Analysis of the Compositional Features and Codon Usage Pattern of Genes Involved in Human Autophagy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval
2.2. Nucleotide Composition
2.3. Relative Synonymous Codon Usage (RSCU)
2.4. Effective Number of Codons (ENC)
2.5. Parity Plot
2.6. Neutrality Plot
2.7. Correspondence Analysis
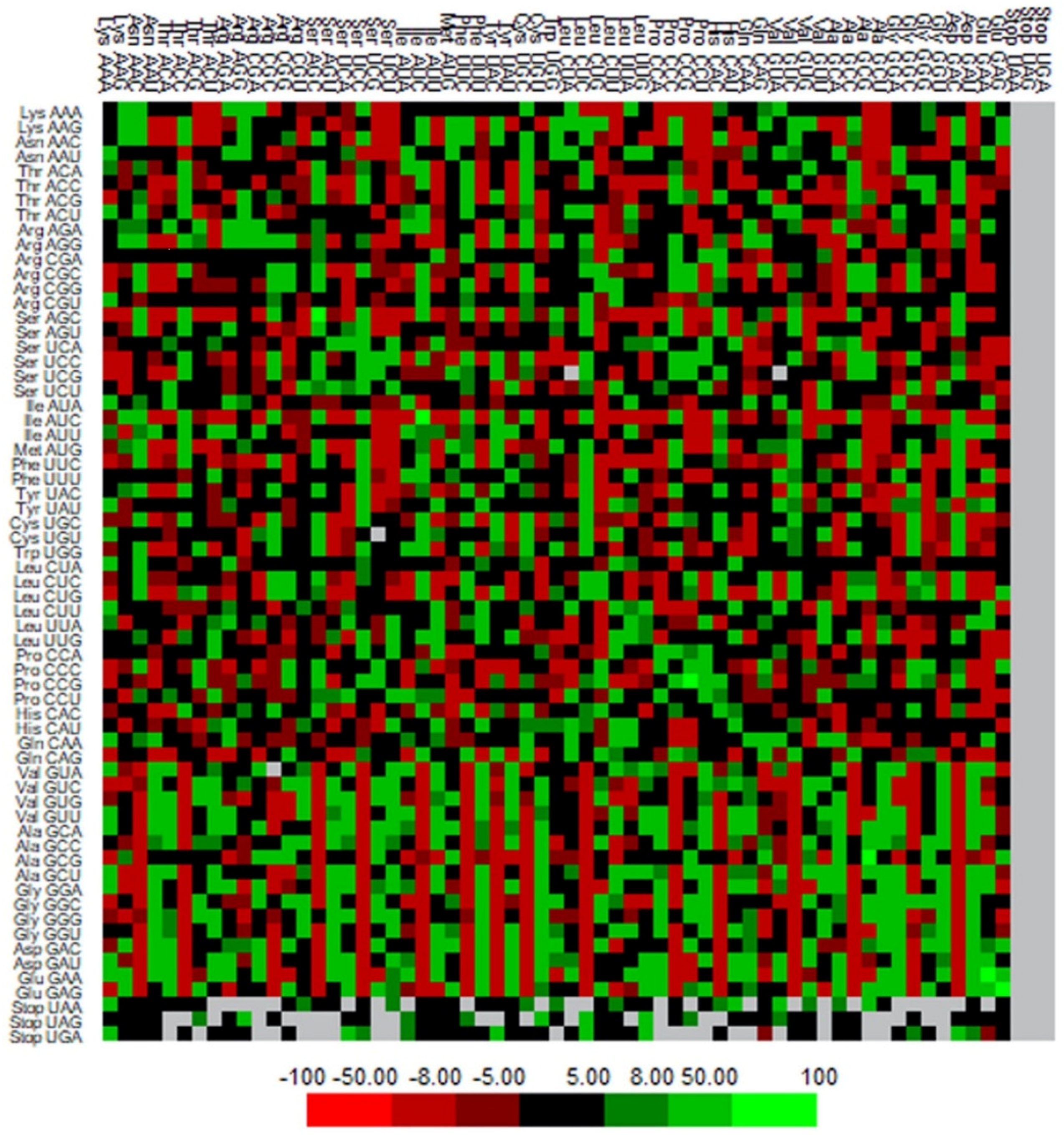
2.8. Codon Context Analysis
2.9. Protein Properties
2.10. Minimum Free Energy (mFE) of mRNA
2.11. Software
3. Results
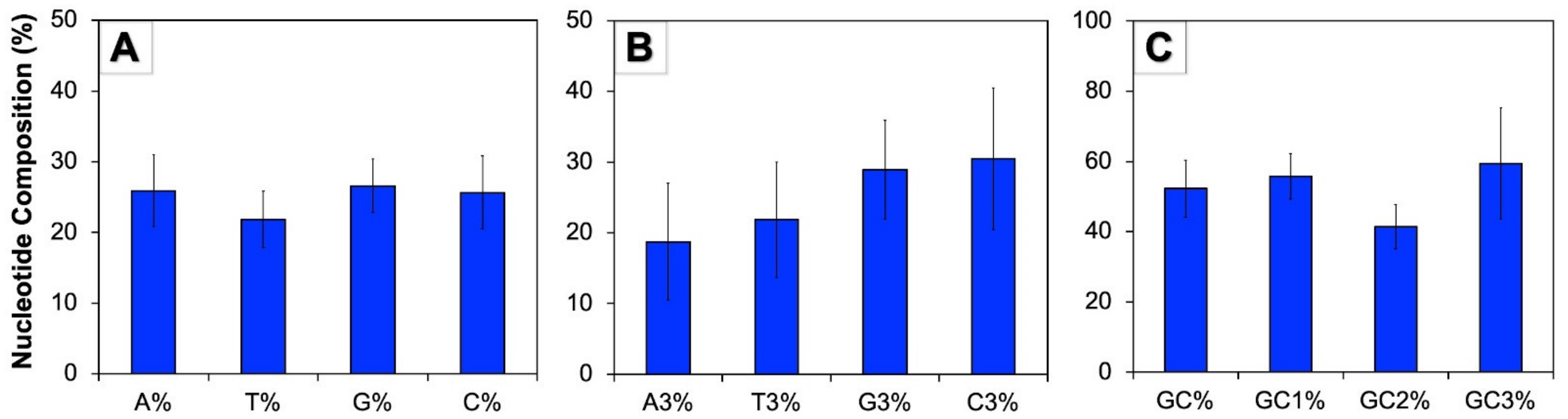
3.1. Human Autophagy Genes Contain Higher GC Content
3.2. Human Autophagy Genes Show Low Codon Usage Bias (CUB)
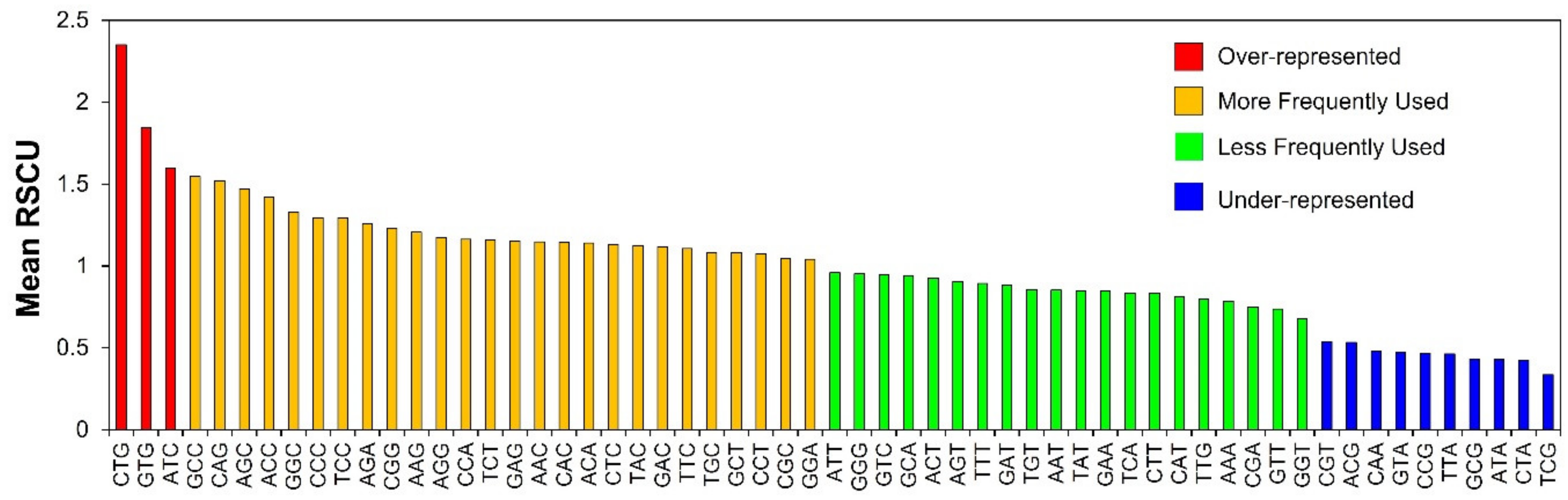
3.3. RSCU Analysis Indicates Stable Genetic Composition of Human Autophagy Genes
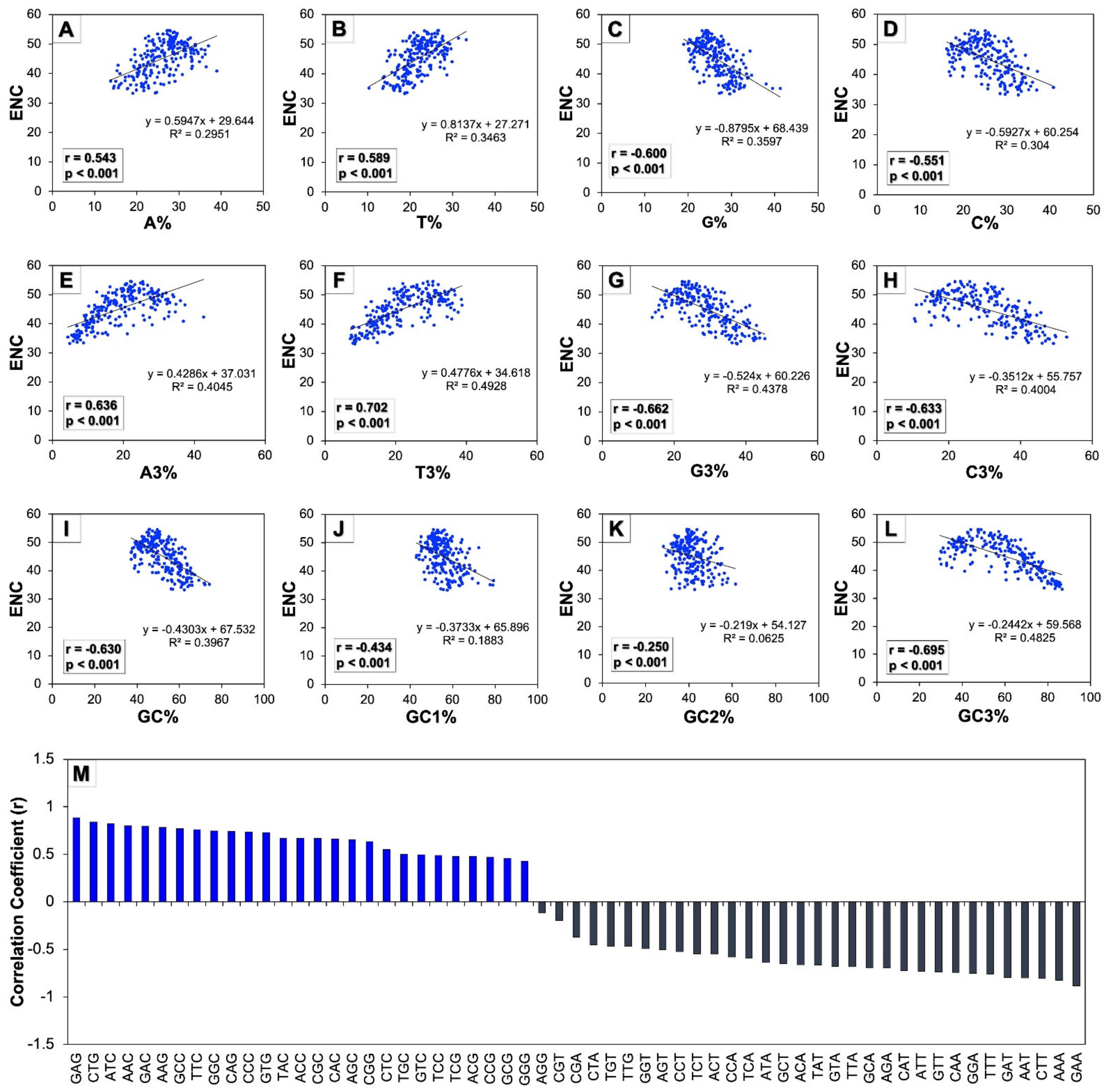
3.4. Base Composition Influences the Codon Usage in Human Autophagy Genes
3.5. Both Natural Selection and Mutation Pressure Determine the Codon Usage in Human Autophagy Genes
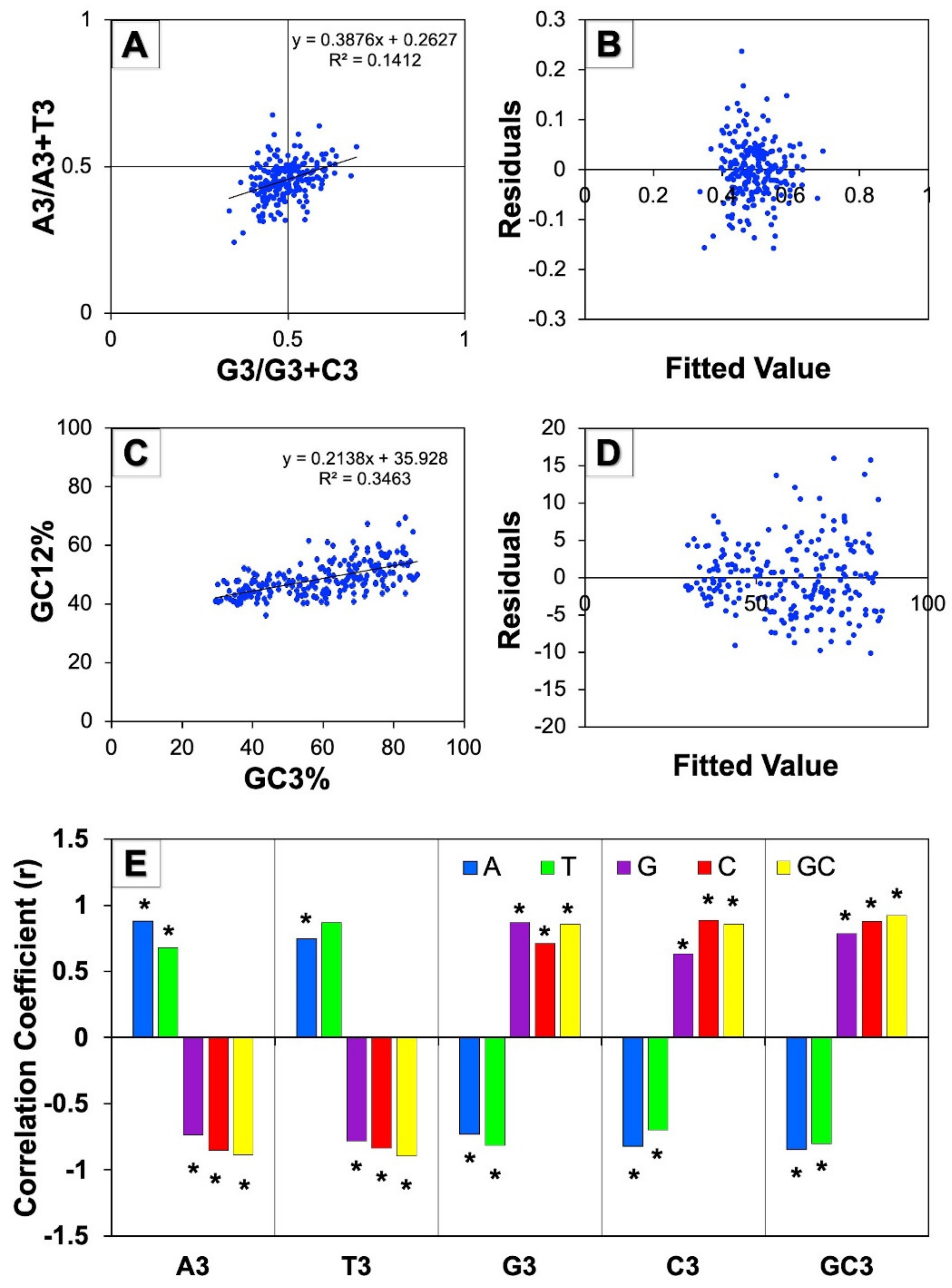
