Lack of Retinoblastoma Protein Shifts Tumor Metabolism from Glycolysis to OXPHOS and Allows the Use of Alternate Fuels
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Cell Lines
2.3. Gene Expression Analysis
2.4. Lentiviral Plasmids and Vectors
2.5. Western Blotting
2.6. Energy Phenotype Assay
2.7. Mitochondrial Stress Assay
2.8. Glycolytic Rate Assay
2.9. Fuel Choice/Mito Fuel Flex Assay
2.10. Metabolite Measurement In Vitro
2.11. Immunofluorescence/Mito Tracker Green
2.12. Statistical Analysis
3. Results
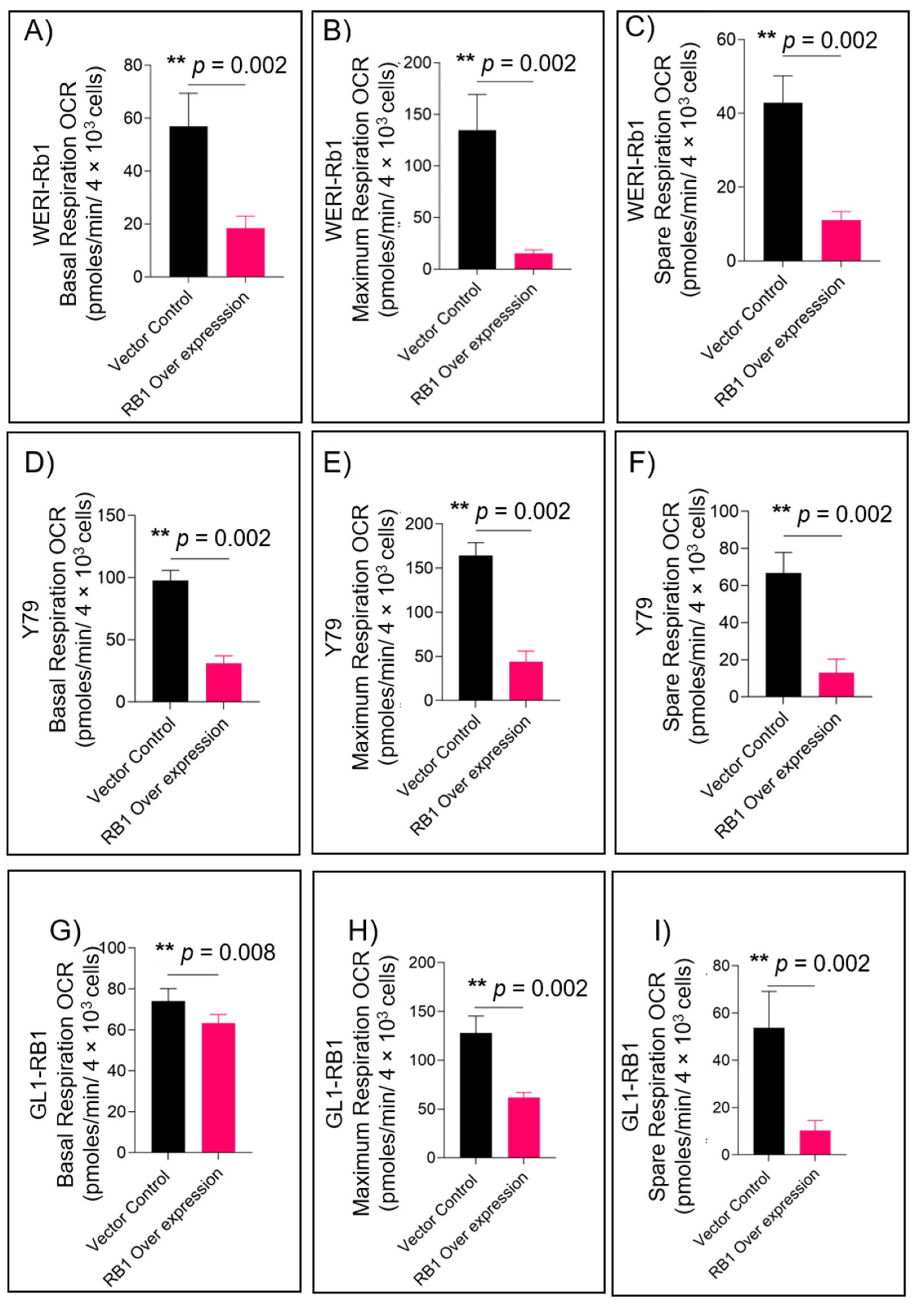
3.1. RB1 Expression Reduces Mitochondrial Respiration in Retinoblastoma Cells
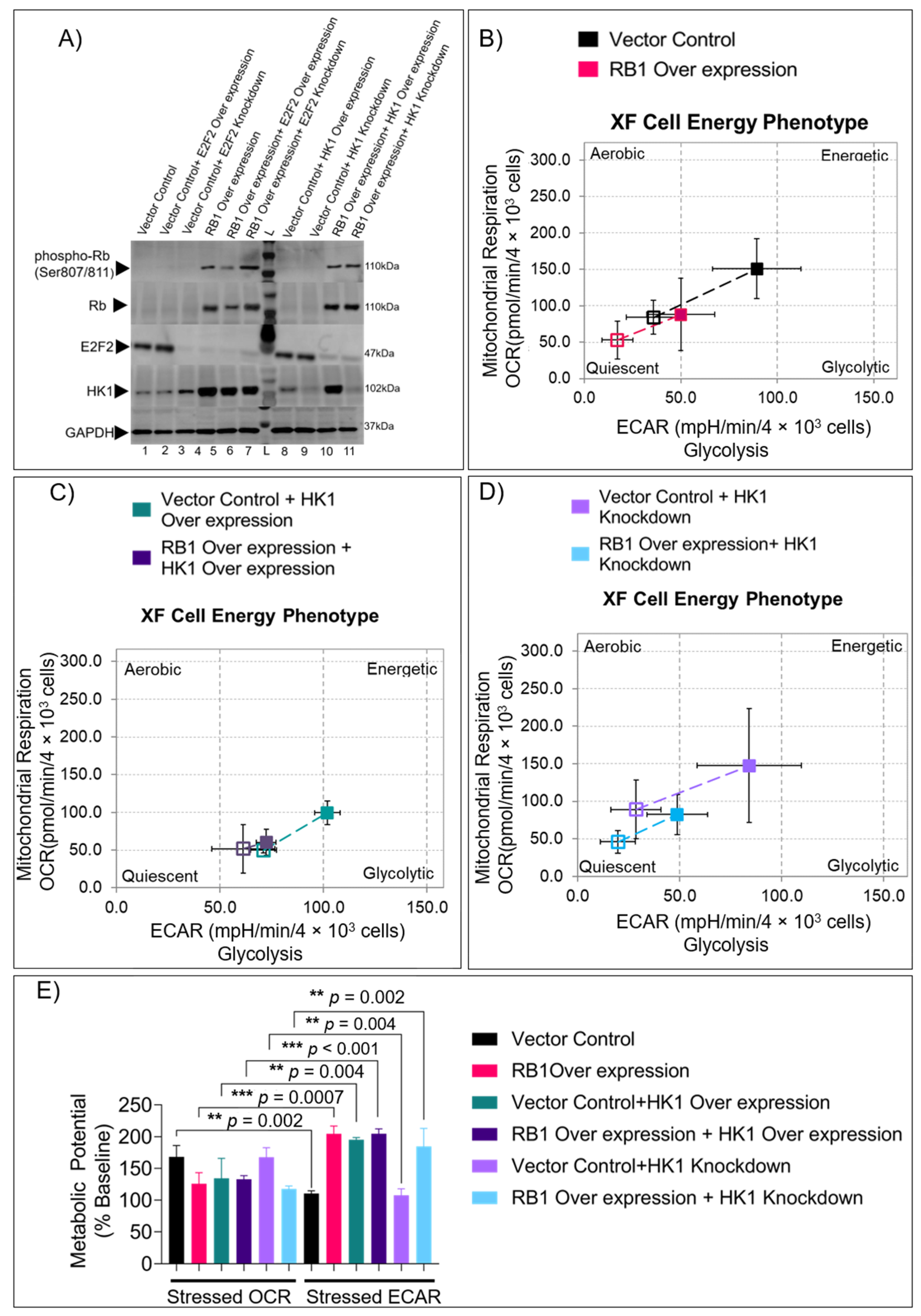
3.2. RB1 and HK1 Complementation in Rb Null Cells Reveal Distinct Energy Profiles
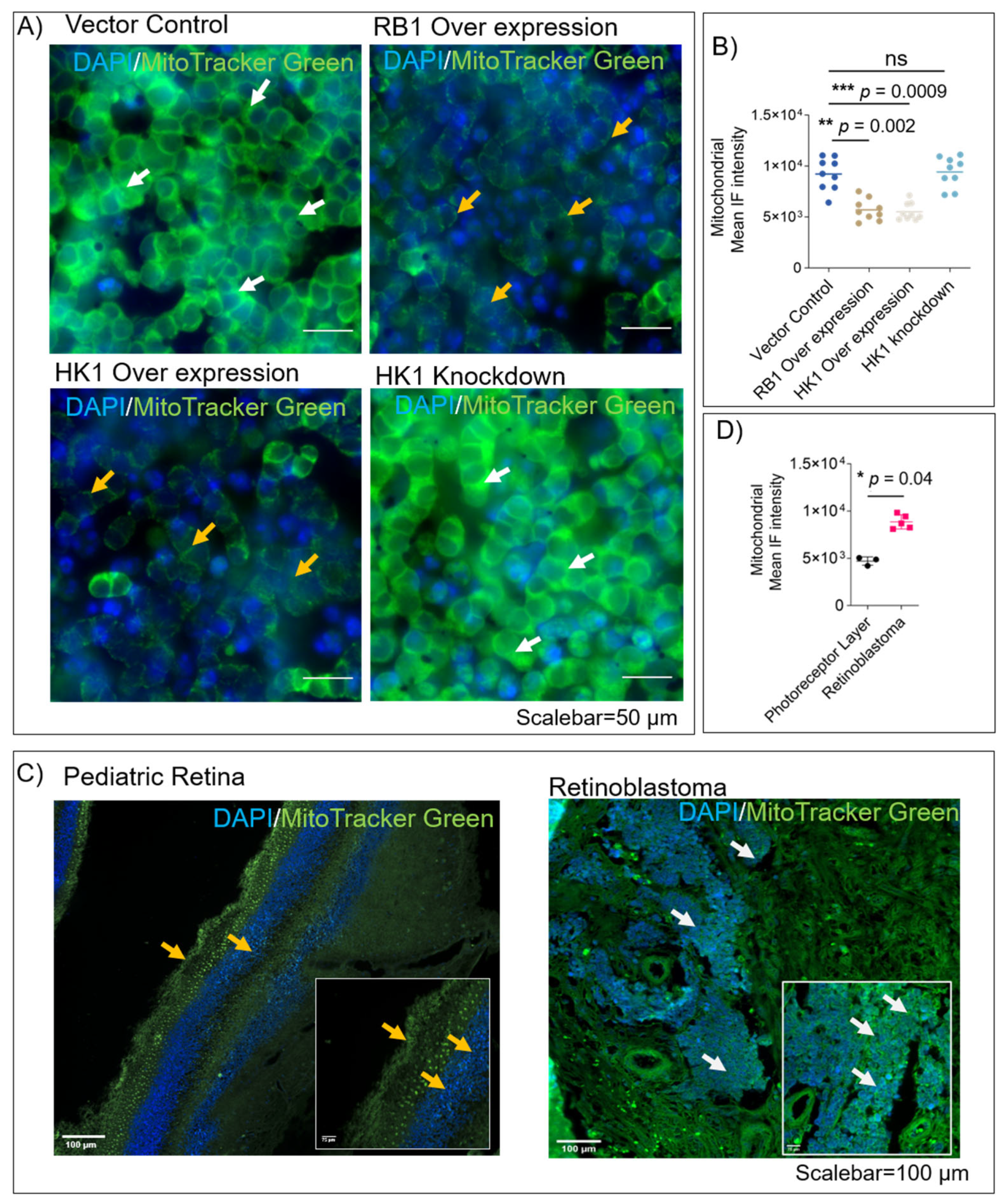
3.3. RB1 and HK1 Augmentation Reduce Mitochondrial Mass in Rb Null Cells
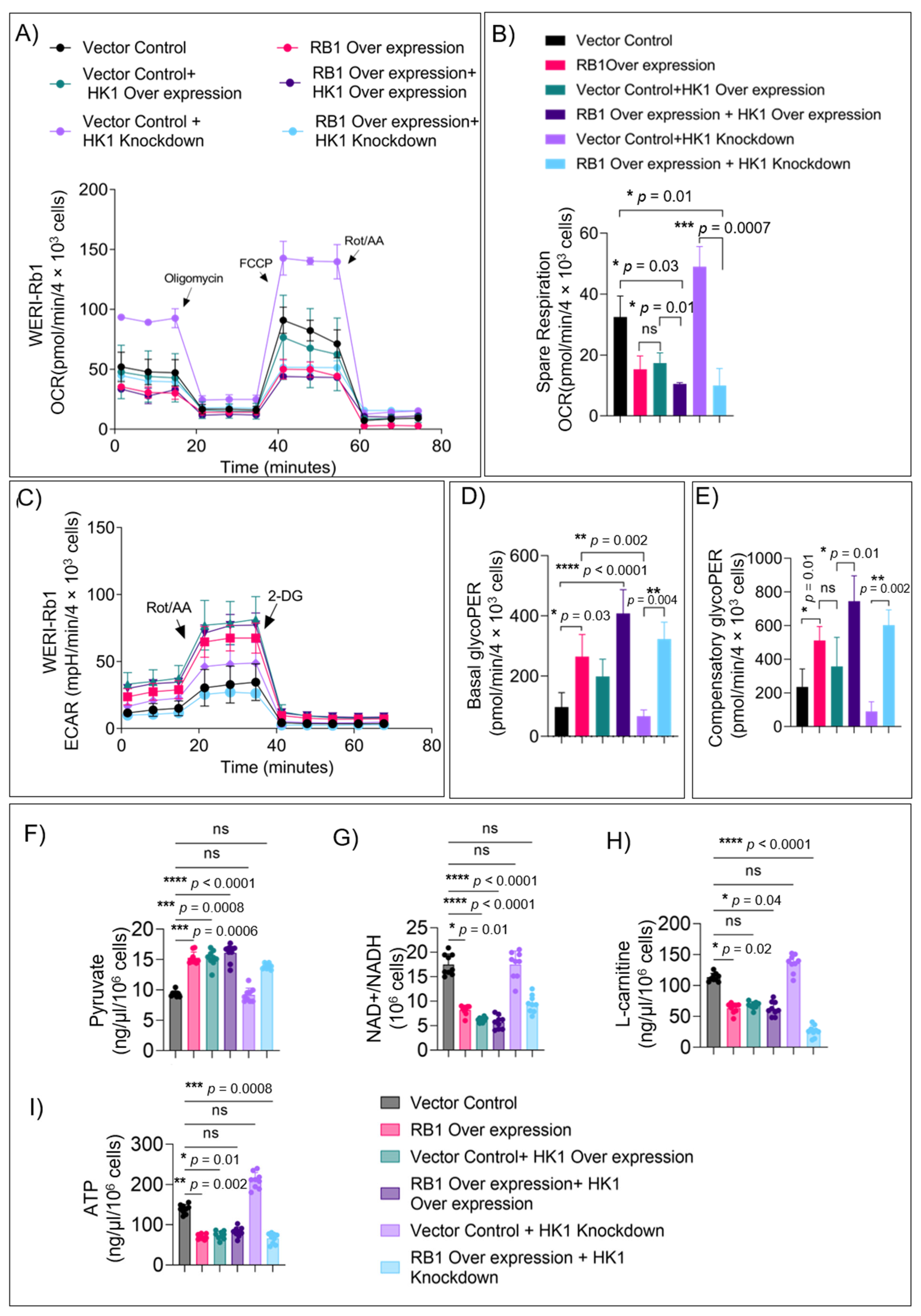
3.4. HK1 and RB1 Expression Induce a Metabolic Switch from Mitochondrial Respiration to Glycolysis
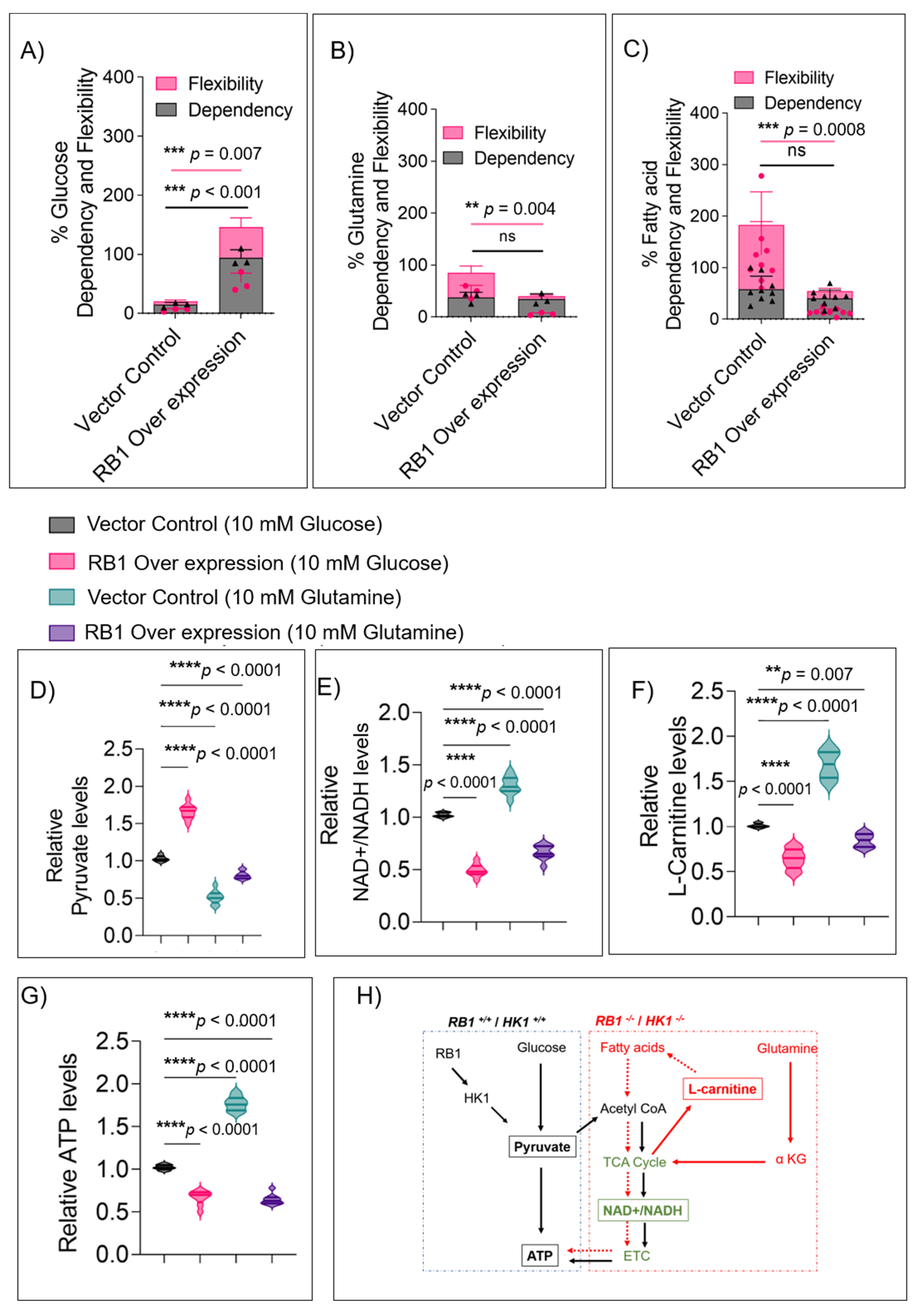
3.5. RB1 Expression Restricts the Usage of Alternate Fuels in Rb Null Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Friend, S.H.; Bernards, R.; Rogelj, S.; Weinberg, R.A.; Rapaport, J.M.; Albert, D.M.; Dryja, T.P. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 1986, 323, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Shields, C.L.; Shields, J.A. Retinoblastoma management: Advances in enucleation, intravenous chemo reduction, and intra-arterial chemotherapy. Curr. Opin. Ophthalmol. 