The Proteome of Neuromelanin Granules in Dementia with Lewy Bodies
Abstract
1. Introduction
2. Materials and Methods
2.1. Disclosure
2.2. Ethical Statement
2.3. Subjects
2.4. Laser Microdissection
2.5. Sample Preparation
2.6. Mass Spectrometric Analysis
2.6.1. Global Mass Spectrometry
2.6.2. Targeted Mass Spectrometry
2.7. Data Analysis and Statistics
2.7.1. Global Mass Spectrometry
2.7.2. Targeted Mass Spectrometry
3. Results
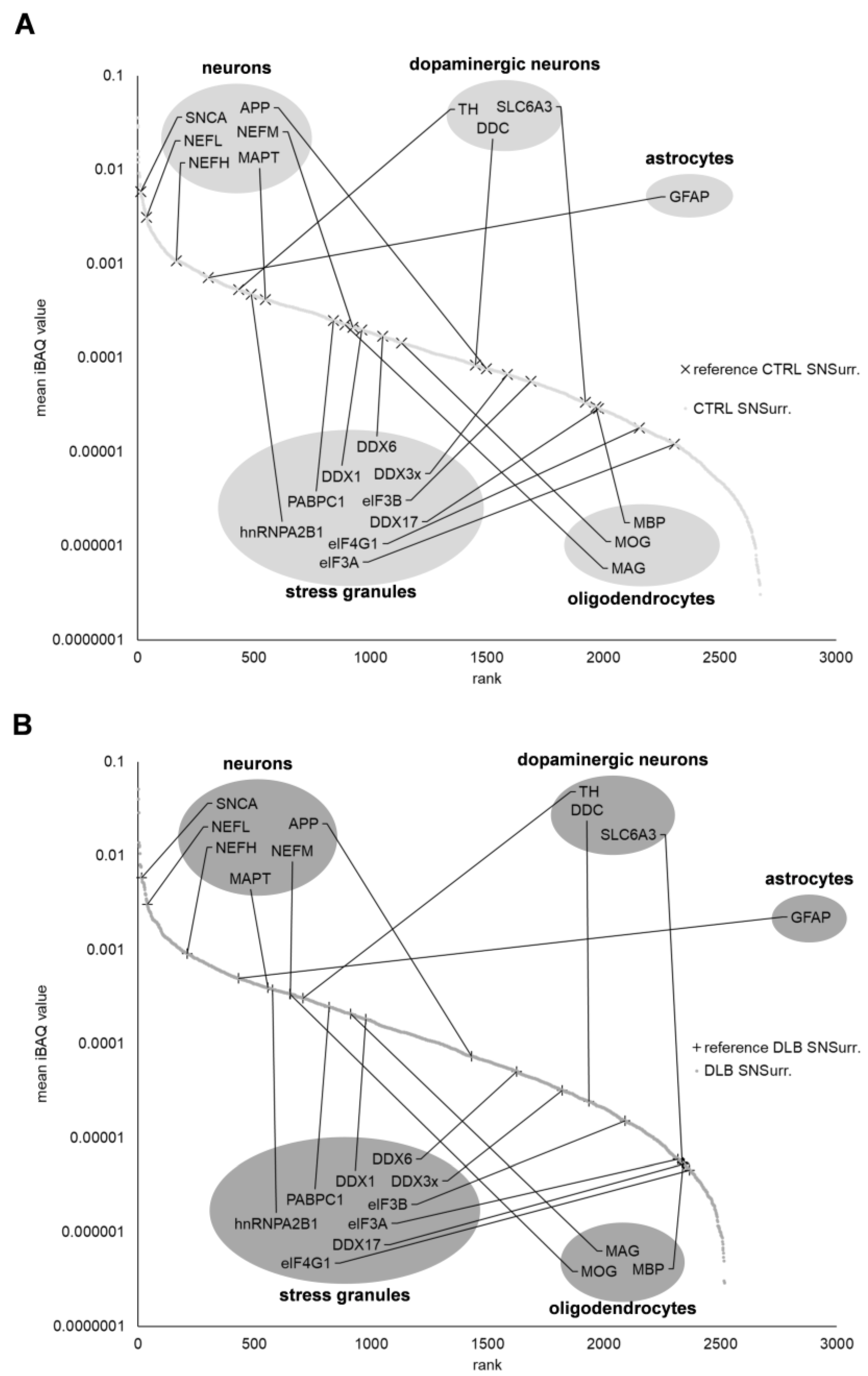
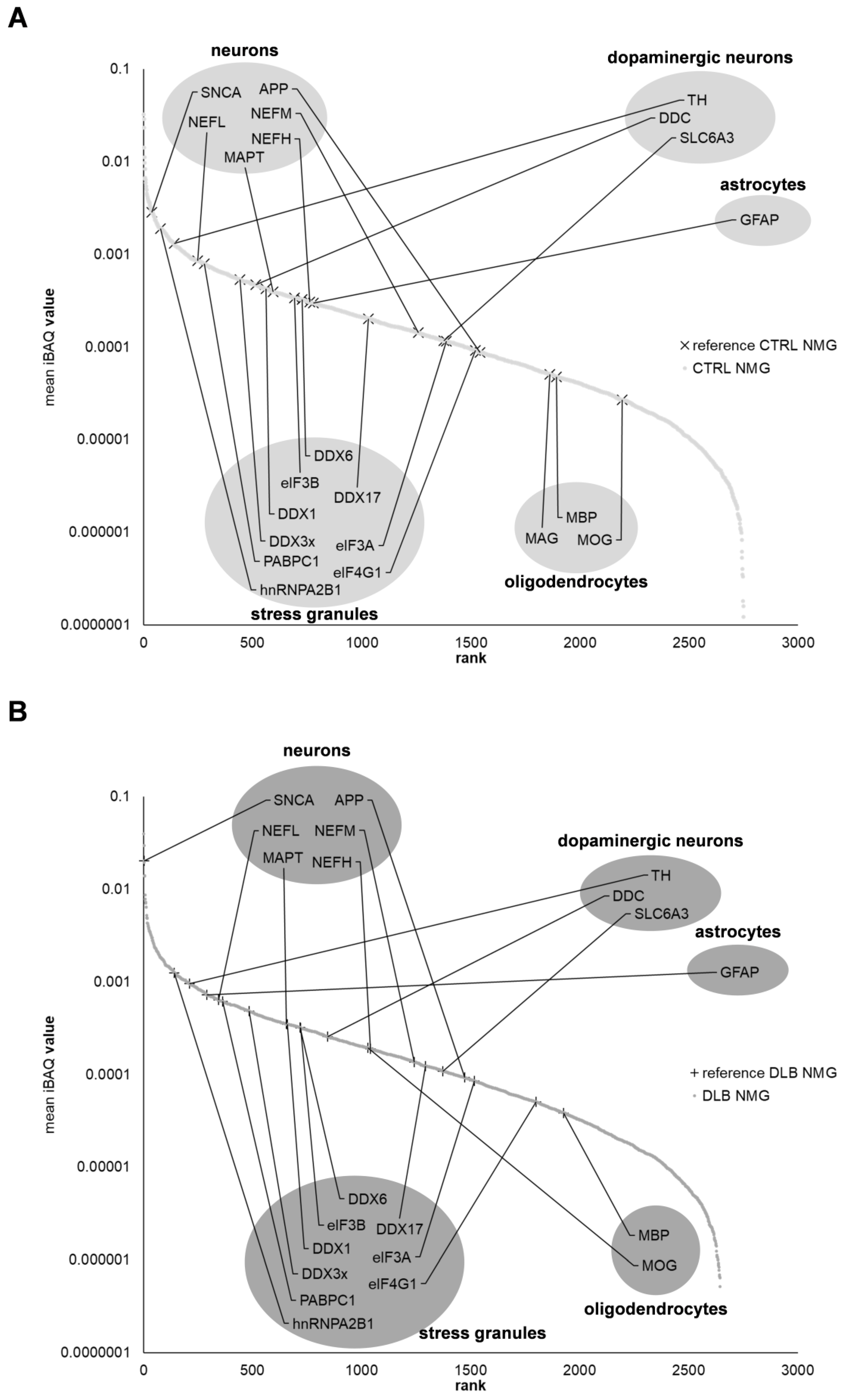
3.1. The Proteome of Neuromelanin Granules and Surrounding Tissue Remains Highly Similar under Neurodegenerative Conditions
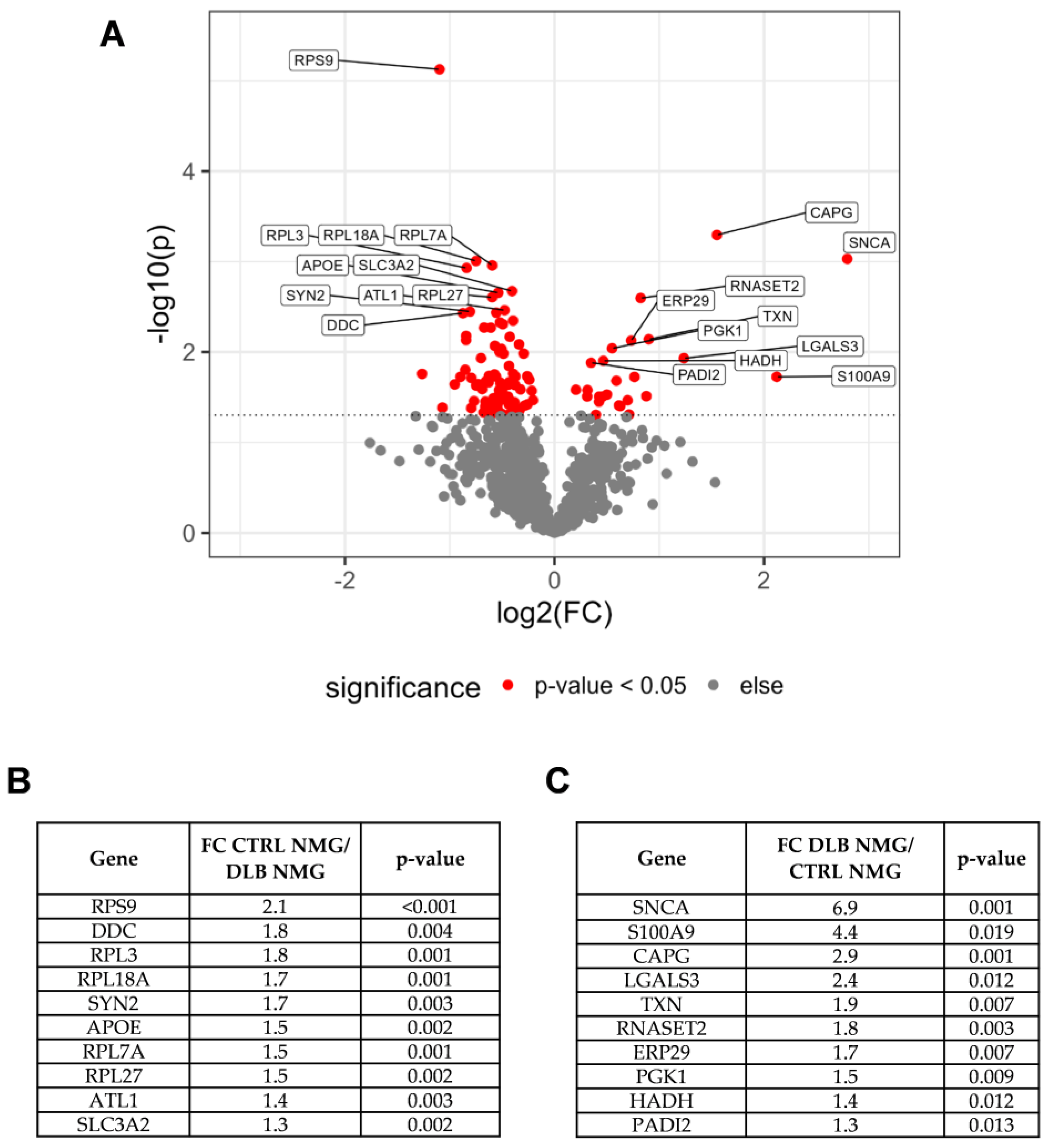
3.2. Dopaminergic Marker Proteins Are Reduced in NMG-Surrounding Tissue of DLB Cases
3.3. Impaired Protein Degradation and Oxidative Stress May Contribute to Neurodegeneration in NMG-Surrounding SN Tissue
3.4. α-Synuclein Is Highly Abundant in NMGs of DLB Cases
3.5. Alpha-Synuclein and Protein S100A9 Are Highly Abundant in NMGs of DLB Cases
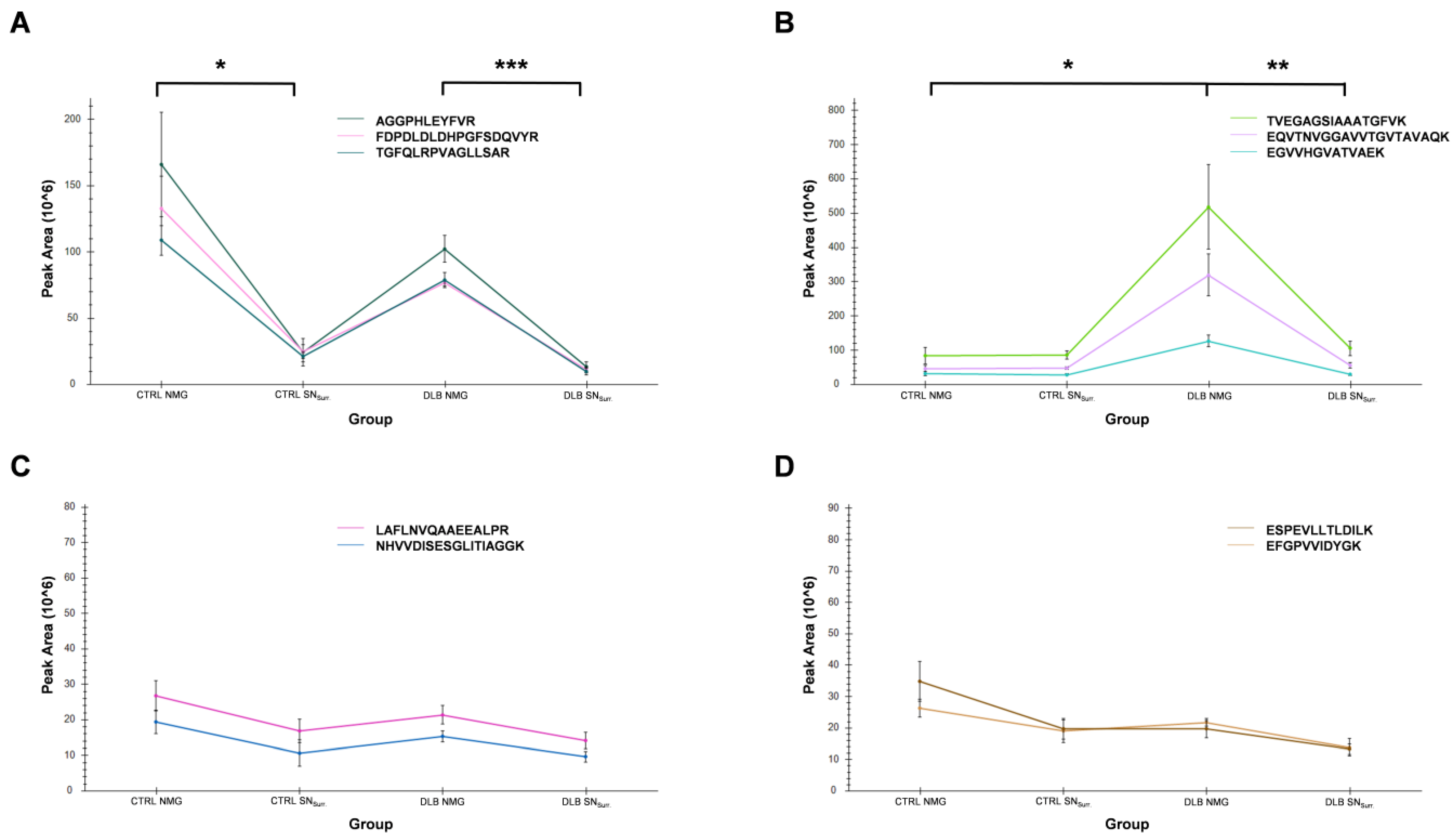
3.6. High Abundances of α-Synuclein in NMGs of DLB Cases Can Verified by Targeted Mass Spectrometry
3.7. Proteins Associated with Endosomes, Lysosomes, Ribosomes and Stress Granules Are Comparably Abundant in NMGs of Both Conditions
4. Discussion
4.1. Proteomic Profiling of NMG Surrounding Tissue Points towards Selective Loss of Dopaminergic Neurons, Response to Oxidative Stress and Impaired Protein Degradation
4.2. α-Synuclein and S100A9 Are Highly Enriched in NMGs of DLB Cases, While the Abundance of Ribosomal and Stress Granule Marker Proteins Are Decreased