3.5.1. Parity Plot Analysis
3.5.2. Neutrality Plot Analysis
3.5.3. Correlation Analysis of the Third Codon Position against Base Composition
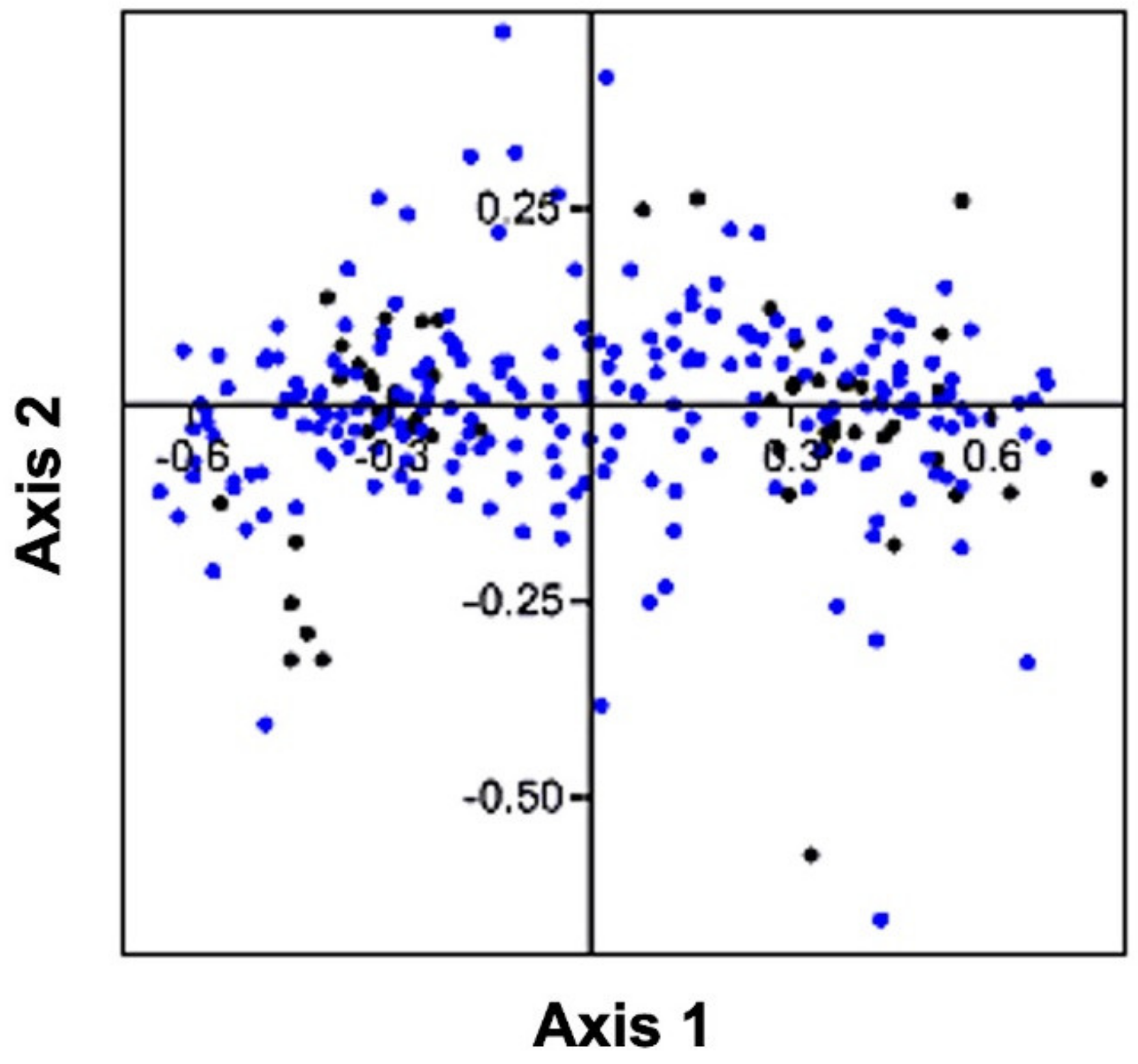
3.6. Correspondence Analysis
3.7. Codon Context Analysis
3.8. Correlation between ENC and Protein Properties
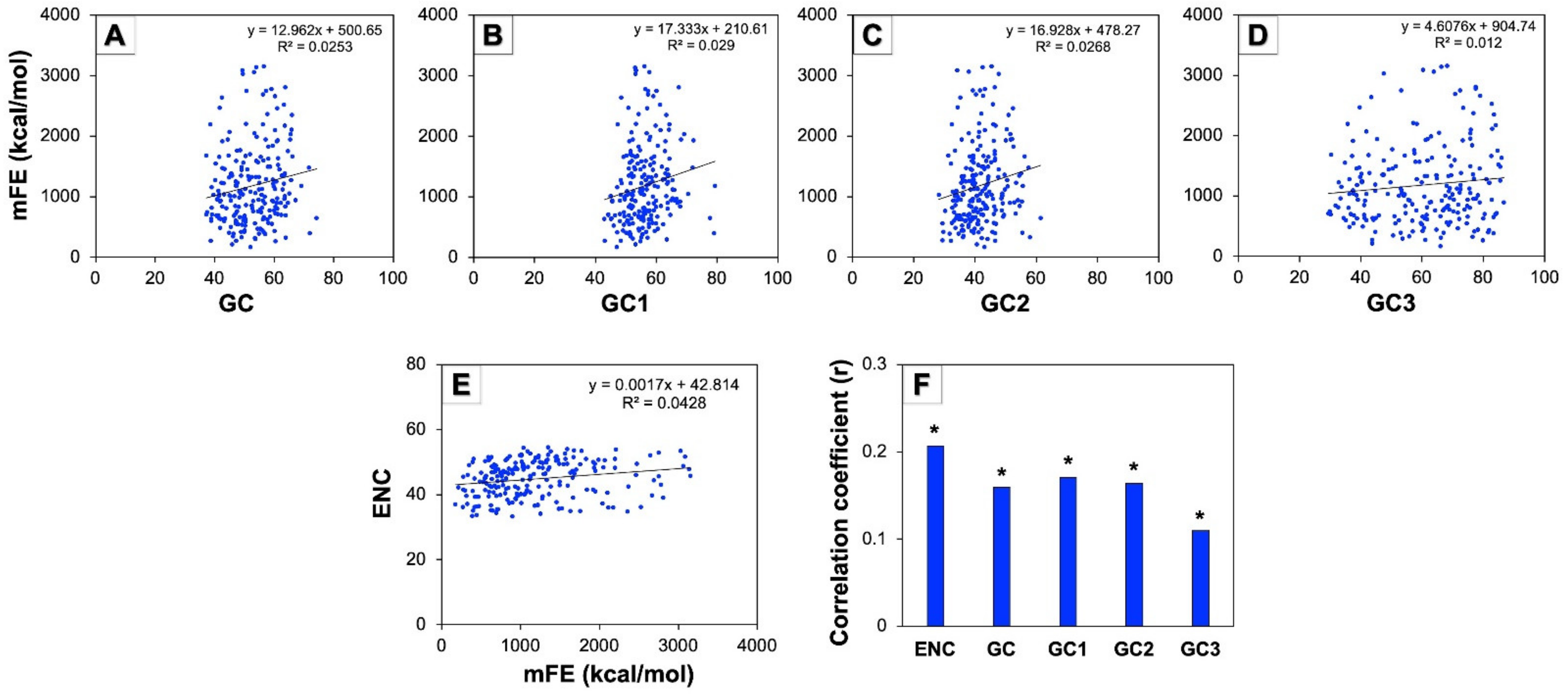
3.9. Minimum Free Energy (mFE) of mRNA and Codon Usage Pattern of Human Autophagy Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of Cells and Tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [Green Version]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Kocaturk, N.M.; Akkoc, Y.; Kig, C.; Bayraktar, O.; Gozuacik, D.; Kutlu, O. Autophagy as a Molecular Target for Cancer Treatment. Eur. J. Pharm. Sci. 2019, 134, 116–137. [Google Scholar] [CrossRef]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-Dependent MTORC1 Association with the ULK1–Atg13–FIP200 Complex Required for Autophagy. MBoC 2009, 20, 1981–1991. [Google Scholar] [CrossRef] [Green Version]
