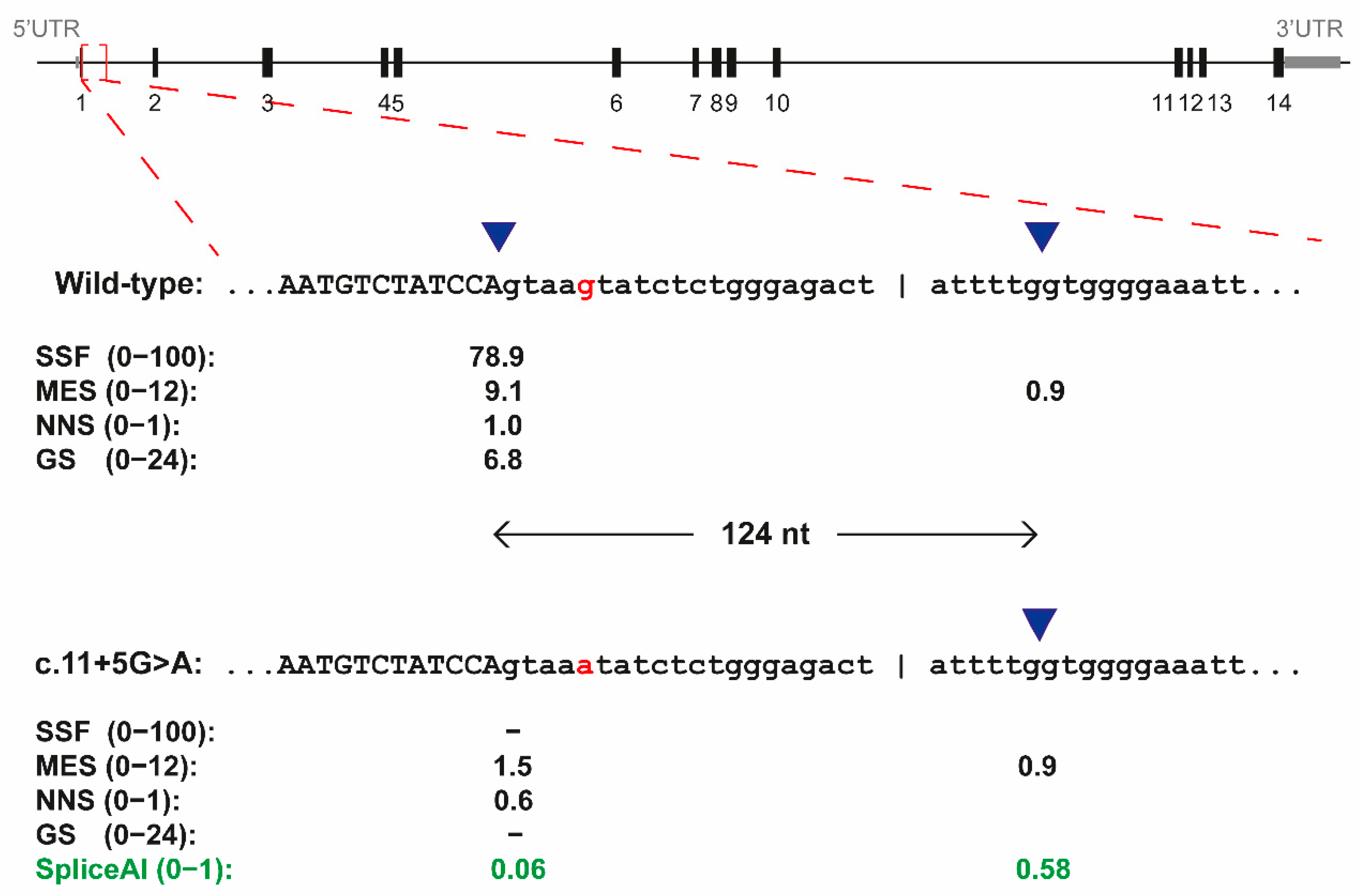
3.1. The c.11+5G>A Variant of RPE65 Results in Exon Elongation In Vitro
3.2. The c.11+5G>A Variant-Mediated Splicing Defect Is Less Noticeable in Patient-Derived iPSCs and Photoreceptor Precursor Cells
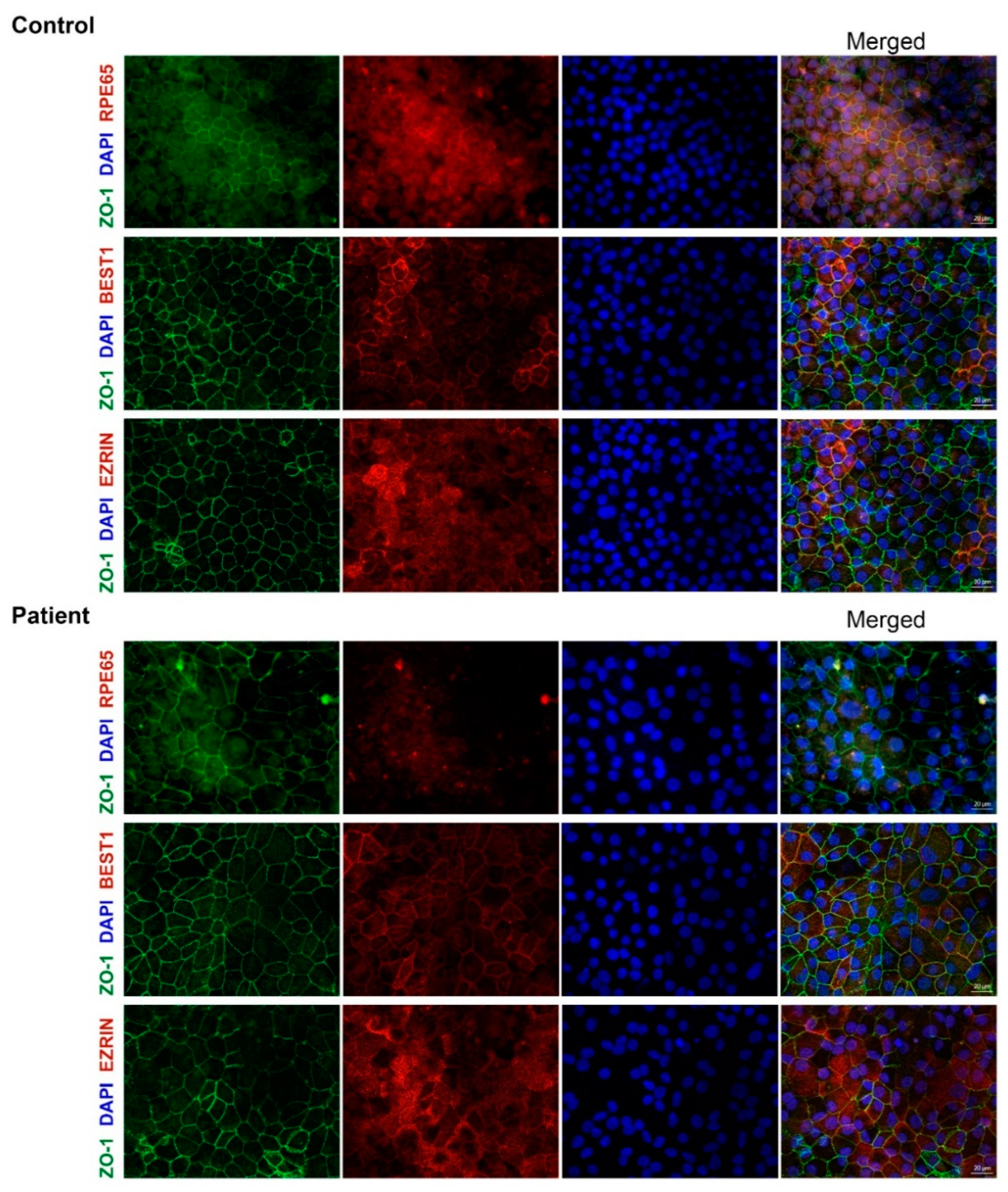
3.3. The Variant c.11+5G>A reduces mRNA Expression Resulting in RPE65 Absence in RPE Cultures
3.4. Long-Read Genome Sequencing Does Not Point Other Causative Variant in The Patient-Derived DNA
3.5. The c.11+5G>A Variant-Mediated mRNA Expression Reduction Is Cell-Context Dependent
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vazquez-Dominguez, I.; Garanto, A.; Collin, R.W.J. Molecular Therapies for Inherited Retinal Diseases-Current Standing, Opportunities and Challenges. Genes 2019, 10, 654. [Google Scholar] [CrossRef] [PubMed]
- Sohocki, M.M.; Daiger, S.P.; Bowne, S.J.; Rodriquez, J.A.; Northrup, H.; Heckenlively, J.R.; Birch, D.G.; Mintz-Hittner, H.; Ruiz, R.S.; Lewis, R.A.; et al. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum. Mutat. 2001, 17, 42–51. [Google Scholar] [CrossRef]
- Strauss, O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005, 85, 845–881. [Google Scholar] [CrossRef] [PubMed]
- Tombran-Tink, J.; Shivaram, S.M.; Chader, G.J.; Johnson, L.V.; Bok, D. Expression, secretion, and age-related downregulation of pigment epithelium-derived factor, a serpin with neurotrophic activity. J. Neurosci. 1995, 15, 4992–5003. [Google Scholar] [CrossRef]
- Hollyfield, J.G.; Varner, H.H.; Rayborn, M.E. Regional variation within the interphotoreceptor matrix from fovea to the retinal periphery. Eye 1990, 4 Pt 2, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, F.; Safa, H.; Finnemann, S.C. Understanding photoreceptor outer segment phagocytosis: Use and utility of RPE cells in culture. Exp. Eye Res. 2014, 126, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Marlhens, F.; Bareil, C.; Griffoin, J.M.; Zrenner, E.; Amalric, P.; Eliaou, C.; Liu, S.Y.; Harris, E.; Redmond, T.M.; Arnaud, B.; et al. Mutations in RPE65 cause Leber’s congenital amaurosis. Nat. Genet. 1997, 17, 139–141. [Google Scholar] [CrossRef]
- Cai, X.; Conley, S.M.; Naash, M.I. RPE65: Role in the visual cycle, human retinal disease, and gene therapy. Ophthalmic Genet. 2009, 30, 57–62. [Google Scholar] [CrossRef]