4.3. NMGs May Form in a Conserved Mechanism including Endosomal, Lysosomal and Ribosomal Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hogan, D.B.; Fiest, K.M.; Roberts, J.I.; Maxwell, C.J.; Dykeman, J.; Pringsheim, T.; Steeves, T.; Smith, E.E.; Pearson, D.; Jetté, N. The Prevalence and Incidence of Dementia with Lewy Bodies: A Systematic Review. Can. J. Neurol. Sci. 2016, 43 (Suppl. S1), S83–S95. [Google Scholar] [CrossRef] [PubMed]
- Owen, A.D.; Schapira, A.H.; Jenner, P.; Marsden, C.D. Indices of oxidative stress in Parkinson’s disease, Alzheimer’s disease and dementia with Lewy bodies. J. Neural Transm. Suppl. 1997, 51, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Lyras, L.; Perry, R.H.; Perry, E.K.; Ince, P.G.; Jenner, A.; Jenner, P.; Halliwell, B. Oxidative damage to proteins, lipids, and DNA in cortical brain regions from patients with dementia with Lewy bodies. J. Neurochem. 1998, 71, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Dalfó, E.; Portero-Otín, M.; Ayala, V.; Martínez, A.; Pamplona, R.; Ferrer, I. Evidence of oxidative stress in the neocortex in incidental Lewy body disease. J. Neuropathol. Exp. Neurol. 2005, 64, 816–830. [Google Scholar] [CrossRef]
- Navarro, A.; Boveris, A.; Bández, M.J.; Sánchez-Pino, M.J.; Gómez, C.; Muntané, G.; Ferrer, I. Human brain cortex: Mitochondrial oxidative damage and adaptive response in Parkinson disease and in dementia with Lewy bodies. Free Radic. Biol. Med. 2009, 46, 1574–1580. [Google Scholar] [CrossRef]
- Spano, M.; Signorelli, M.; Vitaliani, R.; Aguglia, E.; Giometto, B. The possible involvement of mitochondrial dysfunctions in Lewy body dementia: A systematic review. Funct. Neurol. 2015, 30, 151–158. [Google Scholar] [CrossRef]
- Dickson, D.W.; Braak, H.; Duda, J.E.; Duyckaerts, C.; Gasser, T.; Halliday, G.M.; Hardy, J.; Leverenz, J.B.; Del Tredici, K.; Wszolek, Z.K.; et al. Neuropathological assessment of Parkinson’s disease: Refining the diagnostic criteria. Lancet Neurol. 2009, 8, 1150–1157. [Google Scholar] [CrossRef]
