The Ion Channel Gene KCNAB2 Is Associated with Poor Prognosis and Loss of Immune Infiltration in Lung Adenocarcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets and Samples
2.2. GEPIA Database Analysis
2.3. TIMER Database Analysis
2.4. Single-Sample Gene Set Enrichment Analysis
2.5. Kaplan–Meier Plotter Analysis
2.6. UALCAN Database Analysis
2.7. Differentially Expressed Gene Analysis
2.8. Gene Set Enrichment Analysis
2.9. CancerSEA Database Analysis
2.10. Cell Culture
2.11. Cell Transfection
2.12. Cell Proliferation and Invasion Asseys
2.13. Real-Time PCR
2.14. Western Blot
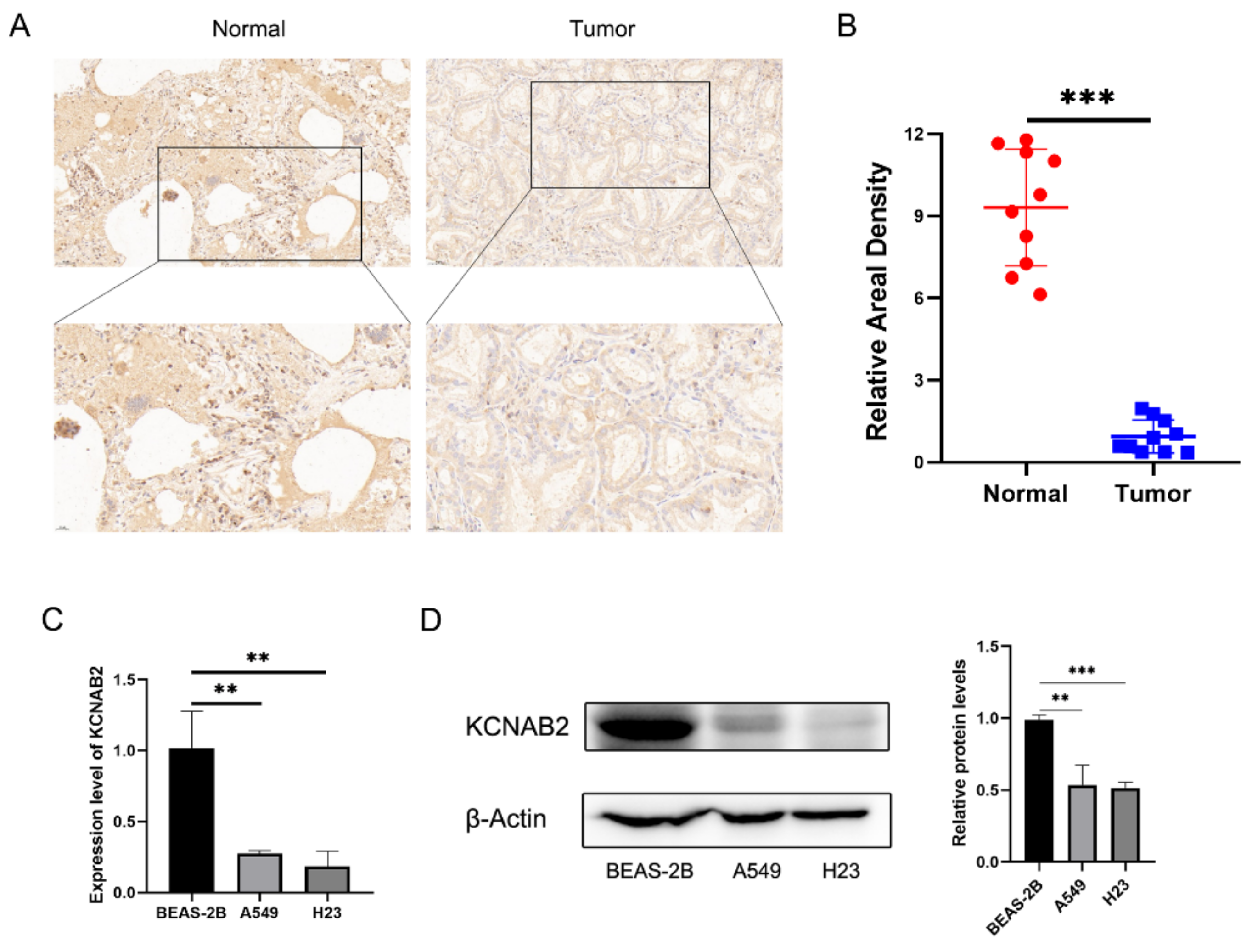
2.15. Immunohistochemical Staining
2.16. Statistical Analyses
3. Results
3.1. KCNAB2 Was Downregulated in LUAD
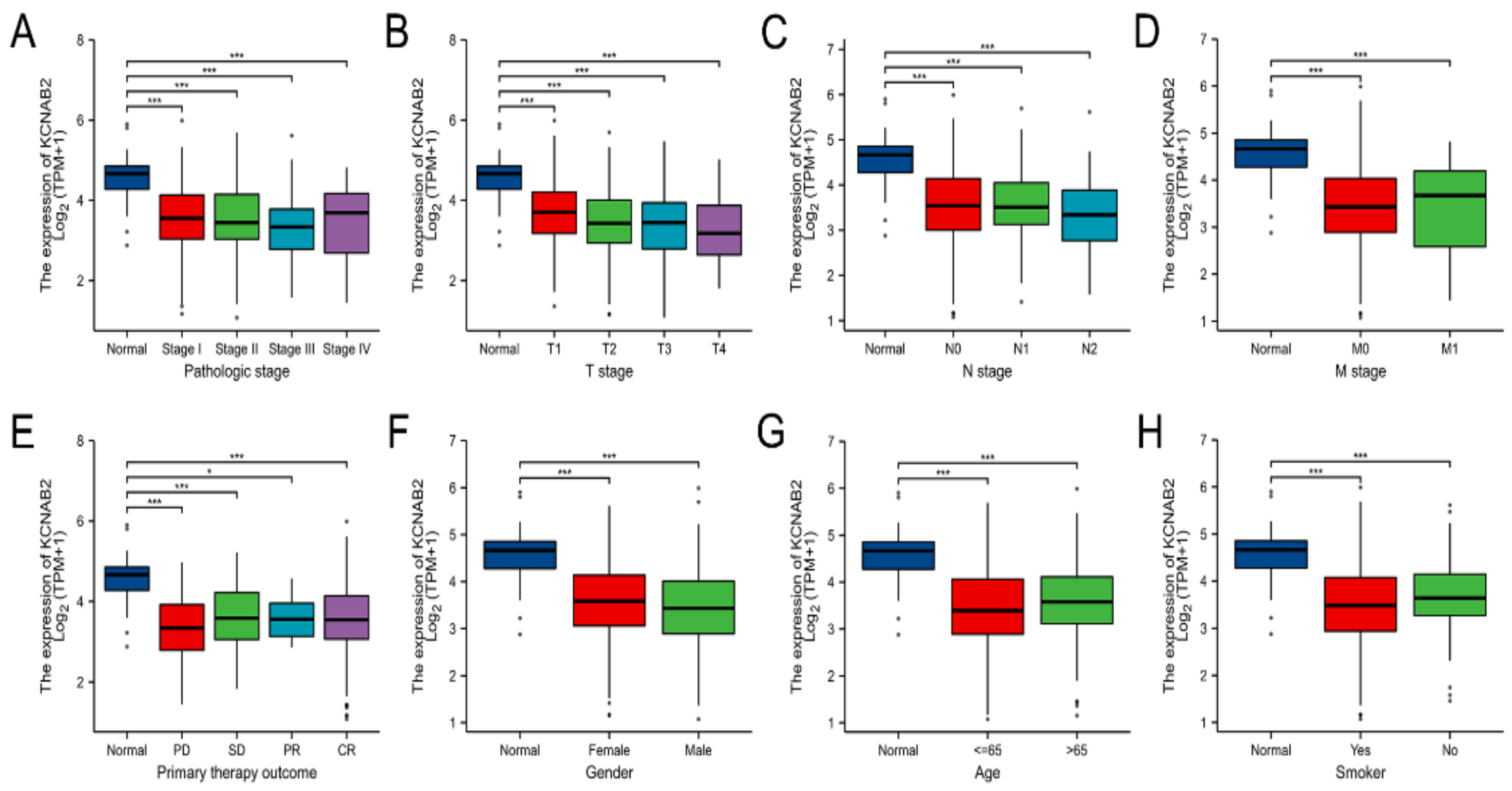
3.2. The Relationship between KCNAB2 Expression and Clinical Parameters in LUAD
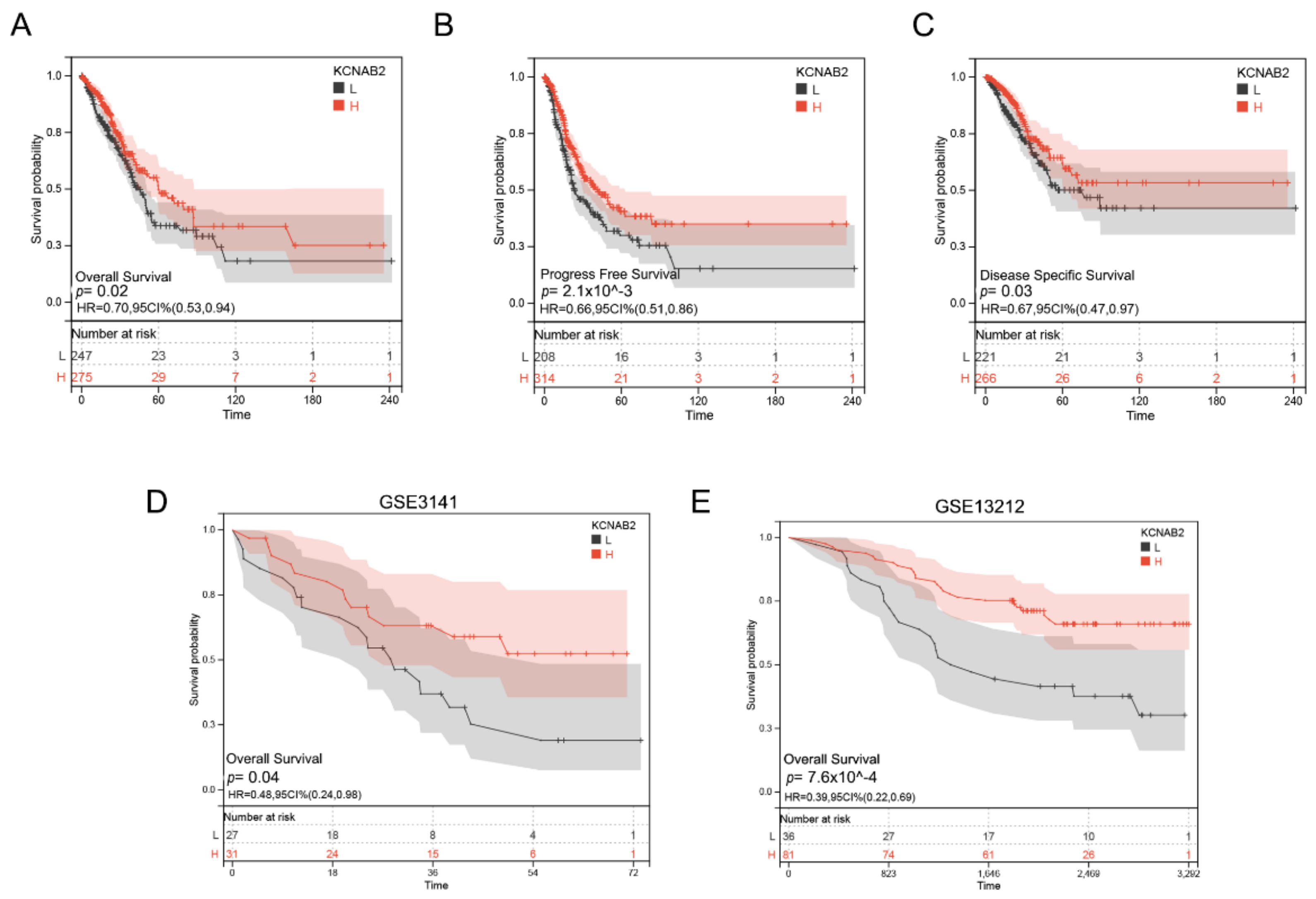
3.3. Decreased KCNAB2 Expression Correlated with Poor Prognosis in LUAD Patients
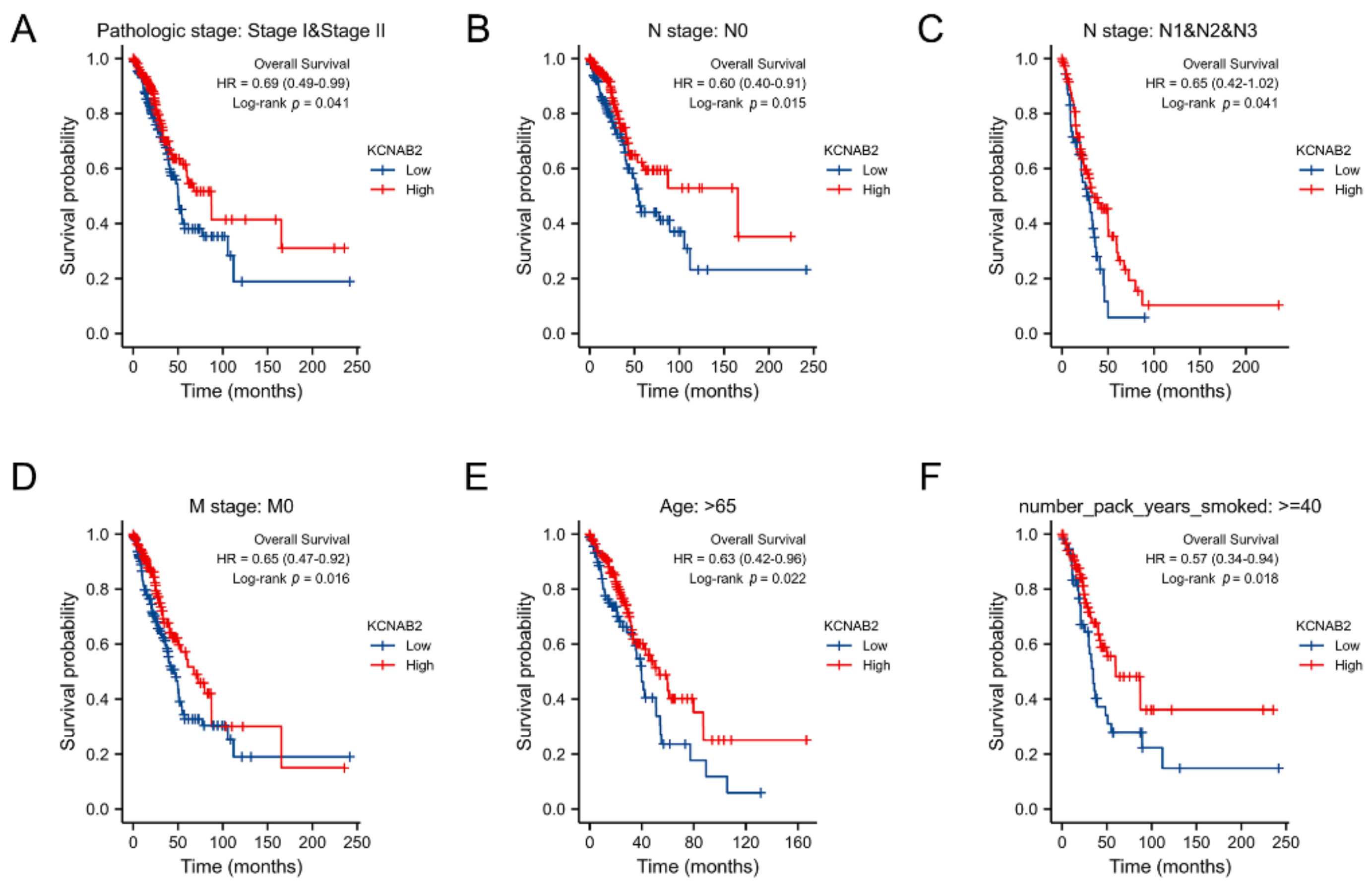
3.4. Survival Analysis of KCNAB2 in Different Clinical Subgroups
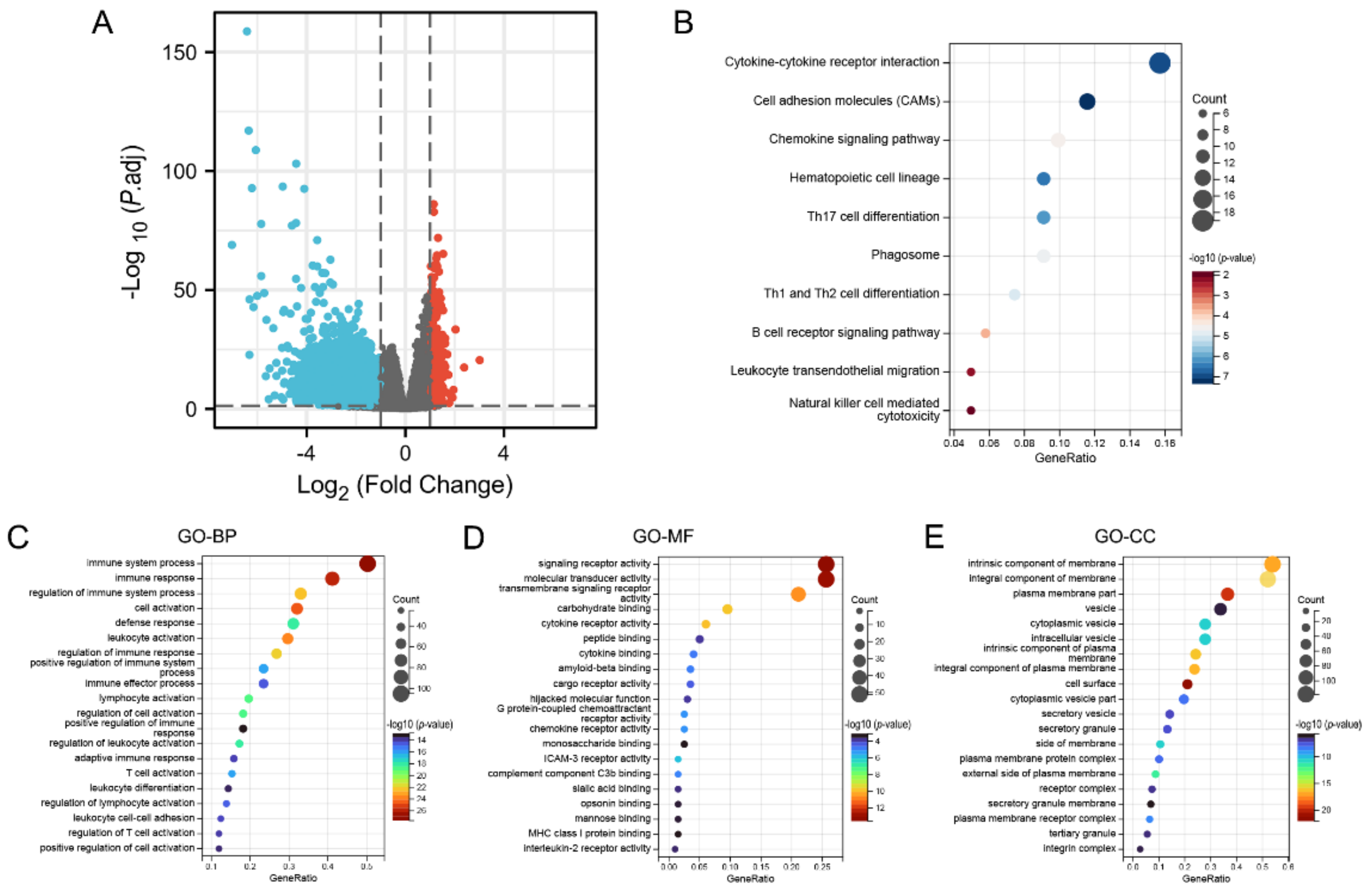
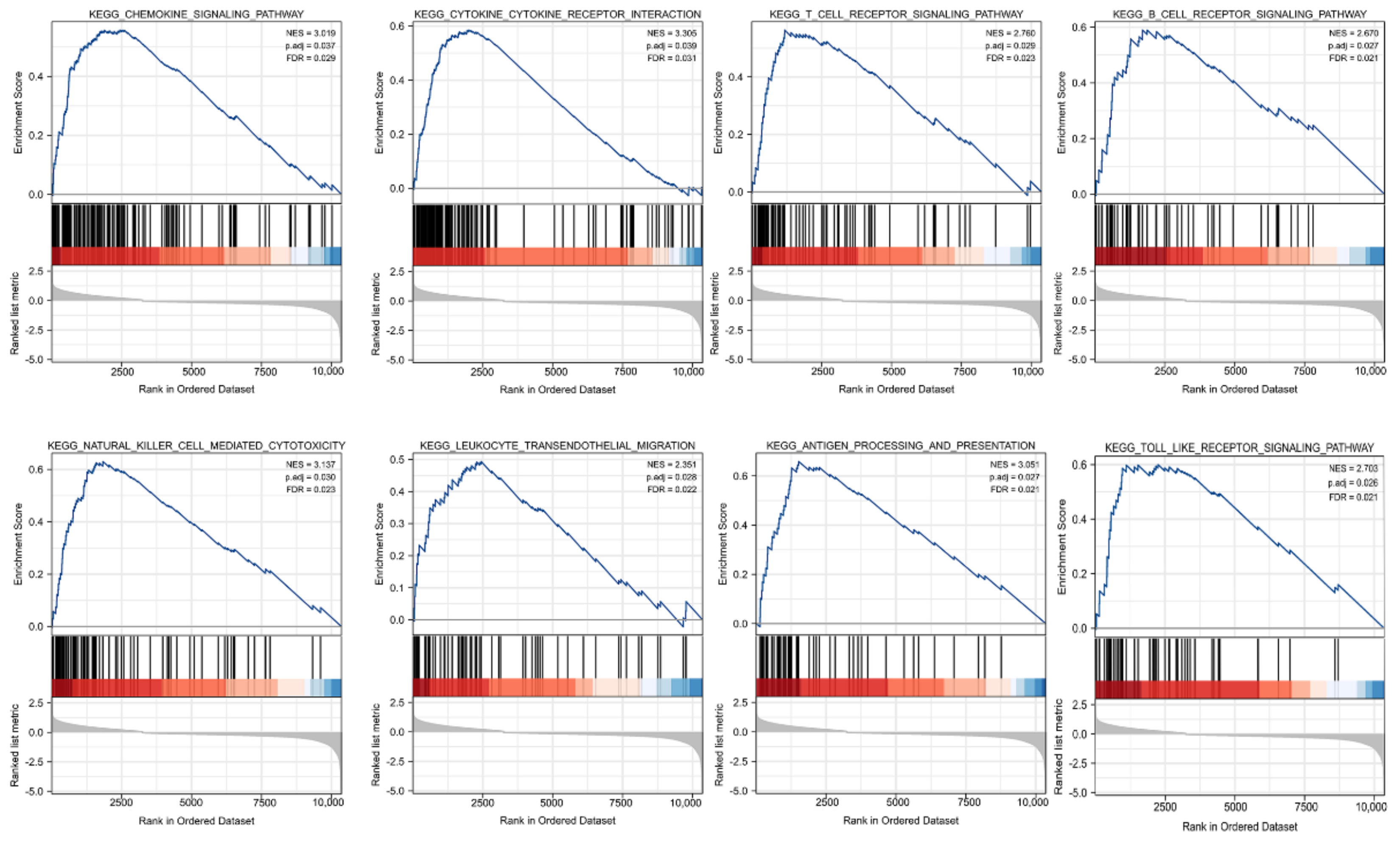
3.5. Identification of DEGs and Functional Enrichment Analysis
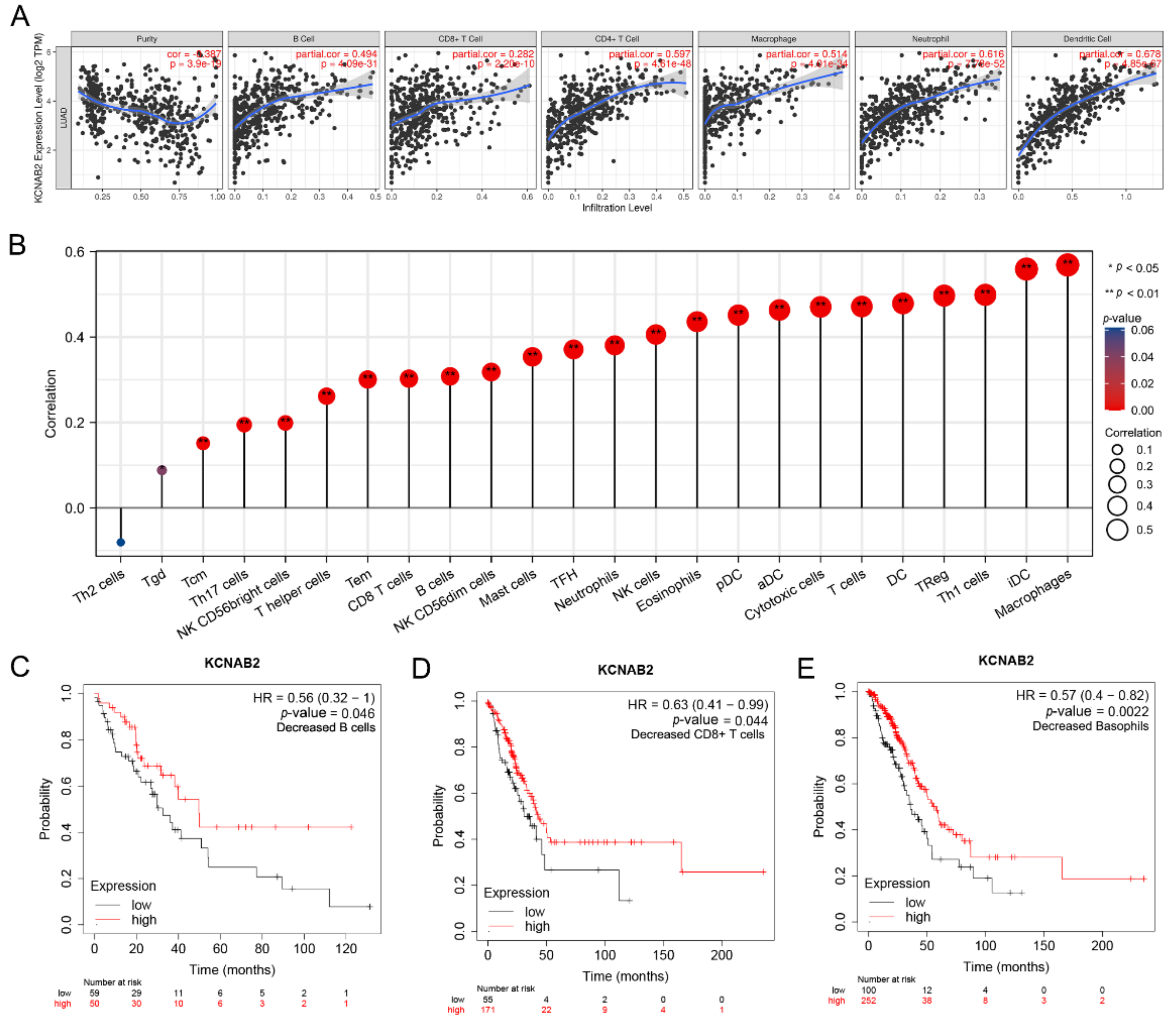
3.6. Immune Cell Infiltration and the Expression of KCNAB2
3.7. The Link between KCNAB2 Expression and Immune Cell Markers
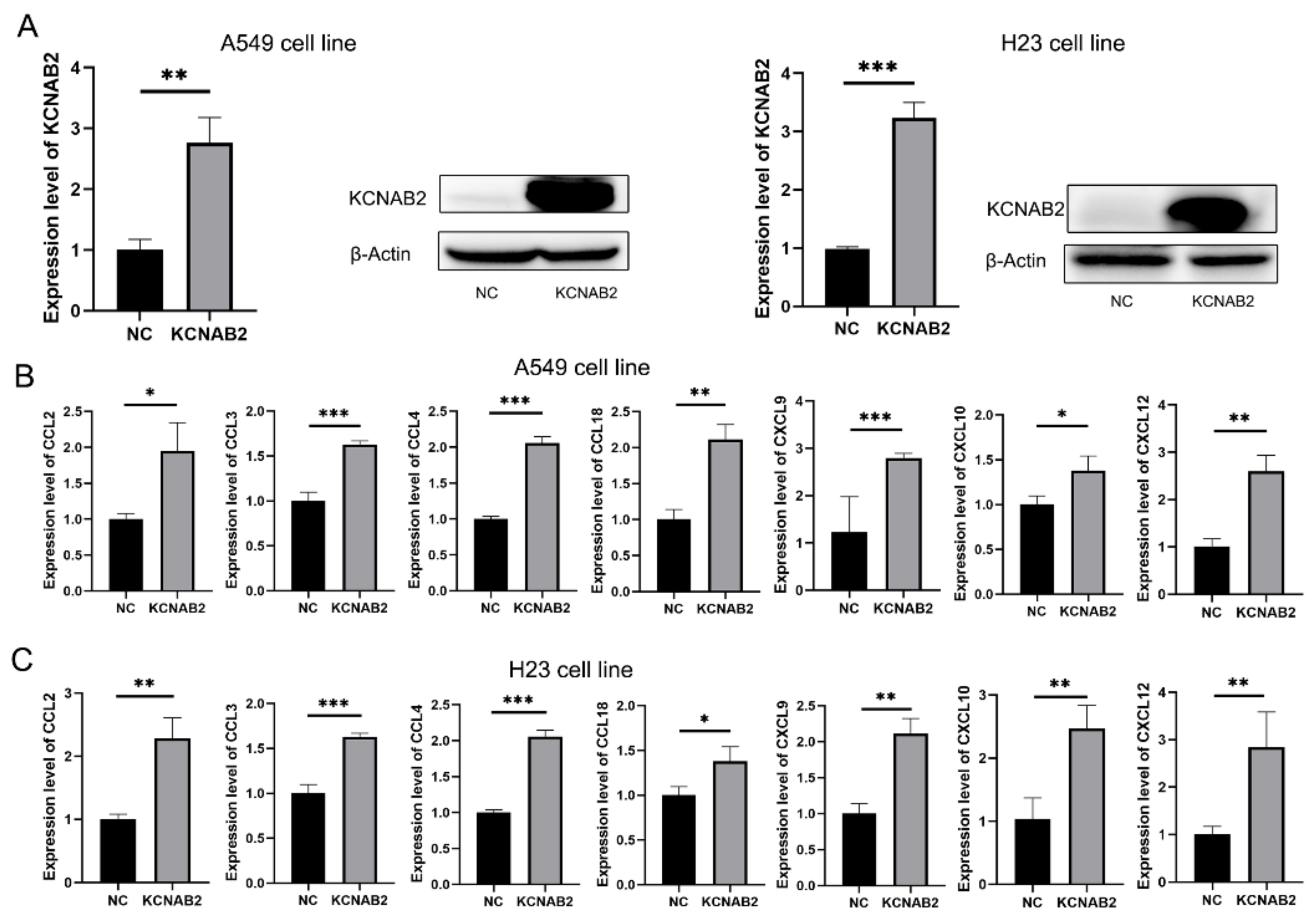
3.8. KCNAB2 Upregulated the Expression of Chemokines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Pasche, B.; Grant, S.C. Non-small cell lung cancer and precision medicine: A model for the incorporation of genomic features into clinical trial design. J. Am. Med. Assoc. 2014, 311, 1975–1976. [Google Scholar] [CrossRef]
