Drug Resistance in Cancers: A Free Pass for Bullying
Abstract
1. Introduction
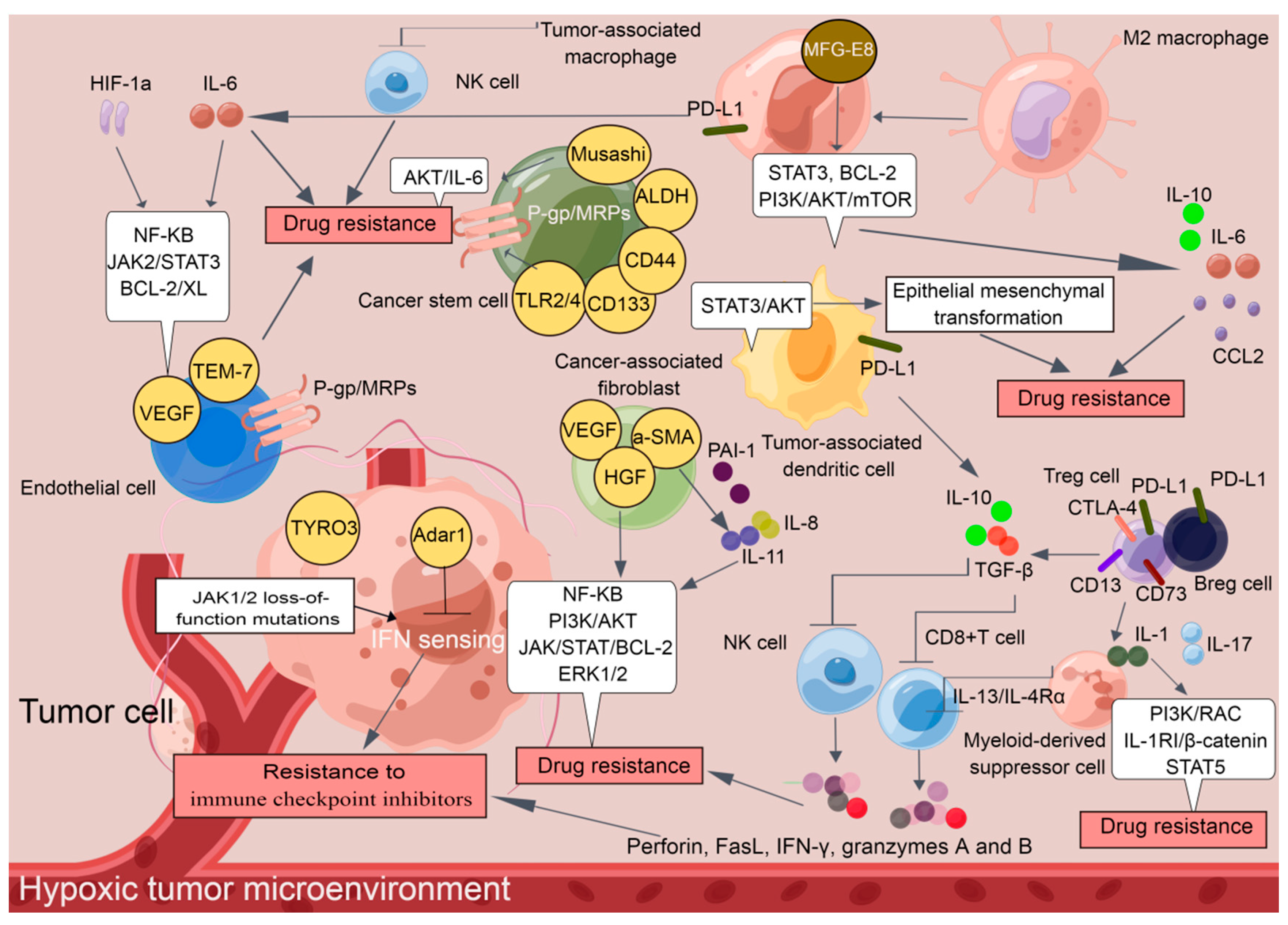
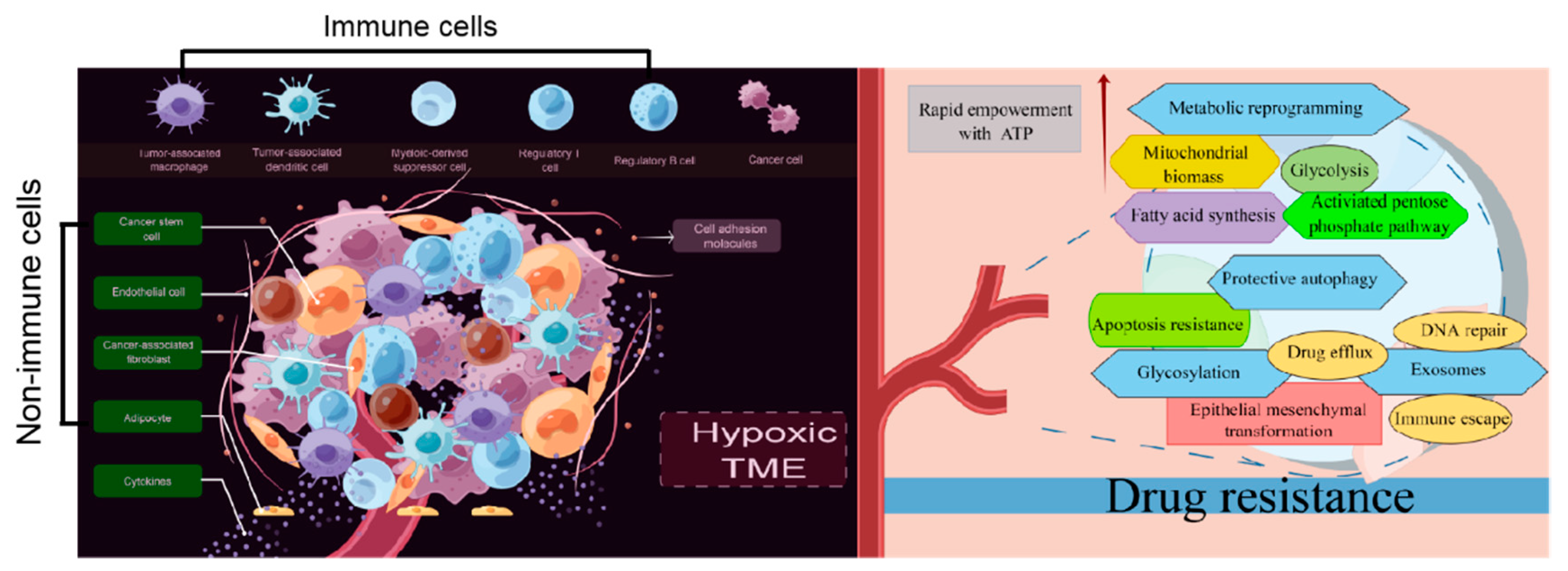
2. TME: A Breeding Ground for Drug Resistance
2.1. Tumor-Associated Immune Cells in the TME
2.1.1. Regulatory T Cells (Tregs) and Regulatory B Cells (Bregs)
2.1.2. Tumor-Associated Macrophages (TAMs)
2.1.3. Myeloid-Derived Suppressor Cells (MDSCs)
2.1.4. Tumor-Associated Dendritic Cells (TADCs)
2.2. Nonimmune Cells in the TME
2.2.1. CAFs
2.2.2. CSCs
2.2.3. Endothelial Cells
2.2.4. Adipocytes
2.3. Cytokines in the TME
2.4. CAMs in the TME
2.5. The Hypoxic TME
3. Exosomes: A Critical Factor in Drug Resistance Development
3.1. Exosome-Mediated Drug Efflux
3.2. Exosome-Mediated Apoptosis Resistance
3.3. Exosome-Mediated DNA Repair
3.4. Exosome-Mediated Immune Escape
3.5. Exosome-Mediated EMT
4. Metabolic Reprogramming: The Genesis of Drug Resistance
4.1. Overstimulation of Glycolysis
4.2. Enhancement of Fatty Acid Metabolism
4.2.1. ATP Citrate Lyase (ACLY)
4.2.2. Fatty Acid Synthase (FASN)
4.2.3. Carnitine Palmitoyltransferase 1 (CPT1)
4.3. Upregulation of Mitochondrial Biomass
4.4. Activation of the Pentose Phosphate Pathway
5. Glycosylation: A Newly Recognized Chapter in the Story of Drug Resistance
5.1. Glycosylation-Mediated Drug Efflux
5.2. Glycosylation-Mediated Apoptosis Resistance
5.3. Other Mechanisms of Glycosylation Action
6. Autophagy: A Novel Force Triggering Drug Resistance
6.1. Protective Autophagy and Drug Resistance
6.2. Protective Autophagy and Apoptosis Resistance
6.3. Autophagy Inhibitors in Clinical Treatment
7. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Wei, G.; Wang, Y.; Yang, G.; Wang, Y.; Ju, R. Recent progress in nanomedicine for enhanced cancer chemotherapy. Theranostics 2021, 11, 6370–6392. [Google Scholar] [CrossRef]
- Cable, J.; Greenbaum, B.; Pe’er, D.; Bollard, C.M.; Bruni, S.; Griffin, M.E.; Allison, J.P.; Wu, C.J.; Subudhi, S.K.; Mardis, E.R.; et al. Frontiers in cancer immunotherapy-a symposium report. Ann. N. Y. Acad. Sci. 2021, 1489, 30–47. [Google Scholar] [CrossRef]
- Lee, Y.T.; Tan, Y.J.; Oon, C.E. Molecular targeted therapy: Treating cancer with specificity. Eur. J. Pharmacol. 2018, 834, 188–196. [Google Scholar] [CrossRef]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int. J. Mol. Sci. 2020, 21, 3233. [Google Scholar] [CrossRef]
- Carneiro, B.A.; El-Deiry, W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef]
- Abad, E.; Graifer, D.; Lyakhovich, A. DNA damage response and resistance of cancer stem cells. Cancer Lett. 2020, 474, 106–117. [Google Scholar] [CrossRef]
- Biswas, A.; De, S. Drivers of dynamic intratumor heterogeneity and phenotypic plasticity. Am. J. Physiol. Cell Physiol. 2021, 320, C750–C760. [Google Scholar] [CrossRef]
- Marusyk, A.; Janiszewska, M.; Polyak, K. Intratumor Heterogeneity: The Rosetta Stone of Therapy Resistance. Cancer Cell 2020, 37, 471–484. [Google Scholar] [CrossRef]
- Quintanal-Villalonga, Á.; Chan, J.M.; Yu, H.A.; Pe’er, D.; Sawyers, C.L.; Sen, T.; Rudin, C.M. Lineage plasticity in cancer: A shared pathway of therapeutic resistance. Nat. Rev. Clin. Oncol. 2020, 17, 360–371. [Google Scholar] [CrossRef]
- Da Silva-Diz, V.; Lorenzo-Sanz, L.; Bernat-Peguera, A.; Lopez-Cerda, M.; Muñoz, P. Cancer cell plasticity: Impact on tumor progression and therapy response. Semin. Cancer Biol. 2018, 53, 48–58. [Google Scholar] [CrossRef]
- Asano, T. Drug Resistance in Cancer Therapy and the Role of Epigenetics. J. Nippon Med. Sch. 2020, 87, 244–251. [Google Scholar] [CrossRef]
- Salgia, R.; Kulkarni, P. The Genetic/Non-genetic Duality of Drug ‘Resistance’ in Cancer. Trends Cancer 2018, 4, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Arneth, B. Tumor Microenvironment. Med. Kaunas 2019, 56, 15. [Google Scholar] [CrossRef]
- Hernández-Camarero, P.; López-Ruiz, E.; Marchal, J.A.; Perán, M. Cancer: A mirrored room between tumor bulk and tumor microenvironment. J. Exp. Clin. Cancer Res. 2021, 40, 217. [Google Scholar] [CrossRef]
- Saleh, R.; Elkord, E. Acquired resistance to cancer immunotherapy: Role of tumor-mediated immunosuppression. Semin. Cancer Biol. 2020, 65, 13–27. [Google Scholar] [CrossRef]
- Nishikawa, H.; Koyama, S. Mechanisms of regulatory T cell infiltration in tumors: Implications for innovative immune precision therapies. J. Immunother. Cancer 2021, 9, e002591. [Google Scholar] [PubMed]
- Li, C.; Jiang, P.; Wei, S.; Xu, X.; Wang, J. Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer 2020, 19, 116. [Google Scholar] [CrossRef]
- Dixon, M.L.; Leavenworth, J.D.; Leavenworth, J.W. Lineage Reprogramming of Effector Regulatory T Cells in Cancer. Front. Immunol. 2021, 12, 717421. [Google Scholar] [CrossRef] [PubMed]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Moreau, J.M.; Velegraki, M.; Bolyard, C.; Rosenblum, M.D.; Li, Z. Transforming growth factor-β1 in regulatory T cell biology. Sci. Immunol. 2022, 7, eabi4613. [Google Scholar] [CrossRef] [PubMed]
- Rana, J.; Biswas, M. Regulatory T cell therapy: Current and future design perspectives. Cell Immunol. 2020, 356, 104193. [Google Scholar] [CrossRef]
- Farhood, B.; Najafi, M.; Mortezaee, K. CD8+ cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell Physiol. 2019, 234, 8509–8521. [Google Scholar] [CrossRef]
- Catalán, D.; Mansilla, M.A.; Ferrier, A.; Soto, L.; Oleinika, K.; Aguillón, J.C.; Aravena, O. Immunosuppressive Mechanisms of Regulatory B Cells. Front. Immunol. 2021, 12, 611795. [Google Scholar] [CrossRef]
- Shalapour, S.; Lin, X.J.; Bastian, I.N.; Brain, J.; Burt, A.D.; Aksenov, A.A.; Vrbanac, A.F.; Li, W.; Perkins, A.; Matsutani, T.; et al. Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature 2017, 551, 340–345. [Google Scholar] [CrossRef]
- Shalapour, S.; Font-Burgada, J.; Di Caro, G.; Zhong, Z.; Sanchez-Lopez, E.; Dhar, D.; Willimsky, G.; Ammirante, M.; Strasner, A.; Hansel, D.E.; et al. Immunosuppressive plasma cells impede T-cell-dependent immunogenic chemotherapy. Nature 2015, 521, 94–98. [Google Scholar] [CrossRef]
- Li, B.; Chan, H.L.; Chen, P. Immune Checkpoint Inhibitors: Basics and Challenges. Curr. Med. Chem. 2019, 26, 3009–3025. [Google Scholar] [CrossRef]
- Fares, C.M.; Van Allen, E.M.; Drake, C.G.; Allison, J.P.; Hu-Lieskovan, S. Mechanisms of Resistance to Immune Checkpoint Blockade, Why Does Checkpoint Inhibitor Immunotherapy Not Work for All Patients? Am. Soc. Clin. Oncol Educ. Book 2019, 39, 147–164. [Google Scholar] [CrossRef]
- Jiang, Z.; Lim, S.O.; Yan, M.; Hsu, J.L.; Yao, J.; Wei, Y.; Chang, S.S.; Yamaguchi, H.; Lee, H.H.; Ke, B.; et al. TYRO3 induces anti-PD-1/PD-L1 therapy resistance by limiting innate immunity and tumoral ferroptosis. J. Clin. Investig. 2021, 131, e139434. [Google Scholar] [CrossRef]
- Ishizuka, J.J.; Manguso, R.T.; Cheruiyot, C.K.; Bi, K.; Panda, A.; Iracheta-Vellve, A.; Miller, B.C.; Du, P.P.; Yates, K.B.; Dubrot, J.; et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 2019, 565, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Zaretsky, J.M.; Garcia-Diaz, A.; Shin, D.S.; Escuin-Ordinas, H.; Hugo, W.; Hu-Lieskovan, S.; Torrejon, D.Y.; Abril-Rodriguez, G.; Sandoval, S.; Barthly, L.; et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N. Engl. J. Med. 2016, 375, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.S.; Zaretsky, J.M.; Escuin-Ordinas, H.; Garcia-Diaz, A.; Hu-Lieskovan, S.; Kalbasi, A.; Grasso, C.S.; Hugo, W.; Sandoval, S.; Torrejon, D.Y.; et al. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discov. 2017, 7, 188–201. [Google Scholar] [CrossRef]
