Cyclin-Dependent Kinases and CTD Phosphatases in Cell Cycle Transcriptional Control: Conservation across Eukaryotic Kingdoms and Uniqueness to Plants
Abstract
1. Introduction
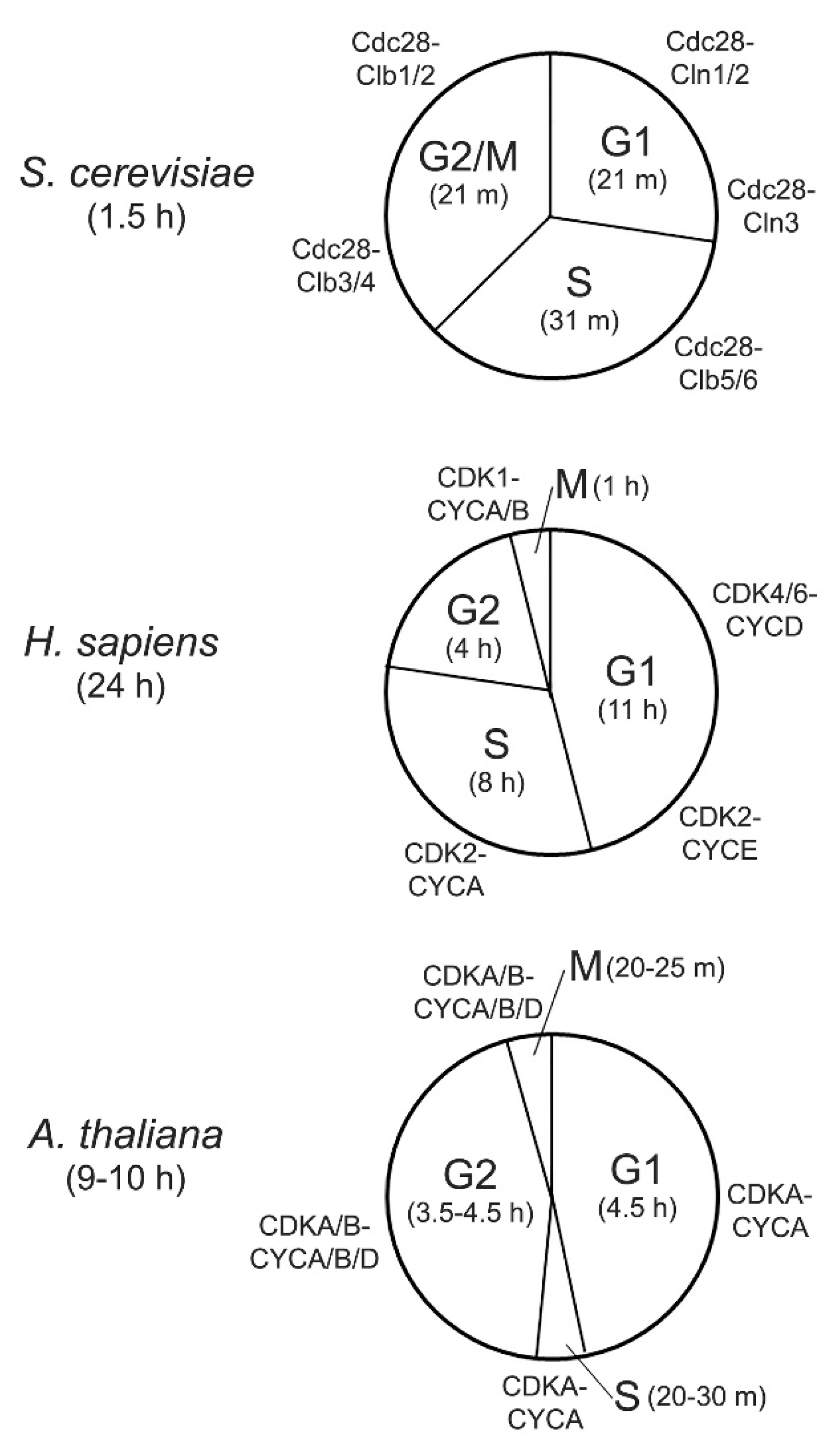
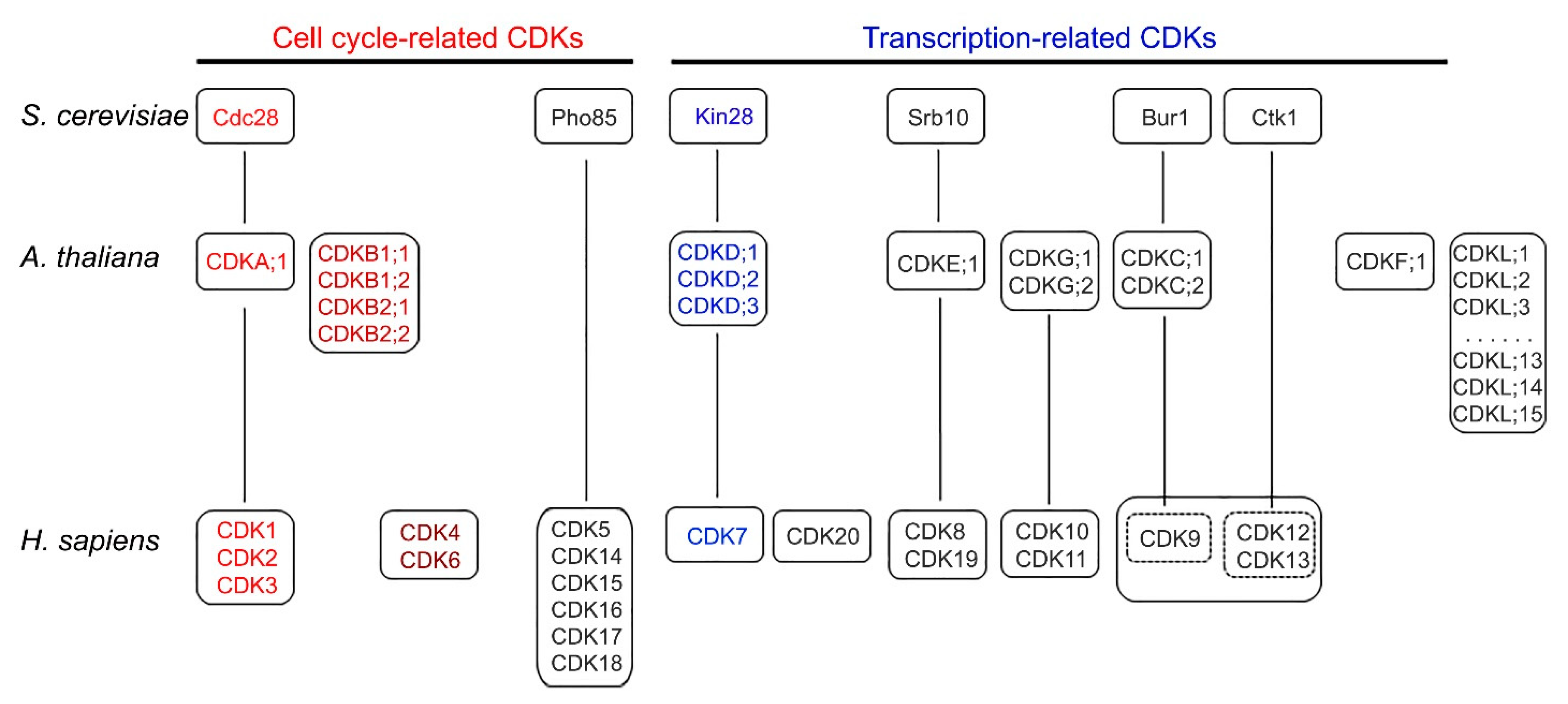
2. Cyclin-Dependent Kinases in Yeast, Human, and Arabidopsis
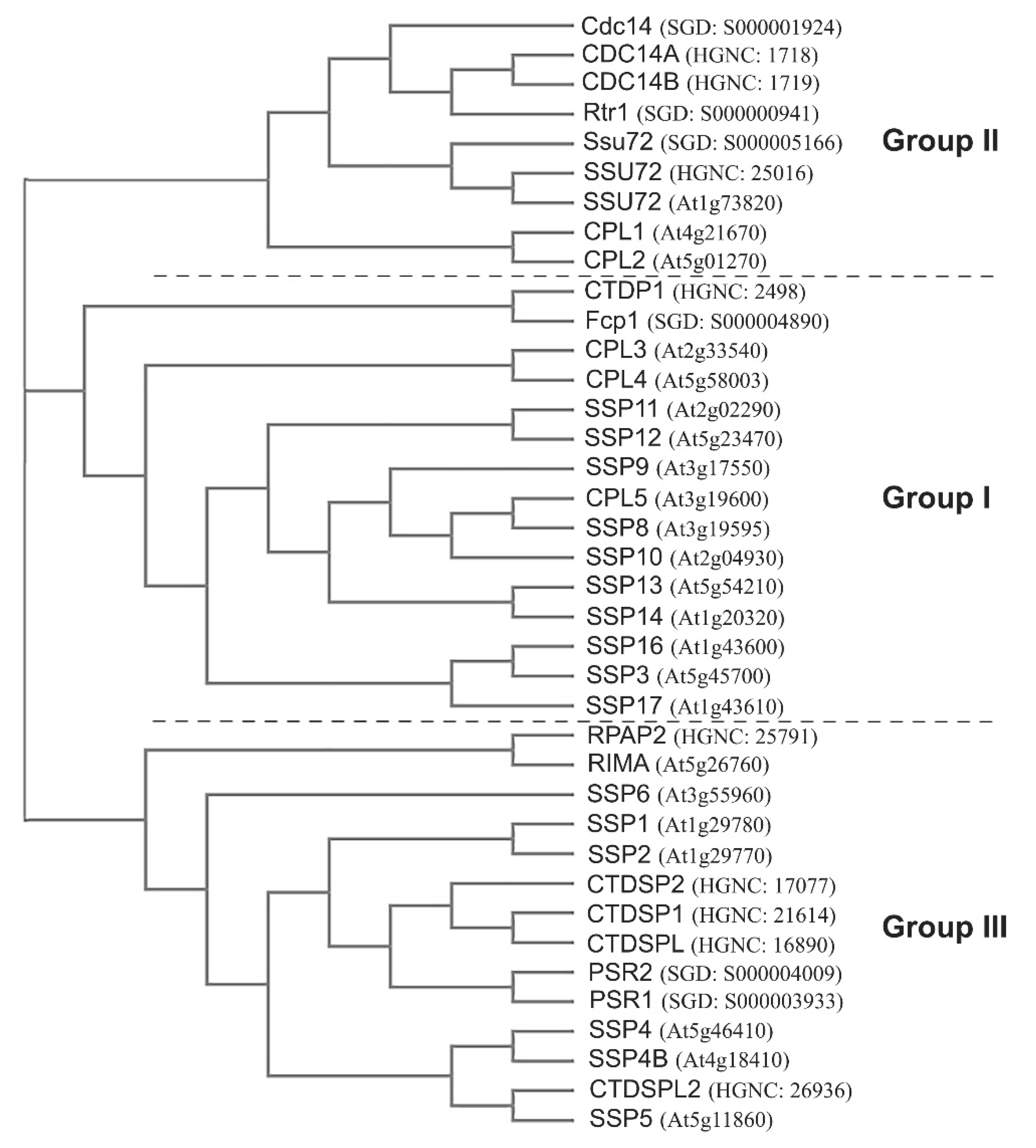
3. Pol II CTD Phosphatases in Yeast, Human, and Arabidopsis
4. Control of Transcription during the Cell Cycle
4.1. Importance of Precise, Global Transcriptional Control in Cell Cycle
4.2. Pol II CTD Phosphorylation Is Controlled by CDKs and CTD Phosphatases
4.3. Transcriptional Control: Global vs. Centralized?
5. Functional Conservation of CDK and CTD Phosphatases
5.1. CDK-Cyclin in Cell Cycle and Transcription
5.2. Substrates RB and E2F
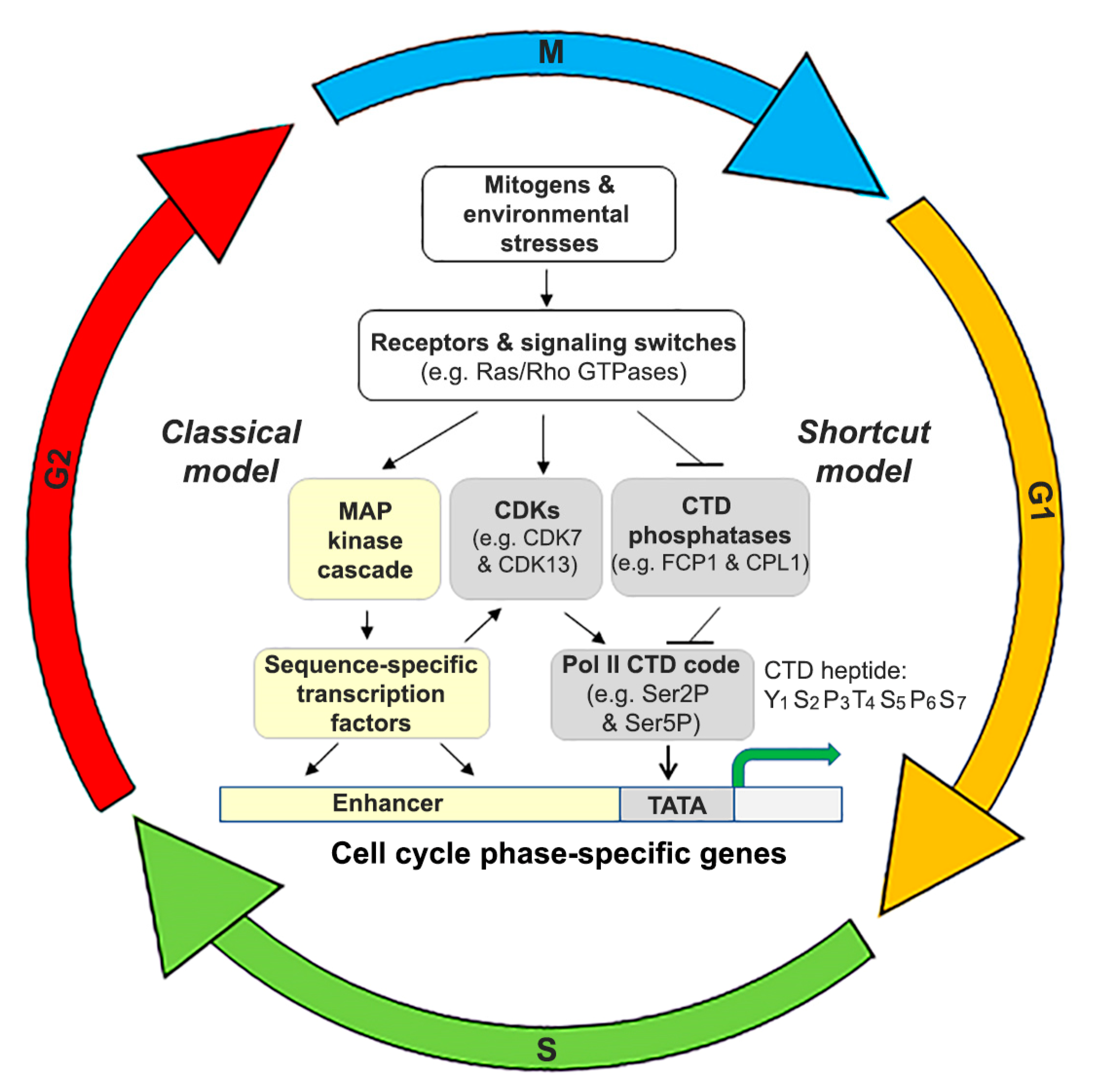
5.3. Substrate Pol II CTD and Its Upstream Regulatory Pathways: Classical vs. Shortcut?
6. Functions of CDKs and CTD Phosphatases Unique to Arabidopsis
6.1. Plant-Specific CDKs
6.2. Plant-Specific CTD Phosphatases
7. Future Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Fischer, M.; Muller, G.A. Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 638–662. [Google Scholar] [CrossRef] [PubMed]
- Sanso, M.; Fisher, R.P. Pause, play, repeat: CDKs push RNAP II’s buttons. Transcription 2013, 4, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef] [PubMed]
- Harashima, H.; Dissmeyer, N.; Schnittger, A. Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 2013, 23, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Brewer, B.J.; Chlebowicz-Sledziewska, E.; Fangman, W.L. Cell cycle phases in the unequal mother/daughter cell cycles of Saccharomyces cerevisiae. Mol. Cell. Biol. 1984, 4, 2529–2531. [Google Scholar] [CrossRef]
- Turner, J.J.; Ewald, J.C.; Skotheim, J.M. Cell size control in yeast. Curr. Biol. 2012, 22, R350–R359. [Google Scholar] [CrossRef]
- Cooper, G.M. The Eukaryotic Cell Cycle. Available online: https://www.ncbi.nlm.nih.gov/books/NBK9876/ (accessed on 1 October 2021).
- Pasternak, T.; Kircher, S.; Palme, K. Estimation of cell cycle kinetics in higher plant root meristem links organ position with cellular fate and chromatin structure. bioRxiv 2021. [Google Scholar] [CrossRef]
- Shimotohno, A.; Aki, S.S.; Takahashi, N.; Umeda, M. Regulation of the Plant Cell Cycle in Response to Hormones and the Environment. Annu. Rev. Plant Biol. 2021, 72, 273–296. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, A.K.; Montessoro, P.D.F.; Fusaro, A.F.; Araujo, B.G.; Hemerly, A.S. Plant CDKs-Driving the Cell Cycle through Climate Change. Plants 2021, 10, 1804. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Zhang, F. Cell Cycle Regulation in the Plant Response to Stress. Front. Plant Sci. 2020, 10, 1765. [Google Scholar] [CrossRef]