- Thomas, A.; Liu, S.V.; Subramaniam, D.S.; Giaccone, G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat. Rev. Clin. Oncol. 2015, 12, 511–526. [Google Scholar] [CrossRef]
- Anagnostou, V.K.; Brahmer, J.R. Cancer immunotherapy: A future paradigm shift in the treatment of non-small cell lung cancer. Clin. Cancer Res. 2015, 21, 976–984. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef]
- Spranger, S.; Gajewski, T.F. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat. Rev. Cancer 2018, 18, 139–147. [Google Scholar] [CrossRef]
- Comes, N.; Bielanska, J.; Vallejo-Gracia, A.; Serrano-Albarrás, A.; Marruecos, L.; Gómez, D.; Soler, C.; Condom, E.; Ramón, Y.; Cajal, S.; et al. The voltage-dependent K(+) channels Kv1.3 and Kv1.5 in human cancer. Front. Physiol. 2013, 4, 283. [Google Scholar] [CrossRef]
- Lin, X.; Wu, J.-F.; Wang, D.-M.; Zhang, J.; Zhang, W.-J.; Xue, G. The correlation and role analysis of KCNK2/4/5/15 in Human Papillary Thyroid Carcinoma microenvironment. J. Cancer 2020, 11, 5162–5176. [Google Scholar] [CrossRef]
- Chen, S.; Wang, C.; Su, X.; Dai, X.; Li, S.; Mo, Z. KCNN4 is a potential prognostic marker and critical factor affecting the immune status of the tumor microenvironment in kidney renal clear cell carcinoma. Transl. Androl. Urol. 2021, 10, 2454–2470. [Google Scholar] [CrossRef] [PubMed]
- Ko, E.-A.; Kim, Y.-W.; Lee, D.; Choi, J.; Kim, S.; Seo, Y.; Bang, H.; Kim, J.-H.; Ko, J.-H. Expression of potassium channel genes predicts clinical outcome in lung cancer. Korean J. Physiol. Pharmacol. 2019, 23, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xiong, Z.-G. Ion channels as targets for cancer therapy. Int. J. Physiol. Pathophysiol. Pharmacol. 2011, 3, 156–166. [Google Scholar] [PubMed]