- Hanada, T.; Noda, N.N.; Satomi, Y.; Ichimura, Y.; Fujioka, Y.; Takao, T.; Inagaki, F.; Ohsumi, Y. The Atg12-Atg5 Conjugate Has a Novel E3-like Activity for Protein Lipidation in Autophagy. J. Biol. Chem. 2007, 282, 37298–37302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsunaga, K.; Saitoh, T.; Tabata, K.; Omori, H.; Satoh, T.; Kurotori, N.; Maejima, I.; Shirahama-Noda, K.; Ichimura, T.; Isobe, T.; et al. Two Beclin 1-Binding Proteins, Atg14L and Rubicon, Reciprocally Regulate Autophagy at Different Stages. Nat. Cell Biol. 2009, 11, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Mauthe, M.; Jacob, A.; Freiberger, S.; Hentschel, K.; Stierhof, Y.-D.; Codogno, P.; Proikas-Cezanne, T. Resveratrol-Mediated Autophagy Requires WIPI-1-Regulated LC3 Lipidation in the Absence of Induced Phagophore Formation. Autophagy 2011, 7, 1448–1461. [Google Scholar] [CrossRef] [PubMed]
- Mehrpour, M.; Esclatine, A.; Beau, I.; Codogno, P. Overview of Macroautophagy Regulation in Mammalian Cells. Cell Res. 2010, 20, 748–762. [Google Scholar] [CrossRef]