- Outeiro, T.F.; Koss, D.J.; Erskine, D.; Walker, L.; Kurzawa-Akanbi, M.; Burn, D.; Donaghy, P.; Morris, C.; Taylor, J.-P.; Thomas, A.; et al. Dementia with Lewy bodies: An update and outlook. Mol. Neurodegener. 2019, 14, 5. [Google Scholar] [CrossRef]
- Zecca, L.; Tampellini, D.; Gerlach, M.; Riederer, P.; Fariello, R.G.; Sulzer, D. Substantia nigra neuromelanin: Structure, synthesis, and molecular behaviour. Mol. Pathol. 2001, 54, 414–418. [Google Scholar]
- Vila, M. Neuromelanin, aging, and neuronal vulnerability in Parkinson’s disease. Mov. Disord. 2019, 34, 1440–1451. [Google Scholar] [CrossRef]
- Marsden, C.D. Pigmentation in the nucleus substantiae nigrae of mammals. J. Anat. 1961, 95, 256–261. [Google Scholar] [PubMed]
- Tribl, F.; Marcus, K.; Meyer, H.E.; Bringmann, G.; Gerlach, M.; Riederer, P. Subcellular proteomics reveals neuromelanin granules to be a lysosome-related organelle. J. Neural Transm. 2006, 113, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Plum, S.; Helling, S.; Theiss, C.; Leite, R.E.P.; May, C.; Jacob-Filho, W.; Eisenacher, M.; Kuhlmann, K.; Meyer, H.E.; Riederer, P.; et al. Combined enrichment of neuromelanin granules and synaptosomes from human substantia nigra pars compacta tissue for proteomic analysis. J. Proteom. 2013, 94, 202–206. [Google Scholar] [CrossRef]
- Plum, S.; Steinbach, S.; Attems, J.; Keers, S.; Riederer, P.; Gerlach, M.; May, C.; Marcus, K. Proteomic characterization of neuromelanin granules isolated from human substantia nigra by laser-microdissection. Sci. Rep. 2016, 6, 37139. [Google Scholar] [CrossRef]