- Villalonga, N.; Ferreres, J.C.; Argilés, J.M.; Condom, E.; Felipe, A. Potassium channels are a new target field in anticancer drug design. Recent Pat. Anti-Cancer Drug Discov. 2007, 2, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Mo, X.; Xia, R.; Jiang, L.; Zhang, C.; Xu, H.; Sun, Q.; Zhou, G.; Zhang, Y.; Wang, Y.; et al. KCNN4 promotes the progression of lung adenocarcinoma by activating the AKT and ERK signaling pathways. Cancer Biomark. 2021, 31, 187–201. [Google Scholar] [CrossRef]
- Lu, X.; Li, K.; Yang, J. Potassium voltage-gated channel subfamily D member 2 induces an aggressive phenotype in lung adenocarcinoma. Neoplasma 2021, 68, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Su, X.; Mo, Z. KCNN4 is a Potential Biomarker for Predicting Cancer Prognosis and an Essential Molecule that Remodels Various Components in the Tumor Microenvironment: A Pan-Cancer Study. Front. Mol. Biosci. 2022, 9, 812815. [Google Scholar] [CrossRef]
- Yang, E.-K.; Alvira, M.R.; Levitan, E.S.; Takimoto, K. Kvβ subunits increase expression of Kv4.3 channels by interacting with their C termini. J. Biol. Chem. 2001, 276, 4839–4844. [Google Scholar] [CrossRef]
- Heilstedt, H.A.; Burgess, D.L.; Anderson, A.E.; Chedrawi, A.; Tharp, B.; Lee, O.; Kashork, C.D.; Starkey, D.E.; Wu, Y.-Q.; Noebels, J.L.; et al. Loss of the potassium channel β-subunit gene, KCNAB2, is associated with epilepsy in patients with 1p36 deletion syndrome. Epilepsia 2001, 42, 1103–1111. [Google Scholar] [CrossRef]