- Miraldi Utz, V.; Coussa, R.G.; Antaki, F.; Traboulsi, E.I. Gene therapy for RPE65-related retinal disease. Ophthalmic Genet. 2018, 39, 671–677. [Google Scholar] [CrossRef]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Evans, K.; Hayden, M.; Heywood, S.; Hussain, M.; Phillips, A.D.; Cooper, D.N. The Human Gene Mutation Database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017, 136, 665–677. [Google Scholar] [CrossRef]
- Cremers, F.P.; van den Hurk, J.A.; den Hollander, A.I. Molecular genetics of Leber congenital amaurosis. Hum. Mol. Genet. 2002, 11, 1169–1176. [Google Scholar] [CrossRef] [PubMed]
- Tsang, S.H.; Sharma, T. Leber Congenital Amaurosis. In Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2018; Volume 1085, pp. 131–137. [Google Scholar] [CrossRef]
- Huang, C.H.; Yang, C.M.; Yang, C.H.; Hou, Y.C.; Chen, T.C. Leber’s Congenital Amaurosis: Current Concepts of Genotype-Phenotype Correlations. Genes 2021, 12, 1261. [Google Scholar] [CrossRef] [PubMed]
- Morimura, H.; Fishman, G.A.; Grover, S.A.; Fulton, A.B.; Berson, E.L.; Dryja, T.P. Mutations in the RPE65 gene in patients with autosomal recessive retinitis pigmentosa or leber congenital amaurosis. Proc. Natl. Acad. Sci. USA 1998, 95, 3088–3093. [Google Scholar] [CrossRef] [PubMed]
- Janecke, A.R.; Verma, A.; Perumalsamy, V.; Shetty, S.; Kulm, M.; Sundaresan, P. Mutational Screening of LCA Genes Emphasizing RPE65 in South Indian Cohort of Patients. PLoS ONE 2013, 8, e73172. [Google Scholar] [CrossRef][Green Version]
- Chen, Y.; Zhang, Q.; Shen, T.; Xiao, X.; Li, S.; Guan, L.; Zhang, J.; Zhu, Z.; Yin, Y.; Wang, P.; et al. Comprehensive mutation analysis by whole-exome sequencing in 41 Chinese families with Leber congenital amaurosis. Investig. Ophthalmol. Vis. Sci. 2013, 54, 4351–4357. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, M.M.; Zack, D.J. Alternative splicing and retinal degeneration. Clin. Genet. 2013, 84, 142–149. [Google Scholar] [CrossRef]
- Aisa-Marin, I.; Garcia-Arroyo, R.; Mirra, S.; Marfany, G. The Alter Retina: Alternative Splicing of Retinal Genes in Health and Disease. Int. J. Mol. Sci. 2021, 22, 1855. [Google Scholar] [CrossRef]
- Ciulla, T.A.; Hussain, R.M.; Berrocal, A.M.; Nagiel, A. Voretigene neparvovec-rzyl for treatment of RPE65-mediated inherited retinal diseases: A model for ocular gene therapy development. Expert Opin. Biol. Ther. 2020, 20, 565–578. [Google Scholar] [CrossRef]