- Vandepoele, K.; Raes, J.; De Veylder, L.; Rouze, P.; Rombauts, S.; Inze, D. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 2002, 14, 903–916. [Google Scholar] [CrossRef]
- Menges, M.; de Jager, S.M.; Gruissem, W.; Murray, J.A. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. Cell Mol. Biol. 2005, 41, 546–566. [Google Scholar] [CrossRef]
- Bang, W.; Kim, S.; Ueda, A.; Vikram, M.; Yun, D.; Bressan, R.A.; Hasegawa, P.M.; Bahk, J.; Koiwa, H. Arabidopsis carboxyl-terminal domain phosphatase-like isoforms share common catalytic and interaction domains but have distinct in planta functions. Plant Physiol. 2006, 142, 586–594. [Google Scholar] [CrossRef]
- Jin, Y.M.; Jung, J.; Jeon, H.; Won, S.Y.; Feng, Y.; Kang, J.S.; Lee, S.Y.; Cheong, J.J.; Koiwa, H.; Kim, M. AtCPL5, a novel Ser-2-specific RNA polymerase II C-terminal domain phosphatase, positively regulates ABA and drought responses in Arabidopsis. New Phytol. 2011, 190, 57–74. [Google Scholar] [CrossRef]
- Tian, Y.; Zheng, H.; Zhang, F.; Wang, S.; Ji, X.; Xu, C.; He, Y.; Ding, Y. PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci. Adv. 2019, 5, eaau7246. [Google Scholar] [CrossRef]
- Munoz, A.; Mangano, S.; Gonzalez-Garcia, M.P.; Contreras, R.; Sauer, M.; De Rybel, B.; Weijers, D.; Sanchez-Serrano, J.J.; Sanmartin, M.; Rojo, E. RIMA-Dependent Nuclear Accumulation of IYO Triggers Auxin-Irreversible Cell Differentiation in Arabidopsis. Plant Cell 2017, 29, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Koiwa, H. Phosphorylation of RNA polymerase II C-terminal domain and plant osmotic-stress responses. In Abiotic Stress Tolerance in Plants: Toward the Improvement of Global Environment and Food; Rai, A.K., Takabe, T., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 47–57. [Google Scholar]
- Feng, Y.; Kang, J.S.; Kim, S.; Yun, D.J.; Lee, S.Y.; Bahk, J.D.; Koiwa, H. Arabidopsis SCP1-like small phosphatases differentially dephosphorylate RNA polymerase II C-terminal domain. Biochem. Biophys. Res. Commun. 2010, 397, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, J.E.; Burkholder, N.T.; Zhang, Y.J. Dephosphorylating eukaryotic RNA polymerase II. Biochim. Biophys. Acta 2016, 1864, 372–387. [Google Scholar] [CrossRef] [PubMed]
- Koiwa, H.; Barb, A.W.; Xiong, L.; Li, F.; McCully, M.G.; Lee, B.H.; Sokolchik, I.; Zhu, J.; Gong, Z.; Reddy, M.; et al. C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc. Natl. Acad. Sci. USA 2002, 99, 10893–10898. [Google Scholar] [CrossRef]
