Accumulation of Glycogen and Upregulation of LEA-1 in C. elegans daf-2(e1370) Support Stress Resistance, Not Longevity
Abstract
1. Introduction
2. Materials and Methods
2.1. C. elegans Strains and Culture Conditions
2.2. RNAi Treatment
2.3. Lifespan Assay
2.4. Stress Resistance Assays
2.4.1. Heat and Oxidative Stress Assays
2.4.2. Osmotic Stress Assay
2.4.3. UV Stress Assay
2.5. Determination of Trehalose and Glycogen Content
2.6. Oil Red O (ORO) Staining and Quantification
2.7. RNAi Efficiency Quantification by Real-Time Quantitative PCR (RT-qPCR)
3. Results
3.1. Accumulation of Glycogen Plays No Role in daf-2(e1370) Longevity
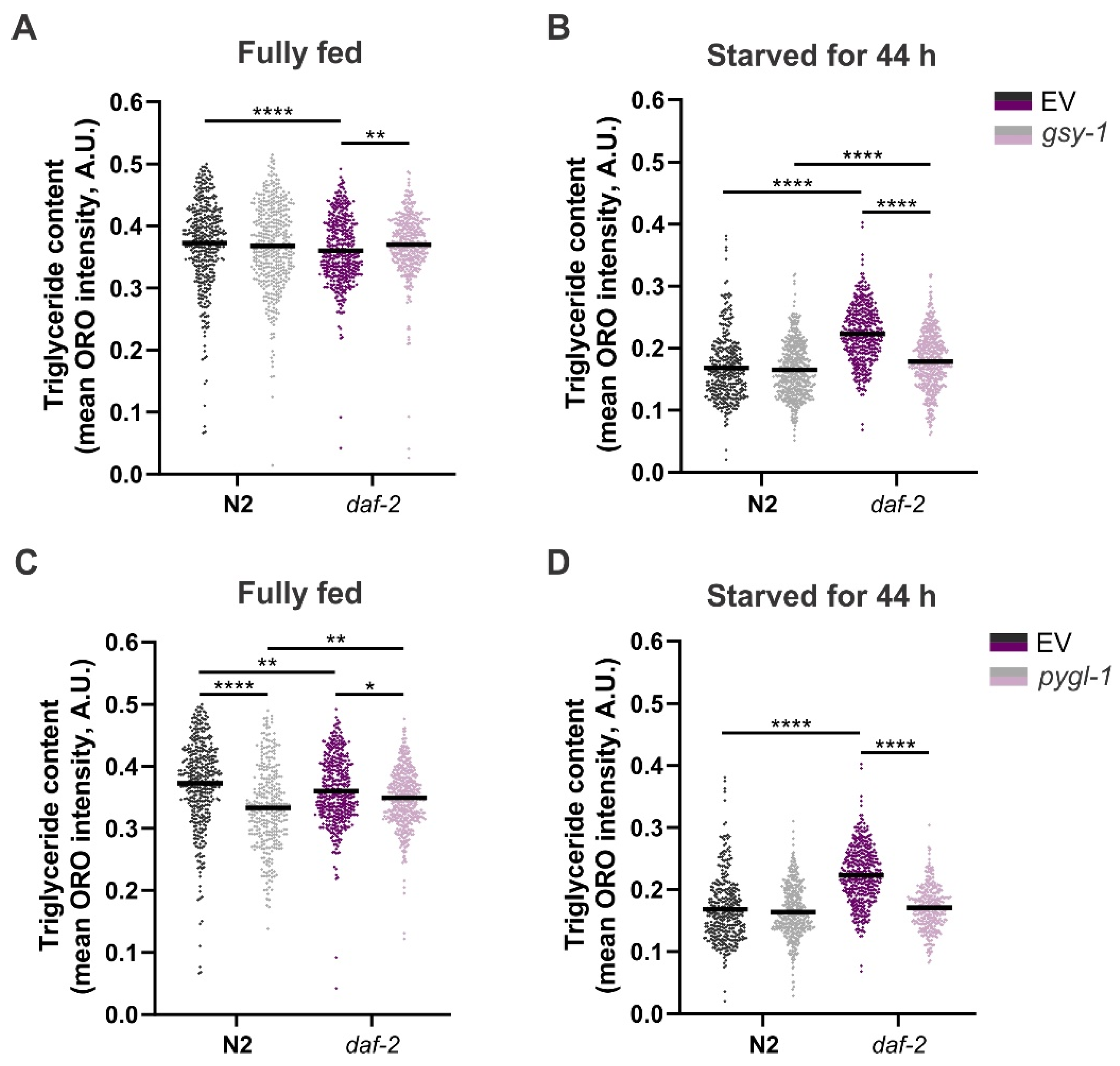
3.2. Gsy-1 and Pygl-1 Knockdowns Affect Glycogen, but Have No Impact on Trehalose Levels in daf-2(e1370) Mutants
3.3. Suppression of Glycogen Synthesis and/or Degradation Reduces Resistance to Osmotic and Heat Stress in daf-2(e1370) Mutants
3.4. Glycogen Is an Important Energy Source during Early Starvation in the daf-2(e1370) Mutant
3.5. lea-1 Is Not Important for daf-2(e1370) Longevity, but Offers Partial Protection against Osmotic, Heat and UV Stress
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kenyon, C.; Chang, J.; Gensch, E.; Rudner, A.; Tabtiang, R. A C. elegans mutant that lives twice as long as wild type. Nature 1993, 366, 461–464. [Google Scholar] [CrossRef]
- Murphy, C.T.; McCarroll, S.A.; Bargmann, C.I.; Fraser, A.; Kamath, R.S.; Ahringer, J.; Li, H.; Kenyon, C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 2003, 424, 277–283. [Google Scholar] [CrossRef]
- McElwee, J.J.; Schuster, E.; Blanc, E.; Thomas, J.H.; Gems, D. Shared Transcriptional Signature in Caenorhabditis elegans Dauer Larvae and Long-lived daf-2 Mutants Implicates Detoxification System in Longevity Assurance. J. Biol. Chem. 2004, 279, 44533–44543. [Google Scholar] [CrossRef] [PubMed]
- Halaschek-Wiener, J.; Khattra, J.S.; McKay, S.; Pouzyrev, A.; Stott, J.M.; Yang, G.S.; Holt, R.A.; Jones, S.J.M.; Marra, M.A.; Brooks-Wilson, A.R.; et al. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 2005, 15, 603–615. [Google Scholar] [CrossRef]
- Depuydt, G.; Xie, F.; Petyuk, V.A.; Smolders, A.; Brewer, H.M.; Camp, D.G.; Smith, R.D.; Braeckman, B.P. LC–MS Proteomics Analysis of the Insulin/IGF-1-Deficient Caenorhabditis elegans daf-2(e1370) Mutant Reveals Extensive Restructuring of Intermediary Metabolism. J. Proteome Res. 2014, 13, 1938–1956. [Google Scholar] [CrossRef]
- Melendez, A. Autophagy Genes Are Essential for Dauer Development and Life-Span Extension in C. elegans. Science 2003, 301, 1387–1391. [Google Scholar] [CrossRef] [PubMed]
- Webb, A.E.; Brunet, A. FOXO transcription factors: Key regulators of cellular quality control. Trends Biochem. Sci. 2014, 39, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Walther, D.M.; Kasturi, P.; Zheng, M.; Pinkert, S.; Vecchi, G.; Ciryam, P.; Morimoto, R.I.; Dobson, C.M.; Vendruscolo, M.; Mann, M.; et al. Widespread Proteome Remodeling and Aggregation in Aging C. elegans. Cell 2015, 161, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Dhondt, I.; Petyuk, V.A.; Cai, H.; Vandemeulebroucke, L.; Vierstraete, A.; Smith, R.D.; Depuydt, G.; Braeckman, B.P. FOXO/DAF-16 Activation Slows Down Turnover of the Majority of Proteins in C. elegans. Cell Rep. 2016, 16, 3028–3040. [Google Scholar] [CrossRef]