- Katzen, F. Gateway® recombinational cloning: A biological operating system. Expert Opin. Drug Discov. 2007, 2, 571–589. [Google Scholar] [CrossRef]
- Reece-Hoyes, J.S.; Walhout, A.J.M. Gateway Recombinational Cloning. Cold Spring Harb. Protoc. 2018, 2018, pdb.top094912. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Dominguez, I.; Kwint, M.; Kroes, H.Y.; Albert, S.; O’Gorman, L.; Gilissen, C.; Cremers, F.P.M.; Collin, R.W.J.; Roosing, S.; Garanto, A. Generation of a patient-derived induced pluripotent cell line (SCTCi016-A) carrying a homozygous variant in RPE65. Stem Cell Res. 2022, 60, 102689. [Google Scholar] [CrossRef] [PubMed]
- Flamier, A.; Barabino, A.; Bernier, G. Differentiation of Human Embryonic Stem Cells into Cone Photoreceptors. Bio-Protocol 2016, 6, e1870. [Google Scholar] [CrossRef]
- Albert, S.; Garanto, A.; Sangermano, R.; Khan, M.; Bax, N.M.; Hoyng, C.B.; Zernant, J.; Lee, W.; Allikmets, R.; Collin, R.W.J.; et al. Identification and Rescue of Splice Defects Caused by Two Neighboring Deep-Intronic ABCA4 Mutations Underlying Stargardt Disease. Am. J. Hum. Genet. 2018, 102, 517–527. [Google Scholar] [CrossRef]
- Garanto, A.; Duijkers, L.; Tomkiewicz, T.Z.; Collin, R.W.J. Antisense Oligonucleotide Screening to Optimize the Rescue of the Splicing Defect Caused by the Recurrent Deep-Intronic ABCA4 Variant c.4539+2001G>A in Stargardt Disease. Genes 2019, 10, 452. [Google Scholar] [CrossRef]
- Khan, M.; Arno, G.; Fakin, A.; Parfitt, D.A.; Dhooge, P.P.A.; Albert, S.; Bax, N.M.; Duijkers, L.; Niblock, M.; Hau, K.L.; et al. Detailed Phenotyping and Therapeutic Strategies for Intronic ABCA4 Variants in Stargardt Disease. Mol. Ther. Nucleic Acids 2020, 21, 412–427. [Google Scholar] [CrossRef]
- Regent, F.; Morizur, L.; Lesueur, L.; Habeler, W.; Plancheron, A.; Ben M’Barek, K.; Monville, C. Automation of human pluripotent stem cell differentiation toward retinal pigment epithelial cells for large-scale productions. Sci. Rep. 2019, 9, 10646. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Poplin, R.; Chang, P.C.; Alexander, D.; Schwartz, S.; Colthurst, T.; Ku, A.; Newburger, D.; Dijamco, J.; Nguyen, N.; Afshar, P.T.; et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018, 36, 983–987. [Google Scholar] [CrossRef]