- Koiwa, H.; Hausmann, S.; Bang, W.Y.; Ueda, A.; Kondo, N.; Hiraguri, A.; Fukuhara, T.; Bahk, J.D.; Yun, D.J.; Bressan, R.A.; et al. Arabidopsis C-terminal domain phosphatase-like 1 and 2 are essential Ser-5-specific C-terminal domain phosphatases. Proc. Natl. Acad. Sci. USA 2004, 101, 14539–14544. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, G.; Chen, Y.; Zhao, Y.; Gao, P.; Liu, B.; Wang, H.; Zheng, Z.L. C-terminal domain (CTD) phosphatase links Rho GTPase signaling to Pol II CTD phosphorylation in Arabidopsis and yeast. Proc. Natl. Acad. Sci. USA 2016, 113, E8197–E8206. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Roman, I.; Munoz-Centeno, M.C. Coupling Between Cell Cycle Progression and the Nuclear RNA Polymerases System. Front. Mol. Biosci. 2021, 8, 691636. [Google Scholar] [CrossRef] [PubMed]
- Hsin, J.P.; Manley, J.L. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012, 26, 2119–2137. [Google Scholar] [CrossRef] [PubMed]
- Egloff, S.; Dienstbier, M.; Murphy, S. Updating the RNA polymerase CTD code: Adding gene-specific layers. Trends Genet. 2012, 28, 333–341. [Google Scholar] [CrossRef]
- Hajheidari, M.; Koncz, C.; Eick, D. Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci. 2013, 18, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Aristizabal, M.J.; Kobor, M.S. A single flexible RNAPII-CTD integrates many different transcriptional programs. Transcription 2016, 7, 50–56. [Google Scholar] [CrossRef][Green Version]
- Zaborowska, J.; Egloff, S.; Murphy, S. The pol II CTD: New twists in the tail. Nat. Struct. Mol. Biol. 2016, 23, 771–777. [Google Scholar] [CrossRef]
- Harlen, K.M.; Churchman, L.S. The code and beyond: Transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 2017, 18, 263–273. [Google Scholar] [CrossRef]
- Schwer, B.; Sanchez, A.M.; Shuman, S. Punctuation and syntax of the RNA polymerase II CTD code in fission yeast. Proc. Natl. Acad. Sci. USA 2012, 109, 18024–18029. [Google Scholar] [CrossRef]
- Jeronimo, C.; Collin, P.; Robert, F. The RNA Polymerase II CTD: The Increasing Complexity of a Low-Complexity Protein Domain. J. Mol. Biol. 2016, 428, 2607–2622. [Google Scholar] [CrossRef]
- Cho, C.Y.; Motta, F.C.; Kelliher, C.M.; Deckard, A.; Haase, S.B. Reconciling conflicting models for global control of cell-cycle transcription. Cell Cycle 2017, 16, 1965–1978. [Google Scholar] [CrossRef]
- Simon, I.; Barnett, J.; Hannett, N.; Harbison, C.T.; Rinaldi, N.J.; Volkert, T.L.; Wyrick, J.J.; Zeitlinger, J.; Gifford, D.K.; Jaakkola, T.S.; et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 2001, 106, 697–708. [Google Scholar] [CrossRef]
- Rahi, S.J.; Pecani, K.; Ondracka, A.; Oikonomou, C.; Cross, F.R. The CDK-APC/C Oscillator Predominantly Entrains Periodic Cell-Cycle Transcription. Cell 2016, 165, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Amoussouvi, A.; Teufel, L.; Reis, M.; Seeger, M.; Schlichting, J.K.; Schreiber, G.; Herrmann, A.; Klipp, E. Transcriptional timing and noise of yeast cell cycle regulators-a single cell and single molecule approach. NPJ Syst. Biol. Appl. 2018, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Andrews, B.; Measday, V. The cyclin family of budding yeast: Abundant use of a good idea. Trends Genet. 1998, 14, 66–72. [Google Scholar] [CrossRef]