2010, 21, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Bosaleh, A.; Sampor, C.; Solernou, V.; Fandino, A.; Dominguez, J.; de Davila, M.T.; Chantada, G.L. Outcome of children with retinoblastoma and isolated choroidal invasion. Arch. Ophthalmol. 2012, 130, 724–729. [Google Scholar] [CrossRef]
- Shields, C.L.; Shields, J.A.; Baez, K.; Cater, J.R.; De Potter, P. Optic nerve invasion of retinoblastoma. Metastatic potential and clinical risk factors. Cancer 1994, 73, 692–698. [Google Scholar] [CrossRef]
- Kohe, S.; Brundler, M.A.; Jenkinson, H.; Parulekar, M.; Wilson, M.; Peet, A.C.; McConville, C.M.; on behalf of the Children’s Cancer and Leukaemia Group (CCLG). Metabolite profiling in retinoblastoma identifies novel clinicopathological subgroups. Br. J. Cancer 2015, 113, 1216–1224. [Google Scholar] [CrossRef]
- Sahoo, S.; Ravi Kumar, R.K.; Nicolay, B.; Mohite, O.; Sivaraman, K.; Khetan, V.; Rishi, P.; Ganesan, S.; Subramanyan, K.; Raman, K.; et al. Metabolite systems profiling identifies exploitable weaknesses in retinoblastoma. FEBS Lett. 2019, 593, 23–41. [Google Scholar] [CrossRef]
- Muranaka, H.; Hayashi, A.; Minami, K.; Kitajima, S.; Kohno, S.; Nishimoto, Y.; Nagatani, N.; Suzuki, M.; Kulathunga, L.A.N.; Sasaki, N.; et al. A distinct function of the retinoblastoma protein in the control of lipid composition identified by lipidomic profiling. Oncogenesis 2017, 6, e350. [Google Scholar] [CrossRef]