- Depuydt, G.; Shanmugam, N.; Rasulova, M.; Dhondt, I.; Braeckman, B.P. Increased Protein Stability and Decreased Protein Turnover in the Caenorhabditis elegans Ins/IGF-1 daf-2 Mutant. J. Gerontol. A Biol. Sci. Med. Sci. 2016, 71, 1553–1559. [Google Scholar] [CrossRef]
- Honda, Y.; Honda, S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 1999, 13, 1385–1393. [Google Scholar] [CrossRef]
- Lithgow, G.J.; White, T.M.; Melov, S.; Johnson, T.E. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc. Natl. Acad. Sci. USA 1995, 92, 7540–7544. [Google Scholar] [CrossRef]
- Scott, B.A.; Avidan, M.S.; Crowder, C.M. Regulation of hypoxic death in C. elegans by the insulin/IGF receptor homolog DAF-2. Science 2002, 296, 2388–2391. [Google Scholar] [CrossRef] [PubMed]
- Garsin, D.A.; Villanueva, J.M.; Begun, J.; Kim, D.H.; Sifri, C.D.; Calderwood, S.B.; Ruvkun, G.; Ausubel, F.M. Long-Lived C. elegans daf-2 Mutants Are Resistant to Bacterial Pathogens. Science 2003, 300, 1921. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Kennedy, S.; Tolonen, A.C.; Ruvkun, G. DAF-16 target genes that control C. elegans life-span and metabolism. Science 2003, 300, 644–647. [Google Scholar] [CrossRef]
- Dong, M.-Q.Q.; Venable, J.D.; Au, N.; Xu, T.; Sung, K.P.; Cociorva, D.; Johnson, J.R.; Dillin, A.; Yates, J.R.; Park, S.K.; et al. Supplementary-Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 2007, 317, 660–663. [Google Scholar] [CrossRef]
- Castro, C.; Krumsiek, J.; Lehrbach, N.J.; Murfitt, S.A.; Miska, E.A.; Griffin, J.L. A study of Caenorhabditis elegans DAF-2 mutants by metabolomics and differential correlation networks. Mol. Biosyst. 2013, 9, 1632–1642. [Google Scholar] [CrossRef]
- Gao, A.W.; Smith, R.L.; van Weeghel, M.; Kamble, R.; Janssens, G.E.; Houtkooper, R.H. Identification of key pathways and metabolic fingerprints of longevity in C. elegans. Exp. Gerontol. 2018, 113, 128–140. [Google Scholar] [CrossRef]
- Fuchs, S.; Bundy, J.G.; Davies, S.K.; Viney, J.M.; Swire, J.S.; Leroi, A.M. A metabolic signature of long life in Caenorhabditis elegans. BMC Biol. 2010, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Frazier, H.N., 3rd; Roth, M.B. Adaptive sugar provisioning controls survival of C. elegans embryos in adverse environments. Curr. Biol. 2009, 19, 859–863. [Google Scholar] [CrossRef]
- Popham, J.D.; Webster, J.M. Aspects of the fine structure of the dauer larva of the nematode Caenorhabditis elegans. Can. J. Zool. 1979, 57, 794–800. [Google Scholar] [CrossRef]
- Wright, D.J.; Grewal, P.; Stolinski, M. Relative Importance of Neutral Lipids and Glycogen as Energy Stores in Dauer Larvae of Two Entomopathogenic Nematodes, Steinernema carpocapsae and Steinernema feltiae. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1997, 118, 269–273. [Google Scholar] [CrossRef]