- Weng, J.; Cao, Y.; Moss, N.; Zhou, M. Modulation of voltage-dependent Shaker family potassium channels by an aldo-keto reductase. J. Biol. Chem. 2006, 281, 15194–15200. [Google Scholar] [CrossRef]
- Tu, J.; Kuang, Z.; Xie, X.; Wu, S.; Wu, T.; Chen, S. Prognostic and predictive value of a mRNA signature in peripheral T-cell lymphomas: A mRNA expression analysis. J. Cell. Mol. Med. 2021, 25, 84–95. [Google Scholar] [CrossRef]
- Garate, J.; Maimo-Barcelo, A.; Bestard-Escalas, J.; Fernandez, R.; Perez-Romero, K.; Martinez, M.A.; Payeras, M.; Lopez, D.; Fernández, J.; Barceló-Coblijn, G. A Drastic Shift in Lipid Adducts in Colon Cancer Detected by MALDI-IMS Exposes Alterations in Specific K(+) Channels. Cancers 2021, 13, 1350. [Google Scholar] [CrossRef] [PubMed]
- Ashton, C.; Rhie, S.K.; Carmichael, J.D.; Zada, G. Role of KCNAB2 expression in modulating hormone secretion in somatotroph pituitary adenoma. J. Neurosurg. 2020, 134, 787–793. [Google Scholar] [CrossRef] [PubMed]
- White, P.S.; Thompson, P.M.; Gotoh, T.; Okawa, E.R.; Igarashi, J.; Kok, M.; Winter, C.; Gregory, S.G.; Hogarty, M.D.; Maris, J.M.; et al. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene 2005, 24, 2684–2694. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef]
- Lánczky, A.; Nagy, Á.; Bottai, G.; Munkácsy, G.; Szabó, A.; Santarpia, L.; Gyorffy, B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 2016, 160, 439–446. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Rudnick, P.A.; Markey, S.P.; Roth, J.; Mirokhin, Y.; Yan, X.; Tchekhovskoi, D.V.; Edwards, N.J.; Thangudu, R.R.; Ketchum, K.A.; Kinsinger, C.R.; et al. A Description of the Clinical Proteomic Tumor Analysis Consortium (CPTAC) Common Data Analysis Pipeline. J. Proteome Res. 2016, 15, 1023–1032. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Havel, J.J.; Chowell, D.; Chan, T.A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 2019, 19, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Maskalenko, N.A.; Zhigarev, D.; Campbell, K.S. Harnessing natural killer cells for cancer immunotherapy: Dispatching the first responders. Nat. Rev. Drug Discov. 2022, 21, 559–577. [Google Scholar] [CrossRef] [PubMed]
- Cui, K.; Hu, S.; Mei, X.; Cheng, M. Innate Immune Cells in the Esophageal Tumor Microenvironment. Front. Immunol. 2021, 12, 654731. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Loibl, S.; Noske, A.; Roller, M.; Müller, B.M.; Komor, M.; Budczies, J.; Darb-Esfahani, S.; Kronenwett, R.; Hanusch, C.; et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 2010, 28, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Teillaud, J.L.; Dieu-Nosjean, M.C. Tertiary Lymphoid Structures: An Anti-tumor School for Adaptive Immune Cells and an Antibody Factory to Fight Cancer? Front. Immunol. 2017, 8, 830. [Google Scholar] [CrossRef]
- Overacre-Delgoffe, A.E.; Chikina, M.; Dadey, R.E.; Yano, H.; Brunazzi, E.A.; Shayan, G.; Horne, W.; Moskovitz, J.M.; Kolls, J.K.; Sander, C.; et al. Interferon-γ Drives Treg Fragility to Promote Anti-tumor Immunity. Cell 2017, 169, 1130–1141.e11. [Google Scholar] [CrossRef]
- Overacre-Delgoffe, A.E.; Vignali, D.A.A. Treg Fragility: A Prerequisite for Effective Antitumor Immunity? Cancer Immunol. Res. 2018, 6, 882–887. [Google Scholar] [CrossRef]