- Zucca, F.A.; Vanna, R.; Cupaioli, F.A.; Bellei, C.; de Palma, A.; Di Silvestre, D.; Mauri, P.; Grassi, S.; Prinetti, A.; Casella, L.; et al. Neuromelanin organelles are specialized autolysosomes that accumulate undegraded proteins and lipids in aging human brain and are likely involved in Parkinson’s disease. NPJ Parkinsons. Dis. 2018, 4, 17. [Google Scholar] [CrossRef]
- Wulf, M.; Barkovits, K.; Schork, K.; Eisenacher, M.; Riederer, P.; Gerlach, M.; Eggers, B.; Marcus, K. Neuromelanin granules of the substantia nigra: Proteomic profile provides links to tyrosine hydroxylase, stress granules and lysosomes. J. Neural Transm. 2022, 129, 1257–1270. [Google Scholar] [CrossRef] [PubMed]
- Duffy, P.E.; Tennyson, V.M. Phase and electron microscopic observations of lewy bodies and melanin granules in the substantia nigra and locus caeruleus in parkinson’s disease. J. Neuropathol. Exp. Neurol. 1965, 24, 398–414. [Google Scholar] [CrossRef]
- Zecca, L.; Costi, P.; Mecacci, C.; Ito, S.; Terreni, M.; Sonnino, S. Interaction of human substantia nigra neuromelanin with lipids and peptides. J. Neurochem. 2000, 74, 1758–1765. [Google Scholar] [CrossRef]
- Tribl, F.; Gerlach, M.; Marcus, K.; Asan, E.; Tatschner, T.; Arzberger, T.; Meyer, H.E.; Bringmann, G.; Riederer, P. “Subcellular proteomics” of neuromelanin granules isolated from the human brain. Mol. Cell. Proteom. 2005, 4, 945–957. [Google Scholar] [CrossRef]
- Sulzer, D.; Mosharov, E.; Talloczy, Z.; Zucca, F.A.; Simon, J.D.; Zecca, L. Neuronal pigmented autophagic vacuoles: Lipofuscin, neuromelanin, and ceroid as macroautophagic responses during aging and disease. J. Neurochem. 2008, 106, 24–36. [Google Scholar] [CrossRef]