- Shpilka, T.; Weidberg, H.; Pietrokovski, S.; Elazar, Z. Atg8: An Autophagy-Related Ubiquitin-like Protein Family. Genome Biol. 2011, 12, 226. [Google Scholar] [CrossRef]
- Crick, F.H.C.; Barnett, L.; Brenner, S.; Watts-Tobin, R.J. General Nature of the Genetic Code for Proteins. Nature 1961, 192, 1227–1232. [Google Scholar] [CrossRef] [PubMed]
- Behura, S.K.; Severson, D.W. Codon Usage Bias: Causative Factors, Quantification Methods and Genome-Wide Patterns: With Emphasis on Insect Genomes. Biol. Rev. 2013, 88, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Zhou, T.; Ma, J.; Sun, X.; Lu, Z. Analysis of Synonymous Codon Usage in SARS Coronavirus and Other Viruses in the Nidovirales. Virus Res. 2004, 101, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Jin, S.; Li, N.; Liu, X.; Wang, H.; Li, J. Comparative and Functional Analysis of Testis-Specific Genes. Biol. Pharm. Bull. 2011, 34, 28–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plotkin, J.B.; Robins, H.; Levine, A.J. Tissue-Specific Codon Usage and the Expression of Human Genes. Proc. Natl. Acad. Sci. USA 2004, 101, 12588–12591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharp, P.M. Codon Usage Bias. In Encyclopedia of Genetics; Elsevier: Amsterdam, The Netherlands, 2001; pp. 402–406. ISBN 978-0-12-227080-2. [Google Scholar]
- Sharp, P.M. Variation in the Strength of Selected Codon Usage Bias among Bacteria. Nucleic Acids Res. 2005, 33, 1141–1153. [Google Scholar] [CrossRef] [PubMed]
- Stenico, M.; Lloyd, A.T.; Sharp, P.M. Codon Usage in Caenorhabditis Elegans: Delineation of Translational Selection and Mutational Biases. Nucleic Acids Res. 1994, 22, 2437–2446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grantham, R.; Gautier, C.; Gouy, M.; Jacobzone, M.; Mercier, R. Codon Catalog Usage Is a Genome Strategy Modulated for Gene Expressivity. Nucleic Acids Res. 1981, 9, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.K.; Bhattacharyya, T.K.; Ghosh, T.C. Synonymous Codon Usage in Lactococcus Lactis: Mutational Bias Versus Translational Selection. J. Biomol. Struct. Dyn. 2004, 21, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Kane, J.F. Effects of Rare Codon Clusters on High-Level Expression of Heterologous Proteins in Escherichia Coli. Curr. Opin. Biotechnol. 1995, 6, 494–500. [Google Scholar] [CrossRef]
- Yang, X.; Luo, X.; Cai, X. Analysis of Codon Usage Pattern in Taenia Saginata Based on a Transcriptome Dataset. Parasites Vectors 2014, 7, 527. [Google Scholar] [CrossRef] [PubMed]
- Bali, V.; Bebok, Z. Decoding Mechanisms by Which Silent Codon Changes Influence Protein Biogenesis and Function. Int. J. Biochem. Cell Biol. 2015, 64, 58–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, R.; Wang, H.; Pu, F.; Feng, X.; Jin, L.; Ma, Z.; Ma, X. Codon Usage Bias in Autophagy-Related Gene 13 in Eukaryotes: Uncovering the Genetic Divergence by the Interplay Between Nucleotides and Codon Usages. Front. Cell. Infect. Microbiol. 2021, 11, 771010. [Google Scholar] [CrossRef] [PubMed]
- Human Autophagy Database (HADb). Available online: http://autophagy.lu/ (accessed on 7 August 2022).