- Possik, E.; Ajisebutu, A.; Manteghi, S.; Gingras, M.-C.; Vijayaraghavan, T.; Flamand, M.; Coull, B.; Schmeisser, K.; Duchaine, T.; van Steensel, M.; et al. FLCN and AMPK Confer Resistance to Hyperosmotic Stress via Remodeling of Glycogen Stores. PLoS Genet. 2015, 11, e1005520. [Google Scholar] [CrossRef]
- Gusarov, I.; Pani, B.; Gautier, L.; Smolentseva, O.; Eremina, S.; Shamovsky, I.; Katkova-Zhukotskaya, O.; Mironov, A.; Nudler, E. Glycogen controls Caenorhabditis elegans lifespan and resistance to oxidative stress. Nat. Commun. 2017, 8, 15868. [Google Scholar] [CrossRef] [PubMed]
- LaMacchia, J.C.; Frazier, H.N.; Roth, M.B. Glycogen Fuels Survival During Hyposmotic-Anoxic Stress in Caenorhabditis elegans. Genetics 2015, 201, 65–74. [Google Scholar] [CrossRef]
- Seo, Y.; Kingsley, S.; Walker, G.; Mondoux, M.A.; Tissenbaum, H.A. Metabolic shift from glycogen to trehalose promotes lifespan and healthspan in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2018, 115, E2791–E2800. [Google Scholar] [CrossRef] [PubMed]
- Stacy, R.A.P.; Aalen, R.B. Identification of sequence homology between the internal hydrophilic repeated motifs of Group 1 late-embryogenesis-abundant proteinsin plants and hydrophilic repeats of the general stress protein GsiB of Bacillus subtilis. Planta 1998, 206, 476–478. [Google Scholar] [CrossRef]
- Honjoh, K.; Matsumoto, H.; Shimizu, H.; Ooyama, K.; Tanaka, K.; Oda, Y.; Takata, R.; Joh, T.; Suga, K.; Miyamoto, T.; et al. Cryoprotective Activities of Group 3 Late Embryogenesis Abundant Proteins from Chlorella vulgaris C-27. Biosci. Biotechnol. Biochem. 2000, 64, 1656–1663. [Google Scholar] [CrossRef]
- Bies-Ethève, N.; Gaubier-Comella, P.; Debures, A.; Lasserre, E.; Jobet, E.; Raynal, M.; Cooke, R.; Delseny, M. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 2008, 67, 107–124. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Lu, H.; Wang, X.; Cai, X.; Zhou, Z.; Zhang, Z.; Salih, H.; Wang, K.; et al. Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 2018, 19, 6. [Google Scholar] [CrossRef]
- Solomon, A.; Salomon, R.; Paperna, I.; Glazer, I. Desiccation stress of entomopathogenic nematodes induces the accumulation of a novel heat-stable protein. Parasitology 2000, 121, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Goyal, K.; Tisi, L.; Basran, A.; Browne, J.; Burnell, A.; Zurdo, J.; Tunnacliffe, A. Transition from natively unfolded to folded state induced by desiccation in an anhydrobiotic nematode protein. J. Biol. Chem. 2003, 278, 12977–12984. [Google Scholar] [CrossRef] [PubMed]
- Gal, T.Z.; Glazer, I.; Koltai, H. An LEA group 3 family member is involved in survival of C. elegans during exposure to stress. FEBS Lett. 2004, 577, 21–26. [Google Scholar] [CrossRef]
- Wu, G.; Zhang, H.; Sun, J.; Liu, F.; Ge, X.; Chen, W.-H.; Yu, J.; Wang, W. Diverse LEA (late embryogenesis abundant) and LEA-like genes and their responses to hypersaline stress in post-diapause embryonic development of Artemia franciscana. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2011, 160, 32–39. [Google Scholar] [CrossRef]