- Engelen, M.; Vanna, R.; Bellei, C.; Zucca, F.A.; Wakamatsu, K.; Monzani, E.; Ito, S.; Casella, L.; Zecca, L. Neuromelanins of human brain have soluble and insoluble components with dolichols attached to the melanic structure. PLoS ONE 2012, 7, e48490. [Google Scholar] [CrossRef] [PubMed]
- Jellinger, K.A. Interaction between α-synuclein and other proteins in neurodegenerative disorders. Sci. World J. 2011, 11, 1893–1907. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, E.; Graybiel, A.M.; Agid, Y.A. Melanized dopaminergic neurons are differentially susceptible to degeneration in Parkinson’s disease. Nature 1988, 334, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, E.C.; Graybiel, A.M.; Agid, Y. Selective vulnerability of pigmented dopaminergic neurons in Parkinson’s disease. Acta Neurol. Scand. Suppl. 1989, 126, 19–22. [Google Scholar] [CrossRef]
- Kastner, A.; Hirsch, E.C.; Lejeune, O.; Javoy-Agid, F.; Rascol, O.; Agid, Y. Is the vulnerability of neurons in the substantia nigra of patients with Parkinson’s disease related to their neuromelanin content? J. Neurochem. 1992, 59, 1080–1089. [Google Scholar] [CrossRef]
- Liang, C.-L.; Nelson, O.; Yazdani, U.; Pasbakhsh, P.; German, D.C. Inverse relationship between the contents of neuromelanin pigment and the vesicular monoamine transporter-2: Human midbrain dopamine neurons. J. Comp. Neurol. 2004, 473, 97–106. [Google Scholar] [CrossRef]
- Zecca, L.; Stroppolo, A.; Gatti, A.; Tampellini, D.; Toscani, M.; Gallorini, M.; Giaveri, G.; Arosio, P.; Santambrogio, P.; Fariello, R.G.; et al. The role of iron and copper molecules in the neuronal vulnerability of locus coeruleus and substantia nigra during aging. Proc. Natl. Acad. Sci. USA 2004, 101, 9843–9848. [Google Scholar] [CrossRef]