- Santamaria, D.; Barriere, C.; Cerqueira, A.; Hunt, S.; Tardy, C.; Newton, K.; Caceres, J.F.; Dubus, P.; Malumbres, M.; Barbacid, M. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 2007, 448, 811–815. [Google Scholar] [CrossRef]
- Satyanarayana, A.; Berthet, C.; Lopez-Molina, J.; Coppola, V.; Tessarollo, L.; Kaldis, P. Genetic substitution of Cdk1 by Cdk2 leads to embryonic lethality and loss of meiotic function of Cdk2. Development 2008, 135, 3389–3400. [Google Scholar] [CrossRef]
- Nowack, M.K.; Harashima, H.; Dissmeyer, N.; Zhao, X.; Bouyer, D.; Weimer, A.K.; De Winter, F.; Yang, F.; Schnittger, A. Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev. Cell 2012, 22, 1030–1040. [Google Scholar] [CrossRef]
- Zhang, J.; Corden, J.L. Identification of phosphorylation sites in the repetitive carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J. Biol. Chem. 1991, 266, 2290–2296. [Google Scholar] [CrossRef]
- Chymkowitch, P.; Eldholm, V.; Lorenz, S.; Zimmermann, C.; Lindvall, J.M.; Bjoras, M.; Meza-Zepeda, L.A.; Enserink, J.M. Cdc28 kinase activity regulates the basal transcription machinery at a subset of genes. Proc. Natl. Acad. Sci. USA 2012, 109, 10450–10455. [Google Scholar] [CrossRef]
- Chymkowitch, P.; Enserink, J.M. The cell cycle rallies the transcription cycle: Cdc28/Cdk1 is a cell cycle-regulated transcriptional CDK. Transcription 2013, 4, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Larochelle, S.; Amat, R.; Glover-Cutter, K.; Sanso, M.; Zhang, C.; Allen, J.J.; Shokat, K.M.; Bentley, D.L.; Fisher, R.P. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat. Struct. Mol. Biol. 2012, 19, 1108–1115. [Google Scholar] [CrossRef]
- Hajheidari, M.; Farrona, S.; Huettel, B.; Koncz, Z.; Koncz, C. CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the C-terminal domain of Arabidopsis RNA polymerase II. Plant Cell 2012, 24, 1626–1642. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhong, X.; Sauane, M.; Zhao, Y.; Zheng, Z.L. Modulation of the Pol II CTD Phosphorylation Code by Rac1 and Cdc42 Small GTPases in Cultured Human Cancer Cells and Its Implication for Developing a Synthetic-Lethal Cancer Therapy. Cells 2020, 9, 621. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowski, N.; Zhang, T.; Rahl, P.B.; Abraham, B.J.; Reddy, J.; Ficarro, S.B.; Dastur, A.; Amzallag, A.; Ramaswamy, S.; Tesar, B.; et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014, 511, 616–620. [Google Scholar] [CrossRef]
- Fisher, R.P. Cdk7: A kinase at the core of transcription and in the crosshairs of cancer drug discovery. Transcription 2019, 10, 47–56. [Google Scholar] [CrossRef]