- Martin, M.; Patterson, M.; Garg, S.; O Fischer, S.; Pisanti, N.; Klau, G.W.; Schöenhuth, A.; Marschall, T. WhatsHap: Fast and accurate read-based phasing. bioRxiv 2016. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Sangermano, R.; Khan, M.; Cornelis, S.S.; Richelle, V.; Albert, S.; Garanto, A.; Elmelik, D.; Qamar, R.; Lugtenberg, D.; van den Born, L.I.; et al. ABCA4 midigenes reveal the full splice spectrum of all reported noncanonical splice site variants in Stargardt disease. Genome Res. 2018, 28, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Garanto, A.; Duijkers, L.; Collin, R.W. Species-dependent splice recognition of a cryptic exon resulting from a recurrent intronic CEP290 mutation that causes congenital blindness. Int. J. Mol. Sci. 2015, 16, 5285–5298. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Cremers, F.P.M. ABCA4-Associated Stargardt Disease. Klin. Mon. Augenheilkd. 2020, 237, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Cornelis, S.S.; Pozo-Valero, M.D.; Whelan, L.; Runhart, E.H.; Mishra, K.; Bults, F.; AlSwaiti, Y.; AlTalbishi, A.; De Baere, E.; et al. Resolving the dark matter of ABCA4 for 1054 Stargardt disease probands through integrated genomics and transcriptomics. Genet. Med. 2020, 22, 1235–1246. [Google Scholar] [CrossRef]
- Zou, G.; Zhang, T.; Cheng, X.; Igelman, A.D.; Wang, J.; Qian, X.; Fu, S.; Wang, K.; Koenekoop, R.K.; Fishman, G.A.; et al. Noncoding mutation in RPGRIP1 contributes to inherited retinal degenerations. Mol. Vis. 2021, 27, 95–106. [Google Scholar]
- Bellingham, J.; Davidson, A.E.; Aboshiha, J.; Simonelli, F.; Bainbridge, J.W.; Michaelides, M.; van der Spuy, J. Investigation of Aberrant Splicing Induced by AIPL1 Variations as a Cause of Leber Congenital Amaurosis. Investig. Ophthalmol. Vis. Sci. 2015, 56, 7784–7793. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bergsma, A.J.; van der Wal, E.; Broeders, M.; van der Ploeg, A.T.; Pim Pijnappel, W.W.M. Alternative Splicing in Genetic Diseases: Improved Diagnosis and Novel Treatment Options. Int. Rev. Cell Mol. Biol. 2018, 335, 85–141. [Google Scholar] [CrossRef]
- Lastella, P.; Surdo, N.C.; Resta, N.; Guanti, G.; Stella, A. In silico and in vivo splicing analysis of MLH1 and MSH2 missense mutations shows exon- and tissue-specific effects. BMC Genom. 2006, 7, 243. [Google Scholar] [CrossRef]