- Zucca, F.A.; Bellei, C.; Giannelli, S.; Terreni, M.R.; Gallorini, M.; Rizzio, E.; Pezzoli, G.; Albertini, A.; Zecca, L. Neuromelanin and iron in human locus coeruleus and substantia nigra during aging: Consequences for neuronal vulnerability. J. Neural Transm. 2006, 113, 757–767. [Google Scholar] [CrossRef]
- Zaccaria, A.; Antinori, P.; Licker, V.; Kövari, E.; Lobrinus, J.A.; Burkhard, P.R. Multiomic Analyses of Dopaminergic Neurons Isolated from Human Substantia Nigra in Parkinson’s Disease: A Descriptive and Exploratory Study. Cell. Mol. Neurobiol. 2021, 42, 2805–2818. [Google Scholar] [CrossRef]
- Petyuk, V.A.; Yu, L.; Olson, H.M.; Yu, F.; Clair, G.; Qian, W.-J.; Shulman, J.M.; Bennett, D.A. Proteomic Profiling of the Substantia Nigra to Identify Determinants of Lewy Body Pathology and Dopaminergic Neuronal Loss. J. Proteome Res. 2021, 20, 2266–2282. [Google Scholar] [CrossRef]
- McKeith, I.G.; Boeve, B.F.; Dickson, D.W.; Halliday, G.; Taylor, J.-P.; Weintraub, D.; Aarsland, D.; Galvin, J.; Attems, J.; Ballard, C.G.; et al. Diagnosis and management of dementia with Lewy bodies: Fourth consensus report of the DLB Consortium. Neurology 2017, 89, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Jellinger, K.A. Is Braak staging valid for all types of Parkinson’s disease? J. Neural Transm. 2019, 126, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Wulf, M.; Barkovits-Boeddinghaus, K.; Sommer, P.; Schork, K.; Eisenacher, M.; Riederer, P.; Gerlach, M.; Kösters, S.; Eggers, B.; Marcus, K. Laser Microdissection-Based Protocol for the LC-MS/MS Analysis of the Proteomic Profile of Neuromelanin Granules. JoVE 2021, 178, e63289. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Michalski, A.; Mann, M. Software lock mass by two-dimensional minimization of peptide mass errors. J. Am. Soc. Mass Spectrom. 2011, 22, 1373–1380. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2020.