- Ganuza, M.; Saiz-Ladera, C.; Canamero, M.; Gomez, G.; Schneider, R.; Blasco, M.A.; Pisano, D.; Paramio, J.M.; Santamaria, D.; Barbacid, M. Genetic inactivation of Cdk7 leads to cell cycle arrest and induces premature aging due to adult stem cell exhaustion. EMBO J. 2012, 31, 2498–2510. [Google Scholar] [CrossRef]
- Simon, M.; Seraphin, B.; Faye, G. KIN28, a yeast split gene coding for a putative protein kinase homologous to CDC28. EMBO J. 1986, 5, 2697–2701. [Google Scholar] [CrossRef]
- Cooper, K. Rb, whi it’s not just for metazoans anymore. Oncogene 2006, 25, 5228–5232. [Google Scholar] [CrossRef]
- Mariconti, L.; Pellegrini, B.; Cantoni, R.; Stevens, R.; Bergounioux, C.; Cella, R.; Albani, D. The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J. Biol. Chem. 2002, 277, 9911–9919. [Google Scholar] [CrossRef]
- Weimer, A.K.; Nowack, M.K.; Bouyer, D.; Zhao, X.; Harashima, H.; Naseer, S.; De Winter, F.; Dissmeyer, N.; Geldner, N.; Schnittger, A. Retinoblastoma related1 regulates asymmetric cell divisions in Arabidopsis. Plant Cell 2012, 24, 4083–4095. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.L. Ras and Rho GTPase regulation of Pol II transcription: A shortcut model revisited. Transcription 2017, 8, 268–274. [Google Scholar] [CrossRef][Green Version]
- Luse, D.S. The RNA polymerase II preinitiation complex: Through what pathway is the complex assembled? Transcription 2013, 5, e27050. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.H.; Jin, Y.; Struhl, K. TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Mol. Cell 2014, 54, 601–612. [Google Scholar] [CrossRef]
- Allen, B.L.; Taatjes, D.J. The Mediator complex: A central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 155–166. [Google Scholar] [CrossRef]
- Chang, Y.W.; Howard, S.C.; Herman, P.K. The Ras/PKA signaling pathway directly targets the Srb9 protein, a component of the general RNA polymerase II transcription apparatus. Mol. Cell 2004, 15, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, P.C.; Hemerly, A.S.; Villarroel, R.; Van Montagu, M.; Inze, D. The Arabidopsis functional homolog of the p34cdc2 protein kinase. Plant Cell 1991, 3, 531–540. [Google Scholar] [CrossRef][Green Version]
- Hirayama, T.; Imajuku, Y.; Anai, T.; Matsui, M.; Oka, A. Identification of two cell-cycle-controlling cdc2 gene homologs in Arabidopsis thaliana. Gene 1991, 105, 159–165. [Google Scholar] [CrossRef]
- Mironov, V.V.; De Veylder, L.; Van Montagu, M.; Inze, D. Cyclin-dependent kinases and cell division in plants-the nexus. Plant Cell 1999, 11, 509–522. [Google Scholar] [CrossRef]
- Umeda, M.; Shimotohno, A.; Yamaguchi, M. Control of cell division and transcription by cyclin-dependent kinase-activating kinases in plants. Plant Cell Physiol. 2005, 46, 1437–1442. [Google Scholar] [CrossRef]