- Baxter, M.A.; Middleton, F.; Cagney, H.P.; Petty, R.D. Resistance to immune checkpoint inhibitors in advanced gastro-oesophageal cancers. Br. J. Cancer 2021, 125, 1068–1079. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, S.; Yuan, R.; Engleman, E.G. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu. Rev. Pathol. 2021, 16, 223–249. [Google Scholar] [CrossRef]
- Ju, S.; Qiu, H.; Zhou, X.; Zhu, B.; Lv, X.; Huang, X.; Li, J.; Zhang, Y.; Liu, L.; Ge, Y.; et al. CD13+CD4+CD25hi regulatory T cells exhibit higher suppressive function and increase with tumor stage in non-small cell lung cancer patients. Cell Cycle 2009, 8, 2578–2585. [Google Scholar] [CrossRef]
- Chen, S.; Wainwright, D.A.; Wu, J.D.; Wan, Y.; Matei, D.E.; Zhang, Y.; Zhang, B. CD73: An emerging checkpoint for cancer immunotherapy. Immunotherapy 2019, 11, 983–997. [Google Scholar] [CrossRef]
- Kong, X. Discovery of New Immune Checkpoints: Family Grows Up. Adv. Exp. Med. Biol. 2020, 1248, 61–82. [Google Scholar]
- Cassetta, L.; Pollard, J.W. Tumor-associated macrophages. Curr. Biol. 2020, 30, R246–R248. [Google Scholar] [CrossRef]
- Jinushi, M.; Chiba, S.; Yoshiyama, H.; Masutomi, K.; Kinoshita, I.; Dosaka-Akita, H.; Yagita, H.; Takaoka, A.; Tahara, H. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc. Natl. Acad. Sci. USA 2011, 108, 12425–12430. [Google Scholar] [CrossRef]
- Yang, C.; He, L.; He, P.; Liu, Y.; Wang, W.; He, Y.; Du, Y.; Gao, F. Increased drug resistance in breast cancer by tumor-associated macrophages through IL-10/STAT3/bcl-2 signaling pathway. Med. Oncol. 2015, 32, 352. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Ji, H.; Niu, X.; Yin, L.; Wang, Y.; Gu, Y.; Wang, J.; Zhou, X.; Zhang, H.; Zhang, Q. Tumor-associated macrophages secrete CC-chemokine ligand 2 and induce tamoxifen resistance by activating PI3K/Akt/mTOR in breast cancer. Cancer Sci. 2020, 111, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Ireland, L.; Santos, A.; Ahmed, M.S.; Rainer, C.; Nielsen, S.R.; Quaranta, V.; Weyer-Czernilofsky, U.; Engle, D.D.; Perez-Mancera, P.A.; Coupland, S.E.; et al. Chemoresistance in Pancreatic Cancer Is Driven by Stroma-Derived Insulin-Like Growth Factors. Cancer Res. 2016, 76, 6851–6863. [Google Scholar] [CrossRef] [PubMed]
- Vari, F.; Arpon, D.; Keane, C.; Hertzberg, M.S.; Talaulikar, D.; Jain, S.; Cui, Q.; Han, E.; Tobin, J.; Bird, R.; et al. Immune evasion via PD-1/PD-L1 on NK cells and monocyte/macrophages is more prominent in Hodgkin lymphoma than DLBCL. Blood 2018, 131, 1809–1819. [Google Scholar] [CrossRef]
- De Beule, N.; De Veirman, K.; Maes, K.; De Bruyne, E.; Menu, E.; Breckpot, K.; De Raeve, H.; Van Rampelbergh, R.; Van Ginderachter, J.A.; Schots, R.; et al. Tumour-associated macrophage-mediated survival of myeloma cells through STAT3 activation. J. Pathol. 2017, 241, 534–546. [Google Scholar] [CrossRef]
- Law, A.M.K.; Valdes-Mora, F.; Gallego-Ortega, D. Myeloid-Derived Suppressor Cells as a Therapeutic Target for Cancer. Cells 2020, 9, 561. [Google Scholar] [CrossRef]
- Gabrilovich, D.I. Myeloid-Derived Suppressor Cells. Cancer Immunol. Res. 2017, 5, 3–8. [Google Scholar] [CrossRef]
- Dysthe, M.; Parihar, R. Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1224, 117–140. [Google Scholar]
- Lindau, D.; Gielen, P.; Kroesen, M.; Wesseling, P.; Adema, G.J. The immunosuppressive tumour network: Myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology 2013, 138, 105–115. [Google Scholar] [CrossRef]
- Bruchard, M.; Mignot, G.; Derangère, V.; Chalmin, F.; Chevriaux, A.; Végran, F.; Boireau, W.; Simon, B.; Ryffel, B.; Connat, J.L.; et al. Chemotherapy-triggered cathepsin B release in myeloid-derived suppressor cells activates the Nlrp3 inflammasome and promotes tumor growth. Nat. Med. 2013, 19, 57–64. [Google Scholar]
- Mendoza-Rodriguez, M.; Arevalo Romero, H.; Fuentes-Panana, E.M.; Ayala-Sumuano, J.T.; Meza, I. IL-1β induces up-regulation of BIRC3, a gene involved in chemoresistance to doxorubicin in breast cancer cells. Cancer Lett. 2017, 390, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Finke, J.; Ko, J.; Rini, B.; Rayman, P.; Ireland, J.; Cohen, P. MDSC as a mechanism of tumor escape from sunitinib mediated anti-angiogenic therapy. Int. Immunopharmacol. 2011, 11, 856–861. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.H.; Lee, K.Y.; Chang, Y.L.; Chan, Y.F.; Kuo, L.W.; Lin, T.Y.; Chung, F.T.; Kuo, C.S.; Yu, C.T.; Lin, S.M.; et al. CD14(+)S100A9(+) monocytic myeloid-derived suppressor cells and their clinical relevance in non-small cell lung cancer. Am. J. Respir. Crit. Care Med. 2012, 186, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, I.R.; Condamine, T.; Lin, C.; Herlihy, S.E.; Garfall, A.; Vogl, D.T.; Gabrilovich, D.I.; Nefedova, Y. Bone marrow PMN-MDSCs and neutrophils are functionally similar in protection of multiple myeloma from chemotherapy. Cancer Lett. 2016, 371, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; Cagnon, L.; Costa-Nunes, C.M.; Baumgaertner, P.; Montandon, N.; Leyvraz, L.; Michielin, O.; Romano, E.; Speiser, D.E. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol. Immunother. 2014, 63, 247–257. [Google Scholar] [CrossRef]
- Tarhini, A.A.; Edington, H.; Butterfield, L.H.; Lin, Y.; Shuai, Y.; Tawbi, H.; Sander, C.; Yin, Y.; Holtzman, M.; Johnson, J.; et al. Immune monitoring of the circulation and the tumor microenvironment in patients with regionally advanced melanoma receiving neoadjuvant ipilimumab. PLoS ONE 2014, 9, e87705. [Google Scholar] [CrossRef]
- Highfill, S.L.; Cui, Y.; Giles, A.J.; Smith, J.P.; Zhang, H.; Morse, E.; Kaplan, R.N.; Mackall, C.L. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci. Transl. Med. 2014, 6, 237ra67. [Google Scholar] [CrossRef]
- Weber, J.; Gibney, G.; Kudchadkar, R.; Yu, B.; Cheng, P.; Martinez, A.J.; Kroeger, J.; Richards, A.; McCormick, L.; Moberg, V.; et al. Phase I/II study of metastatic melanoma patients treated with nivolumab who had progressed after ipilimumab. Cancer Immunol. Res. 2016, 4, 345–353. [Google Scholar] [CrossRef]
- Kvedaraite, E.; Ginhoux, F. Human dendritic cells in cancer. Sci. Immunol. 2022, 7, eabm9409. [Google Scholar] [CrossRef]
- Keirsse, J.; Van Damme, H.; Van Ginderachter, J.A.; Laoui, D. Exploiting tumor-associated dendritic cell heterogeneity for novel cancer therapies. J. Leukoc. Biol. 2017, 102, 317–324. [Google Scholar] [CrossRef]
- Hirohashi, Y.; Sato, N. Tumor-associated dendritic cells: Molecular mechanisms to suppress antitumor immunity. Immunotherapy 2011, 3, 945–947. [Google Scholar] [CrossRef]
- Wang, Y.M.; Qiu, J.J.; Qu, X.Y.; Peng, J.; Lu, C.; Zhang, M.; Zhang, M.X.; Qi, X.L.; Lv, B.; Guo, J.J.; et al. Accumulation of dysfunctional tumor-infiltrating PD-1+ DCs links PD-1/PD-L1 blockade immunotherapeutic response in cervical cancer. Oncoimmunology 2022, 11, 2034257. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.L.; Hung, J.Y.; Tsai, E.M.; Wu, C.Y.; Ho, Y.W.; Jian, S.F.; Yen, M.C.; Chang, W.A.; Hou, M.F.; Kuo, P.L. Benzyl butyl phthalate increases the chemoresistance to doxorubicin/cyclophosphamide by increasing breast cancer-associated dendritic cell-derived CXCL1/GROα and S100A8/A9. Oncol. Rep. 2015, 34, 2889–2900. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.H.; Chen, K.F.; Chou, S.H.; Huang, Y.F.; Wu, C.Y.; Cheng, D.E.; Chen, Y.W.; Yang, C.J.; Hung, J.Y.; Huang, M.S. Lung tumor-associated dendritic cell-derived resistin promoted cancer progression by increasing Wolf-Hirschhorn syndrome candidate 1/Twist pathway. Carcinogenesis 2013, 34, 2600–2609. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.L.; Huang, M.S.; Cheng, D.E.; Hung, J.Y.; Yang, C.J.; Chou, S.H.; Kuo, P.L. Lung tumor-associated dendritic cell-derived amphiregulin increased cancer progression. J. Immunol. 2011, 187, 1733–1744. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tuveson, D.A. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol. Rev. 2021, 101, 147–176. [Google Scholar] [CrossRef]