- Mondal, A.K.; Das, S.K.; Baldini, G.; Chu, W.S.; Sharma, N.K.; Hackney, O.G.; Zhao, J.; Grant, S.F.A.; Elbein, S.C. Genotype and Tissue-Specific Effects on Alternative Splicing of the Transcription Factor 7-Like 2 Gene in Humans. J. Clin. Endocrinol. Metab. 2010, 95, 1450–1457. [Google Scholar] [CrossRef] [PubMed]
- Boulanger, A.; Liu, S.; Henningsgaard, A.A.; Yu, S.; Redmond, T.M. The upstream region of the Rpe65 gene confers retinal pigment epithelium-specific expression in vivo and in vitro and contains critical octamer and E-box binding sites. J. Biol. Chem. 2000, 275, 31274–31282. [Google Scholar] [CrossRef] [PubMed]
- Boulanger, A.; Redmond, T.M. Expression and promoter activation of the Rpe65 gene in retinal pigment epithelium cell lines. Curr. Eye Res. 2002, 24, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Cherry, T.J.; Yang, M.G.; Harmin, D.A.; Tao, P.; Timms, A.E.; Bauwens, M.; Allikmets, R.; Jones, E.M.; Chen, R.; De Baere, E.; et al. Mapping the cis-regulatory architecture of the human retina reveals noncoding genetic variation in disease. Proc. Natl. Acad. Sci. USA 2020, 117, 9001–9012. [Google Scholar] [CrossRef]
- Li, G.; Gao, G.; Wang, P.; Song, X.; Xu, P.; Xie, B.; Zhou, T.; Pan, G.; Peng, F.; Zhang, Q.; et al. Generation and Characterization of Induced Pluripotent Stem Cells and Retinal Organoids From a Leber’s Congenital Amaurosis Patient with Novel RPE65 Mutations. Front. Mol. Neurosci. 2019, 12, 212. [Google Scholar] [CrossRef]
- Al-Ani, A.; Sunba, S.; Hafeez, B.; Toms, D.; Ungrin, M. In Vitro Maturation of Retinal Pigment Epithelium Is Essential for Maintaining High Expression of Key Functional Genes. Int. J. Mol. Sci. 2020, 21, 6066. [Google Scholar] [CrossRef]
- Bennis, A.; Jacobs, J.G.; Catsburg, L.A.E.; Ten Brink, J.B.; Koster, C.; Schlingemann, R.O.; van Meurs, J.; Gorgels, T.; Moerland, P.D.; Heine, V.M.; et al. Stem Cell Derived Retinal Pigment Epithelium: The Role of Pigmentation as Maturation Marker and Gene Expression Profile Comparison with Human Endogenous Retinal Pigment Epithelium. Stem Cell Rev. Rep. 2017, 13, 659–669. [Google Scholar] [CrossRef] [PubMed]