- Shimotohno, A.; Umeda-Hara, C.; Bisova, K.; Uchimiya, H.; Umeda, M. The plant-specific kinase CDKF;1 is involved in activating phosphorylation of cyclin-dependent kinase-activating kinases in Arabidopsis. Plant Cell 2004, 16, 2954–2966. [Google Scholar] [CrossRef] [PubMed]
- Umeda, M.; Bhalerao, R.P.; Schell, J.; Uchimiya, H.; Koncz, C. A distinct cyclin-dependent kinase-activating kinase of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1998, 95, 5021–5026. [Google Scholar] [CrossRef]
- Takatsuka, H.; Umeda-Hara, C.; Umeda, M. Cyclin-dependent kinase-activating kinases CDKD;1 and CDKD;3 are essential for preserving mitotic activity in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2015, 82, 1004–1017. [Google Scholar] [CrossRef] [PubMed]
- Shimotohno, A.; Ohno, R.; Bisova, K.; Sakaguchi, N.; Huang, J.; Koncz, C.; Uchimiya, H.; Umeda, M. Diverse phosphoregulatory mechanisms controlling cyclin-dependent kinase-activating kinases in Arabidopsis. Plant J. Cell Mol. Biol. 2006, 47, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Takatsuka, H.; Ohno, R.; Umeda, M. The Arabidopsis cyclin-dependent kinase-activating kinase CDKF;1 is a major regulator of cell proliferation and cell expansion but is dispensable for CDKA activation. Plant J. Cell Mol. Biol. 2009, 59, 475–487. [Google Scholar] [CrossRef]
- Cavallari, N.; Nibau, C.; Fuchs, A.; Dadarou, D.; Barta, A.; Doonan, J.H. The cyclin-dependent kinase G group defines a thermo-sensitive alternative splicing circuit modulating the expression of Arabidopsis ATU2AF65A. Plant J. Cell Mol. Biol. 2018, 94, 1010–1022. [Google Scholar] [CrossRef]
- Nibau, C.; Gallemi, M.; Dadarou, D.; Doonan, J.H.; Cavallari, N. Thermo-Sensitive Alternative Splicing of FLOWERING LOCUS M Is Modulated by Cyclin-Dependent Kinase G2. Front. Plant Sci. 2019, 10, 1680. [Google Scholar] [CrossRef]
- Kanno, T.; Venhuizen, P.; Wu, M.T.; Chiou, P.; Chang, C.L.; Kalyna, M.; Matzke, A.J.M.; Matzke, M. A Collection of Pre-mRNA Splicing Mutants in Arabidopsis thaliana. Genes Genomes Genet. 2020, 10, 1983–1996. [Google Scholar] [CrossRef]
- Zheng, T.; Nibau, C.; Phillips, D.W.; Jenkins, G.; Armstrong, S.J.; Doonan, J.H. CDKG1 protein kinase is essential for synapsis and male meiosis at high ambient temperature in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2014, 111, 2182–2187. [Google Scholar] [CrossRef]
- Nibau, C.; Dadarou, D.; Kargios, N.; Mallioura, A.; Fernandez-Fuentes, N.; Cavallari, N.; Doonan, J.H. A Functional Kinase Is Necessary for Cyclin-Dependent Kinase G1 (CDKG1) to Maintain Fertility at High Ambient Temperature in Arabidopsis. Front. Plant Sci. 2020, 11, 586870. [Google Scholar] [CrossRef]
- Ma, X.; Qiao, Z.; Chen, D.; Yang, W.; Zhou, R.; Zhang, W.; Wang, M. CYCLIN-DEPENDENT KINASE G2 regulates salinity stress response and salt mediated flowering in Arabidopsis thaliana. Plant Mol. Biol. 2015, 88, 287–299. [Google Scholar] [CrossRef]