- Hu, J.; Locasale, J.W.; Bielas, J.H.; O’Sullivan, J.; Sheahan, K.; Cantley, L.C.; Vander Heiden, M.G.; Vitkup, D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat. Biotechnol. 2013, 31, 522–529. [Google Scholar] [CrossRef]
- Jones, R.A.; Robinson, T.J.; Liu, J.C.; Shrestha, M.; Voisin, V.; Ju, Y.; Chung, P.E.; Pellecchia, G.; Fell, V.L.; Bae, S.; et al. RB1 deficiency in triple-negative breast cancer induces mitochondrial protein translation. J. Clin. Investig. 2016, 126, 3739–3757. [Google Scholar] [CrossRef]
- Zacksenhaus, E.; Shrestha, M.; Liu, J.C.; Vorobieva, I.; Chung, P.E.D.; Ju, Y.; Nir, U.; Jiang, Z. Mitochondrial OXPHOS Induced by RB1 Deficiency in Breast Cancer: Implications for Anabolic Metabolism, Stemness, and Metastasis. Trends Cancer 2017, 3, 768–779. [Google Scholar] [CrossRef]
- Mandigo, A.C.; Yuan, W.; Xu, K.; Gallagher, P.; Pang, A.; Guan, Y.F.; Shafi, A.A.; Thangavel, C.; Sheehan, B.; Bogdan, D.; et al. RB/E2F1 as a Master Regulator of Cancer Cell Metabolism in Advanced Disease. Cancer Discov. 2021, 11, 2334–2353. [Google Scholar] [CrossRef]
- Tarrago-Celada, J.; Cascante, M. Targeting the Metabolic Adaptation of Metastatic Cancer. Cancers 2021, 13, 1641. [Google Scholar] [CrossRef]
