An Imaging and Computational Algorithm for Efficient Identification and Quantification of Neutrophil Extracellular Traps
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Human Neutrophils
2.2. Induction of NET Formation
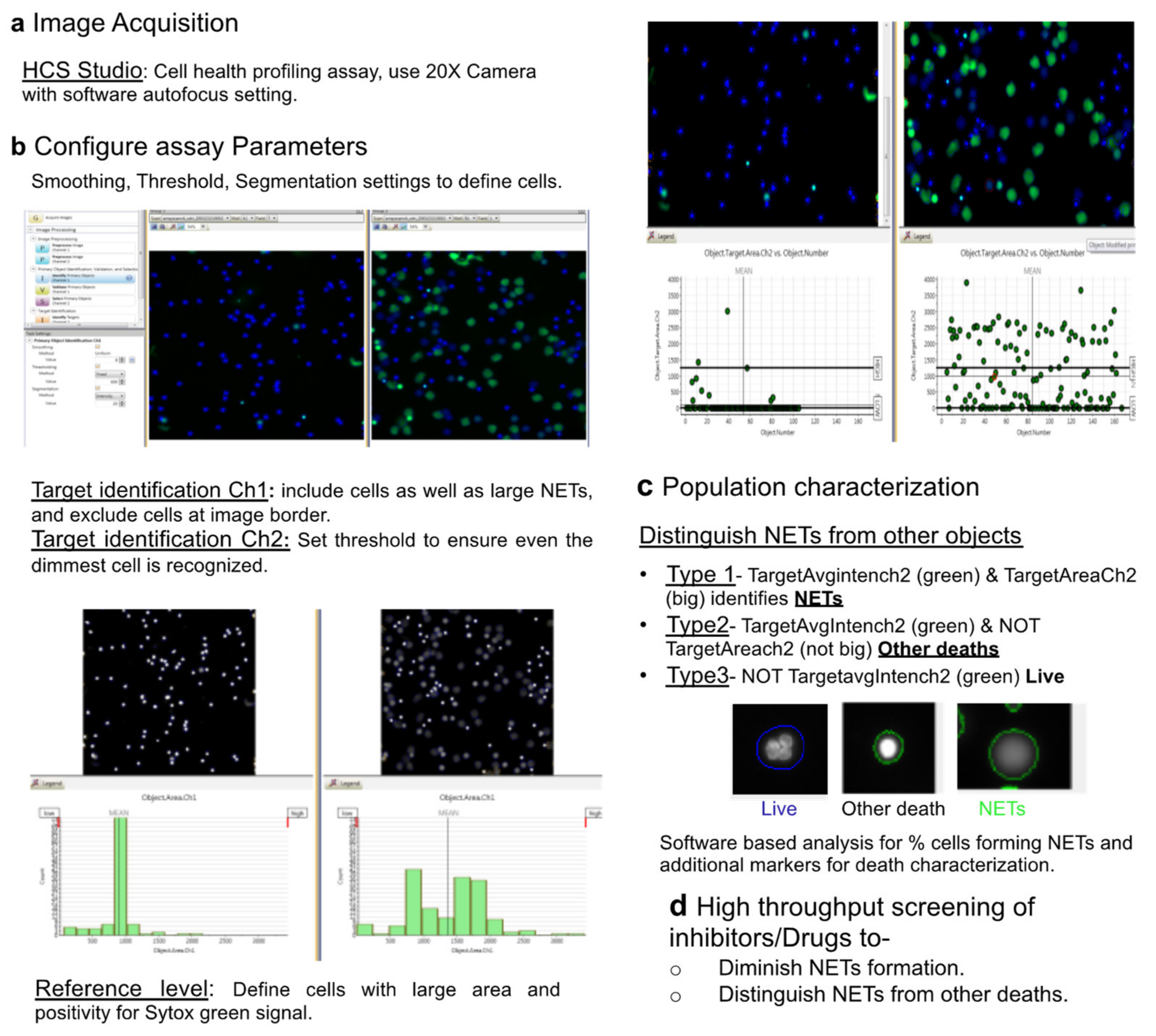
2.3. HCS Studio-Based Acquisition of Images and Identification of NETs
2.4. Automated Image Analysis and Quantification of NETs
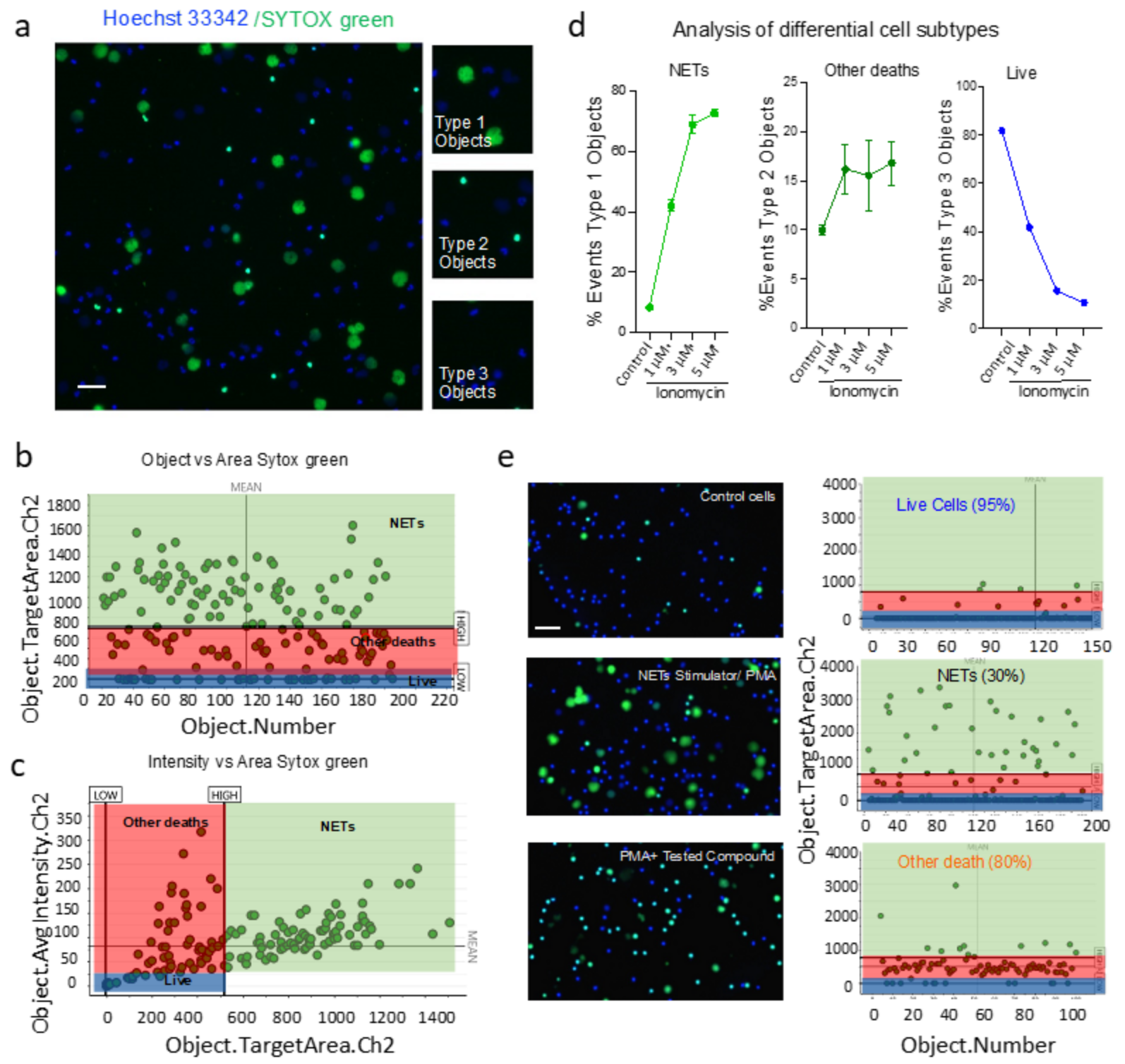
- Type 1 events, i.e., NET forming cells were identified using Target.AvgIntensity.Ch2 AND Target.Area.Ch2 as positive for green and big decondensed nucleus.
- Type 2 events, i.e., green and small cells that were undergoing death through pathways other than NETs were identified using Target.AvgIntensity.Ch2 NOT Target.AreaCh2. parameter.
- Type 3 objects were identified as NOT Target.AvgIntensity.Ch2 for live cells.