- Ton, J.; Jakab, G.; Toquin, V.; Flors, V.; Iavicoli, A.; Maeder, M.N.; Metraux, J.P.; Mauch-Mani, B. Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell 2005, 17, 987–999. [Google Scholar] [CrossRef] [PubMed]
- Slaughter, A.; Daniel, X.; Flors, V.; Luna, E.; Hohn, B.; Mauch-Mani, B. Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 2012, 158, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Lessard, P.; Bouly, J.P.; Jouannic, S.; Kreis, M.; Thomas, M. Identification of cdc2cAt: A new cyclin-dependent kinase expressed in Arabidopsis thaliana flowers. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 1999, 1445, 351–358. [Google Scholar] [CrossRef]
- Li, F.; Cheng, C.; Cui, F.; de Oliveira, M.V.; Yu, X.; Meng, X.; Intorne, A.C.; Babilonia, K.; Li, M.; Li, B.; et al. Modulation of RNA polymerase II phosphorylation downstream of pathogen perception orchestrates plant immunity. Cell Host Microbe 2014, 16, 748–758. [Google Scholar] [CrossRef]
- Fukudome, A.; Aksoy, E.; Wu, X.; Kumar, K.; Jeong, I.S.; May, K.; Russell, W.K.; Koiwa, H. Arabidopsis CPL4 is an essential C-terminal domain phosphatase that suppresses xenobiotic stress responses. Plant J. Cell Mol. Biol. 2014, 80, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Fukudome, A.; Sun, D.; Zhang, X.; Koiwa, H. Salt Stress and CTD PHOSPHATASE-LIKE4 Mediate the Switch between Production of Small Nuclear RNAs and mRNAs. Plant Cell 2017, 29, 3214–3233. [Google Scholar] [CrossRef]
- Fukudome, A.; Goldman, J.S.; Finlayson, S.A.; Koiwa, H. Silencing Arabidopsis CARBOXYL-TERMINAL DOMAIN PHOSPHATASE-LIKE 4 induces cytokinin-oversensitive de novo shoot organogenesis. Plant J. Cell Mol. Biol. 2018, 94, 799–812. [Google Scholar] [CrossRef]
- Li, T.; Natran, A.; Chen, Y.; Vercruysse, J.; Wang, K.; Gonzalez, N.; Dubois, M.; Inze, D. A genetics screen highlights emerging roles for CPL3, RST1 and URT1 in RNA metabolism and silencing. Nat. Plants 2019, 5, 539–550. [Google Scholar] [CrossRef]
- Xiong, L.; Lee, H.; Ishitani, M.; Tanaka, Y.; Stevenson, B.; Koiwa, H.; Bressan, R.A.; Hasegawa, P.M.; Zhu, J.K. Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10899–10904. [Google Scholar] [CrossRef]
- Manavella, P.A.; Hagmann, J.; Ott, F.; Laubinger, S.; Franz, M.; Macek, B.; Weigel, D. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 2012, 151, 859–870. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Wang, B.; Shen, Y.; Wang, H.; Feng, Q.; Shi, H. The arabidopsis RNA binding protein with K homology motifs, SHINY1, interacts with the C-terminal domain phosphatase-like 1 (CPL1) to repress stress-inducible gene expression. PLoS Genet. 2013, 9, e1003625. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Yue, X.; Zeng, H.; Zhu, J. The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 2014, 26, 438–453. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Foley, R.; Casarotto, H.J.; Gao, L.L.; Kamphuis, L.G.; Melser, S.; Singh, K.B. The Arabidopsis RNA Polymerase II Carboxyl Terminal Domain (CTD) Phosphatase-Like1 (CPL1) is a biotic stress susceptibility gene. Sci. Rep. 2018, 8, 13454. [Google Scholar] [CrossRef]