- Märkl, F.; Huynh, D.; Endres, S.; Kobold, S. Utilizing chemokines in cancer immunotherapy. Trends Cancer. 2022, 8, 670–682. [Google Scholar] [CrossRef]
- Vicari, A.P.; Caux, C. Chemokines in cancer. Cytokine Growth Factor Rev. 2002, 13, 143–154. [Google Scholar] [CrossRef]
- Propper, D.J.; Balkwill, F.R. Harnessing cytokines and chemokines for cancer therapy. Nat. Rev. Clin. Oncol. 2022, 19, 237–253. [Google Scholar] [CrossRef]
- Li, H.; Harrison, E.B.; Li, H.; Hirabayashi, K.; Chen, J.; Li, Q.-X.; Gunn, J.; Weiss, J.; Savoldo, B.; Parker, J.S.; et al. Targeting brain lesions of non-small cell lung cancer by enhancing CCL2-mediated CAR-T cell migration. Nat. Commun. 2022, 13, 2154. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Yu, X.; Xue, L.; Ge, X.; Zhao, W.; Peng, W. Intrinsic β-catenin signaling suppresses CD8+ T-cell infiltration in colorectal cancer. Biomed. Pharmacother. 2019, 115, 108921. [Google Scholar] [CrossRef] [PubMed]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Zhao, E.; Kryczek, I.; Zou, W. Th17 cells have stem cell-like features and promote long-term immunity. Oncoimmunology 2012, 1, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Martin-Orozco, N.; Muranski, P.; Chung, Y.; Yang, X.O.; Yamazaki, T.; Lu, S.; Hwu, P.; Restifo, N.P.; Overwijk, W.W.; Dong, C. T helper 17 cells promote cytotoxic T cell activation in tumor immunity. Immunity 2009, 31, 787–798. [Google Scholar] [CrossRef]

| GEO Datasets | Platform | Sample Size | Publication Years | |
|---|---|---|---|---|
| LUAD | Normal | |||
| GSE32863 | GPL6884 | 58 | 58 | 2012 |
| GSE30219 | GPL570 | 85 | 14 | 2013 |
| GSE10072 | GPL96 | 58 | 47 | 2008 |