- Park, D.; Sahai, E.; Rullan, A. SnapShot: Cancer-Associated Fibroblasts. Cell 2020, 181, 486–486.e1. [Google Scholar] [CrossRef]
- Peltanova, B.; Liskova, M.; Gumulec, J.; Raudenska, M.; Polanska, H.H.; Vaculovic, T.; Kalfert, D.; Grega, M.; Plzak, J.; Betka, J.; et al. Sensitivity to Cisplatin in Head and Neck Cancer Cells Is Significantly Affected by Patient-Derived Cancer-Associated Fibroblasts. Int. J. Mol. Sci. 2021, 22, 1912. [Google Scholar] [CrossRef]
- Ma, J.; Song, X.; Xu, X.; Mou, Y. Cancer-Associated Fibroblasts Promote the Chemo-resistance in Gastric Cancer through Secreting IL-11 Targeting JAK/STAT3/Bcl2 Pathway. Cancer Res. Treat. 2019, 51, 194–210. [Google Scholar] [CrossRef]
- Zhai, J.; Shen, J.; Xie, G.; Wu, J.; He, M.; Gao, L.; Zhang, Y.; Yao, X.; Shen, L. Cancer-associated fibroblasts-derived IL-8 mediates resistance to cisplatin in human gastric cancer. Cancer Lett. 2019, 454, 37–43. [Google Scholar] [CrossRef]
- Gonçalves-Ribeiro, S.; Díaz-Maroto, N.G.; Berdiel-Acer, M.; Soriano, A.; Guardiola, J.; Martínez-Villacampa, M.; Salazar, R.; Capellà, G.; Villanueva, A.; Martínez-Balibrea, E.; et al. Carcinoma-associated fibroblasts affect sensitivity to oxaliplatin and 5FU in colorectal cancer cells. Oncotarget 2016, 7, 59766–59780. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Li, Z.; Zhou, J.; Sun, Z.; Bai, C. Response prediction to oxaliplatin plus 5-fluorouracil chemotherapy in patients with colorectal cancer using a four-protein immunohistochemical model. Oncol. Lett. 2019, 18, 2091–2101. [Google Scholar] [CrossRef] [PubMed]
- Che, Y.; Wang, J.; Li, Y.; Lu, Z.; Huang, J.; Sun, S.; Mao, S.; Lei, Y.; Zang, R.; Sun, N.; et al. Cisplatin-activated PAI-1 secretion in the cancer-associated fibroblasts with paracrine effects promoting esophageal squamous cell carcinoma progression and causing chemoresistance. Cell Death Dis. 2018, 9, 759. [Google Scholar] [CrossRef] [PubMed]
- Marusyk, A.; Tabassum, D.P.; Janiszewska, M.; Place, A.E.; Trinh, A.; Rozhok, A.I.; Pyne, S.; Guerriero, J.L.; Shu, S.; Ekram, M.; et al. Spatial Proximity to Fibroblasts Impacts Molecular Features and Therapeutic Sensitivity of Breast Cancer Cells Influencing Clinical Outcomes. Cancer Res. 2016, 76, 6495–6506. [Google Scholar] [CrossRef]
- Mueller, K.L.; Madden, J.M.; Zoratti, G.L.; Kuperwasser, C.; List, K.; Boerner, J.L. Fibroblast-secreted hepatocyte growth factor mediates epidermal growth factor receptor tyrosine kinase inhibitor resistance in triple-negative breast cancers through paracrine activation of Met. Breast Cancer Res. 2012, 14, R104. [Google Scholar] [CrossRef]
- Yu, T.; Yang, G.; Hou, Y.; Tang, X.; Wu, C.; Wu, X.A.; Guo, L.; Zhu, Q.; Luo, H.; Du, Y.E.; et al. Cytoplasmic GPER translocation in cancer-associated fibroblasts mediates cAMP/PKA/CREB/glycolytic axis to confer tumor cells with multidrug resistance. Oncogene 2017, 36, 2131–2145. [Google Scholar] [CrossRef]
- Palomeras, S.; Ruiz-Martínez, S.; Puig, T. Targeting Breast Cancer Stem Cells to Overcome Treatment Resistance. Molecules 2018, 23, 2193. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Z.; Ajani, J.A.; Song, S. Drug resistance and Cancer stem cells. Cell Commun. Signal. 2021, 19, 19. [Google Scholar] [CrossRef]
- Che, F.; Yin, J.; Quan, Y.; Xie, X.; Heng, X.; Du, Y.; Wang, L. TLR4 interaction with LPS in glioma CD133+ cancer stem cells induces cell proliferation.; resistance to chemotherapy and evasion from cytotoxic T lymphocyte-induced cytolysis. Oncotarget 2017, 8, 53495–53507. [Google Scholar] [CrossRef]
- Di Lorenzo, A.; Bolli, E.; Ruiu, R.; Ferrauto, G.; Di Gregorio, E.; Avalle, L.; Savino, A.; Poggio, P.; Merighi, I.F.; Riccardo, F.; et al. Toll-like receptor 2 promotes breast cancer progression and resistance to chemotherapy. Oncoimmunology 2022, 11, 2086752. [Google Scholar] [CrossRef]
- Muñoz-Galván, S.; Carnero, A. Targeting Cancer Stem Cells to Overcome Therapy Resistance in Ovarian Cancer. Cells 2020, 9, 1402. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wu, Z.; Bao, C.; Li, W.; He, H.; Sun, Y.; Chen, Z.; Zhang, H.; Ning, Z. Cancer stem cells in esophageal squamous cell cancer. Oncol. Lett. 2019, 18, 5022–5032. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Dericks, L.; Froment, L.; Boesch, R.; Schmid, R.A.; Karoubi, G. Cisplatin-resistant cells in malignant pleural mesothelioma cell lines show ALDH(high)CD44(+) phenotype and sphere-forming capacity. BMC Cancer 2014, 14, 304. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Bo, Z.; Gong, W.; Guo, Y. Inhibitor of Differentiation 1 (Id1) in Cancer and Cancer Therapy. Int. J. Med. Sci. 2020, 17, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wei, X.; Wu, B.; Xiao, Y.; Yin, M.; Yang, Q. siRNA-mediated knockdown of ID1 disrupts Nanog- and Oct-4-mediated cancer stem cell-likeness and resistance to chemotherapy in gastric cancer cells. Oncol. Lett. 2017, 13, 3014–3024. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, R.; Wu, M.; Smiljanic, S.; Kaskun, O.; Ghannad-Zadeh, K.; Celebre, A.; Isaev, K.; Morrissy, A.S.; Guan, J.; Tong, J.; et al. ID1 Is Critical for Tumorigenesis and Regulates Chemoresistance in Glioblastoma. Cancer Res. 2019, 79, 4057–4071. [Google Scholar] [CrossRef]
- Kudinov, A.E.; Karanicolas, J.; Golemis, E.A.; Boumber, Y. Musashi RNA-Binding Proteins as Cancer Drivers and Novel Therapeutic Targets. Clin. Cancer Res. 2017, 23, 2143–2153. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, J.Y.; Chien, Y.; Yang, Y.P.; Chen, M.T.; Lin, L.T. Overview of the molecular mechanisms of migration and invasion in glioblastoma multiforme. J. Chin. Med. Assoc. 2021, 847, 669–677. [Google Scholar] [CrossRef]
- Legrand, N.; Dixon, D.A.; Sobolewski, C. Stress granules in colorectal cancer: Current knowledge and potential therapeutic applications. World J. Gastroenterol. 2020, 2635, 5223–5247. [Google Scholar] [CrossRef]
- Fang, T.; Lv, H.; Wu, F.; Wang, C.; Li, T.; Lv, G.; Tang, L.; Guo, L.; Tang, S.; Cao, D.; et al. Musashi 2 contributes to the stemness and chemoresistance of liver cancer stem cells via LIN28A activation. Cancer Lett. 2017, 384, 50–59. [Google Scholar] [CrossRef]
- Löblein, M.T.; Falke, I.; Eich, H.T.; Greve, B.; Götte, M.; Troschel, F.M. Dual Knockdown of Musashi RNA-Binding Proteins MSI-1 and MSI-2 Attenuates Putative Cancer Stem Cell Characteristics and Therapy Resistance in Ovarian Cancer Cells. Int. J. Mol. Sci. 2021, 22, 11502. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Molina-Portela, M.D.P.; Parveen, A.; Adler, A.; Adler, C.E.H.; Wang, W.; Ni, M.; Wei, Y.; Atwal, G.; Mohrs, M.; et al. Heterogeneity and chimerism of endothelial cells revealed by single-cell transcriptome in orthotopic liver tumors. Angiogenesis 2020, 23, 581–597. [Google Scholar] [CrossRef] [PubMed]
- Carlson, J.C.; Cantu Gutierrez, M.; Lozzi, B.; Huang-Hobbs, E.; Turner, W.D.; Tepe, B.; Zhang, Y.; Herman, A.M.; Rao, G.; Creighton, C.J.; et al. Identification of diverse tumor endothelial cell populations in malignant glioma. Neuro Oncol. 2021, 23, 932–944. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Zhang, D.; Wu, J.Y.; Xing, K.; Yeo, E.; Li, C.; Zhang, L.; Holland, E.; Yao, L.; Qin, L.; et al. Wnt-mediated endothelial transformation into mesenchymal stem cell-like cells induces chemoresistance in glioblastoma. Sci. Transl. Med. 2020, 12, eaay7522. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y. Adipocyte and lipid metabolism in cancer drug resistance. J. Clin. Investig. 2019, 129, 3006–3017. [Google Scholar] [CrossRef]