2.5. Immunofluorescence Microscopy
2.6. Statistics
3. Results
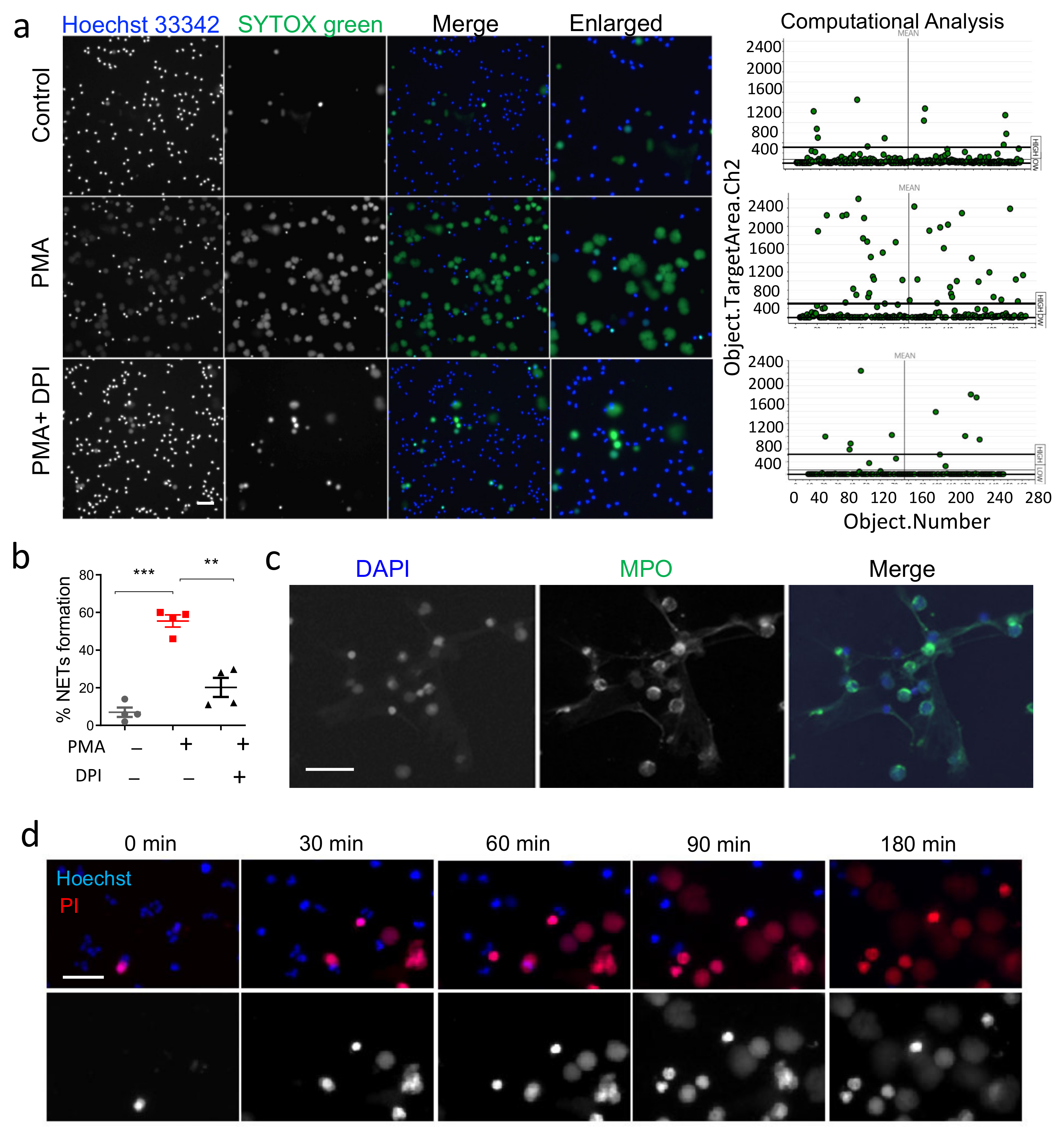
3.1. Validation of HCS-Cellomics Algorithm Using PMA Induced NET Formation
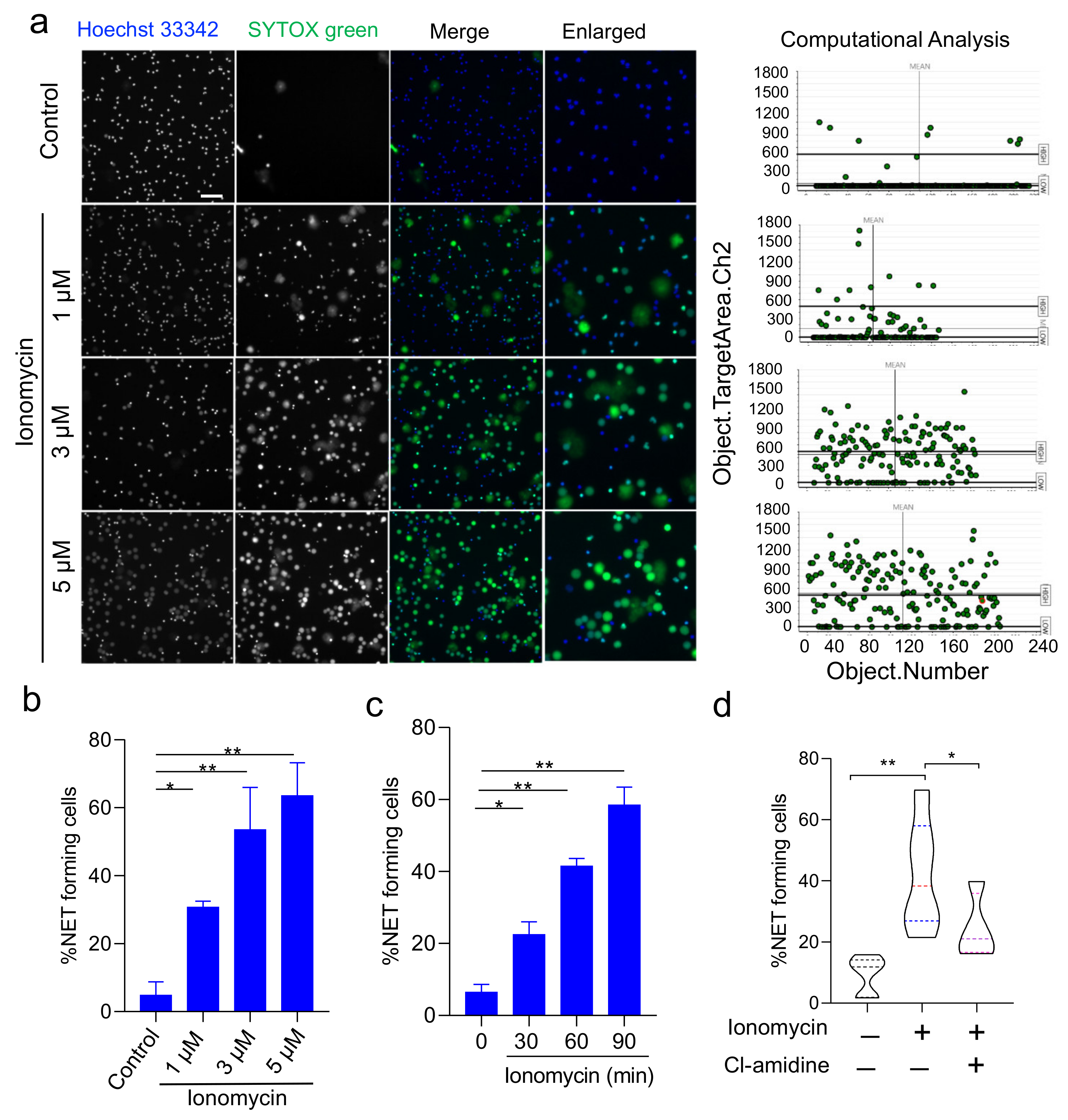
3.2. Validation of HCS-Cellomics Algorithm Using Ionomycin-Induced NET Formation
3.3. HCS-Cellomics Distinguishes NETs from other Forms of Neutrophil Death
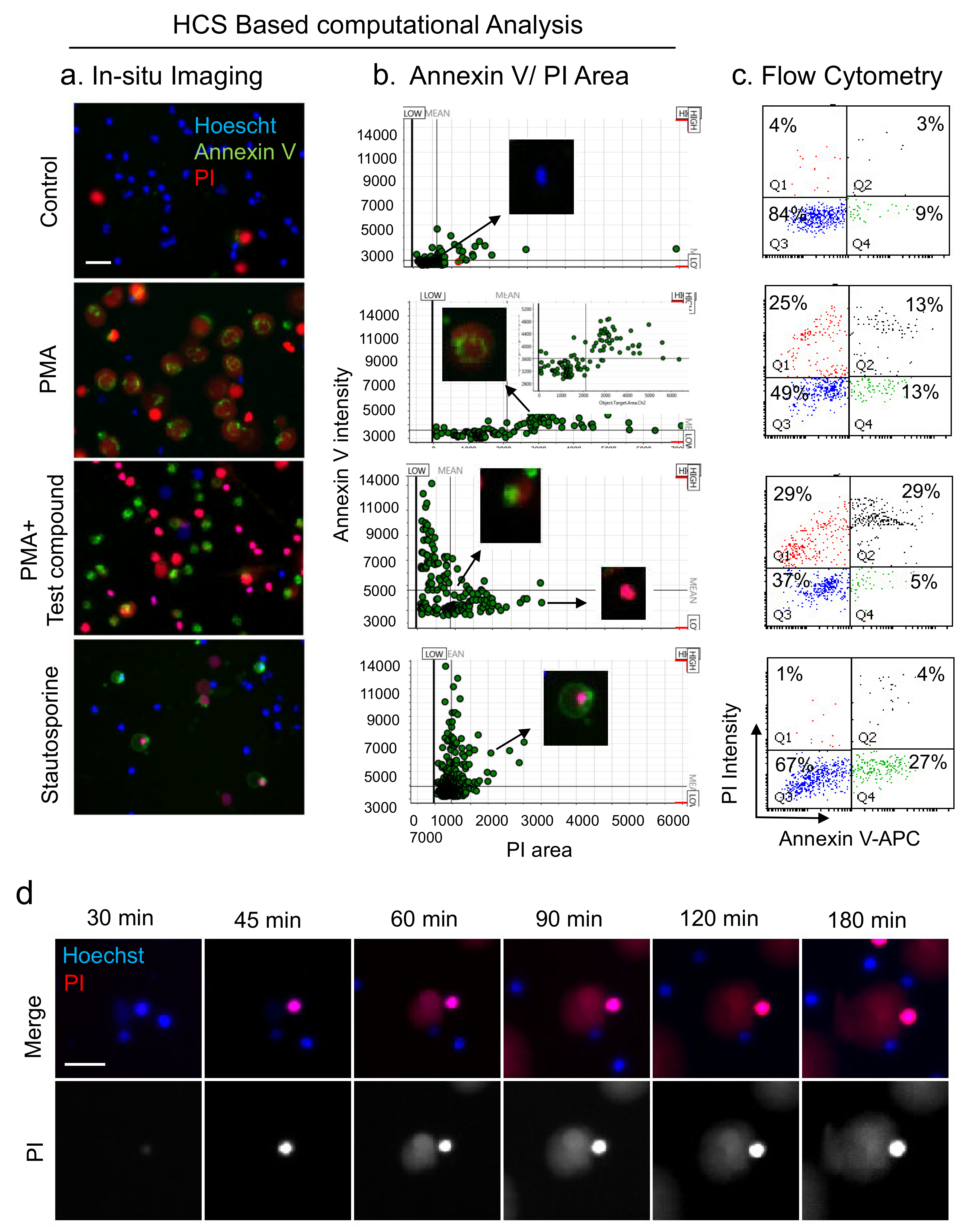
3.4. Analysis of NETs, Apoptosis, and Necrosis with HCS-Cellomics and Flow-Cytometry
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclosure
References
- Nathan, C. Neutrophils and immunity: Challenges and opportunities. Nat. Rev. Immunol. 2006, 6, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, O.E.; Borregaard, N. Neutrophil extracellular traps—The dark side of neutrophils. J. Clin. Investig. 2016, 126, 1612–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Dikshit, M. Metabolic Insight of Neutrophils in Health and Disease. Front. Immunol. 2019, 10, 2099. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Castanheira, F.V.S.; Kubes, P. Neutrophils and NETs in modulating acute and chronic inflammation. Blood 2019, 133, 2178–2185. [Google Scholar] [CrossRef]
- Yipp, B.G.; Petri, B.; Salina, D.; Jenne, C.N.; Scott, B.N.; Zbytnuik, L.D.; Pittman, K.; Asaduzzaman, M.; Wu, K.; Meijndert, H.C.; et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat. Med. 2012, 18, 1386–1393. [Google Scholar] [CrossRef] [Green Version]
- Clark, S.R.; Ma, A.C.; Tavener, S.A.; McDonald, B.; Goodarzi, Z.; Kelly, M.M.; Patel, K.D.; Chakrabarti, S.; McAvoy, E.; Sinclair, G.D.; et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat. Med. 2007, 13, 463–469. [Google Scholar] [CrossRef]
- Fuchs, T.A.; Abed, U.; Goosmann, C.; Hurwitz, R.; Schulze, I.; Wahn, V.; Weinrauch, Y.; Brinkmann, V.; Zychlinsky, A. Novel cell death program leads to neutrophil extracellular traps. J. Cell. Biol. 2007, 176, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Wartha, F.; Beiter, K.; Normark, S.; Henriques-Normark, B. Neutrophil extracellular traps: Casting the NET over pathogenesis. Curr. Opin. Microbiol. 2007, 10, 52–56. [Google Scholar] [CrossRef]