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [PubMed]
- Anger, A.M.; Armache, J.-P.; Berninghausen, O.; Habeck, M.; Subklewe, M.; Wilson, D.N.; Beckmann, R. Structures of the human and Drosophila 80S ribosome. Nature 2013, 497, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Iwata-Ichikawa, E.; Kondo, Y.; Miyazaki, I.; Asanuma, M.; Ogawa, N. Glial cells protect neurons against oxidative stress via transcriptional up-regulation of the glutathione synthesis. J. Neurochem. 1999, 72, 2334–2344. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, D.A.; Moellering, D.R.; Iles, K.E.; Patel, R.P.; Levonen, A.-L.; Wigley, A.; Darley-Usmar, V.M.; Forman, H.J. Cytoprotection against oxidative stress and the regulation of glutathione synthesis. Biol. Chem. 2003, 384, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, K.; Watabe, M.; Nakaki, T. Regulation of neuronal glutathione synthesis. J. Pharmacol. Sci. 2008, 108, 227–238. [Google Scholar] [CrossRef]
- Zhang, C.; Saunders, A.J. An emerging role for Ubiquilin 1 in regulating protein quality control system and in disease pathogenesis. Discov. Med. 2009, 8, 18–22. [Google Scholar]
- Wareham, L.K.; Liddelow, S.A.; Temple, S.; Benowitz, L.I.; Di Polo, A.; Wellington, C.; Goldberg, J.L.; He, Z.; Duan, X.; Bu, G.; et al. Solving neurodegeneration: Common mechanisms and strategies for new treatments. Mol. Neurodegener. 2022, 17, 23. [Google Scholar] [CrossRef]
- Jellinger, K.A. Basic mechanisms of neurodegeneration: A critical update. J. Cell. Mol. Med. 2010, 14, 457–487. [Google Scholar] [CrossRef]
- Moujaber, O.; Mahboubi, H.; Kodiha, M.; Bouttier, M.; Bednarz, K.; Bakshi, R.; White, J.; Larose, L.; Colmegna, I.; Stochaj, U. Dissecting the molecular mechanisms that impair stress granule formation in aging cells. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 475–486. [Google Scholar] [CrossRef]
- Horvath, I.; Iashchishyn, I.A.; Moskalenko, R.A.; Wang, C.; Wärmländer, S.K.T.S.; Wallin, C.; Gräslund, A.; Kovacs, G.G.; Morozova-Roche, L.A. Co-aggregation of pro-inflammatory S100A9 with α-synuclein in Parkinson’s disease: Ex vivo and in vitro studies. J. Neuroinflamm. 2018, 15, 172. [Google Scholar] [CrossRef]
- Werner, T.; Horvath, I.; Wittung-Stafshede, P. Crosstalk Between Alpha-Synuclein and Other Human and Non-Human Amyloidogenic Proteins: Consequences for Amyloid Formation in Parkinson’s Disease. J. Parkinsons. Dis. 2020, 10, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Toleikis, Z.; Ziaunys, M.; Baranauskiene, L.; Petrauskas, V.; Jaudzems, K.; Smirnovas, V. S100A9 Alters the Pathway of Alpha-Synuclein Amyloid Aggregation. Int. J. Mol. Sci. 2021, 22, 7972. [Google Scholar] [CrossRef] [PubMed]
- Toleikis, Z.; Bobrovs, R.; Janoniene, A.; Lends, A.; Ziaunys, M.; Baronaite, I.; Petrauskas, V.; Kitoka, K.; Smirnovas, V.; Jaudzems, K. Interactions between S100A9 and Alpha-Synuclein: Insight from NMR Spectroscopy. Int. J. Mol. Sci. 2022, 23, 6781. [Google Scholar] [CrossRef] [PubMed]
- Halliday, G.M.; Ophof, A.; Broe, M.; Jensen, P.H.; Kettle, E.; Fedorow, H.; Cartwright, M.I.; Griffiths, F.M.; Shepherd, C.E.; Double, K.L. Alpha-synuclein redistributes to neuromelanin lipid in the substantia nigra early in Parkinson’s disease. Brain 2005, 128, 2654–2664. [Google Scholar] [CrossRef]
- Chartier-Harlin, M.-C.; Dachsel, J.C.; Vilariño-Güell, C.; Lincoln, S.J.; Leprêtre, F.; Hulihan, M.M.; Kachergus, J.; Milnerwood, A.J.; Tapia, L.; Song, M.-S.; et al. Translation initiator EIF4G1 mutations in familial Parkinson disease. Am. J. Hum. Genet. 2011, 89, 398–406. [Google Scholar] [CrossRef]