- Tung, S.; Shi, Y.; Wong, K.; Zhu, F.; Gorczynski, R.; Laister, R.C.; Minden, M.; Blechert, A.K.; Genzel, Y.; Reichl, U.; et al. PPAR and fatty acid oxidation mediate glucocorticoid resistance in chronic lymphocytic leukemia. Blood 2013, 122, 969–980. [Google Scholar]
- Dobbs, J.L.; Shin, D.; Krishnamurthy, S.; Kuerer, H.; Yang, W.; Richards-Kortum, R. Confocal fluorescence microscopy to evaluate changes in adipocytes in the tumor microenvironment associated with invasive ductal carcinoma and ductal carcinoma in situ. Int. J. Cancer 2016, 139, 1140–1149. [Google Scholar] [CrossRef]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, F.; Cui, J.Y.; Chen, L.; Chen, Y.T.; Liu, B.W. CAFs enhance paclitaxel resistance by inducing EMT through the IL-6/JAK2/STAT3 pathway. Oncol. Rep. 2018, 39, 2081–2090. [Google Scholar] [CrossRef]
- Yin, Y.; Yao, S.; Hu, Y.; Feng, Y.; Li, M.; Bian, Z.; Zhang, J.; Qin, Y.; Qi, X.; Zhou, L.; et al. The Immune-microenvironment Confers Chemoresistance of Colorectal Cancer through Macrophage-Derived IL6. Clin. Cancer Res. 2017, 23, 7375–7387. [Google Scholar] [CrossRef]
- Wu, W.; Ma, D.; Wang, P.; Cao, L.; Lu, T.; Fang, Q.; Zhao, J.; Wang, J. Potential crosstalk of the interleukin-6-heme oxygenase-1-dependent mechanism involved in resistance to lenalidomide in multiple myeloma cells. FEBS J. 2016, 283, 834–849. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, H.; Momeny, M.; Ghaffari, S.H.; Parsanejad, N.; Poursheikhani, A.; Javadikooshesh, S.; Zarrinrad, G.; Esmaeili, F.; Alishahi, Z.; Sabourinejad, Z.; et al. IL-6/IL-6R pathway is a therapeutic target in chemoresistant ovarian cancer. Tumori 2019, 105, 84–91. [Google Scholar] [CrossRef]
- Zhang, L.; Hannay, J.A.; Liu, J.; Das, P.; Zhan, M.; Nguyen, T.; Hicklin, D.J.; Yu, D.; Pollock, R.E.; Lev, D. Vascular endothelial growth factor overexpression by soft tissue sarcoma cells: Implications for tumor growth, metastasis, and chemoresistance. Cancer Res. 2006, 66, 8770–8778. [Google Scholar] [CrossRef]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease, Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef]
- Apicella, M.; Giannoni, E.; Fiore, S.; Ferrari, K.J.; Fernández-Pérez, D.; Isella, C.; Granchi, C.; Minutolo, F.; Sottile, A.; Comoglio, P.M.; et al. Increased Lactate Secretion by Cancer Cells Sustains Non-cell-autonomous Adaptive Resistance to MET and EGFR Targeted Therapies. Cell Metab. 2018, 28, 848–865.e6. [Google Scholar] [CrossRef]
- Moosavi, F.; Giovannetti, E.; Saso, L.; Firuzi, O. HGF/MET pathway aberrations as diagnostic, prognostic, and predictive biomarkers in human cancers. Crit. Rev. Clin. Lab. Sci. 2019, 56, 533–566. [Google Scholar] [CrossRef]
- Miranda, O.; Farooqui, M.; Siegfried, J.M. Status of Agents Targeting the HGF/c-Met Axis in Lung Cancer. Cancers 2018, 10, 280. [Google Scholar] [CrossRef]
- Smart, J.A.; Oleksak, J.E.; Hartsough, E.J. Cell Adhesion Molecules in Plasticity and Metastasis. Mol. Cancer Res. 2021, 19, 25–37. [Google Scholar] [CrossRef] [PubMed]
- De Abreu Pereira, D.; Sandim, V.; Fernandes, T.F.B.; Almeida, V.H.; Rocha, M.R.; do Amaral, R.J.F.C.; Rossi, M.I.D.; Kalume, D.E.; Zingali, R.B. Proteomic Analysis of HCC-1954 and MCF-7 Cell Lines Highlights Crosstalk between αv and β1 Integrins, E-Cadherin and HER-2. Int. J. Mol. Sci. 2022, 23, 10194. [Google Scholar] [CrossRef] [PubMed]
- Sani, S.; Pallaoro, N.; Messe, M.; Bernhard, C.; Etienne-Selloum, N.; Kessler, H.; Marinelli, L.; Entz-Werle, N.; Foppolo, S.; Martin, S.; et al. Temozolomide-Acquired Resistance Is Associated with Modulation of the Integrin Repertoire in Glioblastoma, Impact of α5β1 Integrin. Cancers 2022, 14, 369. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.K.; Kim, J.S.; Lee, C.G.; Park, Y.S.; Kim, S.D.; Yoon, S.O.; Han, D.H.; Lee, K.Y.; Jeong, M.H.; Jo, W.S. Tumor associated macrophages provide the survival resistance of tumor cells to hypoxic microenvironmental condition through IL-6 receptor-mediated signals. Immunobiology 2017, 222, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Dorayappan, K.D.P.; Wanner, R.; Wallbillich, J.J.; Saini, U.; Zingarelli, R.; Suarez, A.A.; Cohn, D.E.; Selvendiran, K. Hypoxia-induced exosomes contribute to a more aggressive and chemoresistant ovarian cancer phenotype, a novel mechanism linking STAT3/Rab proteins. Oncogene 2018, 37, 3806–3821. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.L.; Wang, C.C.; Lin, Y.J.; Wu, C.P.; Hsieh, C.H. Cycling hypoxia induces chemoresistance through the activation of reactive oxygen species-mediated B-cell lymphoma extra-long pathway in glioblastoma multiforme. J. Transl. Med. 2015, 13, 389. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Vaidya, M. Hypoxia inhibits mesenchymal stem cell proliferation through HIF1α-dependent regulation of P27. Mol. Cell. Biochem. 2016, 415, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Horrée, N.; Gort, E.H.; van der Groep, P.; Heintz, A.P.; Vooijs, M.; van Diest, P.J. Hypoxia-inducible factor 1 alpha is essential for hypoxic p27 induction in endometrioid endometrial carcinoma. J. Pathol. 2008, 214, 38–45. [Google Scholar] [CrossRef]
- Méndez-Blanco, C.; Fondevila, F.; García-Palomo, A.; González-Gallego, J.; Mauriz, J.L. Sorafenib resistance in hepatocarcinoma: Role of hypoxia-inducible factors. Exp. Mol. Med. 2018, 50, 134. [Google Scholar] [CrossRef] [PubMed]
- Wörmann, S.M.; Song, L.; Ai, J.; Diakopoulos, K.N.; Kurkowski, M.U.; Görgülü, K.; Ruess, D.; Campbell, A.; Doglioni, C.; Jodrell, D.; et al. Loss of P53 Function Activates JAK2-STAT3 Signaling to Promote Pancreatic Tumor Growth, Stroma Modification, and Gemcitabine Resistance in Mice and Is Associated With Patient Survival. Gastroenterology 2016, 151, 180–193.e12. [Google Scholar] [CrossRef]
- Modani, S.; Tomar, D.; Tangirala, S.; Sriram, A.; Mehra, N.K.; Kumar, R.; Khatri, D.K.; Singh, P.K. An updated review on exosomes, biosynthesis to clinical applications. J. Drug Target. 2021, 29, 925–940. [Google Scholar] [CrossRef]
- Shedden, K.; Xie, X.T.; Chandaroy, P.; Chang, Y.T.; Rosania, G.R. Expulsion of small molecules in vesicles shed by cancer cells, association with gene expression and chemosensitivity profiles. Cancer Res. 2003, 63, 4331–4337. [Google Scholar]
- Lehuédé, C.; Li, X.; Dauvillier, S.; Vaysse, C.; Franchet, C.; Clement, E.; Esteve, D.; Longué, M.; Chaltiel, L.; Le Gonidec, S.; et al. Adipocytes promote breast cancer resistance to chemotherapy, a process amplified by obesity: Role of the major vault protein (MVP). Breast Cancer Res. 2019, 21, 7. [Google Scholar] [CrossRef]
- Dong, X.; Bai, X.; Ni, J.; Zhang, H.; Duan, W.; Graham, P.; Li, Y. Exosomes and breast cancer drug resistance. Cell Death Dis. 2020, 11, 987. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.Z.; Zhou, H.L.; Tang, S.X.; Jiang, T.; Chen, Q.; Gao, R.; Ding, Y.L. Intercellular transfer of P-glycoprotein mediates the formation of stable multi-drug resistance in human bladder cancer BIU-87 cells. Biol. Open 2019, 8, bio041889. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Qiao, D.; Chen, L.; Xu, M.; Chen, S.; Huang, L.; Wang, F.; Chen, Z.; Cai, J.; Fu, L. Chemotherapeutic drugs stimulate the release and recycling of extracellular vesicles to assist cancer cells in developing an urgent chemoresistance. Mol. Cancer 2019, 18, 182. [Google Scholar] [CrossRef]