- Saitoh, T.; Komano, J.; Saitoh, Y.; Misawa, T.; Takahama, M.; Kozaki, T.; Uehata, T.; Iwasaki, H.; Omori, H.; Yamaoka, S.; et al. Neutrophil extracellular traps mediate a host defense response to human immunodeficiency virus-1. Cell Host Microbe 2012, 12, 109–116. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Weber, A.; Barnes, B.J.; Egeblad, M.; et al. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020, 5, e138999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, P.R.; Palmer, L.J.; Chapple, I.L. Neutrophil extracellular traps as a new paradigm in innate immunity: Friend or foe? Periodontology 2000 2013, 63, 165–197. [Google Scholar] [CrossRef] [PubMed]
- Boeltz, S.; Amini, P.; Anders, H.J.; Andrade, F.; Bilyy, R.; Chatfield, S.; Cichon, I.; Clancy, D.M.; Desai, J.; Dumych, T.; et al. To NET or not to NET:current opinions and state of the science regarding the formation of neutrophil extracellular traps. Cell Death Diffr. 2019, 26, 395–408. [Google Scholar] [CrossRef] [Green Version]
- Masuda, S.; Nakazawa, D.; Shida, H.; Miyoshi, A.; Kusunoki, Y.; Tomaru, U.; Ishizu, A. NETosis markers: Quest for specific, objective, and quantitative markers. Clin. Chim. Acta 2016, 459, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Naccache, P.H.; Fernandes, M.J. Challenges in the characterization of neutrophil extracellular traps: The truth is in the details. Eur. J. Immunol. 2016, 46, 52–55. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, M.J.; Radic, M. Neutrophil extracellular traps: Double-edged swords of innate immunity. J. Immunol. 2012, 189, 2689–2695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keshari, R.S.; Jyoti, A.; Dubey, M.; Kothari, N.; Kohli, M.; Bogra, J.; Barthwal, M.K.; Dikshit, M. Cytokines induced neutrophil extracellular traps formation: Implication for the inflammatory disease condition. PLoS ONE 2012, 7, e48111. [Google Scholar] [CrossRef] [Green Version]
- Caudrillier, A.; Kessenbrock, K.; Gilliss, B.M.; Nguyen, J.X.; Marques, M.B.; Monestier, M.; Toy, P.; Werb, Z.; Looney, M.R. Platelets induce neutrophil extracellular traps in transfusion-related acute lung injury. J. Clin. Investig. 2012, 122, 2661–2671. [Google Scholar] [CrossRef] [PubMed]