- Aksoy, E.; Jeong, I.S.; Koiwa, H. Loss of function of Arabidopsis C-terminal domain phosphatase-like1 activates iron deficiency responses at the transcriptional level. Plant Physiol. 2013, 161, 330–345. [Google Scholar] [CrossRef]
- Yuan, C.; Xu, J.; Chen, Q.; Liu, Q.; Hu, Y.; Jin, Y.; Qin, C. C-terminal domain phosphatase-like 1 (CPL1) is involved in floral transition in Arabidopsis. BMC Genom. 2021, 22, 642. [Google Scholar] [CrossRef]
- Tao, L.Z.; Cheung, A.Y.; Wu, H.M. Plant Rac-like GTPases are activated by auxin and mediate auxin-responsive gene expression. Plant Cell 2002, 14, 2745–2760. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Wen, M.; Nagawa, S.; Fu, Y.; Chen, J.G.; Wu, M.J.; Perrot-Rechenmann, C.; Friml, J.; Jones, A.M.; Yang, Z. Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 2010, 143, 99–110. [Google Scholar] [CrossRef] [PubMed]
| S. cerevisiae | H. sapiens | A. thaliana | Ser-Specificity | |
|---|---|---|---|---|
| CDKs | Kin28 | CDK7 | Ser5, Ser7 | |
| CDKD;1, CDKD;2, CDKD;3 | Ser2, Ser5, Ser7 | |||
| Bur1 | CDK9 | Ser2, Ser5, Ser7 | ||
| CDKC;1, CDKC;2 | Ser2 | |||
| Ctk1 | CDK12, CDK13 | Ser2, Ser5, Ser7 | ||
| CDKF;1 | Ser7 | |||
| Cdc28 | CDKA;1 * | Ser5 | ||
| CDK1 | Ser2, Ser5 | |||
| CTD Phosphatases | Fcp1 | CTDP1 | CPL3 *, CPL4 | Ser2, Ser5 |
| Ssu72 | SSU72 | SSU72 * | Ser5, Ser7 | |
| Rtr1 | RPAP2 | RIMA * | Ser5 | |
| Cdc14 | CDC14A, CDC14B | Ser2, Ser5 | ||
| CTDSP1, CTDSP2, CTDSPL | SSP5 | Ser5 | ||
| SSP4, SSP4B | Ser2, Ser5 | |||
| CPL5 | Ser2 | |||
| CPL1, CPL2 | Ser2, Ser5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Z.-L. Cyclin-Dependent Kinases and CTD Phosphatases in Cell Cycle Transcriptional Control: Conservation across Eukaryotic Kingdoms and Uniqueness to Plants. Cells 2022, 11, 279. https://doi.org/10.3390/cells11020279
Zheng Z-L. Cyclin-Dependent Kinases and CTD Phosphatases in Cell Cycle Transcriptional Control: Conservation across Eukaryotic Kingdoms and Uniqueness to Plants. Cells. 2022; 11(2):279. https://doi.org/10.3390/cells11020279
Chicago/Turabian StyleZheng, Zhi-Liang. 2022. "Cyclin-Dependent Kinases and CTD Phosphatases in Cell Cycle Transcriptional Control: Conservation across Eukaryotic Kingdoms and Uniqueness to Plants" Cells 11, no. 2: 279. https://doi.org/10.3390/cells11020279
APA StyleZheng, Z.-L. (2022). Cyclin-Dependent Kinases and CTD Phosphatases in Cell Cycle Transcriptional Control: Conservation across Eukaryotic Kingdoms and Uniqueness to Plants. Cells, 11(2), 279. https://doi.org/10.3390/cells11020279