- Sun, X.; Wang, M.; Wang, M.; Yu, X.; Guo, J.; Sun, T.; Li, X.; Yao, L.; Dong, H.; Xu, Y. Metabolic Reprogramming in Triple-Negative Breast Cancer. Front. Oncol. 2020, 10, 428. [Google Scholar] [CrossRef]
- Lin, X.; Xiao, Z.; Chen, T.; Liang, S.H.; Guo, H. Glucose Metabolism on Tumor Plasticity, Diagnosis, and Treatment. Front. Oncol. 2020, 10, 317. [Google Scholar] [CrossRef]
- De Berardinis, R.J.; Cheng, T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010, 29, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef]
- Babu, V.S.; Mallipatna, A.; Sa, D.; Dudeja, G.; Kannan, R.; Shetty, R.; Nair, A.P.; Gundimeda, S.; Chaurasia, S.S.; Verma, N.K.; et al. Integrated Analysis of Cancer Tissue and Vitreous Humor from Retinoblastoma Eyes Reveals Unique Tumor-Specific Metabolic and Cellular Pathways in Advanced and Non-Advanced Tumors. Cells 2022, 11, 1668. [Google Scholar] [CrossRef]
- Beemer, F.A.; Vlug, A.M.; Rijksen, G.; Hamburg, A.; Staal, G.E. Characterization of some glycolytic enzymes from human retina and retinoblastoma. Cancer Res. 1982, 42, 4228–4232. [Google Scholar] [PubMed]
- Reynolds, M.R.; Lane, A.N.; Robertson, B.; Kemp, S.; Liu, Y.; Hill, B.G.; Dean, D.C.; Clem, B.F. Control of glutamine metabolism by the tumor suppressor Rb. Oncogene 2014, 33, 556–566. [Google Scholar] [CrossRef]
- De Oliveira, M.P.; Liesa, M. The Role of Mitochondrial Fat Oxidation in Cancer Cell Proliferation and Survival. Cells 2020, 9, 2600. [Google Scholar] [CrossRef] [PubMed]