- MacRae, T.H. Stress tolerance during diapause and quiescence of the brine shrimp, Artemia. Cell Stress Chaperones 2016, 21, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Tunnacliffe, A.; Wise, M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften 2007, 94, 791–812. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The Enigmatic LEA Proteins and Other Hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Goyal, K.; Walton, L.J.; Tunnacliffe, A. LEA proteins prevent protein aggregation due to water stress. Biochem. J. 2005, 388, 151–157. [Google Scholar] [CrossRef]
- Erkut, C.; Vasilj, A.; Boland, S.; Habermann, B.; Shevchenko, A.; Kurzchalia, T. V Molecular Strategies of the Caenorhabditis elegans Dauer Larva to Survive Extreme Desiccation. PLoS ONE 2013, 8, e82473. [Google Scholar] [CrossRef]
- Rasulova, M.; Zečić, A.; Moreno, J.M.M.; Vandemeulebroucke, L.; Dhondt, I.; Braeckman, B.P. Elevated Trehalose Levels in C. elegans daf-2 Mutants Increase Stress Resistance, Not Lifespan. Metabolites 2021, 11, 105. [Google Scholar] [CrossRef]
- Sulston, J.; Hodgkin, J. The Nematode Caenorhabditis Elegans; William, B.W., Ed.; CSH Press: Long Island, NY, USA, 1988. [Google Scholar]
- Kamath, R.S.; Martinez-Campos, M.; Zipperlen, P.; Fraser, A.G.; Ahringer, J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2000, 2, research0002.1. [Google Scholar] [CrossRef]
- Kamath, R. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 2003, 30, 313–321. [Google Scholar] [CrossRef]
- Rual, J.-F.; Ceron, J.; Koreth, J.; Hao, T.; Nicot, A.-S.; Hirozane-Kishikawa, T.; Vandenhaute, J.; Orkin, S.H.; Hill, D.E.; van den Heuvel, S.; et al. Toward Improving Caenorhabditis elegans Phenome Mapping With an ORFeome-Based RNAi Library. Genome Res. 2004, 14, 2162–2168. [Google Scholar] [CrossRef] [PubMed]
- Han, S.K.; Lee, D.; Lee, H.; Kim, D.; Son, H.G.; Yang, J.-S.; Lee, S.-J.V.; Kim, S. OASIS 2: Online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 2016, 7, 56147–56152. [Google Scholar] [CrossRef]
- Benedetto, A.; Bambade, T.; Au, C.; Tullet, J.M.A.; Monkhouse, J.; Dang, H.; Cetnar, K.; Chan, B.; Cabreiro, F.; Gems, D. New label-free automated survival assays reveal unexpected stress resistance patterns during C. elegans aging. Aging Cell 2019, 18, e12998. [Google Scholar] [CrossRef] [PubMed]
- Lamitina, T.; Huang, C.G.; Strange, K. Genome-wide RNAi screening identifies protein damage as a regulator of osmoprotective gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 12173–12178. [Google Scholar] [CrossRef]
- Murakami, S.; Johnson, T.E. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics 1996, 143, 1207–1218. [Google Scholar] [CrossRef]
- Escorcia, W.; Ruter, D.L.; Nhan, J.; Curran, S.P. Quantification of Lipid Abundance and Evaluation of Lipid Distribution in Caenorhabditis elegans by Nile Red and Oil Red O Staining. JoVE 2018, 133, e57352. [Google Scholar] [CrossRef] [PubMed]
- Wählby, C.; Kamentsky, L.; Liu, Z.H.; Riklin-Raviv, T.; Conery, A.L.; O’Rourke, E.J.; Sokolnicki, K.L.; Visvikis, O.; Ljosa, V.; Irazoqui, J.E.; et al. An image analysis toolbox for high-throughput C. elegans assays. Nat. Methods 2012, 9, 714–716. [Google Scholar] [CrossRef] [PubMed]