- Fujioka, S.; Sundal, C.; Strongosky, A.J.; Castanedes, M.C.; Rademakers, R.; Ross, O.A.; Vilariño-Güell, C.; Farrer, M.J.; Wszolek, Z.K.; Dickson, D.W. Sequence variants in eukaryotic translation initiation factor 4-gamma (eIF4G1) are associated with Lewy body dementia. Acta Neuropathol. 2013, 125, 425–438. [Google Scholar] [CrossRef]
- Deng, H.; Wu, Y.; Jankovic, J. The EIF4G1 gene and Parkinson’s disease. Acta Neurol. Scand. 2015, 132, 73–78. [Google Scholar] [CrossRef]


| Group | Sex | Age (In Years) | Ø Age ± SD | PMI (h) | Ø PMI (h) ± SD |
|---|---|---|---|---|---|
| Control (n = 5) | Male | 91 | 78.8 ± 12.2 | 4:00 | 4:23 ± 2:01 |
| Male | 66 | 8:00 | |||
| Female | 82 | 3:20 | |||
| Female | 89 | 3:20 | |||
| Female | 66 | 3:20 | |||
| Control Validation (n = 2) | Male | 79 | - | 11:00 | - |
| Male | 72 | 9:00 | |||
| DLB (n = 5) | Male | 87 | 85.6 ± 1.9 | 14:00 | 7:52 ± 5:49 |
| Female | 85 | 14:20 | |||
| Female | 88 | 2:30 | |||
| Male | 83 | 3:15 | |||
| Female | 85 | 5:20 | |||
| DLB Validation (n = 2) | Female | 93 | - | 1:20 | - |
| Female | 90 | 3:00 |
| Protein | Peptide | m/z | Charge State | RT-Window Mean [±1.5 min] |
|---|---|---|---|---|
| α-synuclein | EGVVHGVATVAEK | 432.57 | 3 | 24.4 |
| EGVVHGVATVAEK | 648.3513 | 2 | 24.3 | |
| EQVTNVGGAVVTGVTAVAQK | 643.3531 | 3 | 66.8 | |
| EQVTNVGGAVVTGVTAVAQK | 964.526 | 2 | 66.8 | |
| TVEGAGSIAAATGFVK | 739.8961 | 2 | 61.0 | |
| Tyrosine hydroxylase | AGGPHLEYFVR | 415.884 | 3 | 43.4 |
| FDPDLDLDHPGFSDQVYR | 712.6605 | 3 | 75.6 | |
| TGFQLRPVAGLLSAR | 529.3106 | 3 | 69.8 | |
| Glycerol-3-phosphate-dehydrogenase | NHVVDISESGLITIAGGK | 603.995 | 3 | 67.3 |
| LAFLNVQAAEEALPR | 821.4516 | 2 | 89.5 | |
| Cytoplasmic dynein 1 heavy chain 1 | ESPEVLLTLDILK | 735.4267 | 2 | 105.6 |
| EFGPVVIDYGK | 612.319 | 2 | 66.1 |
| Gene Name | Marker for | Mean iBAQ Value CTRL | Mean iBAQ Value DLB | Ratio (CTRL/DLB) |
|---|---|---|---|---|
| RAB5C | Endosomes | 0.000525 | 0.000561 | 0.94 |
| RAB7A | Endosomes | 0.001026 | 0.001591 | 0.64 |
| EEA1 | Endosomes | 0.0000349 | 0.0000348 | 1.00 |
| LAMP1 | Lysosomes | 0.000256 | 0.000314 | 0.82 |
| SCARB2 | Lysosomes | 0.000500 | 0.000874 | 0.57 |
| CTSD | Lysosomes | 0.000908 | 0.000631 | 1.44 |
| RPS9 | Ribosomes | 0.004019 | 0.002987 | 1.35 |
| RPS7 | Ribosomes | 0.001508 | 0.001091 | 1.38 |
| RPL3 | Ribosomes | 0.001373 | 0.001115 | 1.23 |
| PABPC1 | Stress Granules | 0.000846 | 0.000654 | 1.29 |
| G3BP1 | Stress Granules | 0.000036 | 0.000029 | 1.25 |
| eIF4G1 | Stress Granules | 0.000097 | 0.000054 | 1.81 |
| List | Number of Included Proteins | Identified in CTRL NMGs (Mean iBAQ > 0) | Identified in DLB NMGs (Mean iBAQ > 0) | Overlap of Proteins Identified in CTRL and DLB NMGs (in %) |
|---|---|---|---|---|
| Endosome | 632 | 148 | 140 | 89 |
| Lysosome | 369 | 97 | 96 | 95 |
| Ribosome | 79 | 67 | 67 | 100 |
| Stress Granule | 41 | 19 | 19 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wulf, M.; Barkovits, K.; Schork, K.; Eisenacher, M.; Riederer, P.; Gerlach, M.; Eggers, B.; Marcus, K. The Proteome of Neuromelanin Granules in Dementia with Lewy Bodies. Cells 2022, 11, 3538. https://doi.org/10.3390/cells11223538
Wulf M, Barkovits K, Schork K, Eisenacher M, Riederer P, Gerlach M, Eggers B, Marcus K. The Proteome of Neuromelanin Granules in Dementia with Lewy Bodies. Cells. 2022; 11(22):3538. https://doi.org/10.3390/cells11223538
Chicago/Turabian StyleWulf, Maximilian, Katalin Barkovits, Karin Schork, Martin Eisenacher, Peter Riederer, Manfred Gerlach, Britta Eggers, and Katrin Marcus. 2022. "The Proteome of Neuromelanin Granules in Dementia with Lewy Bodies" Cells 11, no. 22: 3538. https://doi.org/10.3390/cells11223538
APA StyleWulf, M., Barkovits, K., Schork, K., Eisenacher, M., Riederer, P., Gerlach, M., Eggers, B., & Marcus, K. (2022). The Proteome of Neuromelanin Granules in Dementia with Lewy Bodies. Cells, 11(22), 3538. https://doi.org/10.3390/cells11223538