- Zhang, S.; Hama, Y.; Mizushima, N. The Evolution of Autophagy Proteins—Diversification in Eukaryotes and Potential Ancestors in Prokaryotes. J. Cell Sci. 2021, 134, jcs233742. [Google Scholar] [CrossRef]
- National Centre for Biotechnology Information (NCBI). Available online: http://www.ncbi.nlm.nih.gov (accessed on 8 August 2022).
- Sharp, P.M.; Li, W.-H. An Evolutionary Perspective on Synonymous Codon Usage in Unicellular Organisms. J. Mol. Evol. 1986, 24, 28–38. [Google Scholar] [CrossRef]
- Wright, F. The ‘Effective Number of Codons’ Used in a Gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Satapathy, S.S.; Sahoo, A.K.; Ray, S.K.; Ghosh, T.C. Codon degeneracy and amino acid abundance influence the measures of codon usage bias: Improved Nc and ENCprime measures. Genes Cells 2017, 22, 277–283. [Google Scholar] [CrossRef] [Green Version]
- Agnigarh Web Portal. Available online: http://agnigarh.tezu.ernet.in/~ssankar/cub.php (accessed on 8 August 2022).
- Sueoka, N. Intrastrand Parity Rules of DNA Base Composition and Usage Biases of Synonymous Codons. J. Mol. Evol. 1995, 40, 318–325. [Google Scholar] [CrossRef]
- Sueoka, N. Two Aspects of DNA Base Composition: G+C Content and Translation-Coupled Deviation from Intra-Strand Rule of A=T and G=C. J. Mol. Evol. 1999, 49, 49–62. [Google Scholar] [CrossRef]
- Sueoka, N. Directional Mutation Pressure and Neutral Molecular Evolution. Proc. Natl. Acad. Sci. USA 1988, 85, 2653–2657. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Greenacre, M.J. Theory and Applications of Correspondence Analysis. Biometrics 1986, 42, 223. [Google Scholar] [CrossRef]
- Suzuki, H.; Brown, C.J.; Forney, L.J.; Top, E.M. Comparison of Correspondence Analysis Methods for Synonymous Codon Usage in Bacteria. DNA Res. 2008, 15, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST—PAlaeontological STatistics. Palaeontol. Electron 2001, 4, 1–9. [Google Scholar]
- Moura, G.; Pinheiro, M.; Arrais, J.; Gomes, A.C.; Carreto, L.; Freitas, A.; Oliveira, J.L.; Santos, M.A.S. Large Scale Comparative Codon-Pair Context Analysis Unveils General Rules That Fine-Tune Evolution of MRNA Primary Structure. PLoS ONE 2007, 2, e847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peden, J.F. Analysis of Codon Usage. PhD Thesis, University of Nottingham, Nottingham, UK, 1999. [Google Scholar]
- Kyte, J.; Doolittle, R.F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- GRAVY Calculator. Available online: http://www.gravy-calculator.de/ (accessed on 20 August 2022).
- Kozlowski, L.P. IPC 2.0: Prediction of Isoelectric Point and p K a Dissociation Constants. Nucleic Acids Res. 2021, 49, W285–W292. [Google Scholar] [CrossRef]
- IPC 2.0. Available online: www.ipc2-isoelectric-point.org (accessed on 20 August 2022).
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neubock, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [Green Version]
- RNAfold. Available online: http://rna.tbi.univie.ac.at/ (accessed on 20 August 2022).
- Puigbò, P.; Bravo, I.G.; Garcia-Vallve, S. CAIcal: A Combined Set of Tools to Assess Codon Usage Adaptation. Biol. Direct 2008, 3, 38. [Google Scholar] [CrossRef] [Green Version]
- CAICal Server. Available online: http://genomes.urv.es/CAIcal (accessed on 8 August 2022).