- Wählby, C.; Conery, A.L.; Bray, M.-A.; Kamentsky, L.; Larkins-Ford, J.; Sokolnicki, K.L.; Veneskey, M.; Michaels, K.; Carpenter, A.E.; O’Rourke, E.J. High- and low-throughput scoring of fat mass and body fat distribution in C. elegans. Methods 2014, 68, 492–499. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Hoogewijs, D.; Houthoofd, K.; Matthijssens, F.; Vandesompele, J.; Vanfleteren, J.R. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 2008, 9, 9. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.; Nelson, D.L. Lehninger Principles of Biochemistry, 7th ed.; W.H. Freeman and Company: New York, NY, USA, 2017; ISBN 9781319108243. [Google Scholar]
- Honda, Y.; Tanaka, M.; Honda, S. Trehalose extends longevity in the nematode Caenorhabditis elegans. Aging Cell 2010, 9, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.D.; Tissenbaum, H.A.; Liu, Y.; Ruvkun, G. daf-2, an Insulin Receptor-Like Gene That Regulates Longevity and Diapause in Caenorhabditis elegans. Science 1997, 277, 942–946. [Google Scholar] [CrossRef]
- Ogg, S.; Paradis, S.; Gottlieb, S.; Patterson, G.I.; Lee, L.; Tissenbaum, H.A.; Ruvkun, G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 1997, 389, 994–999. [Google Scholar] [CrossRef]
- Hellerer, T.; Axäng, C.; Brackmann, C.; Hillertz, P.; Pilon, M.; Enejder, A. Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-Stokes Raman scattering (CARS) microscopy. Proc. Natl. Acad. Sci. USA 2007, 104, 14658–14663. [Google Scholar] [CrossRef]
- Fouad, A.D.; Pu, S.H.; Teng, S.; Mark, J.R.; Fu, M.; Zhang, K.; Huang, J.; Raizen, D.M.; Fang-Yen, C. Quantitative Assessment of Fat Levels in Caenorhabditis elegans Using Dark Field Microscopy. G3 2017, 7, 1811–1818. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bretscher, H.; O’Connor, M.B. The Role of Muscle in Insect Energy Homeostasis. Front. Physiol. 2020, 11, 580687. [Google Scholar] [CrossRef] [PubMed]
- Burnell, A.M.; Houthoofd, K.; O’Hanlon, K.; Vanfleteren, J.R. Alternate metabolism during the dauer stage of the nematode Caenorhabditis elegans. Exp. Gerontol. 2005, 40, 850–856. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.J.; Stuart, J.M.; Lund, J.; Kim, S.K. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature 2002, 418, 975–979. [Google Scholar] [CrossRef] [PubMed]
- Hutter, H.; Suh, J. GExplore 1.4: An expanded web interface for queries on Caenorhabditis elegans protein and gene function. Worm 2016, 5, e1234659. [Google Scholar] [CrossRef] [PubMed]
- Gems, D.; Sutton, A.J.; Sundermeyer, M.L.; Albert, P.S.; King, K.V.; Edgley, M.L.; Larsen, P.L.; Riddle, D.L. Two Pleiotropic Classes of daf-2 Mutation Affect Larval Arrest, Adult Behavior, Reproduction and Longevity in Caenorhabditis elegans. Genetics 1998, 150, 129–155. [Google Scholar] [CrossRef]