- Kanlikilicer, P.; Bayraktar, R.; Denizli, M.; Rashed, M.H.; Ivan, C.; Aslan, B.; Mitra, R.; Karagoz, K.; Bayraktar, E.; Zhang, X.; et al. Exosomal miRNA confers chemo resistance via targeting Cav1/p-gp/M2-type macrophage axis in ovarian cancer. EBioMedicine 2018, 38, 100–112. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Qin, Z.; Cai, S.; Yu, L.; Hu, H.; Zeng, S. The role of non-coding RNAs in ABC transporters regulation and their clinical implications of multidrug resistance in cancer. Expert Opin. Drug Metab. Toxicol. 2021, 17, 291–306. [Google Scholar] [CrossRef]
- Takahashi, K.; Yan, I.K.; Wood, J.; Haga, H.; Patel, T. Involvement of extracellular vesicle long noncoding RNA linc-VLDLR, in tumor cell responses to chemotherapy. Mol. Cancer Res. 2014, 12, 1377–1387. [Google Scholar] [CrossRef]
- Ding, C.; Yi, X.; Wu, X.; Bu, X.; Wang, D.; Wu, Z.; Zhang, G.; Gu, J.; Kang, D. Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 2020, 479, 1–12. [Google Scholar] [CrossRef]
- Han, M.; Gu, Y.; Lu, P.; Li, J.; Cao, H.; Li, X.; Qian, X.; Yu, C.; Yang, Y.; Yang, X.; et al. Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol. Cancer 2020, 19, 26. [Google Scholar] [CrossRef]
- Gao, Y.; Li, X.; Zeng, C.; Liu, C.; Hao, Q.; Li, W.; Zhang, K.; Zhang, W.; Wang, S.; Zhao, H.; et al. CD63+ Cancer-Associated Fibroblasts Confer Tamoxifen Resistance to Breast Cancer Cells through Exosomal miR-22. Adv. Sci. Weinh. 2020, 7, 2002518. [Google Scholar] [CrossRef]
- Au Yeung, C.L.; Co, N.N.; Tsuruga, T.; Yeung, T.L.; Kwan, S.Y.; Leung, C.S.; Li, Y.; Lu, E.S.; Kwan, K.; Wong, K.K.; et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat. Commun. 2016, 7, 11150. [Google Scholar] [CrossRef]
- Zhang, Z.; Yin, J.; Lu, C.; Wei, Y.; Zeng, A.; You, Y. Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma. J. Exp. Clin. Cancer Res. 2019, 38, 166. [Google Scholar] [CrossRef]
- Alharbi, M.; Sharma, S.; Guanzon, D.; Lai, A.; Zuñiga, F.; Shiddiky, M.J.A.; Yamauchi, Y.; Salas-Burgos, A.; He, Y.; Pejovic, T.; et al. miRNa signature in small extracellular vesicles and their association with platinum resistance and cancer recurrence in ovarian cancer. Nanomedicine 2020, 28, 102207. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Warshall, C.; Bandyopadhyay, C.; Dutta, D.; Chandran, B. Interactions between exosomes from breast cancer cells and primary mammary epithelial cells leads to generation of reactive oxygen species which induce DNA damage response, stabilization of p53 and autophagy in epithelial cells. PLoS ONE 2014, 9, e97580. [Google Scholar] [CrossRef] [PubMed]
- Hazawa, M.; Tomiyama, K.; Saotome-Nakamura, A.; Obara, C.; Yasuda, T.; Gotoh, T.; Tanaka, I.; Yakumaru, H.; Ishihara, H.; Tajima, K. Radiation increases the cellular uptake of exosomes through CD29/CD81 complex formation. Biochem. Biophys. Res. Commun. 2014, 446, 1165–1171. [Google Scholar] [CrossRef] [PubMed]
- Kok, V.C.; Yu, C.C. Cancer-Derived Exosomes, Their Role in Cancer Biology and Biomarker Development. Int. J. Nanomed. 2020, 15, 8019–8036. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, T.; Taha, E.A.; Calderwood, S.K.; Ono, K. A Novel Model of Cancer Drug Resistance: Oncosomal Release of Cytotoxic and Antibody-Based Drugs. Biology 2020, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Batista, I.A.; Quintas, S.T.; Melo, S.A. The Interplay of Exosomes and NK Cells in Cancer Biology. Cancers 2021, 13, 473. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Xu, X.; Qian, Z.; Zhang, C.; Niu, Y.; Wang, Z.; Sun, J.; Zhang, X.; Yu, Y. The biological functions and clinical applications of exosomes in lung cancer. Cell Mol. Life Sci. 2019, 76, 4613–4633. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; You, Y.; Zhu, X. The Role of Exosomes in the Progression and Therapeutic Resistance of Hematological Malignancies. Front. Oncol. 2022, 12, 887518. [Google Scholar] [CrossRef]
- Zhu, H.; Yu, X.; Zhang, S.; Shu, K. Targeting the Complement Pathway in Malignant Glioma Microenvironments. Front. Cell Dev. Biol. 2021, 9, 657472. [Google Scholar] [CrossRef]
- Lazarova, M.; Steinle, A. The NKG2D axis, an emerging target in cancer immunotherapy. Expert Opin. Ther. Targets 2019, 23, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Fuertes, M.B.; Domaica, C.I.; Zwirner, N.W. Leveraging NKG2D Ligands in Immuno-Oncology. Front. Immunol. 2021, 12, 713158. [Google Scholar] [CrossRef]
- Mhaidly, R.; Mechta-Grigoriou, F. Fibroblast heterogeneity in tumor micro-environment: Role in immunosuppression and new therapies. Semin. Immunol. 2020, 48, 101417. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, R.; Xi, B.; Nie, D.; Xu, H.; Liu, A. Mechanisms of Senescence-Related NKG2D Ligands Release and Immune Escape Induced by Chemotherapy in Neuroblastoma Cells. Front. Cell Dev. Biol. 2022, 10, 829404. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, H.; Choi, Y.J.; Kim, S.Y.; Lee, J.E.; Sung, K.J.; Sung, Y.H.; Pack, C.G.; Jung, M.K.; Han, B.; et al. Exosomal PD-L1 promotes tumor growth through immune escape in non-small cell lung cancer. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pathania, A.S.; Prathipati, P.; Challagundla, K.B. New insights into exosome mediated tumor-immune escape: Clinical perspectives and therapeutic strategies. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188624. [Google Scholar] [CrossRef]
- Ning, T.; Li, J.; He, Y.; Zhang, H.; Wang, X.; Deng, T.; Liu, R.; Li, H.; Bai, M.; Fan, Q.; et al. Exosomal miR-208b related with oxaliplatin resistance promotes Treg expansion in colorectal cancer. Mol. Ther. 2021, 29, 2723–2736. [Google Scholar] [CrossRef]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef]
- Lin, H.; Zhang, L.; Zhang, C.; Liu, P. Exosomal MiR-500a-3p promotes cisplatin resistance and stemness via negatively regulating FBXW7 in gastric cancer. J. Cell Mol. Med. 2020, 24, 8930–8941. [Google Scholar] [CrossRef]
- Shen, M.; Dong, C.; Ruan, X.; Yan, W.; Cao, M.; Pizzo, D.; Wu, X.; Yang, L.; Liu, L.; Ren, X.; et al. Chemotherapy-Induced Extracellular Vesicle miRNAs Promote Breast Cancer Stemness by Targeting ONECUT2. Cancer Res. 2019, 79, 3608–3621. [Google Scholar] [CrossRef]
- Liang, L.; Sun, F.; Wang, H.; Hu, Z. Metabolomics, metabolic flux analysis and cancer pharmacology. Pharmacol. Ther. 2021, 224, 107827. [Google Scholar] [CrossRef]
- Pascale, R.M.; Calvisi, D.F.; Simile, M.M.; Feo, C.F.; Feo, F. The Warburg Effect 97 Years after Its Discovery. Cancers 2020, 12, 2819. [Google Scholar] [CrossRef]
- Abbaszadeh, Z.; Çeşmeli, S.; Biray Avcı, Ç. Crucial players in glycolysis: Cancer progress. Gene 2020, 726, 144158. [Google Scholar] [CrossRef]
- Yang, X.; Lu, Y.; Hang, J.; Zhang, J.; Zhang, T.; Huo, Y.; Liu, J.; Lai, S.; Luo, D.; Wang, L.; et al. Lactate-Modulated Immunosuppression of Myeloid-Derived Suppressor Cells Contributes to the Radioresistance of Pancreatic Cancer. Cancer Immunol. Res. 2020, 8, 1440–1451. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.N.; Luo, Y.; Yang, Y.H.; Fu, J.T.; Geng, X.M.; Shi, J.P.; Yang, J. Lactylation, a Novel Metabolic Reprogramming Code: Current Status and Prospects. Front. Immunol. 2021, 12, 688910. [Google Scholar] [CrossRef]
- Ruprecht, B.; Zaal, E.A.; Zecha, J.; Wu, W.; Berkers, C.R.; Kuster, B.; Lemeer, S. Lapatinib Resistance in Breast Cancer Cells Is Accompanied by Phosphorylation-Mediated Reprogramming of Glycolysis. Cancer Res. 2017, 77, 1842–1853. [Google Scholar] [CrossRef]
- Feng, Y.; Xiong, Y.; Qiao, T.; Li, X.; Jia, L.; Han, Y. Lactate dehydrogenase A, A key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 2018, 7, 6124–6136. [Google Scholar] [CrossRef]
- Xie, H.; Hanai, J.; Ren, J.G.; Kats, L.; Burgess, K.; Bhargava, P.; Signoretti, S.; Billiard, J.; Duffy, K.J.; Grant, A.; et al. Targeting lactate dehydrogenase-a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 2014, 19, 795–809. [Google Scholar] [CrossRef]