- Vyas, S.; Zaganjor, E.; Haigis, M.C. Mitochondria and Cancer. Cell 2016, 166, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Elegheert, J.; Behiels, E.; Bishop, B.; Scott, S.; Woolley, R.E.; Griffiths, S.C.; Byrne, E.F.X.; Chang, V.T.; Stuart, D.I.; Jones, E.Y.; et al. Lentiviral transduction of mammalian cells for fast, scalable and high-level production of soluble and membrane proteins. Nat. Protoc. 2018, 13, 2991–3017. [Google Scholar] [CrossRef]
- Porporato, P.E.; Filigheddu, N.; Pedro, J.M.B.; Kroemer, G.; Galluzzi, L. Mitochondrial metabolism and cancer. Cell Res. 2018, 28, 265–280. [Google Scholar] [CrossRef]
- Kohanim, S.; Palioura, S.; Saeed, H.N.; Akpek, E.K.; Amescua, G.; Basu, S.; Blomquist, P.H.; Bouchard, C.S.; Dart, J.K.; Gai, X.; et al. Acute and Chronic Ophthalmic Involvement in Stevens-Johnson Syndrome/Toxic Epidermal Necrolysis—A Comprehensive Review and Guide to Therapy. II. Ophthalmic Dis. Ocul. Surf. 2016, 14, 168–188. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Alesi, G.N.; Kang, S. Glutaminolysis as a target for cancer therapy. Oncogene 2016, 35, 3619–3625. [Google Scholar] [CrossRef] [PubMed]
- Tanner, L.B.; Goglia, A.G.; Wei, M.H.; Sehgal, T.; Parsons, L.R.; Park, J.O.; White, E.; Toettcher, J.E.; Rabinowitz, J.D. Four Key Steps Control Glycolytic Flux in Mammalian Cells. Cell Syst. 2018, 7, 49–62.e48. [Google Scholar] [CrossRef] [PubMed]