- German, O.L.; Buzzi, E.; Rotstein, N.P.; Rodriguez-Boulan, E.; Politi, L.E. Retinal pigment epithelial cells promote spatial reorganization and differentiation of retina photoreceptors. J. Neurosci. Res. 2008, 86, 3503–3514. [Google Scholar] [CrossRef]
- Sheridan, C.; Boyer, N.P.; Crouch, R.K.; Koutalos, Y. RPE65 and the Accumulation of Retinyl Esters in Mouse Retinal Pigment Epithelium. Photochem. Photobiol. 2017, 93, 844–848. [Google Scholar] [CrossRef]
- Lee, H.; Chung, H.; Arnouk, H.; Lamoke, F.; Hunt, R.C.; Hrushesky, W.J.; Wood, P.A.; Lee, S.H.; Jahng, W.J. Cleavage of the retinal pigment epithelium-specific protein RPE65 under oxidative stress. Int. J. Biol. Macromol. 2010, 47, 104–108. [Google Scholar] [CrossRef]
- Hemati, N.; Feathers, K.L.; Chrispell, J.D.; Reed, D.M.; Carlson, T.J.; Thompson, D.A. RPE65 surface epitopes, protein interactions, and expression in rod- and cone-dominant species. Mol. Vis. 2005, 11, 1151–1165. [Google Scholar] [PubMed]
- Caffe, A.R.; Von Schantz, M.; Szel, A.; Voogd, J.; Van Veen, T. Distribution of Purkinje cell-specific Zebrin-II/aldolase C immunoreactivity in the mouse, rat, rabbit, and human retina. J. Comp. Neurol. 1994, 348, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Garanto, A.; Chung, D.C.; Duijkers, L.; Corral-Serrano, J.C.; Messchaert, M.; Xiao, R.; Bennett, J.; Vandenberghe, L.H.; Collin, R.W. In vitro and in vivo rescue of aberrant splicing in CEP290-associated LCA by antisense oligonucleotide delivery. Hum. Mol. Genet. 2016, 25, 2552–2563. [Google Scholar] [CrossRef] [PubMed]
- Barny, I.; Perrault, I.; Michel, C.; Goudin, N.; Defoort-Dhellemmes, S.; Ghazi, I.; Kaplan, J.; Rozet, J.M.; Gerard, X. AON-Mediated Exon Skipping to Bypass Protein Truncation in Retinal Dystrophies Due to the Recurrent CEP290 c.4723A > T Mutation. Fact or Fiction? Genes 2019, 10, 368. [Google Scholar] [CrossRef] [PubMed]
- Collin, R.W.; den Hollander, A.I.; van der Velde-Visser, S.D.; Bennicelli, J.; Bennett, J.; Cremers, F.P. Antisense Oligonucleotide (AON)-based Therapy for Leber Congenital Amaurosis Caused by a Frequent Mutation in CEP290. Mol. Ther. Nucleic Acids 2012, 1, e14. [Google Scholar] [CrossRef]
- Duijkers, L.; van den Born, L.I.; Neidhardt, J.; Bax, N.M.; Pierrache, L.H.M.; Klevering, B.J.; Collin, R.W.J.; Garanto, A. Antisense Oligonucleotide-Based Splicing Correction in Individuals with Leber Congenital Amaurosis due to Compound Heterozygosity for the c.2991+1655A>G Mutation in CEP290. Int. J. Mol. Sci. 2018, 19, 753. [Google Scholar] [CrossRef]