- Chakraborty, P.K.; Mustafi, S.B.; Xiong, X.; Dwivedi, S.K.D.; Nesin, V.; Saha, S.; Zhang, M.; Dhanasekaran, D.; Jayaraman, M.; Mannel, R.; et al. MICU1 drives glycolysis and chemoresistance in ovarian cancer. Nat. Commun. 2017, 8, 14634. [Google Scholar] [CrossRef]
- Lin, J.; Xia, L.; Liang, J.; Han, Y.; Wang, H.; Oyang, L.; Tan, S.; Tian, Y.; Rao, S.; Chen, X.; et al. The roles of glucose metabolic reprogramming in chemo-and radio-resistance. J. Exp. Clin. Cancer Res. 2019, 38, 218. [Google Scholar] [CrossRef]
- Capelôa, T.; Benyahia, Z.; Zampieri, L.X.; Blackman, M.C.N.M.; Sonveaux, P. Metabolic and non-metabolic pathways that control cancer resistance to anthracyclines. Semin. Cell Dev. Biol. 2020, 98, 181–191. [Google Scholar] [CrossRef]
- Cheng, C.; Xie, Z.; Li, Y.; Wang, J.; Qin, C.; Zhang, Y. PTBP1 knockdown overcomes the resistance to vincristine and oxaliplatin in drug-resistant colon cancer cells through regulation of glycolysis. Biomed. Pharmacother. 2018, 108, 194–200. [Google Scholar] [CrossRef]
- Sukjoi, W.; Ngamkham, J.; Attwood, P.V.; Jitrapakdee, S. Targeting Cancer Metabolism and Current Anti-Cancer Drugs. Adv. Exp. Med. Biol. 2021, 1286, 15–48. [Google Scholar]
- Broadfield, L.A.; Pane, A.A.; Talebi, A.; Swinnen, J.V.; Fendt, S.M. Lipid metabolism in cancer, new perspectives and emerging mechanisms. Dev. Cell 2021, 56, 1363–1393. [Google Scholar] [CrossRef] [PubMed]
- Icard, P.; Wu, Z.; Fournel, L.; Coquerel, A.; Lincet, H.; Alifano, M. ATP citrate lyase, A central metabolic enzyme in cancer. Cancer Lett. 2020, 471, 125–134. [Google Scholar] [CrossRef]
- Zhou, Y.; Bollu, L.R.; Tozzi, F.; Ye, X.; Bhattacharya, R.; Gao, G.; Dupre, E.; Xia, L.; Lu, J.; Fan, F.; et al. ATP citrate lyase mediates resistance of colorectal cancer cells to SN38. Mol. Cancer Ther. 2013, 12, 2782–2791. [Google Scholar] [CrossRef] [PubMed]
- Germain, N.; Dhayer, M.; Boileau, M.; Fovez, Q.; Kluza, J.; Marchetti, P. Lipid Metabolism and Resistance to Anticancer Treatment. Biology 2020, 91, 474. [Google Scholar] [CrossRef]
- Yang, R.; Yi, M.; Xiang, B. Novel Insights on Lipid Metabolism Alterations in Drug Resistance in Cancer. Front. Cell Dev. Biol. 2022, 10, 875318. [Google Scholar] [CrossRef]
- Schcolnik-Cabrera, A.; Chávez-Blanco, A.; Domínguez-Gómez, G.; Taja-Chayeb, L.; Morales-Barcenas, R.; Trejo-Becerril, C.; Perez-Cardenas, E.; Gonzalez-Fierro, A.; Dueñas-González, A. Orlistat as a FASN inhibitor and multitargeted agent for cancer therapy. Expert Opin. Investig. Drugs 2018, 27, 475–489. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.; Duke, G.; Heuer, T.S.; O’Farrell, M.; Wagman, A.S.; McCulloch, W.; Kemble, G. Fatty acid synthase-Modern tumor cell biology insights into a classical oncology target. Pharmacol. Ther. 2017, 177, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Migita, T.; Ruiz, S.; Fornari, A.; Fiorentino, M.; Priolo, C.; Zadra, G.; Inazuka, F.; Grisanzio, C.; Palescandolo, E.; Shin, E.; et al. Fatty acid synthase, a metabolic enzyme and candidate oncogene in prostate cancer. J. Natl. Cancer Inst. 2009, 101, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Karthikeyan, C.; Moorthy, N.S.H.N. Recent Advances in the Development of Fatty Acid Synthase Inhibitors as Anticancer Agents. Mini Rev. Med. Chem. 2020, 20, 1820–1837. [Google Scholar] [CrossRef] [PubMed]
- Al-Bahlani, S.; Al-Lawati, H.; Al-Adawi, M.; Al-Abri, N.; Al-Dhahli, B.; Al-Adawi, K. Fatty acid synthase regulates the chemosensitivity of breast cancer cells to cisplatin-induced apoptosis. Apoptosis 2017, 22, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wang, X.; Du, Y.; Zhang, X.; Ling, Y. FASN Knockdown Inhibited Anoikis Resistance of Gastric Cancer Cells via P-ERK1/2/Bcl-xL Pathway. Gastroenterol. Res. Pract. 2021, 2021, 6674204. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Chen, L.; Zhou, M.; Zhang, J.; Sun, L.; Huang, N.; Bin, J.; Liao, Y.; Liao, W. MACC1 decreases the chemosensitivity of gastric cancer cells to oxaliplatin by regulating FASN expression. Oncol. Rep. 2017, 37, 2583–2592. [Google Scholar] [CrossRef]
- Zhelev, Z.; Aoki, I.; Lazarova, D.; Vlaykova, T.; Higashi, T.; Bakalova, R. A “Weird” Mitochondrial Fatty Acid Oxidation as a Metabolic “Secret” of Cancer. Oxid. Med. Cell Longev. 2022, 2022, 2339584. [Google Scholar] [CrossRef]
- Cacciola, N.A.; Sgadari, M.; Petillo, O.; Margarucci, S.; Martano, M.; Cocchia, N.; Maiolino, P.; Restucci, B. Carnitine palmitoyltransferase 1 A expression profile in canine mammary tumors. Vet. J. 2020, 257, 105453. [Google Scholar] [CrossRef]
- Rios-Colon, L.; Kumar, P.; Kim, S.; Sharma, M.; Su, Y.; Kumar, A.; Singh, S.; Stocks, N.; Liu, L.; Joshi, M.; et al. Carnitine Palmitoyltransferase 1 Regulates Prostate Cancer Growth under Hypoxia. Cancers 2021, 13, 6302. [Google Scholar] [CrossRef]
- Schlaepfer, I.R.; Joshi, M. CPT1A-mediated Fat Oxidation, Mechanisms, and Therapeutic Potential. Endocrinology 2020, 161, bqz046. [Google Scholar] [CrossRef]
- Li, J.; Zhao, S.; Zhou, X.; Zhang, T.; Zhao, L.; Miao, P.; Song, S.; Sun, X.; Liu, J.; Zhao, X.; et al. Inhibition of lipolysis by mercaptoacetate and etomoxir specifically sensitize drug-resistant lung adenocarcinoma cell to paclitaxel. PLoS ONE 2013, 8, e74623. [Google Scholar] [CrossRef] [PubMed]
- Vellinga, T.T.; Borovski, T.; de Boer, V.C.; Fatrai, S.; van Schelven, S.; Trumpi, K.; Verheem, A.; Snoeren, N.; Emmink, B.L.; Koster, J.; et al. SIRT1/PGC1α-Dependent Increase in Oxidative Phosphorylation Supports Chemotherapy Resistance of Colon Cancer. Clin. Cancer Res. 2015, 21, 2870–2879. [Google Scholar] [CrossRef]
- Decker, C.W.; Garcia, J.; Gatchalian, K.; Arceneaux, D.; Choi, C.; Han, D.; Hernandez, J.B. Mitofusin-2 mediates doxorubicin sensitivity and acute resistance in Jurkat leukemia cells. Biochem. Biophys. Rep. 2020, 24, 100824. [Google Scholar] [CrossRef]
- Genovese, I.; Carinci, M.; Modesti, L.; Aguiari, G.; Pinton, P.; Giorgi, C. Mitochondria, Insights into Crucial Features to Overcome Cancer Chemoresistance. Int. J. Mol. Sci. 2021, 22, 4770. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.H.; Pelicano, H.; Zhou, Y.; Carew, J.S.; Feng, L.; Bhalla, K.N.; Keating, M.J.; Huang, P. Inhibition of glycolysis in cancer cells, a novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia. Cancer Res. 2005, 65, 613–621. [Google Scholar] [CrossRef]
- Marcucci, F.; Rumio, C. Glycolysis-induced drug resistance in tumors-A response to danger signals? Neoplasia 2021, 23, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Duanmu, X.; Zeng, L.; Liu, B.; Song, Z. Mitochondrial DNA: Distribution, Mutations, and Elimination. Cells 2019, 8, 379. [Google Scholar] [CrossRef] [PubMed]
- Kopinski, P.K.; Singh, L.N.; Zhang, S.; Lott, M.T.; Wallace, D.C. Mitochondrial DNA variation and cancer. Nat. Rev. Cancer 2021, 21, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Mou, J.J.; Peng, J.; Shi, Y.Y.; Li, N.; Wang, Y.; Ke, Y.; Zhou, Y.F.; Zhou, F.X. Mitochondrial DNA content reduction induces aerobic glycolysis and reversible resistance to drug-induced apoptosis in SW480 colorectal cancer cells. Biomed. Pharmacother. 2018, 103, 729–737. [Google Scholar] [CrossRef]
- Moro, L. Mitochondrial DNA and MitomiR Variations in Pancreatic Cancer, Potential Diagnostic and Prognostic Biomarkers. Int. J. Mol. Sci. 2021, 22, 9692. [Google Scholar] [CrossRef]