- Lande, R.; Ganguly, D.; Facchinetti, V.; Frasca, L.; Conrad, C.; Gregorio, J.; Meller, S.; Chamilos, G.; Sebasigari, R.; Riccieri, V.; et al. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci. Transl. Med. 2011, 3, 73ra19. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.; Kumar, S.; Jyoti, A.; Srinag, B.S.; Keshari, R.S.; Saluja, R.; Verma, A.; Mitra, K.; Barthwal, M.K.; Krishnamurthy, H.; et al. Nitric oxide donors release extracellular traps from human neutrophils by augmenting free radical generation. Nitric Oxide 2010, 22, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Singhal, A.; Dhankani, P.; Mazumder, J.; Adithya, R.; Dikshit, M.; Kumar, S. Rho signaling inhibition mitigates lung injury via targeting neutrophil recruitment and selectin-AKT signaling. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 119122. [Google Scholar] [CrossRef]
- Nakazawa, D.; Desai, J.; Steiger, S.; Muller, S.; Devarapu, S.K.; Mulay, S.R.; Iwakura, T.; Anders, H.J. Activated platelets induce MLKL-driven neutrophil necroptosis and release of neutrophil extracellular traps in venous thrombosis. Cell Death Discov. 2018, 4, 6. [Google Scholar] [CrossRef]
- Papayannopoulos, V. Neutrophil extracellular traps in immunity and disease. Nat. Rev. Immunol. 2018, 18, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Kenny, E.F.; Herzig, A.; Kruger, R.; Muth, A.; Mondal, S.; Thompson, P.R.; Brinkmann, V.; Bernuth, H.V.; Zychlinsky, A. Diverse stimuli engage different neutrophil extracellular trap pathways. Elife 2017, 6, e24437. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Chan, D.W.; Zaal, K.J.; Kaplan, M.J. A High-Throughput Real-Time Imaging Technique To Quantify NETosis and Distinguish Mechanisms of Cell Death in Human Neutrophils. J. Immunol. 2018, 200, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Goosmann, C.; Kuhn, L.I.; Zychlinsky, A. Automatic quantification of in vitro NET formation. Front. Immunol. 2012, 3, 413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraaij, T.; Tengstrom, F.C.; Kamerling, S.W.; Pusey, C.D.; Scherer, H.U.; Toes, R.E.; Rabelink, T.J.; van Kooten, C.; Teng, Y.K. A novel method for high-throughput detection and quantification of neutrophil extracellular traps reveals ROS-independent NET release with immune complexes. Autoimmun. Rev. 2016, 15, 577–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, P.C.; Chicca, I.J.; Ling, M.R.; Wright, H.J.; Cooper, P.R.; Milward, M.R.; Chapple, I.L. Characterization, Quantification, and Visualization of Neutrophil Extracellular Traps. Methods Mol. Biol. 2017, 1537, 481–497. [Google Scholar] [CrossRef] [PubMed]