- Dulla, K.; Aguila, M.; Lane, A.; Jovanovic, K.; Parfitt, D.A.; Schulkens, I.; Chan, H.L.; Schmidt, I.; Beumer, W.; Vorthoren, L.; et al. Splice-Modulating Oligonucleotide QR-110 Restores CEP290 mRNA and Function in Human c.2991+1655A>G LCA10 Models. Mol. Ther. Nucleic Acids 2018, 12, 730–740. [Google Scholar] [CrossRef]
- Gerard, X.; Perrault, I.; Hanein, S.; Silva, E.; Bigot, K.; Defoort-Delhemmes, S.; Rio, M.; Munnich, A.; Scherman, D.; Kaplan, J.; et al. AON-mediated Exon Skipping Restores Ciliation in Fibroblasts Harboring the Common Leber Congenital Amaurosis CEP290 Mutation. Mol. Ther. Nucleic Acids 2012, 1, e29. [Google Scholar] [CrossRef]
- Parfitt, D.A.; Lane, A.; Ramsden, C.M.; Carr, A.J.; Munro, P.M.; Jovanovic, K.; Schwarz, N.; Kanuga, N.; Muthiah, M.N.; Hull, S.; et al. Identification and Correction of Mechanisms Underlying Inherited Blindness in Human iPSC-Derived Optic Cups. Cell Stem Cell 2016, 18, 769–781. [Google Scholar] [CrossRef]
- Bauwens, M.; Garanto, A.; Sangermano, R.; Naessens, S.; Weisschuh, N.; De Zaeytijd, J.; Khan, M.; Sadler, F.; Balikova, I.; Van Cauwenbergh, C.; et al. ABCA4-associated disease as a model for missing heritability in autosomal recessive disorders: Novel noncoding splice, cis-regulatory, structural, and recurrent hypomorphic variants. Genet. Med. 2019, 21, 1761–1771. [Google Scholar] [CrossRef]
- Sangermano, R.; Garanto, A.; Khan, M.; Runhart, E.H.; Bauwens, M.; Bax, N.M.; van den Born, L.I.; Khan, M.I.; Cornelis, S.S.; Verheij, J.; et al. Deep-intronic ABCA4 variants explain missing heritability in Stargardt disease and allow correction of splice defects by antisense oligonucleotides. Genet. Med. 2019, 21, 1751–1760. [Google Scholar] [CrossRef] [PubMed]
- Slijkerman, R.W.; Vache, C.; Dona, M.; Garcia-Garcia, G.; Claustres, M.; Hetterschijt, L.; Peters, T.A.; Hartel, B.P.; Pennings, R.J.; Millan, J.M.; et al. Antisense Oligonucleotide-based Splice Correction for USH2A-associated Retinal Degeneration Caused by a Frequent Deep-intronic Mutation. Mol. Ther. Nucleic Acids 2016, 5, e381. [Google Scholar] [CrossRef] [PubMed]