- Zampieri, L.X.; Grasso, D.; Bouzin, C.; Brusa, D.; Rossignol, R.; Sonveaux, P. Mitochondria Participate in Chemoresistance to Cisplatin in Human Ovarian Cancer Cells. Mol. Cancer Res. 2020, 18, 1379–1391. [Google Scholar] [CrossRef] [PubMed]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, N.; El-Baba, C.; Araji, K.; El-Khoury, R.; Usta, J.; Darwiche, N. The Pentose Phosphate Pathway in Cancer, Regulation and Therapeutic Opportunities. Chemotherapy 2021, 66, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cai, Q.; Zhou, F.; Liu, J.; Jin, X.; Ni, P.; Lu, M.; Wang, G.; Zhang, J. Impaired pentose phosphate pathway in the development of 3D MCF-7 cells mediated intracellular redox disturbance and multi-cellular resistance without drug induction. Redox. Biol. 2018, 15, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Fu, A.; Wu, R.; Wei, N.; Song, K.; Lim, S.; Luo, K.Q. High Expression of G6PD Increases Doxorubicin Resistance in Triple Negative Breast Cancer Cells by Maintaining GSH Level. Int. J. Biol. Sci. 2022, 18, 1120–1133. [Google Scholar] [CrossRef]
- Giacomini, I.; Ragazzi, E.; Pasut, G.; Montopoli, M. The Pentose Phosphate Pathway and Its Involvement in Cisplatin Resistance. Int. J. Mol. Sci. 2020, 21, 937. [Google Scholar] [CrossRef]
- Soltani, M.; Zhao, Y.; Xia, Z.; Ganjalikhani Hakemi, M.; Bazhin, A.V. The Importance of Cellular Metabolic Pathways in Pathogenesis and Selective Treatments of Hematological Malignancies. Front. Oncol. 2021, 11, 767026. [Google Scholar] [CrossRef]
- Cho, E.S.; Cha, Y.H.; Kim, H.S.; Kim, N.H.; Yook, J.I. The Pentose Phosphate Pathway as a Potential Target for Cancer Therapy. Biomol. Ther. Seoul 2018, 26, 29–38. [Google Scholar] [CrossRef]
- Eichler, J. Protein glycosylation. Curr. Biol. 2019, 29, R229–R231. [Google Scholar] [CrossRef]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef]
- Lin, G.; Zhao, R.; Wang, Y.; Han, J.; Gu, Y.; Pan, Y.; Ren, C.; Ren, S.; Xu, C. Dynamic analysis of N-glycomic and transcriptomic changes in the development of ovarian cancer cell line A2780 to its three cisplatin-resistant variants. Ann. Transl. Med. 2020, 8, 289. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Sun, H.; Lu, P.; Ge, C.; Zhang, L.; Li, H.; Zhao, F.; Tian, H.; Zhang, L.; Chen, T.; et al. Tunicamycin potentiates cisplatin anticancer efficacy through the DPAGT1/Akt/ABCG2 pathway in mouse Xenograft models of human hepatocellular carcinoma. Mol. Cancer Ther. 2013, 12, 2874–2884. [Google Scholar] [CrossRef] [PubMed]
- Wojtowicz, K.; Januchowski, R.; Nowicki, M.; Zabel, M. Inhibition of protein glycosylation reverses the MDR phenotype of cancer cell lines. Biomed. Pharmacother. 2015, 74, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.M.; Latorre, A.O.; Hueza, I.M.; Sanches, D.S.; Lippi, L.L.; Gardner, D.R.; Spinosa, H.S. Increased antitumor efficacy by the combined administration of swainsonine and cisplatin in vivo. Phytomedicine 2011, 18, 1096–1101. [Google Scholar] [CrossRef]
- Kanwal, S.; Fardini, Y.; Pagesy, P.; N’tumba-Byn, T.; Pierre-Eugène, C.; Masson, E.; Hampe, C.; Issad, T. O-GlcNAcylation-inducing treatments inhibit estrogen receptor α expression and confer resistance to 4-OH-tamoxifen in human breast cancer-derived MCF-7 cells. PLoS ONE 2013, 8, e69150. [Google Scholar] [CrossRef]
- Pepe, F.; Pagotto, S.; Soliman, S.; Rossi, C.; Lanuti, P.; Braconi, C.; Mariani-Costantini, R.; Visone, R.; Veronese, A. Regulation of miR-483-3p by the O-linked N-acetylglucosamine transferase links chemosensitivity to glucose metabolism in liver cancer cells. Oncogenesis 2017, 6, e328. [Google Scholar] [CrossRef]
- Shen, L.; Dong, X.; Wang, Y.; Qiu, L.; Peng, F.; Luo, Z. β3GnT8 regulates oxaliplatin resistance by altering integrin β1 glycosylation in colon cancer cells. Oncol. Rep. 2018, 39, 2006–2014. [Google Scholar]
- Han, X.; Zhang, X.; Li, H.; Huang, S.; Zhang, S.; Wang, F.; Shi, Y. Tunicamycin enhances the antitumor activity of trastuzumab on breast cancer in vitro and in vivo. Oncotarget 2015, 6, 38912–38925. [Google Scholar] [CrossRef]
- Wu, J.; Chen, S.; Liu, H.; Zhang, Z.; Ni, Z.; Chen, J.; Yang, Z.; Nie, Y.; Fan, D. Tunicamycin specifically aggravates ER stress and overcomes chemoresistance in multidrug-resistant gastric cancer cells by inhibiting N-glycosylation. J. Clin. Cancer Res. 2018, 37, 272. [Google Scholar] [CrossRef]
- Kalra, A.V.; Campbell, R.B. Mucin overexpression limits the effectiveness of 5-FU by reducing intracellular drug uptake and antineoplastic drug effects in pancreatic tumours. Eur. J. Cancer 2009, 45, 164–173. [Google Scholar] [CrossRef]
- Yu, L.G. Cancer cell resistance to anoikis, MUC1 glycosylation comes to play. Cell Death Dis. 2017, 8, e2962. [Google Scholar] [CrossRef] [PubMed]
- Farahmand, L.; Merikhian, P.; Jalili, N.; Darvishi, B.; Majidzadeh-A, K. Significant Role of MUC1 in Development of Resistance to Currently Existing Anti-cancer Therapeutic Agents. Curr. Cancer Drug Targets 2018, 18, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.H.; He, Y.; Hasnain, S.Z.; Wang, R.; Tong, H.; Clarke, D.T.; Lourie, R.; Oancea, I.; Wong, K.Y.; Lumley, J.W.; et al. MUC13 protects colorectal cancer cells from death by activating the NF-κB pathway and is a potential therapeutic target. Oncogene 2017, 36, 700–713. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, D.; Sabri, S.; Xu, Y.; Graham, K.; Bhatnagar, P.; Suresh, M.; Abdulkarim, B. Spontaneous epithelial-mesenchymal transition and resistance to HER-2-targeted therapies in HER-2-positive luminal breast cancer. PLoS ONE 2013, 8, e71987. [Google Scholar] [CrossRef]
- Cheng, S.; Mao, Q.; Dong, Y.; Ren, J.; Su, L.; Liu, J.; Liu, Q.; Zhou, J.; Ye, X.; Zheng, S.; et al. GNB2L1 and its O-GlcNAcylation regulates metastasis via modulating epithelial-mesenchymal transition in the chemoresistance of gastric cancer. PLoS ONE 2017, 12, e0182696. [Google Scholar] [CrossRef]
- Kang, K.A.; Piao, M.J.; Ryu, Y.S.; Kang, H.K.; Chang, W.Y.; Keum, Y.S.; Hyun, J.W. Interaction of DNA demethylase and histone methyltransferase upregulates Nrf2 in 5-fluorouracil-resistant colon cancer cells. Oncotarget 2016, 7, 40594–40620. [Google Scholar] [CrossRef]
- Russo, M.; Russo, G.L. Autophagy inducers in cancer. Biochem. Pharmacol. 2018, 153, 51–61. [Google Scholar] [CrossRef]
- Ferro, F.; Servais, S.; Besson, P.; Roger, S.; Dumas, J.F.; Brisson, L. Autophagy and mitophagy in cancer metabolic remodelling. Semin. Cell Dev. Biol. 2020, 98, 129–138. [Google Scholar] [CrossRef]
- Panigrahi, D.P.; Praharaj, P.P.; Bhol, C.S.; Mahapatra, K.K.; Patra, S.; Behera, B.P.; Mishra, S.R.; Bhutia, S.K. The emerging, multifaceted role of mitophagy in cancer and cancer therapeutics. Semin. Cancer Biol. 2020, 66, 45–58. [Google Scholar] [CrossRef]
- Jin, P.; Jiang, J.; Zhou, L.; Huang, Z.; Nice, E.C.; Huang, C.; Fu, L. Mitochondrial adaptation in cancer drug resistance: Prevalence, mechanisms, and management. J. Hematol. Oncol. 2022, 15, 97. [Google Scholar] [CrossRef]
- Poillet-Perez, L.; Sarry, J.E.; Joffre, C. Autophagy is a major metabolic regulator involved in cancer therapy resistance. Cell Rep. 2021, 36, 109528. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.F.; Wang, X.Y.; Fu, Z.Q.; Peng, Q.H.; Zhang, J.Y.; Ye, F.; Fu, Y.F.; Zhou, C.Y.; Lu, W.G.; Cheng, X.D.; et al. TXNDC17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy 2015, 11, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Zhou, N.; Zhang, D.; Zhang, K.; Zheng, W.; Bao, Y.; Yang, W. PFKFB3 Inhibition Attenuates Oxaliplatin-Induced Autophagy and Enhances Its Cytotoxicity in Colon Cancer Cells. Int. J. Mol. Sci. 2019, 20, 5415. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Loveless, J.; Shay, C.; Teng, Y. Targeting ROS-Mediated Crosstalk Between Autophagy and Apoptosis in Cancer. Adv. Exp. Med. Biol. 2020, 1260, 1–12. [Google Scholar] [PubMed]