- Urban, C.F.; Ermert, D.; Schmid, M.; Abu-Abed, U.; Goosmann, C.; Nacken, W.; Brinkmann, V.; Jungblut, P.R.; Zychlinsky, A. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 2009, 5, e1000639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gavillet, M.; Martinod, K.; Renella, R.; Harris, C.; Shapiro, N.I.; Wagner, D.D.; Williams, D.A. Flow cytometric assay for direct quantification of neutrophil extracellular traps in blood samples. Am. J. Hematol. 2015, 90, 1155–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, S.; Shimizu, S.; Matsuo, J.; Nishibata, Y.; Kusunoki, Y.; Hattanda, F.; Shida, H.; Nakazawa, D.; Tomaru, U.; Atsumi, T.; et al. Measurement of NET formation in vitro and in vivo by flow cytometry. Cytometry A 2017, 91, 822–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Fogg, D.K.; Kaplan, M.J. A novel image-based quantitative method for the characterization of NETosis. J. Immunol. Methods 2015, 423, 104–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moussavi-Harami, S.F.; Mladinich, K.M.; Sackmann, E.K.; Shelef, M.A.; Starnes, T.W.; Guckenberger, D.J.; Huttenlocher, A.; Beebe, D.J. Microfluidic device for simultaneous analysis of neutrophil extracellular traps and production of reactive oxygen species. Integr. Biol. Quant. Biosci. Nano Macro 2016, 8, 243–252. [Google Scholar] [CrossRef] [Green Version]
- Keshari, R.S.; Jyoti, A.; Kumar, S.; Dubey, M.; Verma, A.; Srinag, B.S.; Krishnamurthy, H.; Barthwal, M.K.; Dikshit, M. Neutrophil extracellular traps contain mitochondrial as well as nuclear DNA and exhibit inflammatory potential. Cytometry A 2012, 81, 238–247. [Google Scholar] [CrossRef]
- Korn, K.; Krausz, E. Cell-based high-content screening of small-molecule libraries. Curr. Opin. Chem. Biol. 2007, 11, 503–510. [Google Scholar] [CrossRef]
- Shariff, A.; Kangas, J.; Coelho, L.P.; Quinn, S.; Murphy, R.F. Automated image analysis for high-content screening and analysis. J. Biomol. Screen. 2010, 15, 726–734. [Google Scholar] [CrossRef] [Green Version]
- Steinberg, B.E.; Scott, C.C.; Grinstein, S. High-throughput assays of phagocytosis, phagosome maturation, and bacterial invasion. Am. J. Physiol. Cell Physiol. 2007, 292, C945–C952. [Google Scholar] [CrossRef] [Green Version]