- Robey, R.B.; Hay, N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene 2006, 25, 4683–4696. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Lin, X.; Cai, M.; Zheng, X.; Lian, L.; Fan, D.; Wu, X.; Lan, P.; Wang, J. Overexpression of Hexokinase 1 as a poor prognosticator in human colorectal cancer. Tumor Biol. 2016, 37, 3887–3895. [Google Scholar] [CrossRef]
- Li, Y.; Tian, H.; Luo, H.; Fu, J.; Jiao, Y.; Li, Y. Prognostic Significance and Related Mechanisms of Hexokinase 1 in Ovarian Cancer. OncoTargets Ther. 2020, 13, 11583–11594. [Google Scholar] [CrossRef]
- Lamb, R.; Bonuccelli, G.; Ozsvari, B.; Peiris-Pages, M.; Fiorillo, M.; Smith, D.L.; Bevilacqua, G.; Mazzanti, C.M.; McDonnell, L.A.; Naccarato, A.G.; et al. Mitochondrial mass, a new metabolic biomarker for stem-like cancer cells: Understanding WNT/FGF-driven anabolic signaling. Oncotarget 2015, 6, 30453–30471. [Google Scholar] [CrossRef]
- Xu, X.L.; Singh, H.P.; Wang, L.; Qi, D.L.; Poulos, B.K.; Abramson, D.H.; Jhanwar, S.C.; Cobrinik, D. Rb suppresses human cone-precursor-derived retinoblastoma tumors. Nature 2014, 514, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Reidel, B.; Thompson, J.W.; Farsiu, S.; Moseley, M.A.; Skiba, N.P.; Arshavsky, V.Y. Proteomic profiling of a layered tissue reveals unique glycolytic specializations of photoreceptor cells. Mol. Cell. Proteom. 2011, 10, M110.002469. [Google Scholar] [CrossRef] [PubMed]