| GSE3141 | GPL570 | 58 | 0 | 2005 |
| GSE13213 | GPL6480 | 117 | 0 | 2009 |
| Characteristics | Total (N) | Odds Ratio (OR) | p-Value |
|---|---|---|---|
| T stage (T2, T3, and T4 vs. T1) | 532 | 0.658 (0.522–0.824) | <0.001 |
| N stage (N1, N2, and N3 vs. N0) | 519 | 0.890 (0.714–1.109) | 0.299 |
| M stage (M1 vs. M0) | 386 | 0.982 (0.612–1.592) | 0.940 |
| Pathologic stage (Stage III and Stage IV vs. Stage I and Stage II) | 527 | 0.764 (0.591–0.984) | 0.037 |
| Age (>65 vs. <=65) | 516 | 1.284 (1.043–1.588) | 0.019 |
| Gender (Male vs. Female) | 535 | 0.799 (0.649–0.982) | 0.034 |
| Smoker (Yes vs. No) | 521 | 0.741 (0.546–0.999) | 0.052 |
| Immune Cells | Gene Markers | None | Purity | ||
|---|---|---|---|---|---|
| Correlation | p-Value | Correlation | p-Value | ||
| B cell | CD19 | 0.439 | *** | 0.331 | *** |
| CD79A | 0.371 | *** | 0.257 | *** | |
| T cell (general) | CD3D | 0.471 | *** | 0.346 | *** |
| CD3E | 0.587 | *** | 0.498 | *** | |
| CD2 | 0.59 | *** | 0.499 | *** | |
| CD8+ T cell | CD8A | 0.48 | *** | 0.381 | *** |
| CD8B | 0.394 | *** | 0.306 | *** | |
| Monocyte | CD86 | 0.675 | *** | 0.616 | *** |
| CSF1R | 0.727 | *** | 0.685 | *** | |
| TAM | CCL2 | 0.416 | *** | 0.328 | *** |
| CD68 | 0.674 | *** | 0.636 | *** | |
| IL10 | 0.573 | *** | 0.493 | *** | |
| M1 | IRF5 | 0.627 | *** | 0.587 | *** |
| PTGS2 | -0.123 | * | -0.143 | ** | |
| NOS2 | 0.232 | *** | 0.167 | *** | |
| M2 | CD163 | 0.673 | *** | 0.63 | *** |
| VSIG4 | 0.624 | *** | 0.58 | *** | |
| MS4A4A | 0.628 | *** | 0.572 | *** | |
| Neutrophils | CEACAM8 | 0.286 | *** | 0.289 | *** |
| ITGAM | 0.73 | *** | 0.7 | *** | |