- Douda, D.N.; Khan, M.A.; Grasemann, H.; Palaniyar, N. SK3 channel and mitochondrial ROS mediate NADPH oxidase-independent NETosis induced by calcium influx. Proc. Natl. Acad. Sci. USA 2015, 112, 2817–2822. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, S.M.; Corriden, R.; Nizet, V. How Neutrophils Meet Their End. Trends Immunol. 2020, 41, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Anderton, H.; Wicks, I.P.; Silke, J. Cell death in chronic inflammation: Breaking the cycle to treat rheumatic disease. Nat. Rev. Rheumatol. 2020, 16, 496–513. [Google Scholar] [CrossRef] [PubMed]
- Sawai, H.; Domae, N. Discrimination between primary necrosis and apoptosis by necrostatin-1 in Annexin V-positive/propidium iodide-negative cells. Biochem. Biophys. Res. Commun. 2011, 411, 569–573. [Google Scholar] [CrossRef]
- Leslie, M. New drugs aim to disarm the immune system’s ‘atomic bomb’ cells. Science 2020. [Google Scholar]
- Lefrancais, E.; Mallavia, B.; Zhuo, H.; Calfee, C.S.; Looney, M.R. Maladaptive role of neutrophil extracellular traps in pathogen-induced lung injury. JCI Insight. 2018, 3, e98178. [Google Scholar] [CrossRef] [Green Version]
- Narasaraju, T.; Yang, E.; Samy, R.P.; Ng, H.H.; Poh, W.P.; Liew, A.A.; Phoon, M.C.; van Rooijen, N.; Chow, V.T. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. Am. J. Pathol. 2011, 179, 199–210. [Google Scholar] [CrossRef]
- Barnes, B.J.; Adrover, J.M.; Baxter-Stoltzfus, A. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020, 217, e20200652. [Google Scholar] [CrossRef] [PubMed]
- de Buhr, N.; von Kockritz-Blickwede, M. How Neutrophil Extracellular Traps Become Visible. J. Immunol. Res. 2016, 2016, 4604713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manda-Handzlik, A.; Ostafin, M.; Bystrzycka, W.; Sieczkowska, S.; Moskalik, A.; Demkow, U.; Ciepiela, O. Flow cytometric quantification of neutrophil extracellular traps: Limitations of the methodological approach. Am. J. Hematol. 2016, 91, E9–E10. [Google Scholar] [CrossRef]
- Kumar, S.; Xu, J.; Perkins, C.; Guo, F.; Snapper, S.; Finkelman, F.D.; Zheng, Y.; Filippi, M.D. Cdc42 regulates neutrophil migration via crosstalk between WASp, CD11b, and microtubules. Blood 2012, 120, 3563–3574. [Google Scholar] [CrossRef] [Green Version]
- Chicca, I.J.; Milward, M.R.; Chapple, I.L.C.; Griffiths, G.; Benson, R.; Dietrich, T.; Cooper, P.R. Development and Application of High-Content Biological Screening for Modulators of NET Production. Front. Immunol. 2018, 9, 337. [Google Scholar] [CrossRef] [PubMed]