- Churgin, M.A.; Jung, S.-K.; Yu, C.-C.; Chen, X.; Raizen, D.M.; Fang-Yen, C. Longitudinal imaging of Caenorhabditis elegans in a microfabricated device reveals variation in behavioral decline during aging. Elife 2017, 6, e26652. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.F.; Van Gundy, S.D. Metabolism of Glycogen and Neutral Lipids by Aphelenchus avenae and Caenorhabditis sp. in Aerobic, Microaerobic and Anaerobic Environments. J. Nematol. 1970, 2, 305–315. [Google Scholar] [PubMed]
- Yamada, T.; Habara, O.; Kubo, H.; Nishimura, T. Fat body glycogen serves as a metabolic safeguard for the maintenance of sugar levels in Drosophila. Development 2018, 145, dev158865. [Google Scholar] [CrossRef]
- Chakrabortee, S.; Tripathi, R.; Watson, M.; Kaminski Schierle, G.S.; Kurniawan, D.P.; Kaminski, C.F.; Wise, M.J.; Tunnacliffe, A. Intrinsically disordered proteins as molecular shields. Mol. Biosyst. 2012, 8, 210–219. [Google Scholar] [CrossRef]
- Steponkus, P.L.; Uemura, M.; Joseph, R.A.; Gilmour, S.J.; Thomashow, M.F. Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1998, 95, 14570–14575. [Google Scholar] [CrossRef]
- Gade, V.R.; Traikov, S.; Oertel, J.; Fahmy, K.; Kurzchalia, T. V C. elegans possess a general program to enter cryptobiosis that allows dauer larvae to survive different kinds of abiotic stress. Sci. Rep. 2020, 10, 13466. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, Y. PM2, a group 3 LEA protein from soybean, and its 22-mer repeating region confer salt tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 2005, 331, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ohta, A.; Takagi, M.; Imai, R. Expression of plant group 2 and group 3 lea genes in Saccharomyces cerevisiae revealed functional divergence among LEA proteins. J. Biochem. 2000, 127, 611–616. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H.; Han, J.; Jiang, S.; Geng, X.; Xue, D.; Chen, Y.; Zhang, C.; Zhou, Z.; Zhang, W.; et al. Functional assessment of hydrophilic domains of late embryogenesis abundant proteins from distant organisms. Microb. Biotechnol. 2019, 12, 752–762. [Google Scholar] [CrossRef]
- Hara, M.; Shinoda, Y.; Tanaka, Y.; Kuboi, T. DNA binding of citrus dehydrin promoted by zinc ion. Plant. Cell Environ. 2009, 32, 532–541. [Google Scholar] [CrossRef]
- Boswell, L.C.; Hand, S.C. Intracellular localization of group 3 LEA proteins in embryos of Artemia franciscana. Tissue Cell 2014, 46, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Harris, T.W.; Arnaboldi, V.; Cain, S.; Chan, J.; Chen, W.J.; Cho, J.; Davis, P.; Gao, S.; Grove, C.A.; Kishore, R.; et al. WormBase: A modern Model Organism Information Resource. Nucleic Acids Res. 2020, 48, D762–D767. [Google Scholar] [CrossRef] [PubMed]
- Sarov, M.; Murray, J.I.; Schanze, K.; Pozniakovski, A.; Niu, W.; Angermann, K.; Hasse, S.; Rupprecht, M.; Vinis, E.; Tinney, M.; et al. A Genome-Scale Resource for In Vivo Tag-Based Protein Function Exploration in C. elegans. Cell 2012, 150, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Félix, M.-A.; Braendle, C. The natural history of Caenorhabditis elegans. Curr. Biol. 2010, 20, R965–R969. [Google Scholar] [CrossRef] [PubMed]
- Frézal, L.; Félix, M.-A. C. elegans outside the Petri dish. Elife 2015, 4, e05849. [Google Scholar] [CrossRef] [PubMed]