| CCR7 | 0.567 | *** | 0.477 | *** | |
| Natural killer cell | KIR2DL1 | 0.195 | *** | 0.144 | ** |
| KIR2DL3 | 0.248 | *** | 0.178 | *** | |
| KIR2DL4 | 0.209 | *** | 0.134 | ** | |
| KIR3DL1 | 0.238 | *** | 0.183 | *** | |
| KIR3DL2 | 0.313 | *** | 0.248 | *** | |
| KIR3DL3 | 0.077 | ns | 0.057 | ns | |
| KIR2DS4 | 0.227 | *** | 0.165 | *** | |
| Dendritic cell | HLA-DPB1 | 0.638 | *** | 0.582 | *** |
| HLAD-QB1 | 0.485 | *** | 0.407 | *** | |
| HLA-DRA | 0.573 | *** | 0.503 | *** | |
| HLA-DPA1 | 0.612 | *** | 0.557 | *** | |
| CD1C | 0.384 | *** | 0.316 | *** | |
| NRP1 | 0.203 | *** | 0.172 | *** | |
| ITGAX | 0.804 | *** | 0.777 | *** | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, Y.; Wang, Q.; Liang, J.; Zhang, L.; Zhang, H. The Ion Channel Gene KCNAB2 Is Associated with Poor Prognosis and Loss of Immune Infiltration in Lung Adenocarcinoma. Cells 2022, 11, 3438. https://doi.org/10.3390/cells11213438
Lyu Y, Wang Q, Liang J, Zhang L, Zhang H. The Ion Channel Gene KCNAB2 Is Associated with Poor Prognosis and Loss of Immune Infiltration in Lung Adenocarcinoma. Cells. 2022; 11(21):3438. https://doi.org/10.3390/cells11213438
Chicago/Turabian StyleLyu, Yin, Qiao Wang, Jingtian Liang, Li Zhang, and Hao Zhang. 2022. "The Ion Channel Gene KCNAB2 Is Associated with Poor Prognosis and Loss of Immune Infiltration in Lung Adenocarcinoma" Cells 11, no. 21: 3438. https://doi.org/10.3390/cells11213438
APA StyleLyu, Y., Wang, Q., Liang, J., Zhang, L., & Zhang, H. (2022). The Ion Channel Gene KCNAB2 Is Associated with Poor Prognosis and Loss of Immune Infiltration in Lung Adenocarcinoma. Cells, 11(21), 3438. https://doi.org/10.3390/cells11213438