- Bonifert, T.; Gonzalez Menendez, I.; Battke, F.; Theurer, Y.; Synofzik, M.; Schols, L.; Wissinger, B. Antisense Oligonucleotide Mediated Splice Correction of a Deep Intronic Mutation in OPA1. Mol. Ther. Nucleic Acids 2016, 5, e390. [Google Scholar] [CrossRef] [PubMed]
- Garanto, A.; van der Velde-Visser, S.D.; Cremers, F.P.M.; Collin, R.W.J. Antisense Oligonucleotide-Based Splice Correction of a Deep-Intronic Mutation in CHM Underlying Choroideremia. In Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2018; Volume 1074, pp. 83–89. [Google Scholar] [CrossRef]
- Vazquez-Dominguez, I.; Li, C.H.Z.; Fadaie, Z.; Haer-Wigman, L.; Cremers, F.P.M.; Garanto, A.; Hoyng, C.B.; Roosing, S. Identification of a Complex Allele in IMPG2 as a Cause of Adult-Onset Vitelliform Macular Dystrophy. Investig. Ophthalmol. Vis. Sci. 2022, 63, 27. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez-Domínguez, I.; Duijkers, L.; Fadaie, Z.; Alaerds, E.C.W.; Post, M.A.; van Oosten, E.M.; O’Gorman, L.; Kwint, M.; Koolen, L.; Hoogendoorn, A.D.M.; et al. The Predicted Splicing Variant c.11+5G>A in RPE65 Leads to a Reduction in mRNA Expression in a Cell-Specific Manner. Cells 2022, 11, 3640. https://doi.org/10.3390/cells11223640
Vázquez-Domínguez I, Duijkers L, Fadaie Z, Alaerds ECW, Post MA, van Oosten EM, O’Gorman L, Kwint M, Koolen L, Hoogendoorn ADM, et al. The Predicted Splicing Variant c.11+5G>A in RPE65 Leads to a Reduction in mRNA Expression in a Cell-Specific Manner. Cells. 2022; 11(22):3640. https://doi.org/10.3390/cells11223640
Chicago/Turabian StyleVázquez-Domínguez, Irene, Lonneke Duijkers, Zeinab Fadaie, Eef C. W. Alaerds, Merel A. Post, Edwin M. van Oosten, Luke O’Gorman, Michael Kwint, Louet Koolen, Anita D. M. Hoogendoorn, and et al. 2022. "The Predicted Splicing Variant c.11+5G>A in RPE65 Leads to a Reduction in mRNA Expression in a Cell-Specific Manner" Cells 11, no. 22: 3640. https://doi.org/10.3390/cells11223640
APA StyleVázquez-Domínguez, I., Duijkers, L., Fadaie, Z., Alaerds, E. C. W., Post, M. A., van Oosten, E. M., O’Gorman, L., Kwint, M., Koolen, L., Hoogendoorn, A. D. M., Kroes, H. Y., Gilissen, C., Cremers, F. P. M., Collin, R. W. J., Roosing, S., & Garanto, A. (2022). The Predicted Splicing Variant c.11+5G>A in RPE65 Leads to a Reduction in mRNA Expression in a Cell-Specific Manner. Cells, 11(22), 3640. https://doi.org/10.3390/cells11223640