- Golden, J.W.; Riddle, D.L. The Caenorhabditis elegans dauer larva: Developmental effects of pheromone, food, and temperature. Dev. Biol. 1984, 102, 368–378. [Google Scholar] [CrossRef]
- Cassada, R.C.; Russell, R.L. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 1975, 46, 326–342. [Google Scholar] [CrossRef]
- Klass, M.; Hirsh, D. Non-ageing developmental variant of Caenorhabditis elegans. Nature 1976, 260, 523–525. [Google Scholar] [CrossRef]
- Anderson, G.L. Responses of dauerlarvae of Caenorhabditis elegans (Nematoda: Rhabditidae) to thermal stress and oxygen deprivation. Can. J. Zool. 1978, 56, 1786–1791. [Google Scholar] [CrossRef]
- Xie, M.; Roy, R. AMP-Activated Kinase Regulates Lipid Droplet Localization and Stability of Adipose Triglyceride Lipase in C. elegans Dauer Larvae. PLoS ONE 2015, 10, e0130480. [Google Scholar] [CrossRef]
- Erkut, C.; Penkov, S.; Khesbak, H.; Vorkel, D.; Verbavatz, J.-M.; Fahmy, K.; Kurzchalia, T. V Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr. Biol. 2011, 21, 1331–1336. [Google Scholar] [CrossRef]
- O’Riordan, V.B.; Burnell, A.M. Intermediary metabolism in the dauer larva of the nematode Caenorhabditis elegans—II. The glyoxylate cycle and fatty-acid oxidation. Comp. Biochem. Physiol. Part B Comp. Biochem. 1990, 95, 125–130. [Google Scholar] [CrossRef]
- Erkut, C.; Gade, V.R.; Laxman, S.; Kurzchalia, T. V The glyoxylate shunt is essential for desiccation tolerance in C. elegans and budding yeast. Elife 2016, 5, e13614. [Google Scholar] [CrossRef]
- Mitsumasu, K.; Kanamori, Y.; Fujita, M.; Iwata, K.; Tanaka, D.; Kikuta, S.; Watanabe, M.; Cornette, R.; Okuda, T.; Kikawada, T. Enzymatic control of anhydrobiosis-related accumulation of trehalose in the sleeping chironomid, Polypedilum vanderplanki. FEBS J. 2010, 277, 4215–4228. [Google Scholar] [CrossRef] [PubMed]
- Rozsypal, J.; Koštál, V.; Zahradníčková, H.; Šimek, P. Overwintering strategy and mechanisms of cold tolerance in the codling moth (Cydia pomonella). PLoS ONE 2013, 8, e61745. [Google Scholar] [CrossRef] [PubMed]
- Lamitina, S.T.; Strange, K. Transcriptional targets of DAF-16 insulin signaling pathway protect C. elegans from extreme hypertonic stress. Am. J. Physiol. Physiol. 2005, 288, C467–C474. [Google Scholar] [CrossRef] [PubMed]
- Gal, T.Z.; Solomon, A.; Glazer, I.; Koltai, H. Alterations in the Levels of Glycogen and Glycogen Synthase Transcripts during Desiccation in the Insect-Killing Nematode Steinernema feltiae IS-6. J. Parasitol. 2001, 87, 725–732. [Google Scholar] [CrossRef]
- Dues, D.J.; Andrews, E.K.; Senchuk, M.M.; Van Raamsdonk, J.M. Resistance to Stress Can Be Experimentally Dissociated From Longevity. J. Gerontol. Ser. A 2019, 74, 1206–1214. [Google Scholar] [CrossRef]
- Gsell, M.; Fankl, A.; Klug, L.; Mascher, G.; Schmidt, C.; Hrastnik, C.; Zellnig, G.; Daum, G. A Yeast Mutant Deleted of GPH1 Bears Defects in Lipid Metabolism. PLoS ONE 2015, 10, e0136957. [Google Scholar] [CrossRef][Green Version]