- Okon, I.S.; Coughlan, K.A.; Zhang, M.; Wang, Q.; Zou, M.H. Gefitinib-mediated reactive oxygen specie (ROS) instigates mitochondrial dysfunction and drug resistance in lung cancer cells. J. Biol. Chem. 2015, 290, 9101–9110. [Google Scholar] [CrossRef]
- Xiao, X.; Wang, W.; Li, Y.; Yang, D.; Li, X.; Shen, C.; Liu, Y.; Ke, X.; Guo, S.; Guo, Z. HSP90AA1-mediated autophagy promotes drug resistance in osteosarcoma. J. Exp. Clin. Cancer Res. 2018, 37, 201. [Google Scholar] [CrossRef]
- Liu, Y.; Gu, S.; Li, H.; Wang, J.; Wei, C.; Liu, Q. SNHG16 promotes osteosarcoma progression and enhances cisplatin resistance by sponging miR-16 to upregulate ATG4B expression. Biochem. Biophys. Res. Commun. 2019, 518, 127–133. [Google Scholar] [CrossRef]
- Lin, J.Z.; Wang, W.W.; Hu, T.T.; Zhu, G.Y.; Li, L.N.; Zhang, C.Y.; Xu, Z.; Yu, H.B.; Wu, H.F.; Zhu, J.G. FOXM1 contributes to docetaxel resistance in castration-resistant prostate cancer by inducing AMPK/mTOR-mediated autophagy. Cancer Lett. 2020, 469, 481–489. [Google Scholar] [CrossRef]
- Onorati, A.V.; Dyczynski, M.; Ojha, R.; Amaravadi, R.K. Targeting autophagy in cancer. Cancer 2018, 124, 3307–3318. [Google Scholar] [CrossRef]
- Thorburn, A. Crosstalk between autophagy and apoptosis, Mechanisms and therapeutic implications. Prog. Mol. Biol. Transl. Sci. 2020, 172, 55–65. [Google Scholar]
- Xiong, H.; Ni, Z.; He, J.; Jiang, S.; Li, X.; He, J.; Gong, W.; Zheng, L.; Chen, S.; Li, B.; et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene 2017, 36, 3528–3540. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Zhou, Q.; Yuan, Y.W.; Zhou, L.Q.; Liu, L.; Li, S.H.; Liu, C. METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J. Cancer Res. Clin. Oncol. 2019, 145, 2507–2517. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.R.; Yu, Y.C.; You, S.W.; Li, K.Q.; Tong, X.C.; Chen, S.R.; Chen, E.D.; Lin, X.Z.; Chen, Y.F. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol. Cancer 2017, 16, 174. [Google Scholar]
- Lei, Y.; Tang, L.; Hu, J.; Wang, S.; Liu, Y.; Yang, M.; Zhang, J.; Tang, B. Inhibition of MGMT-mediated autophagy suppression decreases cisplatin chemosensitivity in gastric cancer. Biomed. Pharmacother. 2020, 125, 109896. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Chen, Y.; Shen, Y.; Tantai, J. Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis. 2019, 10, 429. [Google Scholar] [CrossRef]
- Kocaturk, N.M.; Akkoc, Y.; Kig, C.; Bayraktar, O.; Gozuacik, D.; Kutlu, O. Autophagy as a molecular target for cancer treatment. Eur. J. Pharm. Sci. 2019, 134, 116–137. [Google Scholar] [CrossRef]
- Mehnert, J.M.; Mitchell, T.C.; Huang, A.C.; Aleman, T.S.; Kim, B.J.; Schuchter, L.M.; Linette, G.P.; Karakousis, G.C.; Mitnick, S.; Giles, L.; et al. BAMM (BRAF Autophagy and MEK Inhibition in Melanoma): A Phase I/II Trial of Dabrafenib, Trametinib, and Hydroxychloroquine in Advanced BRAFV600-mutant Melanoma. Clin. Cancer Res. 2022, 28, 1098–1106. [Google Scholar] [CrossRef]
- Awada, G.; Schwarze, J.K.; Tijtgat, J.; Fasolino, G.; Kruse, V.; Neyns, B. A lead-in safety study followed by a phase 2 clinical trial of dabrafenib, trametinib and hydroxychloroquine in advanced BRAFV600 mutant melanoma patients previously treated with BRAF-/MEK-inhibitors and immune checkpoint inhibitors. Melanoma Res. 2022, 32, 183–191. [Google Scholar] [CrossRef]
- Zeh, H.J.; Bahary, N.; Boone, B.A.; Singhi, A.D.; Miller-Ocuin, J.L.; Normolle, D.P.; Zureikat, A.H.; Hogg, M.E.; Bartlett, D.L.; Lee, K.K.; et al. A Randomized Phase II Preoperative Study of Autophagy Inhibition with High-Dose Hydroxychloroquine and Gemcitabine/Nab-Paclitaxel in Pancreatic Cancer Patients. Clin. Cancer Res. 2020, 26, 3126–3134. [Google Scholar] [CrossRef]
- Compter, I.; Eekers, D.B.P.; Hoeben, A.; Rouschop, K.M.A.; Reymen, B.; Ackermans, L.; Beckervordersantforth, J.; Bauer, N.J.C.; Anten, M.M.; Wesseling, P.; et al. Chloroquine combined with concurrent radiotherapy and temozolomide for newly diagnosed glioblastoma, a phase IB trial. Autophagy 2021, 17, 2604–2612. [Google Scholar] [CrossRef]

| Anti-Cancer Drugs | Indicated Cancers |
|---|---|
| Chemotherapeutic drugs | |
| Cisplatin | Gastric cancer |
| Breast cancer | |
| Head and neck squamous cell carcinoma | |
| Gastric cancer | |
| Esophageal squamous cell carcinoma | |
| Pleural mesotheliom | |
| Glioblastoma | |
| Osteosarcoma | |
| Ehrlich ascites carcinoma | |
| Hepatocellular carcinoma | |
| Ovarian cancer | |
| Paclitaxel | Breast cancer |
| Glioma | |
| Hepatocellular carcinoma | |
| Osteosarcoma | |
| Ovarian cancer | |
| Lung cancer | |
| Colorectal cancer | |
| Gemcitabine | Pancreatic cancer |
| Hepatocellular carcinoma | |
| Doxorubicin | Gastric cancer |
| Breast cancer | |
| Esophageal cancer | |
| Prostate cancer | |
| Bladder cancer | |
| Osteosarcoma | |
| Prostate cancer | |
| Multiple myeloma | |
| Soft tissue sarcoma | |
| Hepatocellular carcinoma | |
| Colorectal cancer | |
| Etoposide | Gastric cancer |
| Glioma | |
| Irinotecan | Colorectal cancer |
| 5-fluorouracil | Colorectal cancer |
| Gastric cancer | |
| Pancreatic cancer | |
| Esophageal cancer | |
| Hepatocellular carcinoma | |
| Oxaliplatin | Colorectal cancer |
| Gastric cancer | |
| Prostate cancer | |
| Hepatocellular carcinoma | |
| Temozolomide | Glioblastoma/Glioma |
| Oligodendroglioma | |
| Vincristine | Acute lymphoblastic leukemia |
| Gastric cancer | |
| Oral epidermoid carcinoma | |
| Colorectal cancer | |
| Carboplatin | Ovarian cancer |
| Adriamycin | Breast cancer |
| Gastric cancer | |
| Osteosarcoma | |
| Glioma | |
| Burkitt lymphoma | |
| B-cell lymphoma | |
| Cytarabine | Burkitt lymphoma |
| B-cell lymphoma | |
| Staurosporine | Pancreatic cancer |
| Mitomycin C | Hepatocellular carcinoma |
| Camptothecin | Hepatocellular carcinoma |
| Melphalan | Multiple myeloma |
| Cyclophosphamide | Breast cancer |
| Epirubicin | Breast cancer |
| Molecular targeted inhibitors | |
| Sorafenib | Hepatocellular carcinoma |
| Tocilizumab | Ovarian cancer |
| Gefitinib | Lung cancer |
| Glioblastoma | |
| Breast cancer | |
| Trastuzumab | Breast cancer |
| Rituximab | Chronic lymphoblastic leukemia |
| B-cell lymphoma | |
| Cetuximab | Oral squamous cell carcinoma |
| Rapatinib | Breast cancer |
| Sunitinib | Renal cell carcinoma |
| Lapatinib | Breast cancer |
| Afatinib | Lung cancer |
| Bevacizumab | Lung cancer |
| Immune checkpoint inhibitors | |
| Nivolumab | Lung cancer |
| Melanoma | |
| Pembrolizumab | Lung cancer |
| Melanoma | |
| Colorectal cancer | |
| Cervical cancer | |
| Atezolizumab | Lung cancer |
| Ipilimumab | Melanoma |
| Lung cancer | |
| Gastro-esophageal cancer | |
| Tremelimumab | Lung cancer |
| Gastro-esophageal cancer | |
| Tislelizumab | Melanoma |
| Lung cancer | |
| Others | |
| Lenalidomide | Multiple myeloma |
| Bortezomib | Breast cancer |
| Multiple myeloma | |
| Tamoxifen | Breast cancer |
| Dexamethasone | Chronic lymphocytic leukemia |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Li, X.; Guo, Q. Drug Resistance in Cancers: A Free Pass for Bullying. Cells 2022, 11, 3383. https://doi.org/10.3390/cells11213383
Li J, Li X, Guo Q. Drug Resistance in Cancers: A Free Pass for Bullying. Cells. 2022; 11(21):3383. https://doi.org/10.3390/cells11213383
Chicago/Turabian StyleLi, Jing, Xiao Li, and Qie Guo. 2022. "Drug Resistance in Cancers: A Free Pass for Bullying" Cells 11, no. 21: 3383. https://doi.org/10.3390/cells11213383
APA StyleLi, J., Li, X., & Guo, Q. (2022). Drug Resistance in Cancers: A Free Pass for Bullying. Cells, 11(21), 3383. https://doi.org/10.3390/cells11213383