- Ning, W.; Wei, Y.; Gao, L.; Han, C.; Gou, Y.; Fu, S.; Liu, D.; Zhang, C.; Huang, X.; Wu, S.; et al. HemI 2.0: An Online Service for Heatmap Illustration. Nucleic Acids Res. 2022, 50, W405–W411. [Google Scholar] [CrossRef]
- Comeron, J.M.; Aguadé, M. An Evaluation of Measures of Synonymous Codon Usage Bias. J. Mol. Evol. 1998, 47, 268–274. [Google Scholar] [CrossRef]
- Behura, S.K.; Severson, D.W. Comparative Analysis of Codon Usage Bias and Codon Context Patterns between Dipteran and Hymenopteran Sequenced Genomes. PLoS ONE 2012, 7, e43111. [Google Scholar] [CrossRef]
- Deb, B.; Uddin, A.; Chakraborty, S. Composition, Codon Usage Pattern, Protein Properties, and Influencing Factors in the Genomes of Members of the Family Anelloviridae. Arch. Virol. 2021, 166, 461–474. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Gu, W.; Ma, J.; Sun, X.; Lu, Z. Analysis of Synonymous Codon Usage in H5N1 Virus and Other Influenza A Viruses. Biosystems 2005, 81, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Uddin, A. Compositional Features and Codon Usage Pattern of Genes Associated with Anxiety in Human. Mol. Neurobiol. 2020, 57, 4911–4920. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, J.B.; Kudla, G. Synonymous but Not the Same: The Causes and Consequences of Codon Bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [Green Version]
- Mazumder, T.H.; Uddin, A.; Chakraborty, S. Insights into the Nucleotide Composition and Codon Usage Pattern of Human Tumor Suppressor Genes. Mol. Carcinog. 2020, 59, 15–23. [Google Scholar] [CrossRef]
- Nath Choudhury, M.; Uddin, A.; Chakraborty, S. Codon Usage Bias and Its Influencing Factors for Y-Linked Genes in Human. Comput. Biol. Chem. 2017, 69, 77–86. [Google Scholar] [CrossRef]
- Uddin, A.; Mazumder, T.H.; Barbhuiya, P.A.; Chakraborty, S. Similarities and Dissimilarities of Codon Usage in Mitochondrial ATP Genes among Fishes, Aves, and Mammals. IUBMB Life 2020, 72, 899–914. [Google Scholar] [CrossRef]
- Chakraborty, S.; Paul, S.; Nath, D.; Choudhury, Y.; Ahn, Y.; Cho, Y.S.; Uddin, A. Synonymous Codon Usage and Context Analysis of Genes Associated with Pancreatic Cancer. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2020, 821, 111719. [Google Scholar] [CrossRef]
- Chakraborty, S.; Barbhuiya, P.A.; Mazumder, G.A.; Deb, B.; Uddin, A. Compositional Features and Codon Usage Pattern of TP63 Gene. Comput. Biol. Chem. 2019, 83, 107119. [Google Scholar] [CrossRef]
- Chakraborty, S.; Uddin, A.; Mazumder, T.H.; Choudhury, M.N.; Malakar, A.K.; Paul, P.; Halder, B.; Deka, H.; Mazumder, G.A.; Barbhuiya, R.A.; et al. Codon Usage and Expression Level of Human Mitochondrial 13 Protein Coding Genes across Six Continents. Mitochondrion 2018, 42, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Ringnér, M.; Krogh, M. Folding Free Energies of 5′-UTRs Impact Post-Transcriptional Regulation on a Genomic Scale in Yeast. PLoS Comput. Biol. 2005, 1, e72. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jamil, Z.; Uddin, A.; Alam, S.S.M.; Samanta, A.; Altwaijry, N.; Rauf, M.A.; Ali, S.; Khan, M.S.; Asghar, M.N.; Hoque, M. Analysis of the Compositional Features and Codon Usage Pattern of Genes Involved in Human Autophagy. Cells 2022, 11, 3203. https://doi.org/10.3390/cells11203203
Jamil Z, Uddin A, Alam SSM, Samanta A, Altwaijry N, Rauf MA, Ali S, Khan MS, Asghar MN, Hoque M. Analysis of the Compositional Features and Codon Usage Pattern of Genes Involved in Human Autophagy. Cells. 2022; 11(20):3203. https://doi.org/10.3390/cells11203203
Chicago/Turabian StyleJamil, Zarnain, Arif Uddin, Syed Sahajada Mahafujul Alam, Arijit Samanta, Nojood Altwaijry, Mohd Ahmar Rauf, Safdar Ali, Mohd Shahnawaz Khan, Muhammad Nadeem Asghar, and Mehboob Hoque. 2022. "Analysis of the Compositional Features and Codon Usage Pattern of Genes Involved in Human Autophagy" Cells 11, no. 20: 3203. https://doi.org/10.3390/cells11203203
APA StyleJamil, Z., Uddin, A., Alam, S. S. M., Samanta, A., Altwaijry, N., Rauf, M. A., Ali, S., Khan, M. S., Asghar, M. N., & Hoque, M. (2022). Analysis of the Compositional Features and Codon Usage Pattern of Genes Involved in Human Autophagy. Cells, 11(20), 3203. https://doi.org/10.3390/cells11203203