- Petit, L.; Ma, S.; Cipi, J.; Cheng, S.Y.; Zieger, M.; Hay, N.; Punzo, C. Aerobic Glycolysis Is Essential for Normal Rod Function and Controls Secondary Cone Death in Retinitis Pigmentosa. Cell Rep. 2018, 23, 2629–2642. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.B.; Lindsay, K.J.; Du, J. Glucose, lactate, and shuttling of metabolites in vertebrate retinas. J. Neurosci. Res. 2015, 93, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Metallo, C.M.; Gameiro, P.A.; Bell, E.L.; Mattaini, K.R.; Yang, J.; Hiller, K.; Jewell, C.M.; Johnson, Z.R.; Irvine, D.J.; Guarente, L.; et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2011, 481, 380–384. [Google Scholar] [CrossRef] [PubMed]
| ID | Sex | Age at Presentation | Laterality | Clinical Risk | IIRC Group | AJCC Staging |
|---|---|---|---|---|---|---|
| P1 | F | 23 months | Bilateral | Advanced | Group E | cT3b |
| P2 | F | 24 months | Unilateral | Advanced | Group E | cT3b |
| P3 | M | 36 months | Bilateral | Advanced | Group E | cT3b |
| P4 | F | 33 months | Unilateral | Non-advanced Group D | cT2b | 33 months |
| P5 | F | 14 months | Bilateral | Non-advanced Group D | cT2b | 14 months |
| Control 1 | F | 3 months | NA | Cardiac arrest (no ocular complications) | ||
| Control 2 | F | 2 months | NA | Multiple organ dysfunction (no ocular complications) | ||
| Control 3 | M | 6 months | NA | No ocular complication | ||