- Cossins, A.R.; Prosser, C.L. Evolutionary adaptation of membranes to temperature. Proc. Natl. Acad. Sci. USA 1978, 75, 2040–2043. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Kitakaze, T.; Mitani, T.; Furuyashiki, T.; Ashida, H. Enzymatically synthesized glycogen prevents ultraviolet B-induced cell damage in normal human epidermal keratinocytes. J. Clin. Biochem. Nutr. 2020, 67, 36–42. [Google Scholar] [CrossRef]
- Takata, H.; Kajiura, H.; Furuyashiki, T.; Kakutani, R.; Kuriki, T. Fine structural properties of natural and synthetic glycogens. Carbohydr. Res. 2009, 344, 654–659. [Google Scholar] [CrossRef]
- Sokolova, I.M. Energy-Limited Tolerance to Stress as a Conceptual Framework to Integrate the Effects of Multiple Stressors. Integr. Comp. Biol. 2013, 53, 597–608. [Google Scholar] [CrossRef]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef]
- Olvera-Carrillo, Y.; Campos, F.; Reyes, J.L.; Garciarrubio, A.; Covarrubias, A.A. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 2010, 154, 373–390. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.; Boschetti, C.; McGee, B.; Tunnacliffe, A. Trafficking of bdelloid rotifer late embryogenesis abundant proteins. J. Exp. Biol. 2012, 215, 2786–2794. [Google Scholar] [CrossRef] [PubMed]
- Chakrabortee, S.; Boschetti, C.; Walton, L.J.; Sarkar, S.; Rubinsztein, D.C.; Tunnacliffe, A. Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc. Natl. Acad. Sci. USA 2007, 104, 18073–18078. [Google Scholar] [CrossRef] [PubMed]
- Cherkasova, V.; Ayyadevara, S.; Egilmez, N.; Reis, R.S. Diverse Caenorhabditis elegans genes that are upregulated in dauer larvae also show elevated transcript levels in long-lived, aged, or starved adults. J. Mol. Biol. 2000, 300, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Hibshman, J.D.; Goldstein, B. LEA motifs promote desiccation tolerance in vivo. BMC Biol 2021, 19, 263. [Google Scholar] [CrossRef]
- Doonan, R.; McElwee, J.J.; Matthijssens, F.; Walker, G.A.; Houthoofd, K.; Back, P.; Matscheski, A.; Vanfleteren, J.R.; Gems, D. Against the oxidative damage theory of aging: Superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 2008, 22, 3236–3241. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zečić, A.; Dhondt, I.; Braeckman, B.P. Accumulation of Glycogen and Upregulation of LEA-1 in C. elegans daf-2(e1370) Support Stress Resistance, Not Longevity. Cells 2022, 11, 245. https://doi.org/10.3390/cells11020245
Zečić A, Dhondt I, Braeckman BP. Accumulation of Glycogen and Upregulation of LEA-1 in C. elegans daf-2(e1370) Support Stress Resistance, Not Longevity. Cells. 2022; 11(2):245. https://doi.org/10.3390/cells11020245
Chicago/Turabian StyleZečić, Aleksandra, Ineke Dhondt, and Bart P. Braeckman. 2022. "Accumulation of Glycogen and Upregulation of LEA-1 in C. elegans daf-2(e1370) Support Stress Resistance, Not Longevity" Cells 11, no. 2: 245. https://doi.org/10.3390/cells11020245
APA StyleZečić, A., Dhondt, I., & Braeckman, B. P. (2022). Accumulation of Glycogen and Upregulation of LEA-1 in C. elegans daf-2(e1370) Support Stress Resistance, Not Longevity. Cells, 11(2), 245. https://doi.org/10.3390/cells11020245