- van Breda, S.V.; Vokalova, L.; Neugebauer, C.; Rossi, S.W.; Hahn, S.; Hasler, P. Computational Methodologies for the in vitro and in situ Quantification of Neutrophil Extracellular Traps. Front. Immunol. 2019, 10, 1562. [Google Scholar] [CrossRef]
- Coelho, L.P.; Pato, C.; Friaes, A.; Neumann, A.; von Kockritz-Blickwede, M.; Ramirez, M.; Carrico, J.A. Automatic determination of NET (neutrophil extracellular traps) coverage in fluorescent microscopy images. Bioinformatics 2015, 31, 2364–2370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohanty, T.; Sorensen, O.E.; Nordenfelt, P. NETQUANT: Automated Quantification of Neutrophil Extracellular Traps. Front. Immunol. 2017, 8, 1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, J.H.; Schaekel, K.; Gaiser, M.R.; Enk, A.H.; Hadaschik, E.N. Interindividual variation of NETosis in healthy donors: Introduction and application of a refined method for extracellular trap quantification. Exp. Dermatol. 2016, 25, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, T.; Sperandio, M.; Mocsai, A. Neutrophils as emerging therapeutic targets. Nat. Rev. Drug Discov. 2020, 19, 253–275. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singhal, A.; Yadav, S.; Chandra, T.; Mulay, S.R.; Gaikwad, A.N.; Kumar, S. An Imaging and Computational Algorithm for Efficient Identification and Quantification of Neutrophil Extracellular Traps. Cells 2022, 11, 191. https://doi.org/10.3390/cells11020191
Singhal A, Yadav S, Chandra T, Mulay SR, Gaikwad AN, Kumar S. An Imaging and Computational Algorithm for Efficient Identification and Quantification of Neutrophil Extracellular Traps. Cells. 2022; 11(2):191. https://doi.org/10.3390/cells11020191
Chicago/Turabian StyleSinghal, Apurwa, Shubhi Yadav, Tulika Chandra, Shrikant R. Mulay, Anil Nilkanth Gaikwad, and Sachin Kumar. 2022. "An Imaging and Computational Algorithm for Efficient Identification and Quantification of Neutrophil Extracellular Traps" Cells 11, no. 2: 191. https://doi.org/10.3390/cells11020191
APA StyleSinghal, A., Yadav, S., Chandra, T., Mulay, S. R., Gaikwad, A. N., & Kumar, S. (2022). An Imaging and Computational Algorithm for Efficient Identification and Quantification of Neutrophil Extracellular Traps. Cells, 11(2), 191. https://doi.org/10.3390/cells11020191