| Gene | Forward Primer | Reverse Primer | Tm(F/R) |
|---|---|---|---|
| RB1 | TTTGTAACGGGAGTCGGGA | CAGCGAGCTGTGGAGGAG | 54.67/55.89 |
| β-Actin | TCCCTGGAGAAGAGCTACGA | AGGAAGGAAGGCTGGAAGAG | 56.9/55.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suresh Babu, V.; Dudeja, G.; SA, D.; Bisht, A.; Shetty, R.; Heymans, S.; Guha, N.; Ghosh, A. Lack of Retinoblastoma Protein Shifts Tumor Metabolism from Glycolysis to OXPHOS and Allows the Use of Alternate Fuels. Cells 2022, 11, 3182. https://doi.org/10.3390/cells11203182
Suresh Babu V, Dudeja G, SA D, Bisht A, Shetty R, Heymans S, Guha N, Ghosh A. Lack of Retinoblastoma Protein Shifts Tumor Metabolism from Glycolysis to OXPHOS and Allows the Use of Alternate Fuels. Cells. 2022; 11(20):3182. https://doi.org/10.3390/cells11203182
Chicago/Turabian StyleSuresh Babu, Vishnu, Gagan Dudeja, Deepak SA, Anadi Bisht, Rohit Shetty, Stephane Heymans, Nilanjan Guha, and Arkasubhra Ghosh. 2022. "Lack of Retinoblastoma Protein Shifts Tumor Metabolism from Glycolysis to OXPHOS and Allows the Use of Alternate Fuels" Cells 11, no. 20: 3182. https://doi.org/10.3390/cells11203182
APA StyleSuresh Babu, V., Dudeja, G., SA, D., Bisht, A., Shetty, R., Heymans, S., Guha, N., & Ghosh, A. (2022). Lack of Retinoblastoma Protein Shifts Tumor Metabolism from Glycolysis to OXPHOS and Allows the Use of Alternate Fuels. Cells, 11(20), 3182. https://doi.org/10.3390/cells11203182






